Abstract
Clostridium perfringens is a normal gastrointestinal organism that is a reservoir for antibiotic resistance genes and can potentially act as a source from which mobile elements and their associated resistance determinants can be transferred to other bacterial pathogens. Lincomycin resistance in C. perfringens is common and is usually encoded by erm genes that confer macrolide-lincosamide-streptogramin B resistance. In this study we identified strains that are lincomycin resistant but erythromycin sensitive and showed that the lincomycin resistance determinant was plasmid borne and could be transferred to other C. perfringens isolates by conjugation. The plasmid, pJIR2774, is the first conjugative C. perfringens R-plasmid to be identified that does not confer tetracycline resistance. Further analysis showed that resistance was encoded by the lnuP gene, which encoded a putative lincosamide nucleotidyltransferase and was located on tISCpe8, a functional transposable genetic element that was a member of the IS1595 family of transposon-like insertion sequences. This element had significant similarity to the mobilizable lincomycin resistance element tISSag10 from Streptococcus agalactiae. Like tISSag10, tISCpe8 carries a functional origin of transfer within the resistance gene, allowing the element to be mobilized by the conjugative transposon Tn916. The similarity of these elements and the finding that they both contain an oriT-like region support the hypothesis that conjugation may result in the movement of DNA modules that are not obviously mobile since they are not linked to conjugation or mobilization functions. This process likely plays a significant role in bacterial adaptation and evolution.
There has been increasing concern about the emergence of multiply antibiotic-resistant strains of many common bacterial pathogens. The development of multiple resistance phenotypes has already led to compromises in the ability to successfully treat infected patients and to increased treatment costs (15). The emergence of resistant bacteria is often the result of excessive or inappropriate use of antibiotics and the ability of antibiotic resistance genes to be transferred from resistant to susceptible bacteria, either within a bacterial species, between different species within the same genus, or between different genera (14). Different types of mobile genetic elements, including conjugative plasmids, conjugative transposons, mobilizable plasmids, mobilizable transposons, nonconjugative plasmids, and integrons, may contain the resistance genes (14). All of these elements have the ability to mediate the transfer of resistance genes within and between bacterial cells, either independently or cooperatively, which has significant implications for the transfer and evolution of antibiotic resistance, particularly in pathogenic bacterial species.
Clostridium perfringens is a normal gastrointestinal organism that causes food poisoning, necrotic enteritis, and gas gangrene (29). It is a proven reservoir for antibiotic resistance determinants. For example, the catP chloramphenicol resistance determinant, which is located on the Tn4451/Tn4453 family of integrative mobilizable elements in C. perfringens and Clostridium difficile, has been detected in clinical isolates of Neisseria meningitidis (20, 23, 41). Similarly, genetically related variants of the macrolide-lincosamide-streptogramin B (MLS) resistance determinant Erm(B) from C. perfringens have been found in Enterococcus faecalis, Streptococcus agalactiae, and C. difficile (19). It is likely that the C. perfringens determinant is the progenitor of the C. difficile determinant (18, 19, 44). Significantly, both determinants can be transferred into recipient cells by conjugation, although the processes are different (12, 19, 43). The pathogenic clostridia also carry other uncharacterized MLS resistance determinants and can potentially act as a source from which these resistance determinants may be transferred to other bacterial pathogens (10, 18).
Lincomycin belongs to the lincosamide group of antibiotics, which also includes clindamycin. The spectrum of activity of lincosamides predominantly encompasses gram-positive bacteria, and these antimicrobial agents are often used for treatment of infections caused by anaerobic bacteria (45). These antibiotics inhibit protein synthesis by blocking the peptidyltransferase site of the 23S rRNA component of the 50S subunit of the bacterial ribosome (17). Although cross-resistance to MLS antibiotics most commonly involves N6 dimethylation of the A2058 residue of 23S rRNA and is catalyzed by an erm-encoded rRNA methyltransferase (24, 34, 47), specific resistance to the lincosamides is the result of modification and inactivation by a lincosamide nucleotidyltransferase encoded by members of the lnu (previously lin) gene family (5, 34, 45). This type of resistance gene is found in staphylococci and streptococci, where it is often located on plasmids or transposons (5, 45).
Lincomycin resistance in C. perfringens is relatively common, but it is usually conferred as MLS resistance by erm(B) or erm(Q) genes (10, 11). Recent studies have shown that there has been an increase in lincomycin resistance in C. perfringens strains isolated from chickens in Belgium (28). The researchers reported two strains that conferred resistance to lincomycin and carried the lnu(A) or lnu(B) gene, the first such strains reported for C. perfringens.
