Abstract
Growth on N-acetylglucosamine (GlcNAc) produces intracellular N-acetylglucosamine-6-phosphate (GlcNAc6P), which affects the regulation of the catabolism of amino sugars in Escherichia coli in two ways. First, GlcNAc6P is the inducing signal for the NagC repressor, and thus it increases the expression of the enzymes of the nagE-nagBACD operon. Second, it is the allosteric activator of glucosamine-6P (GlcN6P) deaminase, NagB, and thus increases the catalytic capacity of this key enzyme in the metabolism of amino sugars. We showed previously that both the level of expression of the nagB gene and the transport of glucosamine were limiting the growth rate on GlcN (L. I. Álvarez-Añorve et al., J. Bacteriol. 187:2974-2982, 2005). We were unable to conclude if the lack of allosteric activation of wild-type NagB was also contributing to the slower growth rate on GlcN. Using a single-copy plasmid, with a constitutive promoter, we have separated the effects of GlcNAc6P on the NagB protein level and on deaminase activity. We show that over a range of intracellular NagB concentrations it is the quantity of the substrate, GlcN6P, which is limiting growth rather than the concentration of the allosteric activator, GlcNAc6P. On the other hand, the F174A mutant of NagB, which requires higher concentrations of GlcNAc6P for activity in vitro, grew better on GlcN in the presence of GlcNAc6P. However, wild-type NagB behaves as if it is already fully allosterically activated during growth on GlcN, and we present evidence suggesting that sufficient GlcNAc6P for allosteric activation is derived from the recycling of peptidoglycan.
Amino sugars are widely distributed in nature and are valuable nutrients to most organisms. The most commonly found amino sugars are glucosamine (GlcN) and N-acetylglucosamine (GlcNAc). Their most abundant source is chitin, the high-molecular-weight polymer composed of 1-4 β-linked GlcNAc residues found in the cell walls of fungi and the exoskeletons of crustaceans and other arthropods. Amino sugars are both carbon and nitrogen sources but are also essential components of subcellular structures like the peptidoglycan (PG) of bacterial cell walls and the high-molecular-weight glycosaminoglycans and glycoproteins of the extracellular matrix of higher eukaryotes. Enteric Escherichia coli bacteria are likely to encounter host-derived amino sugars during their normal life cycle. The amino sugars, once taken up by the bacteria, can be used both to synthesize the PG and lipopolysaccharides of its cell wall and as an energy source.
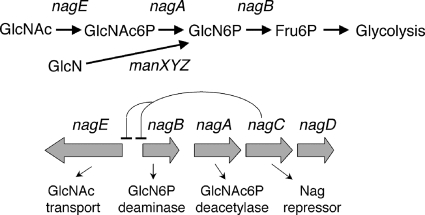
We previously investigated why E. coli grows more slowly on glucosamine than on N-acetylglucosamine. Growth on GlcNAc requires the nagE- or manXYZ-encoded phosphotransferase (PTS) transporters, which produce N-acetylglucosamine-6-phosphate (GlcNAc6P), and the two genes nagA and nagB, which encode GlcNAc6P deacetylase and GlcN6P deaminase, respectively, needed for the metabolism of GlcNAc6P (Fig. 1). Growth on GlcN requires the manXYZ-encoded transporter, producing GlcN6P, and the nagB gene product. We found that growth on GlcN was limited by the quantity of the NagB enzyme, by the amount of the GlcN transporter ManXYZ, and thus by the amount of the substrate for the NagB deaminase, GlcN6P (1). The nagB gene is part of the divergent nagE-nagBACD operon (Fig. 1), and NagB is required for degradation of GlcN6P, whether it is derived from metabolism of GlcNAc6P or by transport of GlcN. NagC is the transcriptional regulator of the operon. The function of NagD, a member of the HAD superfamily, is not known (30), and it is not necessary for the use of GlcNAc or GlcN (25). Expression of the genes nagE and nagBA is strongly induced by growth on GlcNAc but only weakly during growth on GlcN (24).
FIG. 1.
Structure of the divergent nagE-nagBACD operon and function of the encoding genes. GlcNAc enters the cell mostly by the nagE-encoded PTS transporter and is metabolized by the nagA- and nagB-encoded enzymes to fructose-6P. Expression of the genes nagE, nagA, and nagB is repressed by the NagC repressor. GlcN is transported by the manXYZ-encoded PTS transporter and requires just the nagB-encoded GlcN6P deaminase to be converted to Fru6P.
NagB is a homohexameric allosteric enzyme, activated heterotropically by GlcNAc6P or homotropically (due to positive cooperativity) by high concentrations of the substrate, GlcN6P (6). From our previous study, we could not say in the case of the wild-type (wt) enzyme whether the absence of the allosteric activator, GlcNAc6P, during growth on GlcN was also limiting the growth rate. Since GlcNAc6P is both the inducing signal for the NagC repressor of the nag regulon (25) and the allosteric activator of NagB, any increase in the GlcNAc6P concentration should simultaneously increase both the amount of NagB protein (by induction of the operon) and the activity of the NagB enzyme.
