Abstract
Plasmids pRAS3.1 and pRAS3.2 are two closely related, natural variants of the IncQ-2 plasmid family that have identical plasmid backbones except for two differences. Plasmid pRAS3.1 has five 6-bp repeat sequences in the promoter region of the mobB gene and four 22-bp iterons in its oriV region, whereas pRAS3.2 has only four 6-bp repeats and three 22-bp iterons. Plasmid pRAS3.1 was found to have a higher copy number than pRAS3.2, and we show that the extra 6-bp repeat results in an increase in mobB and downstream mobA/repB expression. Placement of repB (primase) behind an arabinose-inducible promoter in trans resulted in an increase in repB expression and an approximately twofold increase in the copy number of plasmids with identical numbers of 22-bp iterons. The pRAS3 plasmids were shown to have a previously unrecognized toxin-antitoxin plasmid stability module within their replicons. The ability of the pRAS3 plasmids to mobilize the oriT regions of two other plasmids of the IncQ-2 family, pTF-FC2 and pTC-F14, suggested that the mobilization proteins pRAS3 are relaxed and can mobilize oriT regions with substantially different sequences. Plasmids pRAS3.1 and pRAS3.2 were highly incompatible with plasmids pTF-FC2 and pTC-F14, and this incompatibility was removed on inactivation of an open reading frame situated downstream of the mobCDE mobilization genes rather than being due to the 22-bp oriV-associated iterons. We propose that the pRAS3 plasmids represent a third, γ incompatibility group within the IncQ-2 family plasmids.
Plasmids of the IncQ family are small (<20 kb), have a broad host range, and are highly promiscuous due to their ability to be mobilized very efficiently by self-transmissible plasmids such as the IncP plasmids. They have been divided into two families, IncQ-1 and IncQ-2, based on the amino acid sequence relatedness of their RepA (helicase), RepB (primase), and RepC (DNA-binding) replication proteins and because the mobilization proteins of the two families are unrelated, consisting of three or five genes, respectively (31). IncQ-1 group plasmids include RSF1010 and the near-identical R1162, pDN1, pIE1107, pIE1115, and pIE1130, while IncQ-2 plasmids include pTF-FC2, pTC-F14, and pRAS3.
IncQ-2 plasmids pRAS3.1 and pRAS3.2 were isolated in Norway from the fish pathogens Aeromonas salmonicida subsp. salmonicida and atypical A. salmonicida, respectively, while investigating plasmids that conferred resistance to tetracycline (21). The two plasmids encode identical replication and mobilization proteins, with the most important differences in the plasmid backbone being that pRAS3.1 has four 22-bp iterons in its oriV region and five 6-bp repeat sequences upstream of its mobB gene, whereas pRAS3.2 has only three iterons and four 6-bp repeat sequences. No biological studies were carried out in the initial report of the pRAS3 plasmids. As a contribution to our studies on the evolution of IncQ plasmids, our longer-term aim is to address the question of why two natural versions of the plasmid exist. Here we report on the major differences in the biology of the two plasmids. In addition, we discovered the presence of repC and mobB genes that were not detected when the sequence of pRAS3 plasmids was previously reported. We also discovered a putative toxin-antitoxin (TA) postsegregational system different from that found in other members of the IncQ plasmids and tested it for functionality.
The IncQ-1 plasmids are subdivided into incompatibility groups α, β, and γ, (31), whereas the IncQ-2 plasmids are subdivided into two incompatibility groups, α and β (14). In this work we also report on the incompatibility between the pRAS3 plasmids and other members of the IncQ-2 plasmid family as well as the IncQ-1 family plasmids. Furthermore, we compare the functional relatedness of the pRAS3 mobilization system with that of previously studied IncQ-2 plasmids.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
Escherichia coli strains, cloning vectors, and plasmid constructs are shown in Table 1. Cultures of E. coli were grown in either Luria-Bertani broth (LB) or on Luria-Bertani agar (LA) plates. The growth medium was supplemented with antibiotics as required at the following concentrations: ampicillin (100 μg ml−1), chloramphenicol (20 μg ml−1), kanamycin (30 μg ml−1), nalidixic acid (35 μg ml−1), streptomycin (35 μg ml−1), tetracycline (10 μg ml−1).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptionb | Reference or source | ||
|---|---|---|---|---|
| Strains | ||||
| DH5α | φ80ΔlacZΔM15 endA1 recA1 gyrA96 thi-1 hsdR17 (rK− mK+) relA1 supE44 deoR Δ(lacZYA-argF)U196 | Promega Corp., Madison, WI | ||
| S17-1 | recA pro hsdR (RP4-2 Tc::Mu Km::Tn7) | 34 | ||
| CSH56 | F−ara Δ(lac pro) supD nalA thi | Cold Spring Harbor Laboratory, Cold Spring Harbor, NY | ||
| EC100D pir+ | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80ΔlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ−rpsL nupG pir+(DHFR) | Epicentre Biotechnologies | ||
| Plasmid vectors | ||||
| EZ-Tn5 | Kmr, R6K γ-ori | Epicentre Biotechnologies | ||
| pACYC177 | Apr Kmr, p15A replicon, cloning vector | 6 | ||
| pBAD28 | Apr Cmr, arabinose-inducible expression vector, pACYC184 replicon | 17 | ||
| pBR322 | Apr Tcr, ColE1 replicon, cloning vector | 4 | ||
| pGEM-T | Apr, T-tailed PCR product cloning vector | Promega Corp., Madison, WI | ||
| pOU82 | Apr, lacZYA, R1 replicon | 15 | ||
| pUC19 | Apr, lacZ′, ColE1 replicon, cloning vector | 41 | ||
| Plasmid constructs | ||||
| pBAD28-mobCDEorf3 | Apr Cmr, 2.7-kb ApaLI-ScaI fragment containing pRAS3.1 mobCDE and orf3 cloned behind PBAD promoter | This study | ||
| pBAD28-mobDEorf3 | Apr Cmr, 2.7-kb HindIII-ScaI fragment from pRAS3.1::mobC containing mobDE and orf3 cloned behind PBAD promoter | This study | ||
| pBAD28-orf3 | Apr Cmr, 1.25-kb PstI-ScaI fragment from pRAS3.1::mobE containing orf3 cloned behind PBAD promoter | This study | ||
| pBAD28-repAC | Apr Cmr, 2.7-kb SalI-StuI fragment containing pRAS3.1 repAC cloned behind PBAD promoter | This study | ||
| pBAD28-repB | Apr Cmr, 1,212-bp PCR fragment containing pRAS3.1 repB (nt position 9891 to 8688)a cloned behind PBAD promoter | This study | ||
| pBAD28-repBAC | Cmr, 3.5-kb PvuI-SphI fragment containing pRAS3.1 pemIK-like and repAC genes cloned into pBAD28-repB after inactivation of the pBAD28 PvuI site | This study | ||
| pBAD28-repC | Apr Cmr, 1,015-bp PCR fragment containing the pRAS3.1 repC (nt position 6903 to 5889)a cloned behind PBAD promoter | This study | ||
| pGEM-OriV3.1 | Apr, 742-bp PCR fragment containing pRAS3.1 oriV (nt position 3123 to 2387)a cloned into pGEM-T | This study | ||
| pIE1108Cm | Cmr, pIE1107 replicon with nonessential oriVa deleted and Str and Kmr genes replaced by Cmr gene | 13 | ||
| pIE1130 | Cmr Kmr Smr Sur, natural 10,687-bp IncQ-like plasmid isolated from piggery manure | 35 | ||
