Abstract
The periodontal pathogen Aggregatibacter actinomycetemcomitans displays on the bacterial surface a nonfimbrial adhesin, EmaA, which is required for collagen binding. In this study, electron tomography was used to characterize the three-dimensional (3D) architecture of this adhesin. The antenna-like surface appendages, corresponding to EmaA, were found to be composed of an ellipsoidal domain capping a rod-like domain that adopts either a straight or a bent conformation at various positions along the length. The most common flexible point along the length of the EmaA appendage was localized 29.4 nm away from the distal end. One-fifth of the appendages were straight and the remaining showed angles distributed between 140° and 170° at this location. Deletion analysis mapped this bend to amino acids 611 to 640 of the protein sequence. The 3D structure of the collagen binding domain of EmaA was generated by alignment and averaging of 9 subvolumes of the adhesin extracted from tomograms. The structure contains three subdomains: a globular structure with a diameter of ∼5 nm and a cylindrical domain (∼4.4 nm by 5.8 nm) separated by a linker region with a diameter of ∼3 nm, followed by a cylindrical domain (∼4.6 nm by 6.6 nm). This is the first 3D structure of a trimeric autotransporter protein of A. actinomycetemcomitans.
Bacterial adhesion to host receptors, a crucial step for colonization and infection, is mediated by fimbrial and nonfimbrial adhesins. These adhesins are proteinaceous appendages displayed on the surface of bacteria and contain the receptor binding domains. Aggregatibacter actinomycetemcomitans, a gram-negative, nonmotile bacterium is found associated with periodontal diseases and other extraoral infections (12, 23, 32, 40). When isolated from the oral cavity, the bacterium exists as a fimbriated form and switches to an afimbriated form upon planktonic subculturing (5, 14). A. actinomycetemcomitans fimbriae mediate the nonspecific adherence of the bacterium to abiotic and organic surfaces and decorate the bacterial surface with long fibrils of 5 to 7 nm in diameter (14, 15). In addition to fimbriae, nonfimbrial adhesins, which mediate the specific binding to host cells and tissues, have been identified in this bacterium (1, 6, 19, 27, 29). Among these nonfimbrial adhesins, only the extracellular matrix protein adhesin A, EmaA, has been visualized forming structures on the bacterial surface by transmission electron microscopy (29).
EmaA is an outer membrane collagen adhesin unique to A. actinomycetemcomitans; however, orthologous proteins exist in other bacterial genera (13, 18, 21, 26, 33, 38). The protein is encoded by a 6-kb gene present in all A. actinomycetemcomitans strains investigated (36). Genetic heterogeneity within the gene exists between different strains, which are based on the serotype of the organism. Based on this heterogeneity, two molecular forms of the protein have been identified: a full-length and an intermediate form. The prototypic or full-length protein exists as a 202-kDa protein and shares 75% amino acid homology with the intermediate form. The intermediate protein form (173 kDa) contains an in-frame 279-amino-acid deletion but maintains collagen binding activity and surface appendages similar to the prototypic form (36).
EmaA is associated with the binding of A. actinomycetemcomitans to both isolated acid-soluble collagen and collagen found in tissues (19, 29, 35, 39). The specificity of EmaA for collagen was demonstrated using a rabbit cardiac valve tissue model (35). Valves with an intact endothelium bound equal amounts of the wild type or emaA isogenic mutants. Removal of the endothelium by trypsin treatment, thereby exposing the underlying collagen, did not affect the level of binding of the mutant. However, the number of wild-type bacteria bound to the exposed collagen was five times the number of mutant bacteria. This represents a 10-fold increase with respect to the number of bacteria bound to the endothelium. The role of EmaA as a virulence determinant in A. actinomycetemcomitans infection was demonstrated in a rabbit endocarditis infection model, in which the wild-type bacterium outcompeted the binding of the mutant 10-fold (35).
Sequence analysis indicates that EmaA belongs to the Oca (oligomeric coiled-coil adhesin) family of autotransporter adhesins (19). Multimers of EmaA oligomerize to form appendages on the bacterial surface and are visible as long rods or antenna-like structures capped by an ellipsoidal domain (29). A strong correlation exists between the translocated region of the protein (head and stalk domains) and the structural features. The head domain, consisting of amino acids 70 to 386, forms the ellipsoidal ending of the appendage, which is essential for collagen binding, while amino acids 387 to 1900 form the stalk domain (39).
Contained within the translocation domain of EmaA are three “neck” sequences, which are conserved in the Oca family protein members (21, 29, 33). These sequences are considered to stabilize the oligomer and transition between β-rolls and coiled-coil regions of the molecule (21, 26). In the EmaA sequence, two “neck” sequences are found within the first 628 amino acids of the protein sequence (19, 29). The third sequence is located in the stalk domain adjacent to the carboxy-terminal membrane anchor domain, which comprises amino acids 1901 to 1965 (19, 29). The membrane anchor domains of three or four monomers are proposed to form β-barrels that are required for pore formation and protein translocation (18, 29, 37).