To separate these two effects of GlcNAc6P on NagB regulation, we have deleted the nagB gene from its position on the chromosome within the inducible nagE-nagBACD operon and placed it on a single-copy plasmid (R1 replicon) under the control of a constitutive plasmid promoter. We have changed the protein levels of NagB within the cell by altering the translational initiation signals by modifying the Shine and Dalgarno (S&D) sequence and including, or not, a C-terminal stem-loop structure to stabilize the nagB transcript. Using these constructs, we have compared growth on GlcN and GlcNAc and tested the effect of mutations, which increased the concentration of either the substrate (GlcN6P) or the allosteric activator (GlcNAc6P). A mutation in the mlc gene, which controls the manXYZ-encoded transporter, increases transport of GlcN two- to threefold (1, 19, 22) and was found to improve the growth rate for all the strains carrying the different plasmid constructs. Mutations which eliminate the nagA-encoded GlcNAc6P deacetylase produce high levels of the allosteric activator GlcNAc6P (1, 25, 33) derived from the recycling of PG (15, 17) but had no effect on the growth rate. These experiments show that it is the supply of substrate that is limiting the growth rate on GlcN and not the absence of the allosteric activator. In fact, during growth on GlcN, NagB behaves as if it is fully active, and we discuss how this could occur.
MATERIALS AND METHODS
Bacterial methods.
Strains used in this work are listed in Table 1. Plasmids expressing nagB were introduced into LAA20 carrying the nagB::cm replacement (1). The nagA::tc and nagC::tc mutations were introduced using P1 lysates grown on LAA44 and LAA43, which also carry the adjacent nagB::cm. Growth rates were measured in minimal morpholineethanesulfonic acid (MOPS) medium with 10 mM GlcN or GlcNAc as described previously (1). Precultures were grown in minimal MOPS medium with glycerol and diluted 1/100 into the test medium. The plasmid experiments were all carried out at 30°C because the pXE1 plasmid carries a thermosensitive replicon. Doubling times (DT) were calculated by linear regression between optical densities measured at 650 nm of 0.08 and 0.8 and are presented as the means of four to eight independent cultures. One-way analysis of variance tests were used to compare growth rates for each NagB construct grown under the six different conditions of growth shown (see Fig. 3). Student's t test was used for pairwise comparisons of different culture conditions with the significance threshold set at 0.05%. Western blotting was carried out as described previously (1). β-Galactosidase activities were measured on MC4100 lysogenized with a λ carrying a nagB-lacZ fusion (MC-B1). Mutations affecting the recycling of PG (nagK::FRTcm, murQ::FRTkan, anmK::FRTcm, nagE::FRTkan, nagZ::cm, ampG::kan [15, 17]) were sequentially introduced into MC-B1 to give MC-B196/7. Antibiotic resistance cassettes surrounded by Flp recombinase target (FRT) sites were removed by transformation with pCP20 carrying the Flp recombinase (7).
TABLE 1.
Bacterial strains used
| Strain | Description | Source or reference |
|---|---|---|
| MC4100 | araD139 Δ(argF-lac)U169 flb5301 deoC1 relA1 rbsR rpsL150 ptsF25 | Lab stock |
| LAA20 | MC4100 nagB::cm | 1 |
| LAA131 | LAA20 pXE(NagB-L1) | This work |
| LAA134 | LAA20 pXE(NagB-L2) | This work |
| LAA135 | LAA20 pXE(NagB-L3) | This work |
| LAA136 | LAA20 pXE(NagB-L4) | This work |
| LAA137 | MC4100 pXE1 | This work |
| LAA178 | LAA20 pXE/NagB F174A | This work |
| LAA43 | MC4100 nagB::cm nagC::tc | 1 |
| LAA44 | MC4100 nagB::cm nagA::tc | 1 |
| IBPC590 | ΔnagE-nagBACD::tc | 23 |
| IBPC1012 | JM101 mlc::tc | 22 |
| KD622 | MC4100 φ(malK-lacZ)1113 (λplacMu50) mlc::Tn10 kan | 8 |
| MC-B1 | MC4100 λRS nagB-lacZ | 17 |
| MC-B196/7 | MC-B1 nagK murQ anmK nagZ ampG nagE | This work |
FIG. 3.
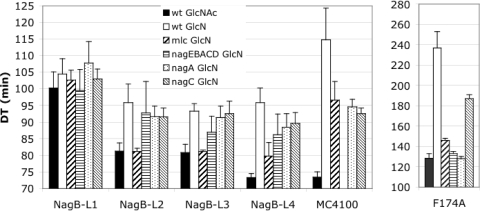
Growth rates of the NagB constructs on GlcN and GlcNAc. LAA20 with pXEI-derived plasmids carrying the NagB-L1, -L2, -L3, and -L4 constructs expressing wt NagB to different levels, NagB-L4 with the F174A mutation, or MC4100 carrying pXE1 was grown on GlcN or GlcNAc. Growth on GlcN was also measured in the presence of the mutations mlc, ΔnagE-nagBACD, nagA, and nagC as indicated. The histogram gives the DT in minutes ± standard deviations and the means of four to eight cultures (note that the scales vary).
Construction of single-copy nagB plasmids.
