Abstract
The opportunistic pathogen Pseudomonas aeruginosa causes a variety of infections in immunocompromised individuals, including individuals with the heritable disease cystic fibrosis. Like the carbon sources metabolized by many disease-causing bacteria, the carbon sources metabolized by P. aeruginosa at the host infection site are unknown. We recently reported that l-alanine is a preferred carbon source for P. aeruginosa and that two genes potentially involved in alanine catabolism (dadA and dadX) are induced during in vivo growth in the rat peritoneum and during in vitro growth in sputum (mucus) collected from the lungs of individuals with cystic fibrosis. The goals of this study were to characterize factors required for alanine catabolism in P. aeruginosa and to assess the importance of these factors for in vivo growth. Our results reveal that dadA and dadX are arranged in an operon and are required for catabolism of l-alanine. The dad operon is inducible by l-alanine, d-alanine, and l-valine, and induction is dependent on the transcriptional regulator Lrp. Finally, we show that a mutant unable to catabolize dl-alanine displays decreased competitiveness in a rat lung model of infection.
A hallmark of successful bacterial pathogens is their ability to replicate within their hosts, where they not only must acquire nutrients for growth but also often compete with commensal microorganisms. Although this basic tenet of bacterial pathogenesis, which was espoused originally by Louis Pasteur in the late nineteenth century (21) and more recently by E. D. Garber and other workers (3, 6), has been recognized for some time, it has generally been overlooked, and the metabolic pathways critical for proliferation in most infection sites are unknown. Basic knowledge regarding metabolic processes utilized by infecting bacteria is of fundamental importance for understanding bacterial pathogenesis and may offer opportunities for development of novel therapeutics. Indeed, interfering with bacterial metabolism in vivo has been efficacious for inhibiting the pathogenesis of several bacterial pathogens. For example, mutants of the poultry pathogen Campylobacter jejuni that are unable to catabolize l-serine display markedly reduced colonization of the chick gut (32). Likewise, loss of threonine utilization by the pulmonary pathogen Legionella pneumophila prevents replication in alveolar macrophages (26).
The gram-negative opportunistic pathogen Pseudomonas aeruginosa is a leading cause of ophthalmic, burn wound, and nosocomial infections and causes chronic pulmonary infections in individuals with cystic fibrosis (CF) (9). In addition, P. aeruginosa colonizes numerous environments outside the host, and its ability to catabolize a wide array of carbon sources likely allows proliferation in these diverse environments. From a host-pathogen perspective, the carbon sources available for growth significantly affect production of extracellular virulence factors and biofilm formation in P. aeruginosa (19, 20, 29). Despite these findings, we have little insight into the molecular mechanism of carbon preference or into the effects that specific carbon sources have on host colonization and proliferation within infection sites.
Previous work in our laboratory determined that P. aeruginosa preferentially catabolizes l-alanine over other carbon sources, including most amino acids, lactic acid, and glucose (19). Interestingly, our laboratory also reported that the mRNA levels of two genes putatively involved in alanine catabolism, dadA and dadX, are highly elevated in a peritoneal rat infection model (17) and during in vitro growth in sputum collected from adult CF lungs (20). Based on the preference for alanine and the induction of genes predicted to be involved in alanine catabolism under these two conditions, we hypothesized that alanine may be an important in vivo carbon source for P. aeruginosa. The goals of the present study were to determine the factors that are critical for P. aeruginosa catabolism of alanine and to assess the importance of alanine catabolism for colonization and proliferation in an in vivo model of infection.
MATERIALS AND METHODS
Bacterial strains and media.
P. aeruginosa wild-type strain UCBPP-PA14 (PA14) (22) and isogenic dadA, dadX, and lrp transposon insertion mutants were obtained from the MGH-Parabiosys:NHLBI Program for Genomic Applications, and the transposon mutants were verified using PCR as described previously (http://pga.mgh.harvard.edu/cgi-bin/pa14/mutants/retrieve.cgi). P. aeruginosa was cultured on tryptic soy agar (TSA), in tryptic soy broth, in brain heart infusion broth (BHI), in morpholinepropanesulfonic acid (MOPS) minimal medium (20) containing 1× Socransky vitamin solution (30) and 20 mM of a specified carbon source, or in synthetic CF sputum medium (19). Bacterial growth was assessed by monitoring the optical density at 600 nm (OD600). All cultures were incubated at 37°C, and liquid cultures were incubated with shaking at 250 rpm. When necessary, antimicrobial agents were used at the following concentrations: carbenicillin, 300 μg/ml; ampicillin, 100 μg/ml; tetracycline, 25 μg/ml for Escherichia coli and 50 μg/ml for P. aeruginosa; and gentamicin, 100 μg/ml. 5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was used at 50 μg/ml.