In the current study we analyzed several multiply antibiotic-resistant isolates of C. perfringens and identified strains that were lincomycin resistant but were susceptible to erythromycin. We characterized these isolates and their lincomycin resistance determinant(s) and showed that resistance could be transferred to other C. perfringens isolates. Detailed analysis of the lincomycin-resistant strain 95-949 showed that resistance was encoded by the lnuP gene, which was located on a transposable genetic element, tISCpe8, that was located on a conjugative plasmid, pJIR2774. This plasmid is the first conjugative C. perfringens R-plasmid to be identified that does not confer tetracycline resistance.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids are listed in Table 1. C. perfringens strains were cultured at 37°C in Trypticase-peptone-glucose broth (36), in brain heart infusion (BHI) broth or on BHI agar (Oxoid), in fluid thioglycolate medium (Difco), or on nutrient agar (35) supplemented with the following antibiotics when appropriate: lincomycin (25 μg/ml), rifampin (rifampicin) (20 μg/ml), nalidixic acid (20 μg/ml), and streptomycin (100 μg/ml). Solid medium containing 1% (vol/vol) potassium chlorate was made by addition of a saturated potassium chlorate solution to the medium before it was dispensed (22). C. perfringens cultures were grown in an atmosphere containing 10% H2, 10% CO2, and 80% N2 at 37°C in anaerobic jars (Oxoid). Escherichia coli strain DB10 (16) was used as the recipient for cloning of the lincomycin resistance gene, and the resultant derivative was cultured aerobically at 37°C in 2×YT medium (30) in the presence of lincomycin (25 μg/ml) and ampicillin (100 μg/ml).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli DB10 | PR7 Lns | 16 |
| C. perfringens strains | ||
| EV3839 | Ems Lnr; wild-type isolate | J. G. Songer |
| UAZ196 | Ems Lnr; wild-type isolate | J. G. Songer |
| 95-949 | Ems Lnr; wild-type isolate; pJIR2774 | J. G. Songer |
| CW504 | Rifr Nalr; conjugation recipient | 37 |
| JIR4 | CW504(pCW3); Tcr | 1 |
| JIR325 | Rifr Nalr; electroporation recipient | 27 |
| JIR4225 | JIR325::Tn916 | 7 |
| JIR4344 | Rifr Nalr Lnr | Transconjugant, 95-949 × CW504 |
| JIR4345 | Rifr Nalr Lnr | Transconjugant, 95-949 × CW504 |
| JIR4394 | Strr Chlr; conjugation recipient | 33 |
| Plasmids | ||
| pUC18 | Apr; cloning vector | 46 |
| pCW3 | Conjugative C. perfringens plasmid; Tcr | 37 |
| pJIR1944 | pUC18 Ω 2.2-kb 95-949 DNA, partial HindIII digest | This study |
| pJIR2774 | Lnr conjugative C. perfringens plasmid | This study |
Lincomycin resistance transfer experiments.
C. perfringens-C. perfringens conjugation experiments were performed as follows. Donor and recipient strains were incubated in fluid thioglycolate broth for 6 h before 100-μl aliquots of the two cultures were mixed by spreading them onto thick BHI agar plates without antibiotics. The mating plates were subsequently incubated anaerobically overnight at 37°C, and the bacterial growth was removed with 2 to 3 ml of dilute (1 in 10) BHI broth, serially diluted, plated onto an appropriate selective medium, and incubated for 24 to 48 h. Donor and recipient controls were included in each conjugation experiment, and donor viable counts were used to calculate the transfer frequency, which was defined as the number of transconjugants per donor cell.
DNA isolation and molecular techniques.
Plasmid DNA from E. coli was obtained using QIAprep spin miniprep columns (Qiagen) according to the manufacturer's instructions. Total DNA and plasmid DNA were obtained from C. perfringens as described previously (25). E. coli (38) and C. perfringens (39) transformations were also performed as described previously. Standard methods for digestion, ligation, and analysis of plasmid DNA and PCR products were used (38). PCR products for nucleotide sequencing were purified from agarose gels with a QIAquick gel extraction kit (Qiagen).
PCR was performed using Taq DNA polymerase (Roche) and 0.5 μM of each primer. Denaturation (94°C for 30 s), annealing (50°C for 30 s), and extension (72°C for 3 to 5 min) steps were carried out for 30 to 35 cycles. DNA sequencing was carried out using a PRISM Ready Reaction DyeDeoxy Terminator cycle sequencing kit (Applied Biosystems) according to the manufacturer's instructions. The oligonucleotide primers used for PCR amplification of DNA or for sequencing were as follows (all 5′-3′): JRP2033 (TAACATTCCGATACCTAAAGA; forward primer for the 3′ end of lnuP), JRP2034 (TACATTGACATTTTGGGCACTC; reverse primer for the 5′ end of tnp), JRP2036 (AAATAAAAAGAATGACCGAAT; forward primer for the 3′ end of tnp), and JRP2037 (GTGGTTGTTTTTATGACTTATT; reverse primer for the 3′ end of tISCpe8, between the end of lnuP and the tISCpe8 right end). Sequence analysis was carried out with an Applied Biosystems 3730S genetic analyzer and Sequencher 3.1 software (Genecodes Corporation). Bioinformatic analyses were performed at the http://www.ncbi.nlm.nih.gov/ and http://npsa-pbil.ibcp.fr) websites and included BLAST, Conserved Domain Database, Clustal W, and helix-turn-helix predictions.
Cloning of the C. perfringens lincomycin resistance gene.
Total DNA extracted from strain 95-949 was subjected to partial HindIII digestion prior to ligation to HindIII-digested pUC18. Ligated products were introduced into E. coli strain DB10 by transformation and incubated at 37°C for 48 h in the presence of lincomycin and ampicillin. The resultant colonies were passaged once on the same selective medium to confirm the antibiotic resistance phenotype prior to plasmid DNA extraction, restriction analysis, and sequencing.