The nagB gene was amplified using the following pairs of oligonucleotides, Nag70-Nag72 (NagB-L1), Nag71-Nag72 (NagB-L2), Nag70-Nag73 (NagB-L3), and Nag71-Nag73 (NagB-L4) (Table 2), and inserted as XbaI-EcoRI fragments into pXE1 to give pXE(NagB-L1), pXE(NagB-L2), pXE(NagB-L3), and pXE(NagB-L4). pXE1 is a single-copy R1 replicon plasmid in which expression of the cloned gene is from a promoter within the λ DNA-derived region of the plasmid expressing the cI857 repressor (16). The whole insert of each NagB plasmid was sequenced with oligonucleotides XE1 and RBP22 (Table 2).
TABLE 2.
Oligonucleotides used
| Oligonucleotide | Function | Sequencea |
|---|---|---|
| NagB70 | 5′ nagB | GACTTCTAGATTACTTATTGAGGGTGAATAATG |
| NagB71 | 5′ nagB with a high-expression S&D sequence | GACTTCTAGAAAAATAAGGAGGAAATAAAATG AGACTGATCCCCCTGAC |
| NagB72 | 3′ nagB | GACTGAATTCAGGAGCCAGGGCAGGGATAA |
| NagB73 | 3′ nagB with stem-loop structure | GACTGAATTCCGTTTAACTGCACATCGATA |
| XE1 | Sequence inserts pXE1 | GGTTGGCTCCAATTATTTGTATATTC |
| RBP22 | Sequence inserts pXE1 | CCGAAAAGTGCCACCTGACGTC |
Restriction sites used for cloning are underlined.
RESULTS AND DISCUSSION
Plasmids expressing fixed amounts of glucosamine-6P deaminase (NagB).
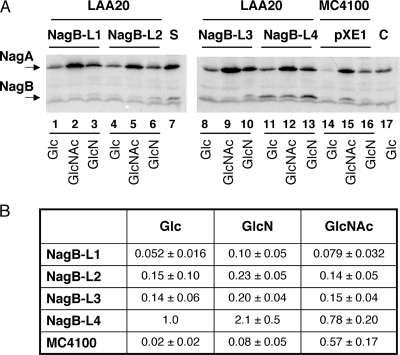
Four constructs were tested expressing various levels of NagB from the pXE1-derived plasmids, called NagB-L1, NagB-L2, NagB-L3, and NagB-L4. NagB-L1 and NagB-L3 have the wt NagB ribosome binding site sequence, while NagB-L2 and NagB-L4 have a high-expression S&D sequence (28). NagB-L3 and NagB-L4 constructs include about 240 bp of DNA downstream of nagB, which carries sequences that can form a stem-loop structure in the transcribed RNA. The 3′ end of the induced nagB transcript was mapped to this region (26), and so it presumably acts as a structure stabilizing the upstream mRNA against 3′ to 5′ exonucleolytic degradation. These plasmids were placed in a strain carrying a deletion of the nagB gene in the chromosome locus (LAA20) (Table 1), and the levels of NagB were tested by Western blotting (Fig. 2). The NagB protein level from the NagB-L4 construct (Fig. 2A, lanes 11 to 13) was about twofold higher than that in MC4100 during growth in GlcNAc (Fig. 2B). The NagB-L1 construct (Fig. 2A, lanes 1 to 3) produces about 5% of the amount of NagB compared to NagB-L4, while the NagB-L2 and -L3 constructs (Fig. 2A, lanes 4 to 6 and 8 to 10) produce two to three times more NagB than NagB-L1 (Fig. 2B). For each plasmid construct, the quantity of NagB in the cell was expected to remain constant during growth on different media, since NagB is expressed from a constitutive plasmid promoter. However, NagB from each construct appeared to be nearly twofold higher in media containing GlcN than in media containing glucose (Glc) or GlcNAc (Fig. 2B). This could be because the plasmid promoter is partially CAP (catabolite gene activator protein) dependent; then expression in GlcNAc and glucose should be lower, since both sugars provoke strong catabolite repression, whereas GlcN does not (9).
FIG. 2.
Western blot of cultures of strains carrying the different NagB-expressing plasmids. (A) LAA20 (ΔnagB::cm) carrying a pXE1-derived plasmid expressing NagB-L1, -L2, -L3, or -L4 or MC4100 carrying the vector plasmid pXE1 was grown in glucose, GlcN, or GlcNAc medium, as indicated, to late exponential phase (A650 = 0.8). Bacteria were chilled, harvested by centrifugation, and lysed by sonication and boiling in sodium dodecyl sulfate sample buffer. Aliquots (0.25 A650 units) were analyzed on 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to Hybond-C membrane, and treated with anti-NagA and anti-NagB as described previously (1). S, standards (100 ng NagA and NagB); C, control LAA20 without any plasmid grown in glucose. Note that NagB is poorly retained on the membranes, resulting in a low signal. The band running just faster than NagB is due to contaminant antibodies reacting with a protein of molecular weight similar to that of NagB and which is equally present in all lanes. (B) NagB protein levels due to each construct and growing on the different media were quantified by using the ImageQuant program of PhosphorImager. For each blot, the levels of NagB were normalized to the level of the NagB protein from the NagB-L4 construct growing in glucose. The numbers are the relative levels ± standard deviations of three or four experiments.