| pOriTF14 | Apr, 203-bp HindIII-NcoI fragment containing pTC-F14 oriT cloned into pUC19 | 40 | ||
| pOriTFC2 | Apr, 208-bp HhaI-HhaI fragment containing pTF-FC2 oriT cloned into pUC19 | 40 | ||
| pOriT-RAS3 | Apr, 196-bp PCR fragment containing pRAS3.1 oriT (nt position 11,820 through 0 to 176)a cloned into pGEM-T | This study | ||
| pOU82-TA | Apr, 731-bp PCR fragment containing pRAS3.1 pemIK-like genes (nt position 8819 to 8088)a cloned into pOU82 | This study | ||
| pR6K.3.1.repCΔ | Kmr, pRAS3.1::tet with repC and tetR truncated by NheI-NheI deletion | This study | ||
| pR6K.3.2.repCΔ | Kmr, pRAS3.2::tet with repC and tetR truncated by NheI-NheI deletion | This study | ||
| pRAS3.1 | Tcr, natural 11,851-bp plasmid isolated from Aeromonas salmonicida subsp. salmonicida with four iterons and five 6-bp repeats | 21 | ||
| pRAS3.1.34 | Tcr, pRAS3.1 derivative with three iterons obtained by random ligation of short iteron fragments after BstEII digestion and four 6-bp repeats from pRAS3.2 by exchange of a 2.9-kb HindIII-PvuI region | This study | ||
| pRAS3.1.35 | Tcr, pRAS3.1 derivative with three iterons obtained by random ligation of short iteron fragments after BstEII digestion | This study | ||
| pRAS3.1Km | Kmr, pRAS3.1 with Tcr replaced by Kmr from pSKm2 at the BamHI-EcoRV sites | This study | ||
| pRAS3.1.44 | Tcr, pRAS3.1 derivative with four 6-bp repeats from pRAS3.2 by exchange of 2.9-kb HindIII-PvuI region | This study | ||
| pRAS3.1::mobC | Kmr Tcr, pRAS3.1 with mobC interrupted by EZ-Tn5 at position 296 | This study | ||
| pRAS3.1::mobD | Kmr Tcr, pRAS3.1 with mobD interrupted by EZ-Tn5 at position 1082 | This study | ||
| pRAS3.1::mobE1 | Kmr Tcr, pRAS3.1 with mobE interrupted by EZ-Tn5 at position 1586 | This study | ||
| pRAS3.1::mobE2 | Kmr Tcr, pRAS3.1 with mobE interrupted by EZ-Tn5 at position 1614 | This study | ||
| pRAS3.1::orf3 | Kmr Tcr, pRAS3.1 with orf3 interrupted by EZ-Tn5 at position 2089 | This study | ||
| pRAS3.1::repB | Kmr Tcr, pRAS3.1 with repB interrupted by EZ-Tn5 at the PvuI site | This study | ||
| pRAS3.1::tetAR | Kmr, pRAS3.1 with tetAR interrupted by EZ-Tn5 at the SphI-SphI sites | This study | ||
| pRAS3.2 | Tcr, natural 11,823-bp plasmid isolated from atypical Aeromonas salmonicida with three iterons and four 6-bp repeats | 21 | ||
| pRAS3.2Km | Kmr, pRAS3.2 with Tcr replaced by Kmr from pSKm2 at the BamHI-EcoRV sites | This study | ||
| pRAS3.2::tetAR | Kmr, pRAS3.2 tetAR interrupted by EZ-Tn5 at the SphI-SphI sites | This study | ||
| pTF-FC2Cm | Cmr, natural pTF-FC2 plasmid with chloramphenicol resistance gene cloned into Tn5467 (called pDR412 in previous manuscripts) | 32 | ||
| pTF-FC2Tet | Tcr, Cmr of pDR412 replaced by Tcr of pACYC184 at the XbaI and EcoRV sites | G. Matcher | ||
| pTC-F14Cm | Cmr, natural pTC-F14 plasmid with Cmr inserted at the BamHI site | 13 | ||
| pTC-F14Km | Kmr, pTC-F14Cm with Cmr replaced by Kmr from Tn5 | 40 | ||
| R6K-OriV3.1 | Kmr, pRAS3.1 oriV from pGEM-OriV3.1 transferred to EZ-Tn5 | This study | ||
| RSF1010K | Kmr, 1,1704 bp of RSF1010 replaced by Tn903 | G. Ziegelin | ||
The nucleotide (nt) positions refer to the positions on pRAS3.1 to which the PCR fragments correspond.
Ap, ampicillin; Cm, chloramphenicol; Km, kanamycin; Tc, tetracycline.
General DNA techniques.
Plasmid preparation, restriction endonuclease digestions, gel electrophoresis, and cloning were carried out using standard methods (2, 33). Where no suitable restriction sites were present, single-strand DNA primers were designed and DNA fragments to be cloned were amplified by PCR. An initial denaturation step of 90 s at 94°C was followed by 30 cycles of denaturation (30 s at 94°C), a variable annealing step, and a standard elongation step (240 s at 72°C). Annealing temperatures were based on the average primer annealing temperature, and extension times were altered as required based on primer sequence (Table 2). PCR was performed in a Sprint temperature cycling system (Hybaid) using the Expand high-fidelity PCR system DNA polymerase (Roche Molecular Biochemicals). The sequences of all constructs that required a PCR step were confirmed by DNA sequencing using the dideoxy chain termination method and an ABI Prism 3100 genetic analyzer.
TABLE 2.
Primers used for cloning, quantitative gene expression, and plasmid copy number assays
| Primer | Sequence |
|---|---|
| E.coli GAPA Fwd | 5′-TGTTAGACGCTGATTACATGG-3′ |
| E.coli GAPA Rev | 5′-CTTTAACGAACATCGGAGTGT-3′ |
| pRAS3A Fwd | 5′-GGAGCCACTATCGACTACG-3′ |
| pRAS3A Rev | 5′-GAAGCAGCCCAGTAGTAGG-3′ |
| pRAS3MobC Fwd | 5′-ACACAACAGAGCAGCTAGA-3′ |
| pRAS3MobC Rev | 5′-TCTGGTCAAGCGTGTATCC-3′ |
| pRAS3MobE Fwd | 5′-GCATCAGCGGAAGCAGCC-3′ |
| pRAS3MobE Rev | 5′-GCCTATCGCACTTCGCC-3′ |
| pRAS3ORF3 Fwd | 5′-CCGTTCGATCTGGTAGACC-3′ |
| pRAS3ORF3 Rev | 5′-GTTCTTCCATGTCTCGACG-3′ |
| pRAS3OriT Fwd | 5′-CTTGCAGGATGAGCCAGAC-3′ |
| pRAS3OriT Rev | 5′-TGGTTGCGGAGTTGACAG-3′ |
| pRAS3OriV Fwd | 5′-GTCGAATTCGTACATTATGTTTCG-3′ |
| pRAS3OriV Rev | 5′-ATAGGTACCAGTCTTTTCCATCC-3′ |
| pRAS3REPB Fwda | 5′-ACGAATTCATGTGCGGGAAG-3′ |
| pRAS3REPB Reva | 5′-TCACTGCAGTGCAACATTGTA-3′ |
| pRAS3REPB2 Fwd | 5′-GCAACTATCAGGCCATCAT-3′ |
| pRAS3REPB2 Rev | 5′-TTGGGCTTGCGGTTCTC-3′ |
| pRAS3REPC2 Reva | 5′-TATCTGCAGCTTGAACAGGTG-3′ |
| pRAS3REPC3 Fwda,b | 5′-TAGAATTCAGGAGGAGGGCTATGACTCAGCAGC-3′ |
| pRAS3SS2 Fwda | 5′-GAATTCAGTGGGAGAAGCTGGAAG-3′ |
| pRAS3SS2 Reva | 5′-GGATCCGGAATGGTGTAGATCGTT-3′ |
| R6KKANR Fwd | 5′-CCATTCTCACCGGATTCAG-3′ |
| R6KKANR Rev | 5′-TCACCGAGGCAGTTCCATA-3′ |
The primer includes an endonuclease restriction site (underlined).
The primer overlaps the start codon (shown in bold) and includes an artificial ribosome binding site (in bold italics).
Reverse transcription-PCR (RT-PCR).
RNA was isolated using the RiboPure RNA isolation kit (Ambion) from mid-logarithmic E. coli DH5α cultures carrying the respective plasmids. The quality of the RNA was assayed on a 1% morpholinepropanesulfonic acid-EDTA agarose gel and quantified using a NanoDrop spectrophotometer. RNA (1 μg) was converted to cDNA using the Transcriptor first-strand cDNA synthesis kit (Roche Diagnostics).
Qualitative gene expression to verify whether orf3 is expressed as part of the mobCDE operon was assayed using the FastStart Taq DNA polymerase (Roche Diagnostics) according to the manufacturer's protocol. Two microliters, or ∼100 ng, of the cDNA was used in each reaction mixture with a final MgCl2 concentration of 2.3 mM. The GC-rich solution was added to a 1× final concentration whenever the pRAS3MOBE forward and reverse primers (Table 1) were used, and 2.5% dimethyl sulfoxide was added to each PCR mixture when the pRAS3MOBC forward or reverse primers were used. The reaction mixtures were subjected to 35 cycles of denaturation, elongation, and extension as described above. The RNA samples were assayed for the presence of contaminating DNA by using ∼400 ng of RNA in control reaction mixtures. PCR products were analyzed on a 1% agarose gel.