DNA and plasmid manipulation.
DNA and plasmids were isolated using standard methods (1). DNA sequencing was performed at the University of Texas at Austin core facility.
RNA methods.
RNA was isolated from P. aeruginosa cultured in MOPS minimal medium containing 20 mM l-alanine and 0.05% yeast extract using RNeasy minicolumns (Qiagen, Santa Clarita, CA). Removal of contaminating DNA (27) and reverse transcription PCR (RT-PCR) (23, 24) were performed as previously described. Primer extension was performed using a fluorescently (6-carboxyfluorescein) labeled primer as previously described (15).
Plasmid construction.
Complementation of P. aeruginosa PA14 mutants was performed using pUCP18 (Table 1), which allows constitutive expression of genes of interest from the lac operon promoter. The lrp, dadA, dadX, and dadAX genes, including their Shine-Dalgarno sequences, were amplified by PCR using an Expand Long Template PCR kit (Roche) from P. aeruginosa PA14 chromosomal DNA using primer combinations listed in Table 2. The resulting amplicons were digested with EcoRI and BamHI and ligated into EcoRI/BamHI-digested pUCP18. Plasmids containing the genes of interest (pLrp, pDadA, pDadX, and pDadAX) were confirmed by restriction digestion and DNA sequencing.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Characteristics | Reference |
|---|---|---|
| E. coli DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U169 deoR [φ80dlac Δ(lacZ)M15] | 25 |
| P. aeruginosa strains | ||
| UCBPP-PA14 | Wild type | 22 |
| lrp mutant | UCBPP-PA14 lrp::Mar2xT7 (Gmr) | 14 |
| dadA mutant | UCBPP-PA14 dadA::Mar2xT7 (Gmr) | 14 |
| dadX mutant | UCBPP-PA14 dadX::Mar2xT7 (Gmr) | 14 |
| PA14_70010 mutant | UCBPP-PA14 PA14_70010::Mar2xT7 (Gmr) | 14 |
| Plasmids | ||
| pUCP18 | E. coli-Pseudomonas Apr shuttle vector | 28 |
| pMP220 | Broad-host-range Tcr transcriptional lacZ fusion vector | 31 |
| pDadAX | pUCP18 with region containing dadA and dadX | This study |
| pDadX | pUCP18 with dadX coding region | This study |
| pLrpRSD | pUCP18 with lrp coding region and modified Shine-Dalgarno sequence | This study |
| pDadA-lacZ | pMP220 with dadA::lacZ transcriptional fusion | This study |
TABLE 2.
Oligonucleotide primer sequences
| Use | Primer | Sequence |
|---|---|---|
| Plasmid construction | ||
| pDadAX | dadAXfor+SD | CTACGAGCTCAAAATAACAAACGTCCGCGGTTC |
| dadAXRevXbaI | TCGGTCTAGATGCCTGCGCAAAGAATTCGGAAAG | |
| pDadX | dadX5′EcoRI | GGGAATTCGCTGAAGAGAGACCCGTCGCC |
| dadX3′BamHI | CGCGGATCCTCAAGCCCCGGAATAGACGCGCGGC | |
| pLrpRSD | Lrp Random SD | GGGAATTCCCAGACNNNNNNCCTCCATCCATGCGTACC |
| 3′lrpBamHI | CGGGATCCTCAATCCGGAACCGGTAGGTCGAGCG | |
| pDadA-lacZ | dadAprom-for | GGGGTACCCGCGAAGGCTCCGCGAGCGCTC |
| dadAprom-rev | GCTCTAGAACGTCGCGGAGGAACCGCGGACG | |
| Primer extension of dadA | CCAAGGACCAGAACTCGCATTG |
pDadA-lacZ was constructed by amplification of the dadA promoter using primers listed in Table 2. The amplified promoter was digested with EcoRI/KpnI and ligated into EcoRI/KpnI-digested pMP220 (31). pMP220 possesses a promoterless lacZ gene with an intact Shine-Dalgarno sequence; thus, ligation of promoters into this vector allows construction of transcriptional fusions of the promoter of interest to lacZ. The resulting plasmid was confirmed by DNA sequencing.