Southern hybridization.
C. perfringens plasmid DNA was digested with EcoRI, subjected to agarose gel electrophoresis, and then transferred to a nylon Hybond H+ membrane (Amersham) (38). Southern hybridization analysis was carried out using standard methods (25). DNA probes were digoxigenin labeled using random PCR labeling according to the manufacturer's instructions (Roche). The blots were hybridized with a probe specific for lnuP, generated using primers JRP2036 and JRP2037.
Transposition experiments.
Suicide plasmid pJIR1944 was introduced into C. perfringens strain JIR325 by electroporation. Cells in which tISCpe8 had moved from pJIR1944 were obtained by selecting for lincomycin resistance. Total DNA was subsequently extracted and subjected to Southern hybridization analysis.
Nucleotide sequence accession numbers.
The GenBank accession number for the intP-dcm region of pJIR2774 is DQ338473, and the GenBank accession number for tISCpe8 is FJ589781.
RESULTS
Lincomycin resistance in C. perfringens is transferable and is plasmid associated.
Analysis of a collection of multiply antibiotic-resistant isolates of C. perfringens revealed three strains that were lincomycin resistant but susceptible to erythromycin. Mixed plate mating showed that two of these strains had the ability to transfer their lincomycin resistance by conjugation. Specifically, when strains UAZ196 and 95-949 were used as donors in mating experiments with recipient strain CW504, transfer frequencies of 3.1 × 10−5 and 1.6 × 10−2 lincomycin-resistant transconjugant per donor cell, respectively, were obtained. In comparison, there was no detectable transfer of lincomycin resistance from strain EV3839 (<1.0 × 10−7 transconjugant per donor cell).
To determine if lincomycin resistance was linked to a mobile extrachromosomal element or if it was located on the chromosome, total genomic DNA was isolated from strain 95-949 and used to transform C. perfringens strain JIR325 to lincomycin resistance. DNA preparations were also obtained for two CW504-derived lincomycin-resistant transconjugants, JIR4344 and JIR4345, which were isolated from mating with 95-949, and used in similar transformation experiments. Transformants were consistently obtained using independently isolated DNA preparations in separate transformation experiments. Several of the resultant independently derived transformants were then used as donors for conjugation with C. perfringens recipient strain JIR4394. Lincomycin resistance was transferred at frequencies similar to those observed for strain 95-949 (data not shown). Taken together, these results suggested that lincomycin resistance was encoded on a conjugative plasmid.
The lincomycin resistance plasmid pJIR2774 is very similar to the prototype C. perfringens conjugative plasmid pCW3.
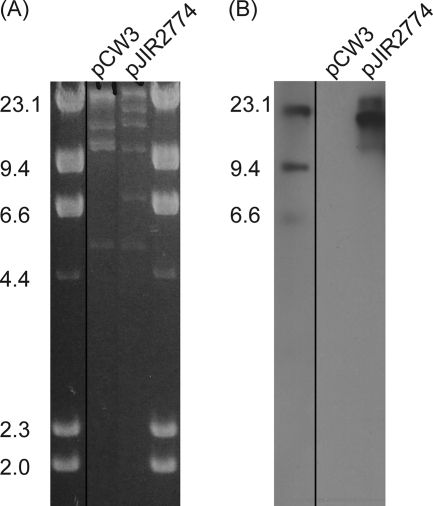
Plasmid DNA was isolated from strain 95-949, and ClaI digests were compared with equivalent digests of pCW3, the prototype conjugative plasmid from C. perfringens (1). The results indicated that a large plasmid, designated pJIR2774, was present in strain 95-949 (Fig. 1A). Southern blotting confirmed that the lincomycin resistance determinant was present on the second largest ClaI fragment of the plasmid (Fig. 1B). As expected, a probe specific for the lincomycin resistance gene carried on pJIR2774 did not hybridize with pCW3 (Fig. 1B). Examination of the restriction digests revealed that pJIR2774 had at least two ClaI fragments in common with pCW3, fragments that were 5,055 bp and 9,744 bp long, as determined using the pCW3 nucleotide sequence (9). These fragments flank one another and correspond to nucleotide positions 4629 to 19428 in pCW3. This region encompasses the rep gene (located at nucleotides 13239 to 14069), which is essential for pCW3 replication and maintenance (9), suggesting that pJIR2774 may utilize a pCW3-like replication mechanism. PCR analysis using primers specific for the pCW3 rep gene confirmed that pJIR2774 contains a similar gene (data not shown).
FIG. 1.
Restriction analysis of pJIR2774. (A) Purified plasmid DNA was digested with ClaI and separated by agarose gel electrophoresis. The sizes of λHindIII standards (in kilobases) are indicated on the left. pCW3 was extracted from strain JIR4, and pJIR2774 was extracted from strain 95-949. (B) Southern hybridization of ClaI-digested pCW3 and pJIR2774. The blot was probed with an lnuP-specific probe.