Quantitative gene expression of the repB and orf3 genes using the pRAS3REPB2 and pRAS3ORF3 primer sets was assayed using a LightCycler as described above. The cDNA was diluted twofold and a total of 50 ng was used in each reaction mixture. The amplification efficiencies were determined as described above using serial dilutions of the cDNA, and the reaction parameters were set as described above for the plasmid copy number determinations. Relative gene expression was determined using the REST analysis tool (30). The R6KkanR primer set was used as a calibrator.
Copy number determinations.
Total genomic DNA was prepared from E. coli DH5α cultures containing the respective plasmids during exponential growth. Cells were grown overnight (in the presence of antibiotics), reinoculated (1/100) into 50 ml of prewarmed LB medium (no antibiotics), and grown while shaking at 37°C to an optical density at 600 nm of ∼0.8. Total genomic DNA was extracted from 1 ml of culture using the QIAamp DNA minikit (Qiagen). The genomic DNA was eluted in 60 μl elution buffer, and the concentration and purity were checked using a Nanodrop spectrophotometer.
Real-time quantitative PCR (qPCR) amplification was performed using a LightCycler (version 2.0) with the LightCycler FastStart DNA master SYBR green I kit (Roche Diagnostics). A total of 4 ng of total DNA was added to each amplification reaction mixture, and the thermal cycling protocol of Lee et al. (22) was followed, except that primer annealing was at 56°C for 4 s and DNA extension was at 72°C for 15 s.
To generate standard curves for plasmid copy number determinations, the gapA amplicon (Table 2) was cloned into pGEM-T(Easy). The pGEM-gapA and pRAS3.1 plasmids were extracted from E. coli DH5α using a Nucleobond AX plasmid DNA purification kit. The concentrations of both DNA samples were determined (six replicates) by using a NanoDrop spectrophotometer. A 10-fold dilution series (100 to 10−5) was set up for each plasmid. Samples were amplified with thermal cycle parameters as specified above, and the threshold cycle (Ct) values were plotted against the number of DNA molecules in each sample. The R2 value for both calibration standard curves was greater than 0.9995. Absolute plasmid copy number was determined by amplification of pRAS3.1 and pRAS3.2 in the same cycle as the calibration curves. The Ct values were used to extrapolate the total amount of chromosome and plasmid present in each sample from the standard curves, using the LightCycler software (version 3.5) according to the calculations of Lee et al. (22). Relative plasmid copy numbers were determined using the same conditions and cycle parameters as described above. All copy numbers were determined relative to pRAS3.1.35 by using the REST analysis tool (30). The same standard curves that were used for absolute copy number determinations were used to calculate the amplification efficiency.
Plasmid copy numbers in the presence of excess replication proteins which were expressed from the PBAD arabinose-inducible promoter of pBAD28 were measured by means of qPCR relative to the same sample with only pBAD28 in trans. Addition of arabinose to Luria-Bertani broth resulted in slow cell growth and proved unnecessary, as rep genes cloned behind the PBAD promoter were sufficiently expressed to be able to complement their respective rep deletion mutants.
Mobilization assay.
E. coli S17.1 donor and E. coli CSH56 recipient cells were cultured separately overnight with appropriate antibiotic selection. Cells were washed three times in phosphate-buffered saline (PBS, pH 7.4) and mixed in a donor-to-recipient ratio of 1:10 or 1:100. One hundred microliters of this mixture was spotted onto an LA plate and incubated at 37°C for 60 min (unless specified otherwise). The agar plug was excised and suspended in 10 ml PBS (pH 7.4) and vigorously shaken, after which 8 ml was collected, pelleted, and resuspended in 1 ml PBS (pH 7.4). Serial dilutions were plated onto donor and transconjugant selective media, and the number of transconjugants per donor was calculated.
Plasmid stability assay.
Stability assays using the pOU82-based test system were performed by growing plasmid-containing E. coli cells without selection in 5 ml LB at 30°C for 4 days, with transfer of ∼1,000 cells to fresh LB at ∼20-generation intervals. Samples taken at ∼20-generation intervals were diluted in PBS (pH 7.4) and plated onto LA plates supplemented with 40 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside and incubated at 37°C. The percentage plasmid loss was determined by calculating the ratio of plasmid-containing (blue) to plasmid-free (white) colonies (15).
Incompatibility assays.
Plasmid-containing E. coli DH5α cells were transformed with a second plasmid and plated on LA plates with antibiotic selection for both plasmids. Single colonies were picked into LB containing appropriate antibiotics and incubated overnight at 30°C. Survival of the plasmids was then tested by removing selection for both plasmids and growing the cells in 5 ml LB for ∼100 generations, with transfer of approximately ∼1,000 cells to fresh medium at ∼20-generation intervals. Finally, 50 colonies were replica plated to antibiotic-containing LA plates to score for plasmid retention. Cells containing individual plasmids were similarly grown and plated as a control to account for spontaneous plasmid loss.
Displacement assays.
Competent E. coli DH5α cells containing a resident plasmid were transformed with a second incoming plasmid and plated on antibiotic-containing medium that selected only for the incoming plasmid. Sixteen colonies were picked and plated onto three sets of solid media, two containing single antibiotics to separately test for the presence of the resident or incoming plasmid and one containing no antibiotics as a control for cell viability. Controls to check for spontaneous loss of the resident plasmids were carried out using the same procedure except that the initial competent E. coli cells containing the resident plasmids were taken through a cycle of growth on solid medium without antibiotic selection before testing for retention of the resident plasmid.
Random knockouts and screening for an incompatibility determinant.
Random knockouts of pRAS3.1 were generated using the EZ-Tn5 transposon system (Epicenter Biotechnologies). The transposon (0.025 pmol) mutagenesis was carried out in vitro using 0.05 pmol pRAS3.1 as per the manufacturer's protocol. The reaction mixture was transformed into electrocompetent E. coli EC100D, and all the colonies were scraped off the plates into 100 ml fresh LB medium and incubated for 1 h at 37°C. The plasmid DNA was purified using the Nucleobond AX plasmid purification kit (Macherey-Nagel).
The bank of random knockouts was screened for a compatible phenotype by transforming 1 ng of the plasmid DNA into electrocompetent E. coli EC100D cells containing a resident pTF-FC2Cm (pTF-FC2 with Cmr gene) plasmid. EZ-Tn5 and pRAS3.1::Tet were used as controls for compatibility and incompatibility, respectively, on double-selective plates. The expression mix was spread on plates containing antibiotic selection for both plasmids, and colonies were replica plated for two rounds of growth on plates selecting for the pRAS3.1 knockouts only. This was followed by selection for cells that still retained pTF-FC2Cm. After restriction analysis of the extracted plasmid DNA, the random knockout plasmids from selected positive colonies were separated from the coresident pTF-FC2Cm plasmid by transforming the extracted DNA into E. coli DH5α and selecting for only the pRAS3.1 knockouts that had allowed pTF-FC2Cm to be retained.
Sequence analysis and bioinformatics.
The DNA sequence previously deposited as pRAS3.1 (accession number AY043298) and pRAS3.2 (accession number AY043299) by L'Abée-Lund and Sørum (21) was analyzed using a variety of software programs but mainly a combination of the Glimmer 2 (www.tigr.org/softlab) (9) and DNAMAN (Lynnon BioSoft) programs. Comparison searches were performed using the gapped BLAST program at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) (1).
RESULTS
Reanalysis of the sequences of pRAS3.1 and pRAS3.2.
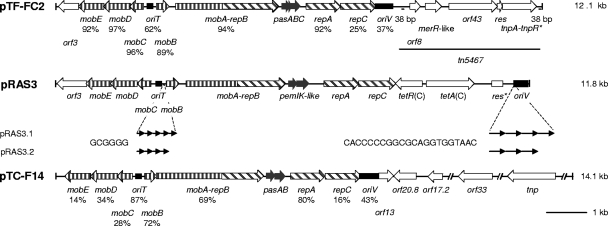
An analysis of the sequences of the pRAS3 plasmids was previously reported by L'Abée-Lund and Sørum (21). Given the high similarity in gene organization and sequence with plasmid pTF-FC2, we were surprised by the apparent absence of mobB and repC genes in the pRAS3 plasmids. When the sequence data were reanalyzed, four genes were identified that were not detected previously. These were the mobB and repC genes, with the amino acid sequences of their products being 87% and 37% identical to the equivalent gene products of pTF-FC2, respectively. In addition two genes were identified for what appears to be a TA postsegregation killing system. The TA genes are distantly related to the previously published pemIK (parDE) (5, 38) and mazEF (chpAI chpAK) (24) systems. A comparison of the pRAS3 plasmids with pTF-FC2 and pTC-F14, the only other two plasmids of the IncQ-2 group identified to date, is shown in Fig. 1. In general, the nucleotide sequences of the backbones of the pRAS3 plasmids are more closely related to pTF-FC2, with the exceptions being the sequences of the oriT and oriV regions, whereas the pRAS3 plasmids were more closely related to pTC-F14. The putative TA system of the pRAS3 plasmids was unrelated to that of either pTF-FC2 or pTC-F14 (which are closely related to each other [8]). However, in all plasmids the TA system is situated in a similar position between the repB and repA genes.