To randomize levels of the leucine-responsive regulatory protein (Lrp) produced from pLrp, pLrpRSD was constructed and selected as follows. In addition to the EcoRI site, the 5′ lrp primer (Table 2) incorporated a six-base randomized sequence (N6) in place of the native lrp Shine-Dalgarno sequence (GGGAGC). Following PCR amplification and ligation into pUCP18, plasmids with randomized lrp Shine-Dalgarno sequences were transformed via electroporation into a P. aeruginosa lrp mutant carrying pDadA-lacZ. Transformants that restored l-alanine-dependent induction of β-galactosidase in pDadA-lacZ were selected, and the DNA sequence of the randomized Shine-Dalgarno sequence was determined.
β-Galactosidase assays.
The β-galactosidase activity of P. aeruginosa carrying pMP220 or pDadA-lacZ was assayed by using the Tropix Galacto-Light Plus chemiluminescent system (Applied Biosystems, Bedford, MA). Strains were grown overnight in synthetic CF sputum medium with 50 μg/ml tetracycline, washed, and subcultured into fresh medium to obtain an OD600 of approximately 0.15. After 2 h of growth, 500 μl of each culture was aliquoted into an Eppendorf tube to which 500 μl Z buffer, 33 μl 0.1% sodium dodecyl sulfate, and 67 μl chloroform were added. After vortexing and centrifugation, assays were performed as described previously (33).
The β-galactosidase activities of a complemented P. aeruginosa lrp mutant carrying pDadA-lacZ and corresponding vector controls were also assayed using the Tropix Galacto-Light Plus system. For these assays, P. aeruginosa strains were grown overnight in MOPS minimal medium with 20 mM succinate and diluted into fresh medium to obtain an OD600 of 0.02. After 4 h of growth, cultures were diluted to obtain an OD600 of 0.15, and individual cultures were then spiked with 5 mM d- or l-alanine or l-valine or with an equal volume of H2O. After 2 h of growth, the β-galactosidase activity was assessed as described above.
BIOLOG analysis.
For BIOLOG analysis, P. aeruginosa wild-type and dadA mutant strains were grown overnight in MOPS minimal medium with 20 mM glucose (17, 20). Cultures were washed twice in MOPS minimal medium (with no carbon source) prewarmed to 37°C, and diluted to obtain an OD600 of 0.005 in 2.4 ml MOPS minimal medium with no carbon source to which 17.6 ml of IF-Oa base, 0.24 ml of a dye mixture, and 3.76 ml of sterile distilled H2O were added. One hundred microliters was added to each well of a BIOLOG PM1 or PM2 plate. The plates were incubated at 37°C, and the absorbance at 595 nm was measured with a Varioskan microplate reader (Thermo Scientific, Rockford, IL) every 24 h for 72 h to monitor substrate oxidation.
In vitro competition assays.
Overnight BHI cultures of wild-type P. aeruginosa or the P. aeruginosa dadA mutant were diluted to obtain an OD600 of 0.02 in fresh BHI and grown for 4 h to mid-exponential phase. Then 5 × 104 CFU of each strain was added to 5 ml of BHI or MOPS minimal medium containing 20 mM l-alanine. At 0 and 24 h, cultures were serially diluted and plated in duplicate onto TSA to determine the total numbers of bacteria (wild-type P. aeruginosa and the dadA mutant) and onto TSA containing 100 μg/ml gentamicin to determine the numbers of P. aeruginosa dadA mutant bacteria. The numbers of P. aeruginosa wild-type cells were calculated by subtracting the numbers of P. aeruginosa dadA mutant bacteria from the total number of bacteria.
In vivo competition assays.
Wild-type P. aeruginosa and the dadA mutant were incorporated into agar beads and introduced into the lungs of male Sprague-Dawley rats as described previously (4, 7, 11-13, 35) in accordance with the requirements of the ethics committee for animal treatment. After 7 days, the lungs were removed from sacrificed rats, and homogenized tissues were plated in triplicate onto Mueller-Hinton agar (which recovers both wild-type and dadA mutant bacteria) and Mueller-Hinton agar containing 100 μg/ml gentamicin (which recovers dadA mutant bacteria) to determine the number of CFU. The in vivo competitive index (CI) was then determined for each animal. The CI was determined by dividing the output ratio of mutant CFU to wild-type CFU by the input ratio of mutant CFU to wild-type CFU (2, 8). The reported CIs were calculated by determining the geometric mean for animals in the same group. Mann-Whitney sum tests were performed using GraphPad Prism 5 software.