Expression of NagA from the chromosomal nagE-nagBACD operon in MC4100 is induced slightly by growth on GlcN and strongly by growth on GlcNAc (Fig. 2A, lanes 14 to 16). The cm cassette, which has replaced nagB in LAA20, carries a promoter which leads to some expression of the downstream gene nagA during growth on glucose (Fig. 2A, compare NagA levels in glucose from LAA20 in lanes 1, 4, 8, and 11 with that from MC4100 in lane 14). NagA levels are also slightly higher in LAA20 during growth on GlcN or GlcNAc than in MC4100 during growth in the same medium, presumably corresponding to the sum of the expression from the cm promoter and that coming from the GlcN- or GlcNAc-induced nagB promoter. In conclusion, expression of NagA was still induced by growth on GlcNAc, whereas the NagB protein level from the plasmid was essentially insensitive to the composition of the medium and did not increase during growth on GlcNAc.
Growth rates on glucosamine and N-acetylglucosamine with fixed amounts of NagB.
LAA20 (ΔnagB::cm) carrying the four plasmid constructs expressing NagB was grown on Glc, GlcN, and GlcNAc, and growth rates were compared to those of MC4100 carrying the vector plasmid (pXE1). All constructs grew equally on glucose (DT = 70 min ± 2.5 min), whereas the growth rates on GlcN and GlcNAc varied (Fig. 3). The growth pattern of MC4100, using the chromosomally encoding nagB gene and carrying the pXE1 plasmid at 30°C, was similar to that observed previously for MC4100 at 37°C (1). Growth was rather slow on GlcN (DT = 115 min) but almost as fast on GlcNAc (DT = 73 min) as on glucose. Mutations in mlc, nagA, or nagC all improved the growth rate of MC4100 on GlcN (see below).
The growth rates of the different NagB plasmid constructs on GlcN and GlcNAc varied. The NagB-L1 construct grew at similar rates on GlcN and GlcNAc (DT = 105 and 101 min), whereas the NagB-L2, -L3, and -L4 constructs grew somewhat faster on GlcN (DT = 93 to 96 min) and more rapidly on GlcNAc (DT = 81 min for NagB-L2 and -L3 and 73 min for NagB-L4) (Fig. 3). We interpret the fact that the growth rates of the NagB-L1 construct are the same on GlcN and GlcNAc to mean that for this plasmid it is the amount of NagB protein that is limiting growth on either medium. GlcNAc, which supplies higher concentrations of the substrate GlcN6P as well as the allosteric activator, GlcNAc6P, did not improve the growth rate of NagB-L1 compared to GlcN. Likewise, the mlc mutation, which improves the growth rate of MC4100 (by derepressing the ManXYZ transporter and thus increasing the substrate concentration), did not have any significant effect on the DT of NagB-L1. This is consistent with the hypothesis that the level of NagB protein in construct NagB-L1 is the growth-limiting factor, as discussed below.
On the other hand, NagB-L2, -L3, and -L4 grew better on GlcNAc than on GlcN (a P value of <6 × 10−4 for each pairwise combination for each construct), even though the quantity of NagB from each construct should be identical in GlcN and GlcNAc and in fact was higher in the GlcN-grown cultures (Fig. 2B). Thus, something other than the quantity of the NagB protein is limiting growth on GlcN compared to that on GlcNAc for these three constructs. The limiting factor could be a lack of substrate or a lack of allosteric activation.
Effect of increasing levels of GlcN6P, the substrate for NagB.
Introduction of an mlc mutation improved the growth rates of NagB-L2, -L3, and -L4 on GlcN, producing the same growth rate (DT = 80 min) for all three (P = 0.8; one-way analysis of variance), despite the fact that there was significantly more NagB protein from the NagB-L4 construct than from the other two. This implies that it is the amount of the substrate, GlcN6P, which is determining the growth rate on GlcN in all three constructs. An mlc mutation increases the expression of the GlcN transporter (encoded by manXYZ) two- to threefold and hence should increase the intracellular concentration of GlcN6P by a similar factor (1, 19, 20, 22). On the other hand, growth on GlcNAc induces expression of the chromosomal nagE and nagBA genes about 20-fold (21, 24) and could be expected to produce a greater increase in the concentration of GlcN6P than the mlc mutation. In the case of NagB-L4, when the NagB protein level is considerably higher the extra GlcN6P coming from growth on GlcNAc together with the higher level of NagB protein resulted in a faster growth rate on GlcNAc. The mlc mutation had no effect on NagB-L1, whose growth rate stayed the same as that on GlcN, consistent with the hypothesis that it is the amount of the NagB protein which is limiting growth rates of this strain and not the GlcN6P concentration.
Effect of mutations increasing levels of GlcNAc6P, the allosteric activator of NagB.