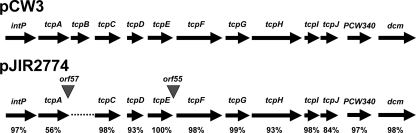
Comparative PCR walking was subsequently performed with total genomic DNA isolated from strains carrying pCW3 and pJIR2774, as previously described for other C. perfringens conjugative plasmids (9). These experiments utilized 10 overlapping primer pairs that together encompassed the 11 transfer-related genes located on pCW3: intP and tcpA to tcpJ. The results showed that all of these genes were present in pJIR2774, with the exception of tcpB (Fig. 2). The differences between pCW3 and pJIR2774 were confirmed by complete nucleotide sequencing of the region of pJIR2774 from intP to dcm. Two small open reading frames (ORFs), orf57 and orf55, which encoded hypothetical proteins, were identified between tcpA and tcpC and between tcpE and tcpF, respectively. The amino acid sequences of the pJIR2774 tcp-encoded products were aligned with the amino acid sequences of the corresponding pCW3-derived proteins, which showed that the encoded products were very closely related (Fig. 2). Overall, when the transfer region of pJIR2774 was compared to that of pCW3, a very high degree of amino acid sequence identity was observed, suggesting that the mechanisms of conjugative DNA transfer are likely to be very similar in pJIR2774 and pCW3. Furthermore, when these results were combined with the results of the comparative restriction analysis of pCW3 and pJIR2774, it appeared that the entire region from nucleotide 4,629 to nucleotide 40,030 (of a total of 47,263 bp) in pCW3 is present in pJIR2774. This analysis suggests that the similarities between these two plasmids extend well beyond the transfer-related region, although pJIR2774 is clearly larger than pCW3 and does not encode tetracycline resistance.
FIG. 2.
Comparative analysis of the region from intP to dcm of pCW3 and pJIR2774. The percentages indicate the levels of amino acid sequence identity between the orthologous proteins encoded by pCW3 (accession number DQ366035) (9) and pJIR2774 (accession number DQ338473).
A lincosamide nucleotidyltransferase is responsible for lincomycin resistance.
Total genomic DNA was isolated from strain 95-949 and digested with HindIII, and the lincomycin resistance determinant was cloned into pUC18, generating pJIR1944. Sequence analysis of the insert identified a gene whose product had 60% identity to LnuC from S. agalactiae (accession number AY928180). LnuC confers lincomycin resistance since it is a lincosamide nucleotidyltransferase that converts the antibiotic to a nontoxic form (5). The high level of sequence identity suggested that the pJIR2774 lincomycin resistance gene in C. perfringens strain 95-948, which we designated lnuP, conferred resistance by a similar mechanism. PCR analysis showed that strain UAZ196 also carried lnuP (data not shown), but the lincomycin resistance determinant from EV3839 could not be identified.
Lincomycin resistance is associated with an IS1595-like transposable element.
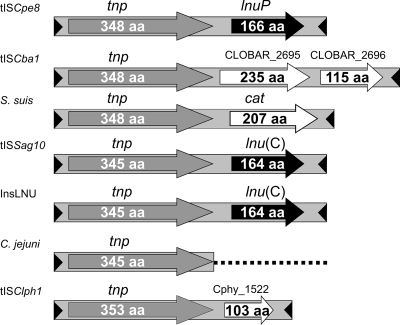
Sequence analysis of pJIR1944 suggested that the lnuP gene was located on a putative mobile element that contained two potential genes, tnp encoding a potential transposase and lnuP (Fig. 3). The genetic organization of this element was almost identical to that of tISSag10 from S. agalactiae (4), which is a mobilizable and transposable member of the IS1595 family of transposon-like insertion elements (42). Accordingly, the C. perfringens strain 95-949-derived element was designated tISCpe8. PCR analysis showed that the lnuP gene in strain UAZ196 was also located on a tISCpe8-like element (data not shown). Comparative analysis of the putative 348-amino-acid transposase encoded by tISCpe8 showed that it had significant sequence identity to putative transposases from Clostridium bartlettii (99% identity; encoded by tISCba1), Clostridium phytofermentans (32% identity; encoded by tISClph1), S. agalactiae (36% identity; encoded by tISSag10), and Campylobacter jejuni (36% identity) (Fig. 3). The other related tnp genes were located on similar elements or remnants of such elements (Fig. 3).
FIG. 3.
Comparison of the genetic organizations of orthologous gene regions. The genetic map of the lincomycin resistance transposon tISCpe8 shows the locations of the putative transposase gene (tnp) and lincomycin resistance gene (lnuP), their direction of transcription, and the inverted repeats located at the ends of the element (arrowheads). The numbers of amino acids (aa) in the encoded proteins are indicated. The elements related to tISCpe8 include tISSag10 from S. agalactiae (5). The remaining elements were identified predominantly by searches of existing genome sequences and included tISCba1 from C. bartlettii (accession number NZ_ABEZ02000022), an element from S. suis (YP_003028723.1), an element from C. jejuni (AY701528) (32), InsLNU from H. parasuis (NC_012661.1), and tISClph1 from C. phytofermentans (NC_010001). Related transposase genes are indicated by gray arrows, conserved inverted repeats are indicated by arrowheads, and related resistance genes are indicated by black arrows. Open arrows indicate ORFs with unknown functions and the chloramphenicol acetyltransferase gene (cat).