RESULTS AND DISCUSSION
Two genes in E. coli, dadA and dadX, encode proteins that are critical for catabolism of l-alanine to pyruvate (Fig. 1A). DadX is an alanine racemase that converts l-alanine to the d-alanine isomer, and DadA is a component of alanine dehydrogenase that oxidatively deaminates d-alanine to pyruvate and ammonia (5). Examination of the P. aeruginosa PA14 genome sequence revealed two genes with high levels of homology to the DadX and DadA genes. PA14_69990 (PA5302 in P. aeruginosa PAO1) putatively encodes a protein with 48% identity (with an E value of <10−82 as determined using BLASTp) to DadX from E. coli K-12, and PA14_70040 (PA5304 in P. aeruginosa PAO1) putatively encodes a protein with 62% identity (with an E value of <10−158 as determined using BLASTp) to DadA from E. coli K-12. It is noteworthy that a P. aeruginosa gene also encodes a second alanine racemase, Alr; however, as in E. coli, alr has a low level of constitutive expression in P. aeruginosa and Alr is used primarily for d-alanine production for cell wall biosynthesis instead of alanine catabolism (34).
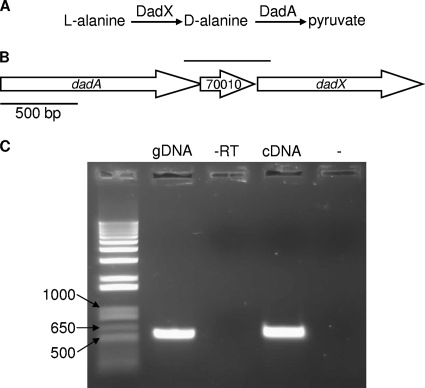
FIG. 1.
Organization and characterization of the dad operon in P. aeruginosa. (A) Reaction scheme for l-alanine and d-alanine degradation. (B) Chromosomal region of the P. aeruginosa dad operon. The dadA, PA14_70010, and dadX genes are indicated by arrows, and the line above the arrows indicates the region amplified in the RT-PCR experiments described below. (C) RT-PCR amplification of the region indicated by the line above the dad operon in panel B. Lane gDNA, P. aeruginosa PA14 chromosomal DNA template positive control; lane −RT, negative control template derived from RT of P. aeruginosa RNA with no reverse transcriptase added; lane cDNA, cDNA template synthesized from P. aeruginosa RNA; lane −, no-template control. The numbers on the left indicate the sizes of standards (in base pairs). The expected length of the amplicon is 566 bp.
Genes involved in specific catabolic pathways are often arranged in operons in bacteria, and this is the case for dadA and dadX in E. coli (34). Examination of the P. aeruginosa PA14 genome sequence revealed that dadA and dadX likely comprise an operon with a third gene, PA14_70010, which encodes a putative endoribonuclease-translation inhibitor (Fig. 1B). To test this prediction, RT-PCR was performed using RNA isolated from P. aeruginosa grown in an alanine-containing medium. The results indicate that, as predicted, an operon encompassing dadA, PA14_70010, and dadX is present in P. aeruginosa (Fig. 1C). This operon structure is supported by microarray data indicating that dadA, dadX, and PA14_70010 are coregulated in PA14 (17, 20) (data not shown).
To assess the role of the dad operon in growth with alanine, dadA, PA14_70010, and dadX transposon mutants were obtained from the randomized, nonredundant library of P. aeruginosa PA14 mutants (14). Mutants in this library contain single-site MAR2xT7 transposon insertions. MAR2xT7 is a mariner transposon that contains aacC1 that is expressed from a constitutive promoter and confers gentamicin resistance. Insertional inactivation of dadA eliminated the ability of P. aeruginosa to grow using l-alanine or d-alanine as a sole source of carbon and energy, while growth on the small acid succinate was not altered (Table 3). As expected, inactivation of dadX also eliminated growth on l-alanine but not growth on d-alanine (Table 3). Growth of both mutants could be restored by addition of the inactivated genes in trans, although complementation of the dadA mutant required expression of dadA and dadX. This was likely due to polar effects of the dadA MAR2xT7 transposon insertion, which inserted into dadA such that the constitutive aacC1 promoter was oriented opposite the direction of dadAX transcription. It is noteworthy that MAR2xT7 insertional inactivation of PA14_70010 had no effect on growth with d- or l-alanine (Table 3).
TABLE 3.
Growth of P. aeruginosa mutants
| P. aeruginosa strain | Growth with carbon sourcesa
|
||
|---|---|---|---|
| Succinate | d-Alanine | l-Alanine | |
| Wild type/pUCP18 | + | + | + |
| dadA mutant/pUCP18 | + | − | − |
| dadA mutant/pDadAX | + | + | + |
| dadX mutant/pUCP18 | + | + | − |
| dadX mutant/pDadX | + | + | + |
| PA14_70010 mutant | + | + | + |
| lrp mutant/pUCP18 | + | − | − |
| lrp mutant/pLrpRSD | + | + | + |
Growth was assessed at 48 h. +, growth greater than that of the no-carbon control.