To investigate if the presence of the allosteric activator, GlcNAc6P, enhanced growth rates on GlcN, we tested the effect of mutations nagA and ΔnagE-nagBACD. Both of these mutations result in high internal concentrations of GlcNAc6P due to the loss of the nagA-encoded GlcNAc6P deacetylase (17, 25, 33). High levels of GlcNAc6P, the inducer for NagC, normally cause derepression of the nagE-nagBACD operons as well as permit allosteric activation of NagB. Using the plasmid constructs, we eliminated the effect of GlcNAc6P on NagB protein levels and expected to see only effects due to changes in the enzymatic activity of NagB. However, the presence of a nagA mutation did not improve growth rates on GlcN or had only a small effect which was not significantly different from that of a nagC mutation (P values of >0.09, 0.9, 0.6, and 0.7 for pairwise comparisons of nagA and nagC mutant strains with the four constructs) (Fig. 3). A nagC mutation improved growth on GlcN when nagB was expressed from its normal chromosomal locus in MC4100 but had a much smaller effect or no effect on growth of the strains carrying the four NagB constructs on GlcN (Fig. 3). Growth of the nagA and nagC mutants of NagB-L2, -L3, and -L4 was always distinctly slower than that of the mlc derivatives. The fact that for these three constructs the mlc mutation has a greater effect on growth rates than the nagA mutation demonstrates that it is the supply of substrate which is limiting the growth rate and not the lack of allosteric activator. We also tested the combination of an mlc mutation with ΔnagE-nagBACD, nagA, or nagC mutations to see if allosteric activation by GlcNAc6P was detectable when the substrate level was increased by the mlc mutation. In all cases, the growth rates of the double mutants were the same as that of the single mlc mutant (data not shown), confirming that under these conditions it is the rate of transport of GlcN which is limiting growth on GlcN.
When NagB was expressed from its natural location in the nagE-nagBACD operon (MC4100 pXE1) (Fig. 3), the mlc, nagA, and nagC mutations all improved growth on GlcN at 30°C, as seen previously at 37°C (1), but the relative effects of mlc and nagA and/or nagC mutations were slightly different: at 30°C, mlc was almost as effective as nagA or nagC. An in vitro kinetic analysis has shown that the NagB deaminase is still allosteric at 30°C, although the cooperativity, as measured by the Hill coefficient, is slightly lower than at 37°C (3). Thus, use of the lower temperature should not affect our ability to detect allosteric activation of NagB.
Allosteric activation of the NagB F174A mutant.
As a control to verify that allosteric activation by GlcNAc6P was detectable at 30°C with the plasmid constructs, we tested the F174A mutation of NagB. This mutant form of deaminase is essentially inactive in vitro without the allosteric activator, GlcNAc6P, to stabilize the active site conformation (5). We cloned the F174A mutation into the NagB-L4 plasmid construction to give NagB-F174A and tested growth on GlcN in the presence of the different mutations in trans (Fig. 3). As expected, the presence of the nagA or ΔnagE-nagBACD mutation produced significantly better growth rates (DT = 130 min) than the wt strain on GlcN (DT = 236 min) or the nagC mutation (DT = 187 min) on GlcN. So in the context of the F174A mutation, allosteric regulation of deaminase is detectable for NagB cloned into the single-copy plasmids. Interestingly, the mlc mutation also appreciably increased the growth rate of F174A (DT = 146 min), suggesting that the substrate concentration is also limiting growth. (The reason for the improvement in growth produced by the nagC mutation is not known. We note that the nagC mutation also affected the NagB-L4 construct slightly. It is perhaps an indication that some other NagC-controlled gene is also affecting growth on GlcN.)
Growth on GlcN produces GlcNAc6P via PG recycling.
The absence of any effect of the nagA mutation on growth on GlcN over a range of NagB enzyme concentrations implies that the wt NagB enzyme is already fully active and cannot be further allosterically activated by higher concentrations of GlcNAc6P (unlike the F174A mutant, which requires higher concentrations of GlcNAc6P for full activation) (see above). GlcN6P deaminase is subject to positive cooperativity (homotropic activation) by substrate binding, but this requires concentrations of GlcN6P of >5 mM (in vitro), which are unlikely to be attained within the cell during growth on GlcN. The fact that growth on GlcN does increase expression of the nagE-nagBACD operon (Fig. 2; Table 3) (24) is evidence that some of the GlcN6P is converted to GlcNAc6P, the inducer of the NagC repressor. Thus, we considered ways in which the GlcN6P could be converted to GlcNAc6P. No enzymes are known which can acetylate GlcN6P in E. coli. GlcN6P has an essential function in bacteria as a precursor in the formation of PG of the cell wall. PG is a dynamic material, undergoing continuous synthesis and degradation during cell growth and division. The PG recycling process involves a set of dedicated enzymes to hydrolyze the PG, transport muropeptides into the cell, and then degrade them to GlcNAc6P and GlcN6P (reviewed in reference 15). We have recently shown that NagE and ManXYZ PTS transporters also contribute to the recycling process and the pool of GlcNAc6P (17). Introduction of mutations, which prevented the majority of the recycling of the PG (strain MC-B196/7) (Table 3), considerably reduced the expression of nagB-lacZ, indicating that the intracellular concentration of GlcNAc6P (the inducing signal for the NagC repressor) has been reduced, which is consistent with the idea that GlcNAc6P is generated via PG recycling. The DT on GlcN also seemed to increase just slightly, consistent with the idea that a reduction in the GlcNAc6P concentration reduces NagB activity (Table 3). The manXYZ-encoded PTS transporter is also implicated in PG recycling (17), but we could not eliminate this gene because it is required for GlcN uptake. A low level of GlcNAc6P generated by PG recycling via ManXYZ could explain why nagB-lacZ β-galactosidase activities do not fall to the background level observed during growth on glycerol.