FIG. 1.
Comparison of the genetic maps of the pRAS3 plasmids with pTF-FC2 and pTC-F14. Percentages below the plasmid backbone genes of pTF-FC2 and pTC-F14 indicate the percent amino acid sequence identity of the gene product with that of the pRAS3 plasmids. Percentages below the oriT and oriV regions indicate nucleotide sequence identity. Plasmids pRAS3.1 and pRAS3.2 have different numbers of 6-bp repeats and 22-bp iterons, while the nucleotide sequence of each repeat or iteron is identical, as indicated below pRAS3.
Copy numbers of pRAS3.1 and pRAS3.2.
The absolute and relative plasmid copy numbers (PCN) of pRAS3.1 and pRAS3.2 were determined in E. coli DH5α by quantitative real-time PCR using the chromosomal gapA gene as a calibration standard. The copy number of pRAS3.1 was found to be 45 ± 13 (n = 11) plasmids per chromosome and that for pRAS3.2 was 30 ± 5 (n = 4). This large difference in copy number was surprising, as pRAS3.2 has three oriV-associated 22-bp iterons while pRAS3.1 has four of these identical 22-bp iterons, and one would expect the plasmid with fewer iterons to have the higher copy number. Furthermore, other IncQ-2 family plasmids have been reported to have a considerably lower PCN of 12 to 16 plasmids per chromosome (10, 36), which is two- to threefold lower than determined for pRAS3.2 and pRAS3.1, respectively. The PCN of the well-known cloning vector pBR322 has been determined to be ∼18 plasmids per chromosome based on both absolute and relative quantification real-time PCR methods (22). As the tetracycline resistance gene of pBR322 is identical to that of pRAS3.2, the PCN of pBR322 could be quantified using the same primer sets. It was therefore included in an additional set of real-time PCR assays as a copy number control. The relative PCN of pRAS3.1 and pRAS3.2 were 2.3- and 1.7-fold higher than that of pBR322, respectively. Based on a copy number of 18 for pBR322, this is equal to PCN values of approximately 41 for pRAS3.1 and 30 for pRAS3.2.
Reason for the difference in copy number between pRAS3.1 and pRAS3.2.
The observation that pRAS3.1 with four 22-bp oriV-associated iterons had a higher copy number than pRAS3.2 with three 22-bp iterons was unexpected. We therefore investigated the effect on PCN of the additional 6-bp CCCCCG repeat upstream of the mobB gene of pRAS3.1. The repeats are located 6 bp upstream of a putative ribosomal binding site of the mobB gene (Fig. 2). Three derivatives of pRAS3.1 (five 6-bp repeats and four 22-bp iterons) were constructed to isolate the effects of the 6-bp repeats and 22-bp iterons on PCN (Table 1). When the number of 22-bp iterons in pRAS3.1 was decreased from four to three (pRAS3.1.35), the calculated PCN increased from approximately 41 to 59 copies (Table 3). The five 6-bp repeats in this construct were then exchanged for those in pRAS3.2 (Table 1) to give a construct still with three 22-bp iterons but now with only four 6-bp repeats (pRAS3.1.34). If the number of 6-bp repeats had an influence on PCN, the PCN of pRAS3.1.34 should fall to that of pRAS3.2, as these two plasmids have equal 6-bp repeat and 22-bp iteron copy numbers. This is what was found, as the relative PCN of pRAS3.1.34 decreased by approximately 47% to 31 copies, the same as that of pRAS3.2. When the number of 22-bp iterons in this lower-copy-number plasmid was increased from three to four (pRAS3.1.44) the PCN decreased further to approximately 23 copies. Therefore, the presence of a fifth 6-bp repeat resulted in the PCN of a plasmid with four 22-bp iterons to increase by 1.77-fold (pRAS3.1.44 compared with pRAS3.1) and a plasmid with three 22-bp iterons increased by 1.89-fold (pRAS3.1.34 compared with pRAS3.1.35). A decrease in the number of 22-bp iterons from four to three also resulted in an increase in PCN. This was 1.45-fold in a plasmid with five 6-bp repeats (pRAS3.1 compared with pRAS3.1.35) and 1.35-fold in a plasmid with four 6-bp repeats (pRAS3.1.44 compared with pRAS3.1.34).
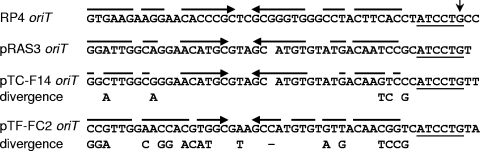
FIG. 2.
The intergenic sequence between the oriT and mobB of pRAS3.1, showing the position of the 6-bp repeats. The oriT is underlined and the imperfect inverted repeat within the oriT is indicated by broken inverted arrows. The conserved hexameric nick site is indicated in bold with a vertical arrow indicating the putative nick position. The 6-bp CCCCCG repeats are labeled 1 to 5. The first repeat consists of only 5 bp, as it lacks a cytosine base. A putative promoter with a near-consensus −35 region and a weak −10 region is shown in bold italics and is separated by a 17-bp spacer.
TABLE 3.
Effects of the number of 6-bp repeats and 22-bp iterons on plasmid copy number
| Plasmid construct | No. of 6-bp repeats | No. of 22-bp iterons | Relative plasmid copy no.b | Calculated plasmid copy no.c |
|---|---|---|---|---|
| pRAS3.2 | 4 | 3 | 0.51 ± 0.09 | 30 ± 5 |
| pRAS3.1 | 5 | 4 | 0.69 ± 0.062 | 41 ± 4 |
| pRAS3.1.35a | 5 | 3 | 1.0 | 59 |
| pRAS3.1.34 | 4 | 3 | 0.53 ± 0.002 | 31 ± 1 |
| pRAS3.1.44 | 4 | 4 | 0.39 ± 0.037 | 23 ± 2 |
pRAS3.1.35 served as the reference for the determination of relative copy numbers and standard deviations.
The number of replicates for relative copy number determinations was four to six, and a P value of 0.001 was obtained for each qPCR experiment.
Plasmid numbers were calculated to the nearest whole plasmid.
These results suggested that it was the presence of an additional 6-bp repeat upstream of the mobB gene that resulted in a higher PCN and raised the question of how the extra 6-bp repeat exerted this effect on PCN. The most obvious possibility was that the 6-bp repeat affected the level of expression of mobB as well as the downstream mobA/repB genes (Fig. 2). To test this, the levels of expression of mobB, mobA/repB, and the divergently transcribed mobCDE and orf3 operon were determined for pRAS3.1 and compared with pRAS3.2 by using qPCR. To carry out the comparison, an R6K oriV and kanamycin resistance gene (EZ-Tn5) were cloned into the tetAR genes of both pRAS3.1 and pRAS3.2, whereafter the native replicons were inactivated through truncation of the repC and tetR genes. This ensured that both plasmids had the same copy number and allowed the relative levels of gene expression to be determined. The expression of mobA/repB in the case of a pRAS3.1 equivalent (five 6-bp repeats; pR6K.3.1.repCΔ) was approximately twofold higher (2.0 ± 0.9; n = 12; P = 0.048) relative to a pRAS3.2 equivalent (four 6-bp repeats; pR6K.3.2.repCΔ). In contrast, expression in the opposite direction (mobCDE-orf3) was the same (1.1 ± 0.5; n = 4; P = 0.875) for both pRAS3.1 and pRAS3.2 equivalents. These results suggested that the reason for the increase in PCN of pRAS3.1 compared to pRAS3.2 was that the additional 6-bp repeat resulted in an increase in mobB-mobA/repB expression and that increased expression of repB was the actual cause.
Effect of increased repBAC expression on plasmid copy number.