Amino acid dehydrogenases can potentially affect growth on a variety of amino acids other than the primary substrate. To test the possibility that DadA is important for oxidation of additional amino acids in P. aeruginosa, the abilities of wild-type P. aeruginosa and the dadA mutant to oxidize an array of potential carbon sources were assessed using BIOLOG phenotype microarrays. As expected, the dadA mutant exhibited a reduced ability to oxidize d- and l-alanine, as well as the l-alanine-containing dipeptide l-alanyl glycine and the l-alanine derivative l-alaninamide (Table 4). The dadA mutant also oxidized l-uridine to a lesser extent and showed increased oxidation of l-serine (Table 4). Although oxidation of carbon sources in addition to carbon sources containing alanine was impacted by loss of dadA, the most severe phenotypes were observed with alanine and alanine derivatives.
TABLE 4.
Carbon source oxidation by wild-type P. aeruginosa and the dadA mutant
| Carbon source | Oxidation (A595)a
|
|
|---|---|---|
| Wild type | dadA mutant | |
| d-Alanine | 0.38, 0.35 | 0.08, 0.09 |
| l-Alanine | 1.16, 1.20 | 0.09, 0.08 |
| l-Alanyl glycine | 0.60, 0.67 | 0.12, 0.09 |
| l-Alaninamide | 0.61, 0.54 | 0.09, 0.10 |
| l-Serine | 0.15, 0.16 | 0.63, 0.47 |
| l-Uridine | 0.56, 0.57 | 0.13, 0.12 |
Carbon source oxidation in BIOLOG phenotype microarrays was determined by measuring the A595 at 48 h postinoculation. The two numbers in each cell are values from two independent experiments. Only substrates for which there was at least a threefold change in each experiment are shown.
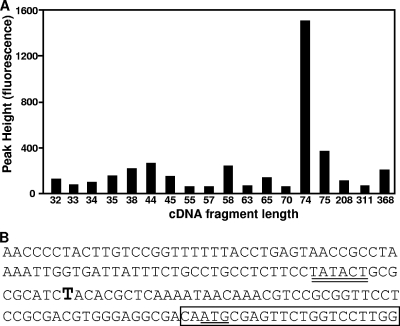
As dadA and dadX in P. aeruginosa are induced during growth in a rat peritoneal model and in sputum collected from CF lungs (17, 20), we were interested in defining and characterizing the promoter region controlling transcription of these genes. To identify the promoter region, primer extension was used to map the transcriptional start site upstream of dadA. A prominent 74-bp cDNA was detected (Fig. 2A) that mapped the transcriptional start site to the thymidine located 54 bases upstream of the dadA translational start site (Fig. 2B). Examination of the promoter region revealed a putative −10 DNA sequence (TATACT) but no clear −35 sequence.
FIG. 2.
Identification of the P. aeruginosa dad operon promoter by primer extension analysis. Primer extension was performed using a fluorescently (6-carboxyfluorescein) labeled primer as previously described (15). (A) Sizes of primer extension cDNA fragments detected and fluorescence values for the fragments. The fluorescence value indicates the relative quantity of the cDNA product. (B) Promoter region of the dad operon based on primer extension data. The transcriptional start site is indicated by bold type, the dadA translational site is underlined, the putative −10 is double underlined, and the target sequence of the primer extension oligonucleotide primer is enclosed in a box.
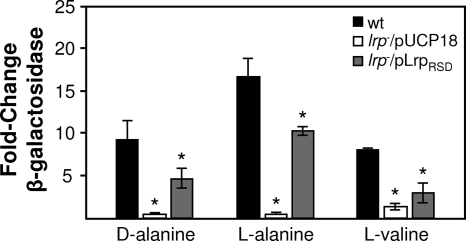
In E. coli, dadAX is inducible by leucine as well as d- and l-alanine (16). To examine regulation of the dad genes in P. aeruginosa, the promoter region of dadAX was ligated into the lacZ transcriptional fusion vector pMP220 (31) to create pDadA-lacZ. pDadA-lacZ was transformed into P. aeruginosa, and the β-galactosidase activity was monitored in the presence and absence of potential amino acid inducers. Addition of l-alanine, d-alanine, and l-valine resulted in significant increases in dad operon transcription (Fig. 3) while all other amino acids, including leucine, had no significant effect on dad transcription (data not shown).
FIG. 3.