To precisely delineate the ends of tISCpe8, inverse PCR was used to generate PCR products that encompassed the ends of the transposon and flanking DNA from strain 95-949 and two CW504-derived lincomycin-resistant transconjugants, JIR4344 and JIR4345, which were isolated from mating with 95-949. The flanking DNA sequences were then used to generate primers that were used to amplify the insertion sites from 95-459 and CW504. The PCR products were then sequenced. It was predicted that the sequence of the tISCpe8 insertion site in 95-949 would be the same as the sequence of the regions flanking tISCpe8 in pJIR1944. Unexpectedly, the insertion site in pJIR1944 corresponded to an intergenic region located between aprX, which encodes a putative intracellular serine proteinase, and a sequence encoding a predicted hypothetical protein, CPE2609, located in a chromosomal region of the parent of JIR325, strain 13 (40), not to the plasmid-derived sequence. In addition, the sequences of the PCR products representing the insertion sites were very different from the sequence of pJIR1944 (Fig. 4A), suggesting that there must more than one insertion site in 95-949. This suggestion was confirmed by Southern hybridization analysis, which showed that multiple copies of tISCpe8 were present in this strain (data not shown). It was also expected that the tISCpe8 insertion sites in strains JIR4344 and JIR4345 would also match that of 95-949, since these transconjugants inherited pJIR2774 from this parent strain. However, these insertion sites were different from each other and from those of 95-949 and pJIR1944. This result suggested that tISCpe8 was an active transposon that moves very readily. We postulated that upon inheriting pJIR2774, tISCpe8 transposed in the new host, resulting in multiple genomic copies and locations; this hypothesis was confirmed by Southern hybridization analysis (data not shown). Note that none of these insertion sites matched pCW3-derived sequences; they all were chromosomal sequences. For example, the sequence derived from strain JIR4343 was identified as another chromosomal intergenic insertion, and in JIR4344 insertion occurred toward the end of the thiM gene, which is located in the middle of a putative operon.
FIG. 4.
Comparison of related sequences. (A) Regions flanking tISCpe8 elements. The insertion sites sequenced include the sites from pJIR1944, strain 95-949, and two CW504-derived transconjugants, JIR4344 and JIR4345, which were isolated after mating with 95-949. Underlining indicates the first 8 bp of the direct repeat, bold italics indicates the other 8 bp that constitute the direct repeat, the arrows below the sequences indicate the direct repeat sequences, and the triangle indicates the position of the inserted transposon. In the absence of the transposon only one of the 8-bp sequences is present; after transposon insertion the same 8 bp is present at both ends of the transposon. (B) Alignment of inverted repeats at the left (IRL) and right (IRR) ends of the tISCpe8, tISSag10, InsLNU (H. parasuis), tISClph1 (C. phytofermentans), tISCba1 (C. bartlettii), and S. suis sequences and at the left end of the C. jejuni upstream transposase sequence. (C) Alignment of putative oriT sites of tISCpe8 and tISSag10.
The finding that tISCpe8 had different insertion sites in the various C. perfringens derivatives allowed precise delineation of this element, which was found to consist of 1,964 bp and to include 24-bp imperfect inverted repeats (22 of 24 bp conserved) at the termini (Fig. 4B). tISCpe8 appears to duplicate an 8-bp target sequence at the site of insertion, like other members of the ISPna2 group in the IS1595 family (42). Furthermore, insertional specificity appears to involve an AT-rich target site (Fig. 4A). The ends of the closely related tISSag10 element and other ISPna2 elements (42) are very similar to the ends of tISCpe8 (Fig. 4B). A similar sequence was also identified immediately upstream of the related putative transposase gene from C. jejuni plasmid pCG8245, as well as in C. phytofermentans and C. bartlettii, again upstream of putative transposase genes (Fig. 4B). In C. jejuni the right end of the putative element could not be identified.
The first 1,250 bp of tISCpe8 exhibited 98% and 95% identity with tISCba1 from C. bartlettii and an isoform from Streptococcus suis (Fig. 3), respectively, and the last 130 bp of these elements exhibited 90% and 87% identity to tISCpe8, respectively. The transposase gene is at the 5′ end of all three elements. However, despite this marked similarity, the passenger gene(s) present in each element is different. tISCpe8 contains lnuP downstream of the tnp gene, whereas tISCba1 contains two additional ORFs with unknown functions and the putative S. suis element contains a chloramphenicol acetyltransferase gene.
tISCpe8 is capable of transposition in C. perfringens.