To confirm that the additional repB expression resulted in an increased PCN as well as to determine whether the products of the repA and repC genes affected PCN, the repC, repAC, and repB genes were cloned behind the PBAD promoter of the vector pBAD28. The arabinose-inducible repB construct was placed in E. coli(pRAS3.1.34) cells and the copy number was compared with the same cells containing the pBAD28 vector only. With the repB-containing construct in trans, the PCN of pRAS3.1.34 was increased approximately 2.2-fold ± 0.53-fold (n = 4; P = 0.001) relative to the vector control. This is approximately equal to the 1.96-fold difference in PCN between plasmids differing only in their number of 6-bp repeats, such as pRAS3.2 (three iterons; four 6-bp repeats) and pRAS3.1.35 (three iterons; five 6-bp repeats), as shown in Table 3. Therefore, the additional repB expression raised the copy number of a plasmid which contained four 6-bp repeats to approximately the same as that of a plasmid that had five 6-bp repeats.
To examine the effects of additional repC or repAC gene products, plasmid pRAS3.1.35 was used, as this plasmid with five 6-bp repeats already had high levels of repB expression and it was possible that the repC or repAC gene products were limiting. Expression of repC in trans did not result in a change in PCN (1.02 ± 0.22; n = 4; P = 0.874), whereas expression of repAC (pBAD28-repAC) resulted in an approximately 30% reduction in PCN (0.67 ± 0.13; n = 4; P = 0.008). This reduction in PCN due to overexpression of repAC was consistent with that reported by Matcher and Rawlings (25).
Comparison of the mobilization frequencies of pRAS3.1 and pRAS3.2.
As the mobilization frequencies of pRAS3.1 and pRAS3.2 were not reported in the study by L'Abée-Lund and Sørum (21), we determined the mobilization frequency between E. coli S17.1 donor and E. coli CSH56 recipient cells. This was found to be 0.032 ± 0.014 for pRAS3.1 and 0.021 ± 0.013 for pRAS3.2. Experiments using plasmids with different numbers of 6-bp repeats but identical 22-bp iteron numbers, as well as plasmids with identical 6-bp repeats but different 22-bp iteron numbers, showed that neither had any significant effect on the mobilization frequencies. Thus, although increased transcription in the direction mobB-mobA/repB compared with mobCDEorf3 as a result of the extra 6-bp repeat had an effect on plasmid replication, it did not have a marked effect on plasmid mobilization.
Functional relatedness of mobilization regions within the IncQ-2 plasmids.
Prior to plasmid conjugation a relaxase cleaves one of the DNA strands at the origin of transfer (oriT), forming a covalent protein-DNA complex in the donor cell that is transferred to the recipient. In the case of plasmid F, the relaxase-oriT recognition has a high degree of structural and sequence specificity (12, 26). In contrast, the oriT region of the IncQ-1 plasmid R1162 is small, structurally simple, and can accommodate base pair changes without a complete loss of function (3, 20, 27). Since the five-protein mobilization operon of the IncQ-2 plasmids has more in common with the IncP plasmids than with the three mobilization protein operons of IncQ-1 plasmids, we tested whether the relaxase-oriT recognition of the pRAS3 plasmids was relaxed like that of R1162 or specific like plasmid F and related plasmids. The amino acid sequences of the mob gene products of the pRAS3 plasmids were closely related to that of pTF-FC2, with an average amino acid sequence identity of over 90% for the five mob gene products (Fig. 1). This contrasted with pTC-F14, where the average amino acid sequence identity was approximately 25% for the MobCDE genes and 70% for the MobAB genes. However, when the nucleotide sequences of the oriT regions were compared (Fig. 3), the oriT of the pRAS3 plasmids was considerably more related to that of pTC-F14 (87% identity) than to pTF-FC2 (62% identity). We therefore tested whether the oriT of the pRAS3 plasmids was able to be mobilized by E. coli S17.1 cells containing the conjugative plasmid RP4 by the products of the mob genes of pTF-FC2 and pTC-F14. A 196-bp fragment from pRAS3.1 containing the oriT was PCR amplified and cloned into the nonmobilizable pUC19 vector to produce pOriT-RAS3, and the sequence was confirmed by DNA sequencing. Plasmid pOriT-RAS3 was mobilized at the saturation frequency within 60 min of mating by E. coli S17.1 when either pRAS3.1 or pRAS3.2 was coresident, while no transconjugants were obtained in the absence of coresident pRAS3 plasmids. When either pTF-FC2Cm or pTC-F14Cm was coresident instead of the pRAS3 plasmids, no transconjugants were obtained. However, in the reverse experiments, coresident pRAS3.1 plasmid was able to mobilize pUC19 vector containing the pTF-FC2 and pTC-F14 oriTs at frequencies of 1.20 (±0.44) × 10−1 and 3.42 (±1.64) × 10−4 transconjugants per donor in 60 min, respectively. The ability of Mob proteins of pRAS3 to mobilize the oriTs pTF-FC2 and pTC-F14 suggested that like R1162 these oriTs are also relaxed and that the Mob proteins pRAS3 can mobilize oriTs with very different sequences.
FIG. 3.
Alignment of oriT regions of IncQ-2 plasmids and the IncPα plasmid RP4, showing the sequence divergence that could be tolerated by the Mob proteins of plasmid pRAS3 while they were still able to mobilize DNA from an oriT. A vertical arrow indicates the relaxase nic site at which single-stranded cleavage takes place as determined for plasmid RK2/RP4 (29).
The toxin-antitoxin system on pRAS3 is functional.
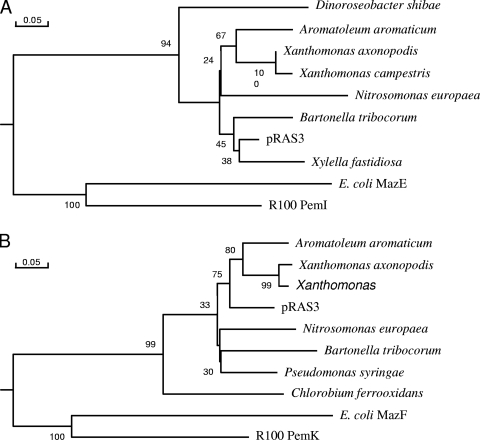
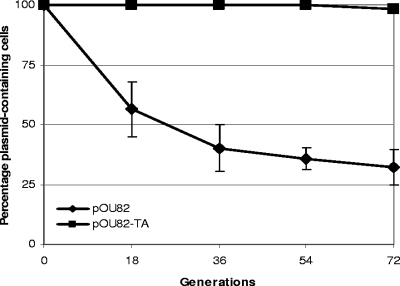
Careful analysis of the sequence of pRAS3.1 and pRAS3.2 revealed the presence of two small tandem ORFs encoding proteins of 74 and 108 amino acids (aa), respectively, situated between repB and repA. BLAST analysis of the proteins from these ORFs indicated that they were most closely related to pairs of proteins of similar size from adjacent ORFs detected in genome sequencing data obtained from a number of other bacteria, such as Xanthomonas campestris, Xanthomonas axonopodis, Aeromatoleum aromaticum, and Nitrosomonas europaea (Fig. 4A and B). These proteins are related to toxin-antitoxin postsegregational killing plasmid stability systems and are listed as being either PemIK-like or MazEF-like, although they group in a cluster well separated from either of these protein pairs. No members of the clusters shown in Fig. 4A and B, besides the distantly related PemIK and MazEF, have been tested for toxin-antitoxin activity. Using PCR we amplified the two ORFs from pRAS3.1 and cloned them into the segregationally unstable test plasmid pOU82, to give plasmid pOU82-TA. When grown without plasmid selection for 72 generations, approximately 98% of E. coli DH5α cells retained pOU82-TA, whereas only 35% of cells retained pOU82 (Fig. 5). This enhanced plasmid stability suggests that the other untested proteins shown in Fig. 4A and B are also likely to be toxin-antitoxin pairs.
FIG. 4.