P. aeruginosa dad operon is inducible by d-alanine, l-alanine, and l-valine and is controlled by Lrp. The β-galactosidase activities of wild-type P. aeruginosa (wt), the lrp mutant with the complementation vector (lrp−/pUCP18), and the genetically complemented lrp mutant (lrp−/pLrpRSD) carrying pDadA-lacZ were assessed in the presence and absence of d-alanine, l-alanine, and l-valine. The changes were determined by dividing the average β-galactosidase values for pDadA-lacZ with the amino acid inducer by the average β-galactosidase values without the amino acid inducer. The values are the averages of two separate experiments performed in triplicate, and the error bars indicate standard deviations. *, P < 0.05, as determined by Student's t test.
In E. coli, Lrp is a transcriptional regulator of the dad operon (10, 18, 36, 37). E. coli Lrp is a global regulator of the feast/famine regulatory protein family that binds DNA either specifically or nonspecifically, alone or with other transcriptional regulators (such as the cyclic AMP receptor protein). Lrp both transcriptionally activates and represses dadAX in E. coli through binding of activating sites and removal from repressor sites in the presence of the inducers d-alanine, l-alanine, and leucine (37). Examination of the P. aeruginosa genome sequence revealed the presence of eight putative feast/famine regulatory protein family members. One of these proteins, encoded by PA14_70080 (PA5308 in P. aeruginosa PAO1), exhibits 48% identity (with an E value of <10−82 as determined using BLASTp) to Lrp from E. coli K-12 and is located approximately 3.6 kb upstream of the dad operon on the P. aeruginosa PA14 chromosome. To determine whether Lrp is a transcriptional regulator of the dad operon in P. aeruginosa, a P. aeruginosa PA14 MAR2xT7 lrp insertion mutant was obtained from the PA14NR library and transformed with the dadA::lacZ reporter plasmid, pDadA-lacZ, and β-galactosidase activity was monitored in the presence and absence of the dadA inducers d-alanine, l-alanine, and l-valine. Insertional inactivation of lrp significantly reduced β-galactosidase activity in the presence of all three inducers, and induction could be significantly restored by expression of lrp in trans (Fig. 2). These results, correlated with Genechips microarray experiments (data not shown), reveal that, in contrast to the situation in E. coli, Lrp is an activator of dad operon transcription in P. aeruginosa and d-alanine, l-alanine, and l-valine are coinducers. As expected, the lrp mutant was also defective for growth with d- or l-alanine, and growth could be restored by addition of lrp in trans (Table 3). It should be noted that for lrp complementation, lrp containing a modified Shine-Dalgarno sequence (modified from the native sequence GGGAGC to TCCCTC) was used to reduce expression from the multicopy pUCP18-derived complementation plasmid.
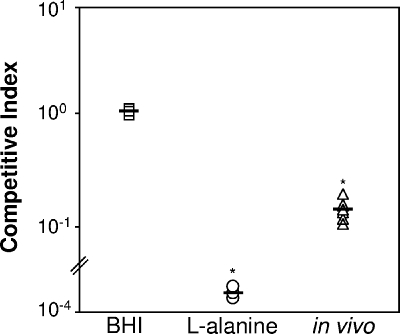
To determine if the ability to catabolize alanine was important for P. aeruginosa proliferation in vivo, we assessed the ability of the dadA mutant to compete with wild-type P. aeruginosa in a rat model of infection. The model utilized was an agar bead lung infection model in which agar beads containing both wild-type P. aeruginosa and the dadA mutant were introduced into the lungs of male Sprague-Dawley rats. This model has been used extensively to examine the ability of P. aeruginosa to proliferate in the rat lung (7, 11-13, 35). Experiments examining competition between wild-type P. aeruginosa and the dadA mutant were also performed in vitro to determine if the dadA mutant has a generalized competition defect. As expected, the dadA mutant exhibited no competitive disadvantage when it was cultured in a complex laboratory medium (BHI agar), but a 3- to 4-log competitive disadvantage was observed for the dadA mutant when it was grown in MOPS minimal medium with l-alanine as the primary energy source (Fig. 4). Interestingly, the dadA mutant exhibited a >3-fold reduction in competitive fitness compared to wild-type P. aeruginosa in the rat lung (Fig. 4).
FIG. 4.