The detection of several different insertion sites suggested that tISCpe8 readily transposes in C. perfringens. To obtain direct evidence for transposition, the tISCpe8-containing suicide plasmid pJIR1944 was introduced into C. perfringens strain JIR325 by electroporation. Several lincomycin-resistant derivatives of JIR325 were isolated in independent transformation experiments. PCR analysis confirmed that these isolates carried tISCpe8 independent of pJIR1944, implying that the element had transposed to a different genetic location in the new host strain. Southern blotting confirmed this conclusion and also suggested that there may be a hotspot for tISCpe8 insertion in JIR325. However, in other C. perfringens hosts, such as CW504 and 95-949, multiple insertions were found (data not shown). Two copies of tISCpe8 were also observed in a JIR325 transconjugant carrying pJIR2774, the plasmid on which tISCpe8 was initially carried (data not shown). These results confirm that tISCpe8 is a functional transposon that is capable of transposition in C. perfringens.
tISCpe8 contains a functional oriT site.
tISSag10 has a functional origin of transfer located in the 3′ end of the lnuC gene, allowing the element to be mobilized by the conjugative transposon Tn916 (4). A similar oriT-like region was found in the equivalent region of the lnuP gene of tISCpe8 (Fig. 4C). To determine if this site was functional, we introduced tISCpe8 into the chromosome of JIR4225 (JIR325::Tn916) by electroporation with pJIR1944. Subsequent mating of the resultant transformants showed that Tn916 could mobilize tISCpe8 into recipient strain JIR4394. All of the resultant lincomycin-resistant transconjugants were susceptible to tetracycline, whereas selection for tetracycline resistance did not yield any lincomycin-resistant transconjugants. These results confirmed that tISCpe8 was not genetically linked to Tn916 in these isolates and provided evidence that tISCpe8 carried a functional oriT site.
DISCUSSION
Conjugative plasmids from C. perfringens have been shown to confer tetracycline resistance (2, 3, 9, 26), enterotoxin production (13, 31), and ɛ-toxin production (21). All of these plasmids are closely related to the prototype tetracycline resistance plasmid pCW3 and carry the same tcp conjugation locus. The lincomycin resistance plasmid identified in this study, pJIR2774, is also a member of this conjugative plasmid family and is the first conjugative C. perfringens resistance plasmid that does not confer tetracycline resistance. Similarly, the tISCpe8 lincomycin resistance element carried on pJIR2774 is the first insertion element-like resistance element shown to transpose in C. perfringens. The only other antibiotic resistance transposons detected in C. perfringens confer chloramphenicol resistance and are members of the Tn4451 family of integrative mobilizable elements, the excision and insertion of which are mediated by a large serine recombinase (6).
The similarity between pJIR2774 and pCW3 includes the rep region, suggesting that the mechanisms of plasmid replication are the same in these plasmids. Furthermore, 10 of the 11 genes in the tcp conjugation region of pCW3 are also present in pJIR2774, including the tcpA, tcpF, and tcpH genes, which are essential for the conjugative transfer of pCW3 (9, 33). The pJIR2774 tcpA gene can complement a tcpA mutant of pCW3 and restore conjugative transfer (33). The tcpB gene is not present in pJIR2774 and also is not present in several other pCW3-like plasmids, including the conjugative tetracycline resistance plasmid pJIR26 (9) and the β-toxin plasmid pJGS1495 (9, 33), which supports the previous conclusion that tcpB is not required for conjugation (33). Our analysis of pJIR2774 provides further evidence that conjugative plasmids from C. perfringens have the same mechanism of conjugative transfer (9, 21) and are derived from a common ancestor, most likely a Tn916-like conjugative transposon (8), but have diverged to the extent that they carry an array of other genes, which now includes a lincomycin resistance gene.
tISCpe8, tISSag10 (4, 5), and a putative element from Haemophilus parasuis have very similar genetic organizations (Fig. 3), and each element carries only two genes, a gene encoding a putative transposase and a lincomycin resistance gene. They are all members of the IS1595 family of transposon-like insertion elements and belong to the ISPna2 group (42). They appear to duplicate 8-bp sequences upon insertion and encode closely related transposases. These enzymes have several common regions that are important for their function, including a zinc finger motif, a putative helix-turn-helix motif, and a catalytic tetrad (42). Each element is also flanked by imperfect inverted repeats that are 24 and 25 bp long, with 50% identity. These regions are similar to other sequences located just upstream of similar transposase genes (Fig. 4B). The observation that different resistance genes are located downstream of the transposase genes in these elements (Fig. 3) suggests that there may be a mechanism by which gene cassettes can be inserted into or deleted from this group of elements, but there is no evidence for any integron-like structures.
tISCpe8 is capable of movement both within the original host strain and within recipient strains following DNA transfer by conjugation. Transposition has been demonstrated in C. perfringens, either by the presence of multiple insertions after conjugative transfer or following electroporation-mediated introduction of the element, in association with either pJIR2774 or the clostridial suicide plasmid pJIR1944.
The genetic and functional similarity between tISCpe8 and tISSag10 suggests that these elements originated from a common ancestor and that DNA transfer may have occurred between C. perfringens and S. agalactiae, possibly via intermediate hosts. Perhaps a progenitor transposon was disseminated either by a promiscuous conjugative plasmid or by mobilization by an oriT site within the element itself. Evidence for the latter scenario is provided by studies of tISSag10 that showed that this element contains a functional oriT site and can be mobilized by Tn916 (4). We have shown that tISCpe8 has a similar oriT site and also can be mobilized by Tn916. These data reinforce the notion that conjugation may be responsible for the movement of genetic elements that are not obviously mobile since they do not contain conjugation or mobilization genes and that this process plays a very significant role in bacterial adaptation and evolution.
Acknowledgments
Research at Monash University was supported by grants from the Monash University Small Grants Scheme, the Australian National Health and Medical Research Council, and the Australian Research Council. Research at the University of Arizona was partially supported by Alpharma Ltd.