Phylogenies of toxin-antitoxin proteins of pRAS3 and comparison with closely related proteins as well as the more distantly related PemIK and MazEF proteins. (A) Antitoxins were as follows: Aromatoleum aromatium, CAI08016; Bartonella tribocorum, CAK00897; Dinoroseobacter shibae, YP_001541878; Nitrosomonas europaea ATCC 19718, CAD85218; Xanthomonas axonopolis pv. citri strain 306, NP_644761; Xanthomonas campestris pv. vesicatoria strain 85-10, CAJ19793; Xylella fastidiosa Ann-1, ZP_00682677; E. coli MG1165 MazE, AAA69293; plasmid R100 PemI, P13975. (B) Toxins were as follows: Aromatoleum aromatium, CAI08015; Bartonella tribocorum, CAK00896; Chlorobium ferrooxidans, EAT59633; Nitrosomonas europaea ATCC 19718, CAD85217; Pseudomonas syringae pv. phaseolicola, AAZ37969; Xanthomonas axonopolis pv. citri strain 306, NP_644760; Xanthomonas campestris pv. vesicatoria strain 85-10, CAJ19792; E. coli MG1165 MazF, AAA69292; plasmid R100 PemK, P13976.
FIG. 5.
Loss of the low-copy-number test plasmid, pOU82, with and without the PemIK-like TA genes from pRAS3 in the absence of plasmid selection.
Compatibility of pRAS3.1 and pRAS3.2 with other IncQ plasmids.
We tested whether plasmids pRAS3.1 and pRAS3.2 were compatible with different IncQ-1 and IncQ-2 plasmids. Plasmids RSF1010 (16), pIE1108 (35), and pIE1130 (37) were used as representatives of the α, β, and γ incompatibility groups of InQ-1 plasmids, respectively. The incompatibility assay involved the placement of the two test plasmids into a single E. coli DH5α host cell and then testing for how many cells retained both plasmids after approximately 34 generations in the absence of selection. When present on their own, plasmids RSF1010K, pIE1108Cm, and pIE1130 were slightly less stable after 34 generations (94 to 100% retention) than pRAS3.1 or pRAS3.2 (100% retention). When either pRAS3.1 or pRAS3.2 was coresident with plasmid RSF1010K, pIE1108Cm, or pIE1130, the stability of each plasmid was indistinguishable from that when it was present in the E. coli host on its own.
When testing for the compatibility of pRAS3.1 and pRAS3.2 with pTF-FC2 and pTC-F14, members of the IncQ-2 α and β incompatibility groups, respectively, a different compatibility assay had to be used. The pRAS3.1 and pRAS3.2 plasmids were so highly incompatible with either pTF-FC2Cm or pTC-F14Cm that we were unable to isolate E. coli host cells containing two test plasmids. When attempts were made to transform competent E. coli pRAS3.1- or pRAS3.2-containing cells with either pTF-FC2Cm or pTC-F14Cm, no transformants were isolated, although the competent cells could be readily transformed with other plasmids, such as pUC19 and pACYC177. In the reciprocal experiments with either pTF-FC2Cm or pTC-F14Cm being resident in the competent cells prior to transformation with either pRAS3.1 or pRAS3.2, successful transformation by the incoming plasmid was achieved but the resident plasmid was immediately displaced.
Search for the source of strong plasmid incompatibility.
Experiments were carried out in an attempt to identify the reason for this strong incompatibility. A functional 751-bp pRAS3.1 oriV region containing the four 22-bp iteron DNA was cloned into the E. coli pGEM-T vector to give pGEM-OriV3.1. However, this construct was fully compatible with either the pTF-FC2 or pTC-F14 replicons and therefore the iterons and associated DNA were not the cause of the strong incompatibility. The pRAS3.1 repC, repAC, and repB genes were cloned behind the PBAD promoter of the pBAD28 expression vector and shown to be expressed by their ability to complement pRAS3.1 repC or repB mutants. Again, none of these clones displaced pTF-FC2 or pTC-F14. Next, the region containing the repBAC genes, including the genes for the pemIK-like TA system, was cloned behind the PBAD promoterto give pBAD28-repBAC. This plasmid could support the replication of the 751-bp pRAS3.1 oriV region when cloned into a R6K vector (EZ-Tn5), which was unable to replicate in E. coli DH5α unless pBAD28-repBAC was present in trans. This showed that the repBAC fragment was functional, but this fragment also did not displace the pTF-FC2 or pTC-F14 replicons.
As plasmid incompatibility did not appear to be associated with the replicon region, we tested whether we could knock out whatever was responsible for incompatibility using the EZ-Tn5 transposon mutagenesis system. A bank of random mutants of pRAS3.1 was generated and screened for mutants that did not displace E. coli containing a resident pTF-FC2Cm plasmid. Approximately 5% of mutants displayed a compatible phenotype by selection on plates for both plasmids. All 200 compatible pRAS3.1 mutant plasmids tested retained pTF-FC2Cm after two rounds of growth from a single cell to a colony when selecting for only the pRAS3.1 mutants. Restriction endonuclease analysis using BamHI and SalI for 48 of the 200 mutants indicated that the mutations fell into five groups, with all transposons located within a 1.75-kb region. Nucleotide sequencing of a representative of each group indicated that the five insertions were evenly spaced within the region containing the mobCDE genes and a previously unreported downstream ORF (called orf3), with one insertion in each gene and two in mobE. To determine whether orf3 is expressed from the same transcript as mobCDE, mRNA was isolated from E. coli DH5α(pRAS3.1) cells and analyzed by RT-PCR. Primer sets to mobC and orf3 or to mobE and orf3 gave positive amplification products of the predicted sizes, while a lack of amplification products in experiments in which reverse transcriptase was omitted indicated that amplification products were not due to DNA contamination (data not shown). We concluded that orf3 is expressed as part of the mobCDE operon. Since orf3 is the last gene in the series of four genes, we predicted that insertion into the upstream mob genes presumably affected expression of orf3 and that it was orf3 that was probably responsible for plasmid incompatibility.
Role of orf3.
orf3 is 753 bp long and encodes a predicted protein of 250 aa that is preceded by a putative ribosome-binding site (GGAGG) 5 bp upstream of the ATG start. A BLAST analysis against the nonredundant NCBI protein database indicated two strong hits of 98 and 97% amino acid identity along the entire length of a hypothetical protein (240 aa) from an uncultured bacterium and to ORFX (163 aa) of plasmid Rms149 from Pseudomonas aeruginosa (18). Rms149 is a 57-kb IncP-6 plasmid that has a different replicon from IncQ plasmids but a 5.6-kb mobilization region that is very similar to that of the pRAS3 plasmids and pTF-FC2. Like the IncQ-2 plasmids this mobilization region includes a mobB followed by a mobA/repB gene fusion and the divergent mobCDE genes and has between 86 and 100% amino acid identity to the corresponding gene products of pRAS3 and pTF-FC2. The orfX of Rms149 is in the same location as orf3, immediately downstream of mobE, but appears to have been truncated by the insertion of Tn1012. Plasmid pTF-FC2 has an orf4 encoding a 270-aa product in a similar location to orf3 and orfX, but this ORF is unrelated in nucleotide or predicted amino acid sequence.
Despite the Tn1012 insertion in orfX, the 5.6-kb mobilization region of Rms149 remained functional (18), but without an untruncated orfX the effect of orfX on mobilization frequency could not be tested. We therefore tested whether the insertion of EZ-Tn5 into orf3 of pRAS3.1, which was used to indentify orf3 as the cause of incompatibility, affected the mobilization frequency. Construct pRAS3.1::tet was mobilized at a frequency of 12.44 ± 3.95 transconjugants per donor, while pRAS3::orf3 was mobilized twofold lower, at 6.04 ± 4.33 transconjugants per donor. We concluded that like mobD and mobE, orf3 plays a minor role in mobilization frequency. To determine whether the product of orf3 was responsible for the strong plasmid incompatibility, mobDE-orf3 and orf3 were cloned behind the PBAD promoter. The genes were shown to be expressed by using RT-PCR and primers specific to the mobE and orf3 genes. However, when these constructs were placed in trans with pTF-FC2Tet (or pTC-F14Km) they did not cause plasmid incompatibility. Plasmid pTF-FC2Tet was, however, displaced when the entire operon (pBAD28-mobCDEorf3) was placed in trans, and the reason for strong incompatibility requires further investigation.
DISCUSSION
The low nucleotide sequence identity between the repC genes of the pRAS3 plasmids with that of pTF-FC2 and pTC-F14, the other members of the IncQ-2 plasmid group, was unexpected. The iteron-binding protein, RepC, is essential for IncQ plasmid replication and is generally the most conserved Rep protein within the IncQ plasmid family (31). The other genes of the plasmid backbone (mobEDCBA and repBA) of pRAS3 and pTF-FC2 are highly conserved (Fig. 1), and it is unlikely that differences in the repC genes are due to a higher mutation rate. It is more likely that the repC gene of the pRAS3 plasmids has been acquired by gene swapping with some as-yet-unidentified IncQ-like plasmid that is different from any so far discovered.