In vivo competition and in vitro competition of wild-type P. aeruginosa and the dadA mutant. Equal numbers of the wild type and the dadA mutant were combined and used to inoculate BHI or MOPS minimal medium containing l-alanine and a vitamin solution (30) or were incorporated into agar beads and introduced into the lungs of male Sprague-Dawley rats (in vivo). Bacteria were quantified for starting inocula at the onset of competition, after 24 h of coculture in vitro, and for homogenized lung tissue 7 days postinfection by dilution plating onto growth media with and without gentamicin to determine the total numbers of bacteria (growth with no gentamicin) and the numbers of dadA mutant bacteria (growth with gentamicin). The ratio of the mutant to the wild type after competition was divided by the ratio of the mutant to the wild type at the onset of competition to determine the CI. Average CIs are indicated by horizontal lines (n = 3 in vitro and n = 7 in vivo). *, P < 0.02, as determined by the Mann-Whitney sum test.
An important goal of this study was to test the hypothesis that alanine is an important carbon source for P. aeruginosa during in vivo growth. This hypothesis was based on the observation that alanine is a preferred in vitro carbon source in P. aeruginosa (19) and on the observation that genes involved in alanine catabolism are induced during growth in a peritoneal rat model of infection and in sputum collected from the lungs of individuals with CF (17, 19, 20). Although the dadA mutant displayed a relatively modest reduction (3-fold) in competitive fitness in the agar bead model, it is interesting that despite its ability to grow with a wide array of carbon sources, loss of a single catabolic enzyme involved in alanine utilization reduced the competitive fitness of P. aeruginosa in vivo. These results support the idea that even in metabolically diverse bacteria, such as P. aeruginosa, inactivation of specific carbon catabolic pathways can impact in vivo proliferation.
Acknowledgments
This work was funded by a grant from the Cystic Fibrosis Foundation (to M.W.), by the National Science Foundation (award 0650600 to M.W.), and by a minority supplement to NSF award 0650600 (to A.L. and K.B.). P.J.B. was supported by a Research Opportunity Award, and M.L.B. was supported by a postdoctoral fellowship from the Cystic Fibrosis Foundation. M.W. is a Burroughs Wellcome Investigator in the Pathogenesis of Infectious Disease.
Footnotes
Published ahead of print on 7 August 2009.
REFERENCES
- 1.Ausubel, F. M. 2002. Short protocols in molecular biology: a compendium of methods from Current Protocols in Molecular Biology, 5th ed. Wiley, New York, NY.
- 2.Beuzon, C. R., and D. W. Holden. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 3:1345-1352. [DOI] [PubMed] [Google Scholar]
- 3.Brown, S. A., K. L. Palmer, and M. Whiteley. 2008. Revisiting the host as a growth medium. Nat. Rev. Microbiol. 6:657-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cash, H. A., D. E. Woods, B. McCullough, W. G. Johanson, Jr., and J. A. Bass. 1979. A rat model of chronic respiratory infection with Pseudomonas aeruginosa. Am. Rev. Respir. Dis. 119:453-459. [DOI] [PubMed] [Google Scholar]
- 5.Franklin, F. C., and W. A. Venables. 1976. Biochemical, genetic, and regulatory studies of alanine catabolism in Escherichia coli K12. Mol. Gen. Genet. 149:229-237. [DOI] [PubMed] [Google Scholar]
- 6.Garber, E. D. 1960. The host as a growth medium. Ann. N. Y. Acad. Sci. 88:1187-1194. [DOI] [PubMed] [Google Scholar]
- 7.Gooderham, W. J., S. L. Gellatly, F. Sanschagrin, J. B. McPhee, M. Bains, C. Cosseau, R. C. Levesque, and R. E. Hancock. 2009. The sensor kinase PhoQ mediates virulence in Pseudomonas aeruginosa. Microbiology 155:699-711. [DOI] [PubMed] [Google Scholar]
- 8.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 45:1389-1406. [PMC free article] [PubMed] [Google Scholar]
- 9.Hoiby, N. 1998. Pseudomonas in cystic fibrosis: past, present, and future. Cystic Fibrosis Trust, London, United Kingdom.