TABLE 3.
Effect of peptidoglycan recycling on expression of nagBa
| Strain | Description | Glycerol
|
GlcN
|
||
|---|---|---|---|---|---|
| nagB-lacZ (U) | DT (min) | nagB-lacZ (U) | DT (min) | ||
| MC-B1 | wt | 69 ± 5 | 68 ± 5 | 229 ± 25 | 101 ± 4 |
| MC-B196/7 | nagK murQ anmK nagZ ampG nagE | 61 ± 4 | 64 ± 5 | 119 ± 19 | 110 ± 3 |
β-Galactosidase activities (Miller units) were measured in minimal MOPS medium with 0.2% glycerol or 0.2% GlcN at 37°C. MC-B1 is MC4100 with a nagB-lacZ fusion on a λ lysogen. MC-B196/7 is MC-B1 into which mutations nagK, murQ, anmK, nagZ, ampG, and nagE have been sequentially introduced to reduce recycling of peptidoglycan and formation of intracellular GlcNAc6P. DT (min) were calculated by linear regression between A650 values of 0.08 to 0.8. Results are the means of four independent cultures.
Concluding remarks.
In summary, our results show that over a physiological range of concentrations of NagB in the cell, it is the level of the substrate, GlcN6P, rather than the presence of the allosteric effector, GlcNAc6P, which determines the growth rate on GlcN. In our original study (1), we found that the major limiting effect during growth on GlcN was the amount of the NagB protein due to a much lower level of induction of the chromosomal nagE-nagBACD operon during growth on GlcN compared to growth on GlcNAc. We also observed that growth improved when the amount of the GlcN transporter increased, and thus the rate of uptake of the substrate increased. In this work, we show that at a fixed level of NagB protein it is the amount of the substrate, determined by the expression of the GlcN transporter, which is limiting the growth rate on GlcN. Contrary to our expectations, we have failed to detect any effect of increased concentrations of GlcNAc6P on the activity of NagB deaminase in vivo under conditions when its protein levels were fixed at a low level. This is true whether GlcNAc6P is derived from transport of GlcNAc or is due to the presence of a nagA mutation. We also present evidence that the low level of induction of the nagE-nagBACD operon during growth on GlcN is due to generation of GlcNAc6P from PG recycling.
The in vitro kinetic data for NagB deaminase are consistent with the Monod, Wyman, and Changeux model of allosteric activation (12) where the enzyme can exist in two extreme forms, one with low affinity for the substrate (T form) and an activated form with high affinity for the substrate (R form). The calculated Km for the substrate, GlcN6P, to the T form of the enzyme (i.e., in the absence of the allosteric activator, GlcNAc6P) is so high (22 mM) that the T form appears to be practically inactive (4). Substrate binding does produce positive cooperativity (homotropic activation), but this requires concentrations of GlcN6P >3 to 5 mM to see any significant activity in vitro (4, 6). Since our results strongly suggest that GlcN6P concentrations are limiting growth, it seems unlikely that GlcN6P could attain in vivo the concentrations required in vitro. On the other hand, the Km for GlcN6P binding to the GlcNAc6P-saturated R form is 0.55 mM (4). Since there is no effect of GlcNAc6P (derived from either growth on GlcNAc or the presence of a nagA mutation) on growth on GlcN when the amount of the enzyme is fixed, as in the case of the plasmid constructs studied here, it appears as if the NagB enzyme is already fully allosterically active during growth on GlcN. The most likely explanation of this paradox is that growth on GlcN generates sufficient GlcNAc6P to allosterically activate the enzyme. The dissociation constant (Kdis) for GlcNAc6P of the wt enzyme measured in vitro is 35 μM to the R state. The F174A mutant enzyme has a higher Kdis for GlcNAc6P (139 μM) (5), which could explain why its activity is enhanced by growth on GlcNAc or by the presence of a nagA mutation in vivo.
The results presented here imply that growth on GlcN increases the GlcNAc6P level such that it is sufficient for full allosteric activation of wt NagB but not for F174A and produces only partial inactivation of the NagC repressor, as shown by the low-level induction of the nagB-lacZ fusion. Moreover, we show that the increase in GlcNAc6P is dependent upon the presence of genes involved in PG recycling. One interpretation of this observation is that growth on GlcN, by supplying de novo GlcN6P, slows down the reuse of GlcNAc6P derived from PG recycling so that GlcNAc6P starts to accumulate. This could be the case if NagA was subject to product inhibition by GlcN6P. Inhibition of NagA by GlcN6P was observed in vitro but only at rather high (>4 mM GlcN6P) concentrations (29).