In several other plasmids the total number of oriV-associated iterons in a cell has been shown to affect plasmid copy number (7, 23, 39), and our expectation was that pRAS3.1 with four 22-bp oriV-associated iterons would have a lower copy number than pRAS3.2, with three 22-bp iterons. Unexpectedly, we found that pRAS3.1 had a copy number considerably greater than pRAS3.2 despite of it having more iterons. This increase in copy number was due to the number of 6-bp repeats in the intergenic region between the mobCBE-orf3 and the mobB-mobA/repB operons, with the higher-copy-number pRAS3.1 plasmid having five 6-bp repeats while plasmid pRAS3.2 had four 6-bp repeats. The additional 6-bp repeat resulted in an increase in expression of the mobB-mobA/repB operon and it was the increase in expression of the RepB primase that resulted in a higher plasmid copy number. The altered promoter could either increase the efficiency of initiation of mobB-mobA/repB transcription or decrease autorepression by RepB (14). Swapping the region with the repeats between pRAS3.1 and pRAS3.2 resulted in a corresponding change in copy number. We further demonstrated that the level of transcription of the mobB-mobA/repB operon was increased approximately twofold in the presence of five 6-bp repeats. An increase in PCN as a result of increased repB expression was confirmed by placement of a repB gene in trans under the control of an arabinose-inducible promoter. This resulted in a 2.2-fold increase in copy number of a plasmid containing four 6-bp repeats to approximately the level of a plasmid containing five 6-bp repeats. This provided strong evidence that the copy number of pRAS3.1 was affected by both the number of 22-bp oriV-associated iterons and the level of transcription of the repB gene.
The 30% reduction in pRAS3 copy number that occurred on overexpression of repAC from a PBAD vector promoter is different from the result obtained by Haring et al. (19), who reported that the copy number of the IncQ-1 plasmid, RSF1010, was increased sixfold upon overexpression of repAC. However, the small reduction in copy number of pRAS3 is similar to the reduction in copy number that occurred on overexpression of repAC in the case of the related plasmid, pTF-FC2 (25). We observed that when repAC from either pRAS3 or pTF-FC2 was expressed from the PBAD promoter, the E. coli host cells were slow growing and clearly stressed. Whether this might have affected the copy number is unknown.
Demonstration that the pRAS3 plasmids contain a TA system means that all of the IncQ-2 plasmids so far discovered contain a toxin-antitoxin plasmid stability module situated within the replicon in exactly the same position between the repB and repA genes. However, although the pas toxin-antitoxin systems of pTF-FC2 and pTC-F14 are related to each other and are deep-branching members of the E. coli relBE family, they are totally unrelated to the toxin-antitoxin system of the pRAS3 plasmids. As the repB and repA genes of pRAS3 plasmids are highly related to pTF-FC2 and pTC-F14 and flank the unrelated TA modules (Fig. 1), this implies that the TA modules of the pRAS3 plasmids were acquired independently of pTF-FC2 and pTC-F14. The observation that IncQ-2 plasmid replicons have acquired different TA systems located in the same position suggests that there may be a biological reason for their occurrence in that exact position. Matcher and Rawlings (25) have presented evidence that a strong, autoregulated promoter such as that provided by a TA system within the replicon of pTF-FC2 confers on the plasmid the ability to rapidly respond to a fall in copy number. This is because a transient burst of expression of the TA genes results in a related increase in expression of the downstream repAC genes. IncQ plasmids appear not to have an active partitioning system, and a fall in copy number might occur on cell division when one daughter cell could receive many more copies of a plasmid than the other. Similarly, a strongly expressed, autoregulated TA system would allow the rapid expression of repAC on arrival in a recipient cell following conjugation.
To date plasmids pRAS3.1, pRAS3.2, pTF-FC2, and pTC-F14 are the only reported representatives of the IncQ-2 plasmids. Previous work demonstrated that pTF-FC2 and pTC-F14 are fully compatible and therefore were placed in the IncQ-2 incompatibility groups α and β, respectively. When we tested the incompatibility of pRAS3.1 and pRAS3.2 against representatives of the IncQ-1 α, β, and γ incompatibility groups, they were fully compatible but were violently incompatible with other members of the IncQ-2 plasmids. Plasmid incompatibility is understood as being the inability of two plasmids to coexist in the same host in the absence of selective pressure and implies that they belong to the same incompatibility group (28). This is clearly not the case with pRAS3 plasmids and pTC-FC2 and pTC-F14, as the strong incompatibility observed would imply that they belong to the same incompatibility group. Strong incompatibility appeared to be due to a phenomenon associated with orf3 that is still not fully understood. If, however, the region containing orf3 was interrupted, then the replicon of pRAS3.1 was compatible with pTF-FC2 and pTC-F14. This suggests that the pRAS3 plasmids form a third, γ plasmid incompatibility grouping within the IncQ-2 plasmids. Support for this proposal is that the nucleotide sequence of the 22-bp oriV-associated iterons and the protein sequences of the RepC iteron-binding proteins of the pRAS3 plasmids are very different from pTF-FC2 or pTC-F14.
In previous work, plasmid pTF-FC2 was mobilized at saturation frequency by the RP4 conjugative plasmid that had been integrated into the chromosome of E. coli S17.1, while pTC-F14 was mobilized at a frequency 3,500-fold lower (40). This difference in mobilization frequencies was due to differences in the MobD and MobE proteins, because when the genes for these proteins from pTF-FC2 were provided in trans, the mobilization frequency of pTC-F14 was raised to near saturation levels. The hypothesis for the differences in mobilization frequency was that pTF-FC2 and pTC-F14 had been adapted for efficient mobilization by two different conjugative plasmids and that the very different amino acid sequences of mobCDE gene products between the two plasmids were responsible for this. The pRAS3 plasmids have mobCDE genes that are closely related to those of pTF-FC2 (Fig. 1) and appeared to be more efficiently mobilized by RP4 than even pTF-FC2 was. Mating of the pRAS3 plasmids was so efficient that the mating time had to be shortened from 60 to 30 min and the donor-to-recipient ratio decreased from 1:10 to 1:100 to prevent mating from reaching saturation so that the mating frequency could be calculated. The finding that the pRAS3 plasmids could mobilize the oriTs of pTF-FC2 and pTC-F14 even though they have different sequences from the pRAS3 plasmids (Fig. 3) is interesting in the context of the work of Meyer (27). He showed that the IncQ-1 plasmid R1162 can initiate transfer from a 19-bp locus that is partly degenerate in sequence and that such sites are likely to occur by chance in a bacterial chromosome. R1162-dependent transfer of chromosomal DNA from such a potential oriT was demonstrated, and it was pointed out that this might indicate a previously unrecognized potential for the exchange of bacterial DNA. The relaxed nature of the relaxase and oriT interaction of the pRAS3 plasmids suggests that these plasmids also have the potential to mobilize chromosomal DNA from cryptic oriT-like sequences. The observations that the nucleotide sequence of the oriT of the pRAS3 plasmids is 25% more identical to pTC-F14 but that the pRAS3 plasmids do not complement the mobilization of pTC-F14 as well as pTF-FC2 suggest that conservation of specific base pairs within the oriT probably affects the mobilization frequency more than the general level of oriT sequence identity.
IncQ family plasmids are highly promiscuous, and related plasmids have been found in very different environments. An illustration of this was obtained while searching for genes with homology to those present on the pRAS3 plasmids. A genomic island containing tetracycline resistance genes has been isolated from seven isolates of the obligate intracellular human and animal pathogen Chlamydia suis (11). The tetracycline resistance genes were associated with a plasmid that is almost identical to pRAS3.2. Approximately 10.1 kb of plasmid-like DNA is present, the nucleotide sequence of which is 99% identical to pRAS3.2 and contains large stretches of perfect identity. The biggest difference is that a 1.7-kb fragment containing the mobA/repB gene fusion extending 64 bp into the antitoxin gene is missing. Although the toxin gene remains, this is not likely to be expressed, as its promoter lies upstream of the antitoxin gene and within the missing region. There is an 8-bp deletion in the region of the operator of the divergent tetA and tetR genes as well as a deletion in the oriV region, such that only one 22-bp iteron remains. The presence of pRAS3-like plasmids in an obligately intracellular parasite such as Chlamydia suis illustrates the remarkable promiscuity of IncQ family plasmids and their ability to participate in the horizontal gene pool.