- 10.Janes, B. K., and R. A. Bender. 1998. Alanine catabolism in Klebsiella aerogenes: molecular characterization of the dadAB operon and its regulation by the nitrogen assimilation control protein. J. Bacteriol. 180:563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kukavica-Ibrulj, I., A. Bragonzi, M. Paroni, C. Winstanley, F. Sanschagrin, G. A. O'Toole, and R. C. Levesque. 2008. In vivo growth of Pseudomonas aeruginosa strains PAO1 and PA14 and the hypervirulent strain LESB58 in a rat model of chronic lung infection. J. Bacteriol. 190:2804-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kukavica-Ibrulj, I., and R. C. Levesque. 2008. Animal models of chronic lung infection with Pseudomonas aeruginosa: useful tools for cystic fibrosis studies. Lab. Anim. 42:389-412. [DOI] [PubMed] [Google Scholar]
- 13.Kukavica-Ibrulj, I., F. Sanschagrin, A. Peterson, M. Whiteley, B. Boyle, J. Mackay, and R. C. Levesque. 2008. Functional genomics of PycR, a LysR family transcriptional regulator essential for maintenance of Pseudomonas aeruginosa in the rat lung. Microbiology 154:2106-2118. [DOI] [PubMed] [Google Scholar]
- 14.Liberati, N. T., J. M. Urbach, S. Miyata, D. G. Lee, E. Drenkard, G. Wu, J. Villanueva, T. Wei, and F. M. Ausubel. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 103:2833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lloyd, A. L., B. J. Marshall, and B. J. Mee. 2005. Identifying cloned Helicobacter pylori promoters by primer extension using a FAM-labelled primer and GeneScan analysis. J. Microbiol. Methods 60:291-298. [DOI] [PubMed] [Google Scholar]
- 16.Lobocka, M., J. Hennig, J. Wild, and T. Klopotowski. 1994. Organization and expression of the Escherichia coli K-12 dad operon encoding the smaller subunit of d-amino acid dehydrogenase and the catabolic alanine racemase. J. Bacteriol. 176:1500-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mashburn, L. M., A. M. Jett, D. R. Akins, and M. Whiteley. 2005. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J. Bacteriol. 187:554-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathew, E., J. Zhi, and M. Freundlich. 1996. Lrp is a direct repressor of the dad operon in Escherichia coli. J. Bacteriol. 178:7234-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palmer, K. L., L. M. Aye, and M. Whiteley. 2007. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol. 189:8079-8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer, K. L., L. M. Mashburn, P. K. Singh, and M. Whiteley. 2005. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J. Bacteriol. 187:5267-5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pasteur, L. 1881. On the germ theory. Science 2:420-422. [DOI] [PubMed] [Google Scholar]
- 22.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 23.Ramsey, M. M., and M. Whiteley. 2009. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc. Natl. Acad. Sci. USA 106:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramsey, M. M., and M. Whiteley. 2004. Pseudomonas aeruginosa attachment and biofilm development in dynamic environments. Mol. Microbiol. 53:1075-1087. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russell (ed.). 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Sauer, J. D., M. A. Bachman, and M. S. Swanson. 2005. The phagosomal transporter A couples threonine acquisition to differentiation and replication of Legionella pneumophila in macrophages. Proc. Natl. Acad. Sci. USA 102:9924-9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109-121. [DOI] [PubMed] [Google Scholar]
- 29.Shrout, J. D., D. L. Chopp, C. L. Just, M. Hentzer, M. Givskov, and M. R. Parsek. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62:1264-1277. [DOI] [PubMed] [Google Scholar]
- 30.Socransky, S. S., J. L. Dzink, and C. M. Smith. 1985. Chemically defined medium for oral microorganisms. J. Clin. Microbiol. 22:303-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spaink, H. P., R. J. H. Okker, C. A. Wijffelman, E. Pees, and B. J. J. Lugtenberg. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 9:27-39. [DOI] [PubMed] [Google Scholar]
- 32.Velayudhan, J., M. A. Jones, P. A. Barrow, and D. J. Kelly. 2004. l-Serine catabolism via an oxygen-labile l-serine dehydratase is essential for colonization of the avian gut by Campylobacter jejuni. Infect. Immun. 72:260-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wild, J., J. Hennig, M. Lobocka, W. Walczak, and T. Klopotowski. 1985. Identification of the dadX gene coding for the predominant isozyme of alanine racemase in Escherichia coli K12. Mol. Gen. Genet. 198:315-322. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, S., Y. Chen, E. Potvin, F. Sanschagrin, R. C. Levesque, F. X. McCormack, and G. W. Lau. 2005. Comparative signature-tagged mutagenesis identifies Pseudomonas factors conferring resistance to the pulmonary collectin SP-A. PLoS Pathog. 1:259-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhi, J., E. Mathew, and M. Freundlich. 1998. In vitro and in vivo characterization of three major dadAX promoters in Escherichia coli that are regulated by cyclic AMP-CRP and Lrp. Mol. Gen. Genet. 258:442-447. [DOI] [PubMed] [Google Scholar]
- 37.Zhi, J., E. Mathew, and M. Freundlich. 1999. Lrp binds to two regions in the dadAX promoter region of Escherichia coli to repress and activate transcription directly. Mol. Microbiol. 32:29-40. [DOI] [PubMed] [Google Scholar]