We thank Michael Chandler and Patricia Siguier for helpful discussions.
Footnotes
Published ahead of print on 14 August 2009.
REFERENCES
- 1.Abraham, L. J., and J. I. Rood. 1985. Cloning and analysis of the Clostridium perfringens tetracycline resistance plasmid, pCW3. Plasmid 13:155-162. [DOI] [PubMed] [Google Scholar]
- 2.Abraham, L. J., and J. I. Rood. 1985. Molecular analysis of transferable tetracycline resistance plasmids from Clostridium perfringens. J. Bacteriol. 161:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abraham, L. J., A. J. Wales, and J. I. Rood. 1985. Worldwide distribution of the conjugative Clostridium perfringens tetracycline resistance plasmid, pCW3. Plasmid 14:37-46. [DOI] [PubMed] [Google Scholar]
- 4.Achard, A., and R. Leclercq. 2007. Characterization of a small mobilizable transposon, MTnSag1, in Streptococcus agalactiae. J. Bacteriol. 189:4328-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Achard, A., C. Villers, V. Pichereau, and R. Leclercq. 2005. New lnu(C) gene conferring resistance to lincomycin by nucleotidylation in Streptococcus agalactiae UCN36. Antimicrob. Agents Chemother. 49:2716-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams, V., D. Lyras, K. Farrow, and J. Rood. 2002. The clostridial mobilisable transposons. Cell. Mol. Life Sci. 59:2033-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Awad, M. M., and J. I. Rood. 1997. Isolation of α-toxin, θ-toxin and κ-toxin mutants of Clostridium perfringens by Tn916 mutagenesis. Microb. Pathog. 22:275-284. [DOI] [PubMed] [Google Scholar]
- 8.Bannam, T. L., P. K. Crellin, and J. I. Rood. 1995. Molecular genetics of the chloramphenicol-resistance transposon Tn4451 from Clostridium perfringens: the TnpX site-specific recombinase excises a circular transposon molecule. Mol. Microbiol. 16:535-551. [DOI] [PubMed] [Google Scholar]
- 9.Bannam, T. L., W. L. Teng, D. Bulach, D. Lyras, and J. I. Rood. 2006. Functional identification of conjugation and replication regions of the tetracycline resistance plasmid pCW3 from Clostridium perfringens. J. Bacteriol. 188:4942-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berryman, D. I., M. Lyristis, and J. I. Rood. 1994. Cloning and sequence analysis of ermQ, the predominant macrolide-lincosamide-streptogramin B resistance gene in Clostridium perfringens. Antimicrob. Agents Chemother. 38:1041-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berryman, D. I., and J. I. Rood. 1989. Cloning and hybridization analysis of ermP, a macrolide-lincosamide-streptogramin B resistance determinant from Clostridium perfringens. Antimicrob. Agents Chemother. 33:1346-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berryman, D. I., and J. I. Rood. 1995. The closely related ermB-ermAM genes from Clostridium perfringens, Enterococcus faecalis (pAMβ1), and Streptococcus agalactiae (pIP501) are flanked by variants of a directly repeated sequence. Antimicrob. Agents Chemother. 39:1830-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brynestad, S., M. R. Sarker, B. A. McClane, P. E. Granum, and J. I. Rood. 2001. Enterotoxin plasmid from Clostridium perfringens is conjugative. Infect. Immun. 69:3483-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601-610. [DOI] [PubMed] [Google Scholar]
- 15.Cassell, G. 1997. Emergent antibiotic resistance: health risks and economic impact. FEMS Immunol. Med. Microbiol. 18:271-274. [DOI] [PubMed] [Google Scholar]
- 16.Datta, N., R. W. Hedges, D. Becker, and J. Davies. 1974. Plasmid-determined fusidic acid resistance in Enterobacteriaceae. J. Gen. Microbiol. 83:191-196. [DOI] [PubMed] [Google Scholar]
- 17.Douthwaite, S. 1992. Interaction of the antibiotics clindamycin and lincomycin with Escherichia coli 23S ribosomal RNA. Nucleic Acids Res. 20:4717-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrow, K. A., D. Lyras, and J. I. Rood. 2001. Genomic analysis of the erythromycin resistance element Tn5398 from Clostridium difficile. Microbiology 147:2717-2728. [DOI] [PubMed] [Google Scholar]
- 19.Farrow, K. A., D. Lyras, and J. I. Rood. 2000. The macrolide-lincosamide-streptogramin B resistance determinant from Clostridium difficile 630 contains two erm(B) genes. Antimicrob. Agents Chemother. 44:411-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galimand, M., G. Gerbaud, M. Guibourdenche, J. Y. Riou, and P. Courvalin. 1998. High-level chloramphenicol resistance in Neisseria meningitidis. N. Engl. J. Med. 339:868-874. [DOI] [PubMed] [Google Scholar]