We began this work as part of a study aimed at answering the question of why two versions of what are two highly similar plasmids existed. Although this question has not yet been fully addressed, we have established that the effect of the additional 6 bp upstream of the mobB-mobA/repB operon in pRAS3.1 was to raise plasmid copy number while the effect of the additional 22-bp oriV-associated iteron was to partly reduce the copy number (though this reduction was not down to the level of pRAS3.2). During evolution, plasmid variants would be expected to compete for space in the cytoplasm of the host but in such a way that they do not place an evolutionary significant additional metabolic burden on their host cells. For plasmids that do not have an active partitioning mechanism (such as the pRAS3 plasmids), one would predict that the ideal plasmid copy number would be a compromise between a copy number high enough to minimize plasmid loss that might occur on host cell division but not one so high that the plasmid-associated metabolic burden makes the host noncompetitive. In addition, there may also be selection for a plasmid to evolve in such a way that it is not easily displaced by its sister variant. Future studies will include the construction of a number of plasmid variants that will enable us to address this question.
Acknowledgments
We thank Trine L'Abée-Lund for the gift of plasmids pRAS3.1 and pRAS3.2.
This work was supported by grants from the National Research Foundation (Pretoria, South Africa) and BHP Billiton Johannesburg Technology Centre (Randburg, South Africa).
Footnotes
Published ahead of print on 14 August 2009.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1993. Current protocols in molecular biology. Wiley Interscience, New York, NY.
- 3.Becker, E. C., and R. J. Meyer. 2003. Relaxed specificity of the R1162 nickase: a model for evolution of a system for conjugative mobilization of plasmids. J. Bacteriol. 185:3538-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 5.Bravo, A., G. de Torrontegui, and R. Diaz. 1987. Identification of components of a new stability sysyem of plasmid R1, ParD, that is close to the origin of replication of this plasmid Mol. Gen. Genet. 210:101-110. [DOI] [PubMed] [Google Scholar]
- 6.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chattoraj, D. 2000. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol. Microbiol. 37:467-476. [DOI] [PubMed] [Google Scholar]
- 8.Deane, S. M., and D. E. Rawlings. 2004. Interaction between the plasmid addiction stability systems of two related broad-host-range plasmids. J. Bacteriol. 186:2123-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorrington, R. A., and D. E. Rawlings. 1989. Identification and sequence of the basic replication region of a broad-host-range plasmid isolated from Thiobacillus ferrooxidans. J. Bacteriol. 171:2735-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dugan, J., D. D. Rockey, L. Jones, and A. A. Andersen. 2004. Tetracycline resistance in Chlamydia suis mediated by genomic islands inserted into the chlamydial inv-like gene. Antimicrob. Agents Chemother. 48:3989-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fekete, R., and L. Frost. 2000. Mobilization of chimeric oriT plasmids by F and R100-01: role of relaxosome formation in defining plasmid specificity. J. Bacteriol. 182:4022-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner, M. N., S. M. Deane, and D. E. Rawlings. 2001. Isolation of a new broad-host-range IncQ-like plasmid, pTC-F14, from the acidophilic bacterium Acidithiobacillus caldus and analysis of the plasmid replicon. J. Bacteriol. 183:3303-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardner, M. N., and D. E. Rawlings. 2004. Evolution of compatible replicons of the related IncQ-like plasmids, pTC-F14 and pTF-FC2. Microbiology 150:1797-1808. [DOI] [PubMed] [Google Scholar]
- 15.Gerdes, K., J. E. Larsen, and S. Molin. 1985. Stable inheritance of plasmid R1 requires two different loci. J. Bacteriol. 161:292-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerry, P., J. Van Embden, and S. Falkow. 1974. Molecular nature of two nonconjugative plasmids carrying drug resistance genes. J. Bacteriol. 117:619-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guzman, L., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haines, A. S., K. Jones, M. Cheung, and C. M. Thomas. 2005. The IncP-6 plasmid Rms149 consists of a small mobilizable backbone with multiple large insertions. J. Bacteriol. 187:4728-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haring, V., P. Scholtz, E. Scherzinger, J. Frey, K. Derbyshire, G. Hatfull, N. S. Willetts, and M. Bagdasarian. 1985. Protein RepC is involved in copy number control of the broad host range plasmid RSF1010. Proc. Natl. Acad. Sci. USA 82:60690-60694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jandle, S., and R. Meyer. 2006. Stringent and relaxed recognition of oriT by related systems for plasmid mobilization: implications for horizontal gene transfer. J. Bacteriol. 188:499-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.L'Abée-Lund, T., and H. Sørum. 2002. A global non-conjugative Tet C plasmid, pRAS3, from Aeromonas salmonicida. Plasmid 47:172-181. [DOI] [PubMed] [Google Scholar]
- 22.Lee, C., J. Kim, S. G. Shin, and S. Hwang. 2006. Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J. Biotechnol. 123:273-280. [DOI] [PubMed] [Google Scholar]
- 23.Lin, L.-S., Y.-J. Kim, and R. J. Meyer. 1987. The 20 bp, directly repeated DNA sequence of broad host range plasmid R1162 exerts incompatibility in vivo and inhibits R1162 DNA replication in vitro. Mol. Gen. Genet. 208:390-397. [DOI] [PubMed] [Google Scholar]
- 24.Masuda, Y., K. Miyakawa, Y. Nishimura, and E. Ohtsubo. 1993. chpA and chpB, Escherichia coli homologs of the pem locus responsible for stable maintenance of plasmid R100. J. Bacteriol. 175:6850-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matcher, G. M., and D. E. Rawlings. 2009. The effect of the location of the proteic post-segregational stability system within the replicon of plasmid pTF-FC2 on the fine regulation of plasmid replication. Plasmid 62:98-107. [DOI] [PubMed] [Google Scholar]
- 26.Matson, S. W., and B. Morton. 1991. Escherichia coli DNA helicase I catalyzes a site- and strand-specific nicking reaction at the F plasmid oriT. J. Biol. Chem. 226:16232-16237. [PubMed] [Google Scholar]
- 27.Meyer, R. 2009. The R1162 mob proteins can promote conjugative transfer from cryptic origins in the bacterial chromosome. J. Bacteriol. 191:1574-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novick, R. P. 1987. Plasmid incompatibility. Microbiol. Rev. 51:381-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pansegrau, W., G. Ziegelin, and E. Lanka. 1988. The origin of conjugative IncP plasmid transfer: interaction of plasmid encoded products and the nucleotide sequence at the relaxation site. Biochim. Biophys. Acta 951:365-374. [DOI] [PubMed] [Google Scholar]
- 30.Pfaffl, M. W., G. W. Horgan, and L. Dempfle. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rawlings, D. E., and E. Tietze. 2001. Comparative biology of IncQ and IncQ-like plasmids. Microbiol. Mol. Biol. Rev. 65:481-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rawlings, D. E., I. M. Pretorius, and D. R. Woods. 1984. Expression of a Thiobacillus ferrooxidans origin of replication in Escherichia coli. J. Bacteriol. 158:737-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-790. [Google Scholar]
- 35.Smalla, K., H. Heuer, A. Götz, D. Niemeyer, E. Krögerrechlenfort, and E. Tietze. 2000. Exogenous isolation of antibiotic resistance plasmids from piggery manure slurries reveals a high prevalence and diversity of IncQ-like plasmids. Appl. Environ. Microbiol. 66:4854-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, A. G. S., and D. E. Rawlings. 1997. The poison-antidote system of the broad host-range Thiobacillus ferroxidans plasmid pTF-FC2. Mol. Microbiol. 25:961-970. [DOI] [PubMed] [Google Scholar]
- 37.Tietze, E., H. Tschape, and W. Voigt. 1989. Characterization of new resistance plasmids belonging to incompatibility group Q. J. Basic Microbiol. 10:695-706. [DOI] [PubMed] [Google Scholar]
- 38.Tsuchimoto, S., H. Ohtsubo, and E. Ohtsubo. 1988. Two genes, pemK and pemI, responsible for stable maintenance of the resistance plasmid R100. J. Bacteriol. 170:1461-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsutsui, H., A. Fujiyama, T. Murotsu, and K. Matsabara. 1983. Role of nine repeating sequences of the mini-F genome for expression of F-specific incompatibility phenotype and copy number control. J. Bacteriol. 155:337-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Zyl, L. J., S. M. Deane, and D. E. Rawlings. 2003. Analysis of the mobilization region of the broad host-range IncQ-like plasmid pTC-F14 and its ability to interact with a related plasmid, pTF-FC2. J. Bacteriol. 185:6104-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequence of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]