As NagB is an allosteric enzyme, binding of the substrate and activator are coupled reactions so that relatively small changes in one or other or both can shift the enzyme toward the active R state. If growth on GlcN does fully activate GlcN6P deaminase, then we can ask what is the purpose of the allosteric regulation and under what conditions is the enzyme not fully active. In the absence of an exogenous supply of an amino sugar, the enzyme GlcN6P synthase (GlmS) is essential. Expression of GlmS is controlled both transcriptionally by NagC, acting as an activator in the absence of exogenous amino sugars (18), and by an elaborate posttranscriptional mechanism, implicating RNA processing and two small RNAs and a probable GlcN6P sensor protein (10, 27, 31), which demonstrates the importance of controlling GlcN6P production. We could speculate that one function of the allosteric regulation is to keep the activity of NagB very low in the absence of all external supplies of amino sugars so that GlmS-synthesized GlcN6P is not immediately degraded by NagB in a futile cycle.
Glucosamine-6P deaminases are ubiquitous. In most organisms, both prokaryotes and eukaryotes, the enzymes are orthologues of the E. coli NagB, with a conserved catalytic triad of readily identifiable amino acids (13). GlcN6P deaminases have been purified from several different sources and found to exist in different quaternary structures. In some cases (e.g., the enzyme from dogs [11] and two cloned human enzymes [2], LIA-A and MLC [unpublished data]), the purified enzymes have been shown to be hexameric and to be allosterically activated by GlcNAc6P like the E. coli version. Other NagB orthologues exist as dimers (e.g., from Candida albicans [14]) or monomers (e.g., from Bacillus subtilis [32]) and are nonallosteric. The abundance of conserved allosteric enzymes, particularly in mammals, suggests that the allosteric regulation plays an important role in the cellular function of these enzymes.
Acknowledgments
We are very grateful to Florent Busi for help with data analysis and to Annie Kolb, Harald Putzer, and Mathias Springer for comments on the manuscript.
This work was supported by funds from the CNRS and Université of Paris 7—Denis Diderot (to UPR9073, Paris, France) and from CONACYT (to M.L.C.) (Mexico) and was partly financed by a CNRS-CONACYT international cooperation (project no. 18330).
Footnotes
Published ahead of print on 21 August 2009.
REFERENCES
- 1.Álvarez-Añorve, L. I., M. Calcagno, and J. Plumbridge. 2005. Why does Escherichia coli grow more slowly on glucosamine than on N-acetylglucosamine? Effects of enzyme levels and allosteric activation of GlcN6P deaminase (NagB) on growth rates. J. Bacteriol. 187:2974-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arreola, R., B. Valderrama, M. L. Morante, and E. Horjales. 2003. Two mammalian glucosamine-6-phosphate deaminases: a structural and genetic study. FEBS Lett. 551:63-70. [DOI] [PubMed] [Google Scholar]
- 3.Bustos-Jaimes, I., and M. Calcagno. 2001. Allosteric transition and substrate binding are entropy driven in glucosamine-6-phosphate deaminase from Escherichia coli. Arch. Biochem. Biophys. 394:156-160. [DOI] [PubMed] [Google Scholar]
- 4.Bustos-Jaimes, I., M. Ramírez-Costa, L. D. Anda-Aguilar, P. Hinjosa-Ocaña, and M. Calcagno. 2005. Evidence for two different mechanisms triggering the change in quaternary structure of the allosteric enzyme, glucosamine-6-phosphate deaminase. Biochemistry 44:1127-1135. [DOI] [PubMed] [Google Scholar]
- 5.Bustos-Jaimes, I., A. Sosa-Peinado, E. Rudiño-Piñera, E. Horjales, and M. L. Calcagno. 2002. On the role of the conformational flexibility of the active site lid on the allosteric kinetics of glucosamine-6-phosphate deaminase. J. Mol. Biol. 319:183-189. [DOI] [PubMed] [Google Scholar]
- 6.Calcagno, M., P. J. Campos, G. Mulliert, and J. Suastegui. 1984. Purification, molecular and kinetic properties of glucosamine-6-phosphate isomerase (deaminase) from E. coli. Biochim. Biophys. Acta 787:165-173. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One step inactivation of chromosomal genes in Escherichia coli K12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decker, K., J. Plumbridge, and W. Boos. 1998. Negative transcriptional regulation of a positive regulator: the expression of malT, encoding the transcriptional activator of the maltose regulon of Escherichia coli, is negatively controlled by Mlc. Mol. Microbiol. 27:381-390. [DOI] [PubMed] [Google Scholar]
- 9.Dobrogosz, W. J. 1968. Effect of amino sugars on catabolite repression in Escherichia coli. J. Bacteriol. 95:578-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalamorz, F., B. Reichenbach, W. Marz, B. Rak, and B. Görke. 2007. Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA and involves the novel protein YhbJ in Escherichia coli. Mol. Microbiol. 65:1518-1533. [DOI] [PubMed] [Google Scholar]
- 11.Lara-Lemus, R., C. A. Libreros-Minotta, M. M. Altamirano, and M. L. Calcagno. 1992. Purification and characterisation of glucosamine-6-phosphate deaminase from dog kidney cortex. Arch. Biochem. Biophys. 297:213-220. [DOI] [PubMed] [Google Scholar]