- 21.Hughes, M. L., R. Poon., V. Adams, S. Sayeed, S. Saputo, F. A. Uzal, B. A. McClane, and J. I. Rood. 2007. Epsilon toxin plasmids of Clostridium perfringens type D are conjugative. J. Bacteriol. 189:7531-7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johanesen, P. A., D. Lyras, T. L. Bannam, and J. I. Rood. 2001. Transcriptional analysis of the tet(P) operon from Clostridium perfringens. J. Bacteriol. 183:7110-7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jorgensen, J. H., S. A. Crawford, and K. R. Fiebelkorn. 2005. Susceptibility of Neisseria meningitidis to 16 antimicrobial agents and characterization of resistance mechanisms affecting some agents. J. Clin. Microbiol. 43:3162-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leclercq, R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482-492. [DOI] [PubMed] [Google Scholar]
- 25.Lyras, D., and J. Rood. 2000. Transposition of Tn4451 and Tn4453 involves a circular intermediate that forms a promoter for the large resolvase, TnpX. Mol. Microbiol. 38:588-601. [DOI] [PubMed] [Google Scholar]
- 26.Lyras, D., and J. I. Rood. 1996. Genetic organization and distribution of tetracycline resistance determinants in Clostridium perfringens. Antimicrob. Agents Chemother. 40:2500-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyristis, M., A. E. Bryant, J. Sloan, M. M. Awad, I. T. Nisbet, D. L. Stevens, and J. I. Rood. 1994. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol. Microbiol. 12:761-777. [DOI] [PubMed] [Google Scholar]
- 28.Martel, A., L. A. Devriese, K. Cauwerts, K. De Gussem, A. Decostere, and F. Haesebrouck. 2004. Susceptibility of Clostridium perfringens strains from broiler chickens to antibiotics and anticoccidials. Avian Pathol. 33:3-7. [DOI] [PubMed] [Google Scholar]
- 29.McClane, B. A., D. M. Lyerly, J. S. Moncrief, and T. D. Wilkins. 2000. Enterotoxic clostridia: Clostridium perfringens type A and Clostridium diffcile, p. 531-562. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, DC.
- 30.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 31.Miyamoto, K., D. J. Fisher, J. Li, S. Sayeed, S. Akimoto, and B. A. McClane. 2006. Complete sequencing and diversity analysis of the enterotoxin-encoding plasmids in Clostridium perfringens type A non-food-borne human gastrointestinal disease isolates. J. Bacteriol. 188:1585-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nirdnoy, W., C. J. Mason, and P. Guerry. 2005. Mosaic structure of a multiple-drug-resistant, conjugative plasmid from Campylobacter jejuni. Antimicrob. Agents Chemother. 49:2454-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parsons, J. A., T. L. Bannam, R. J. Devenish, and J. I. Rood. 2007. TcpA, an FtsK/SpoIIIE homolog, is essential for transfer of the conjugative plasmid pCW3 from Clostridium perfringens. J. Bacteriol. 189:7782-7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts, M. C. 2008. Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol. Lett. 282:147-159. [DOI] [PubMed] [Google Scholar]
- 35.Rood, J. I. 1983. Transferable tetracycline resistance in Clostridium perfringens strains of porcine origin. Can. J. Microbiol. 29:1241-1246. [DOI] [PubMed] [Google Scholar]
- 36.Rood, J. I., E. A. Maher, E. B. Somers, E. Campos, and C. L. Duncan. 1978. Isolation and characterization of multiply antibiotic-resistant Clostridium perfringens strains from porcine feces. Antimicrob. Agents Chemother. 13:871-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rood, J. I., V. N. Scott, and C. L. Duncan. 1978. Identification of a transferable resistance plasmid (pCW3) from Clostridium perfringens. Plasmid 1:563-570. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 39.Scott, P. T., and J. I. Rood. 1989. Electroporation-mediated transformation of lysostaphin-treated Clostridium perfringens. Gene 82:327-333. [DOI] [PubMed] [Google Scholar]
- 40.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shultz, T. R., J. W. Tapsall, P. A. White, C. S. Ryan, D. Lyras, J. I. Rood, E. Binotto, and C. J. Richardson. 2003. Chloramphenicol-resistant Neisseria meningitidis containing catP isolated in Australia. J. Antimicrob. Chemother. 52:856-859. [DOI] [PubMed] [Google Scholar]
- 42.Siguier, P., L. Gagnevin, and M. Chandler. 2009. The new IS1595 family, its relation to IS1 and the frontier between insertion sequences and transposons. Res. Microbiol. 160:232-241. [DOI] [PubMed] [Google Scholar]
- 43.Spigaglia, P., F. Barbanti, and P. Mastrantonio. 2007. Detection of a genetic linkage between genes coding for resistance to tetracycline and erythromycin in Clostridium difficile. Microb. Drug Res. 13:90-95. [DOI] [PubMed] [Google Scholar]
- 44.Spigaglia, P., and P. Mastrantonio. 2002. Analysis of macrolide-lincosamide-streptogramin B (MLS(B)) resistance determinant in strains of Clostridium difficile. Microb. Drug Res. 8:45-53. [DOI] [PubMed] [Google Scholar]
- 45.Spizek, J., J. Novotna, and T. Rezanka. 2004. Lincosamides: chemical structure, biosynthesis, mechanism of action, resistance, and applications. Adv. Appl. Microbiol. 56:121-154. [DOI] [PubMed] [Google Scholar]
- 46.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 47.Weisblum, B. 1995. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 39:577-585. [DOI] [PMC free article] [PubMed] [Google Scholar]