- 12.Monod, J., J. Wyman, and J. Changeux. 1965. On the nature of the allosteric transitions a plausible model. J. Mol. Biol. 12:88-118. [DOI] [PubMed] [Google Scholar]
- 13.Montero-Morán, G. M., S. Lara-González, L. I. Álvarez-Añorve, J. A. Plumbridge, and M. L. Calcagno. 2001. On the multiple functional roles of the active site histidine in catalysis and allosteric regulation of Escherichia coli glucosamine 6-phosphate deaminase. Biochemistry 40:10187-10196. [DOI] [PubMed] [Google Scholar]
- 14.Natarajan, K., and A. Datta. 1993. Molecular cloning and analysis of the NAG1 cDNA coding for glucosamine-6-phosphate deaminase from Candida albicans. J. Biol. Chem. 268:9206-9214. [PubMed] [Google Scholar]
- 15.Park, J. T., and T. Uehara. 2008. How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol. Mol. Biol. Rev. 72:211-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pennetier, C., L. Domínguez-Ramírez, and J. Plumbridge. 2008. Different regions of Mlc and NagC, homologous transcriptional repressors controlling expression of the glucose and N-acetylglucosamine phosphotransferase systems in Escherichia coli, are required for inducer signal recognition. Mol. Microbiol. 67:364-377. [DOI] [PubMed] [Google Scholar]
- 17.Plumbridge, J. 2009. An alternate route for recycling of N-acetylglucosamine from peptidoglycan involves the N-acetylglucosamine phosphotransferase system in E. coli. J. Bacteriol. 191:5641-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plumbridge, J. 1995. Co-ordinated regulation of aminosugar biosynthesis and degradation: the NagC repressor acts as an activator for the transcription of the glmUS operon and requires two separated NagC binding sites. EMBO J. 14:3958-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plumbridge, J. 1998. Control of the expression of the manXYZ operon in Escherichia coli: Mlc is a negative regulator of the mannose PTS. Mol. Microbiol. 27:369-381. [DOI] [PubMed] [Google Scholar]
- 20.Plumbridge, J. 2002. Regulation of gene expression in the PTS in Escherichia coli: the role and interactions of Mlc. Curr. Opin. Microbiol. 5:187-193. [DOI] [PubMed] [Google Scholar]
- 21.Plumbridge, J. 1996. How to achieve constitutive expression of a gene within an inducible operon: the example of the nagC gene of Escherichia coli. J. Bacteriol. 178:2629-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plumbridge, J., and E. Vimr. 1999. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine and N-acetylneuraminic acid by Escherichia coli. J. Bacteriol. 181:47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plumbridge, J. A. 1992. A dominant mutation in the gene for the Nag repressor of Escherichia coli that renders the nag regulon uninducible. J. Gen. Microbiol. 138:1011-1017. [DOI] [PubMed] [Google Scholar]
- 24.Plumbridge, J. A. 1990. Induction of the nag regulon of Escherichia coli by N-acetylglucosamine and glucosamine: role of the cyclic AMP-catabolite activator protein complex in expression of the regulon. J. Bacteriol. 172:2728-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plumbridge, J. A. 1991. Repression and induction of the nag regulon of Escherichia coli K12: the roles of nagC and nagA in maintenance of the uninduced state. Mol. Microbiol. 5:2053-2062. [DOI] [PubMed] [Google Scholar]
- 26.Plumbridge, J. A. 1989. Sequence of the nagBACD operon in Escherichia coli K12 and pattern of transcription within the nag regulon. Mol. Microbiol. 3:506-515. [DOI] [PubMed] [Google Scholar]
- 27.Reichenbach, B., A. Maes, F. Kalamorz, E. Hajnsdorf, and B. Görke. 2008. The small RNA GlmY acts upstream of the sRNA GlmZ in the activation of glmS expression and is subject to regulation by polyadenylation in Escherichia coli. Nucleic Acids Res. 36:2570-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ringquist, S., S. Shinedling, D. Barrick, L. Green, J. Binkley, G. Stormo, and L. Gold. 1992. Translational initiation in Escherichia coli: sequences within the ribosome-binding site. Mol. Microbiol. 6:1219-1229. [DOI] [PubMed] [Google Scholar]
- 29.Souza, J.-M., J. A. Plumbridge, and M. L. Calcagno. 1997. N-Acetyl-d-glucosamine-6-phosphate deacetylase from Escherichia coli: purification and molecular and kinetic characterization. Arch. Biochem. Biophys. 340:338-346. [DOI] [PubMed] [Google Scholar]
- 30.Tremblay, L., D. Dunaway-Mariano, and K. Allen. 2006. Structure and activity analyses of Escherichia coli K-12 NagD provide insight into the evolution of biochemical function in the haloalkanoic acid dehalogenase superfamily. Biochemistry 45:1183-1193. [DOI] [PubMed] [Google Scholar]
- 31.Urban, J., and J. Vogel. 2008. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 6:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vincent, F., G. Davies, and J. Brannigan. 2005. Structure and kinetics of a monomeric glucosamine-6 phosphate deaminase. Missing link of the NagB superfamily? J. Biol. Chem. 280:19649-19655. [DOI] [PubMed] [Google Scholar]
- 33.White, R. J. 1968. Control of aminosugar metabolism in Escherichia coli and isolation of mutants unable to degrade amino sugars. Biochem. J. 106:847-858. [DOI] [PMC free article] [PubMed] [Google Scholar]