The translocated domain of EmaA has been subjected to a two-dimensional (2D) study by transmission electron microscopy, and the overall dimensions of the EmaA appendages have been determined by the analysis of a large number of micrographs (29). The ellipsoidal ending shows diameters of 2.8 by 4.6 nm, and the stalk domain, which is at least 150 nm long, has a diameter of 4.1 nm. Several conformations of the stalk domain were present in the micrographs: either straight or containing a bend at 29.2 nm from the distal end. This bend position was correlated with amino acids localized between the first two neck sequences (29).
In this study, electron tomography was used to characterize the 3D structure of the EmaA appendages of A. actinomycetemcomitans in situ. The functional domain of EmaA was found to be composed of three distinct subdomains followed by a long stalk domain. Distinct regions of the molecule were identified that provide flexibility for the molecule and allow for the deformation or bending of the adhesin. A correlation between these flexible regions and specific amino acids in the sequence was ascertained.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids are listed in Table 1. A. actinomycetemcomitans strains were grown statically in trypticase soy broth (Becton Dickinson, Sparks, MD) supplemented with 0.6% yeast extract (TSBYE; Becton Dickinson). One bacterial colony was inoculated in broth and grown in a humidified, 10% CO2 atmosphere at 37°C for 16 h. One-milliliter cultures were subsequently diluted to 10 ml with fresh TSBYE broth and incubated for 2 to 3 h until mid-log phase (optical density at 495 nm [OD495], 0.2 to 0.4). Escherichia coli strains were inoculated in Luria-Bertani medium (USB Co., Cleveland, OH) and grown at 37°C with agitation.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| A. actinomycetemcomitans strains | ||
| VT1169 WT | Afimbriated strain with spontaneous rifampin- and nalidixic acid-resistant strain derived from strain SUNY465 | 20 |
| WT/emaA+ | Wild-type strain transformed with plasmid pKM9, containing the full-length emaA gene and 500-bp putative promoter region, to overexpress EmaA | This study |
| emaA mutant | Allelic replacement mutant with antibiotic cassette coding for resistance to spectinomycin inserted into unique HindIII site within the emaA gene | 19 |
| emaA mutant/pKM1 | emaA mutant strain transformed with plasmid pKM1 | 29 |
| Complemented strains | emaA mutant strain complemented with plasmid pKM9 | 29 |
| morC mutant | Allelic replacement mutant with antibiotic cassette coding for resistance to spectinomycin inserted at 5′ end of morC | 9 |
| morC/emaA+ | morC mutant strain transformed with plasmid pKM9 to overexpress EmaA | This study |
| E. coli DH10B | F−endA1 recA1 galU galK deoR nupG rpsL ΔlacX74 φ80lacZΔM15 araD139 Δ(ara leu)7697 mcrA Δ(mrr-hsdRMS-mcrBC)λ− | 11 |
| Plasmids | ||
| pKM1 | pDMG4 with aminoglycoside phosphotransferase gene (kan) replacing spectinomycin adenyltransferase gene (aad9) | 29 |
| pKM9 | emaA gene with 500-bp upstream sequence of the start codon cloned into pKM1 | 29 |
| pKMSmaI1504 | pKM9 with SmaI site introduced 1,504 bp downstream of start codon of emaA | This study |
| pKM1603C | pKM9 with adenine at bp 1603 mutated to a cytosine | This study |
| pKMSmaI1603 | pKM9 with SmaI site introduced 1,603 bp downstream of start codon of emaA derived from pKM1603C | This study |
| pKMSmaI1831 | pKM9 with SmaI site introduced 1,831 bp downstream of start codon of emaA | This study |
| pKMSmaI1924 | pKM9 with SmaI site introduced 1,924 bp downstream of start codon of emaA | This study |
| pKMΔ502-534 | In-frame deletion of bp 1504 to 1602 of coding sequence of the emaA gene engineered using pKMSmaI1504 and pKMSmaI1603 | This study |
| pKMΔ535-610 | In-frame deletion of bp 1603 to 1830 of coding sequence of the emaA gene engineered using pKMSmaI1603 and pKMSmaI1831 | This study |
| pKMΔ611-641 | In-frame deletion of bp 1831 to 1923 of coding sequence of the emaA gene engineered using pKMSmaI1831 and pKMSmaI1924 | This study |
The plasmid pKM9, containing the full-length emaA gene and 500 bp upstream of the start methionine, was constructed as described previously (29). pKM1, the empty shuttle plasmid, was transformed into the emaA mutant strain (19) as a negative control for the collagen binding assays. The emaA mutant strain transformed with pKM9 was designated as the complemented strain. All of the emaA in-frame deletion mutations were constructed using pKM9 as template. In-frame deletion mutants were constructed based on a site-directed mutagenesis strategy using the QuikChange XL kit (Stratagene, La Jolla, CA) as described previously (39). SmaI restriction sites were generated within the specific sequences of emaA using the overlapping mutagenesis primers listed in Table 2. The strains containing these plasmids were propagated in medium containing 50 μg/ml kanamycin.
TABLE 2.
Oligonucleotides used for generating SmaI sites in the emaA gene
| Oligonucleotide | Sequence (5′ to 3′)a | Positionb (bp) |
|---|---|---|
| SmaI1504f | CCGATAGTAATTACGACAATCCCGGGGCTAAAGCAGTAGGTTC | 1483-1526 |
| SmaI1504r | GAACCTACTGCTTTAGCCCCGGGATTGTCGTAATTACTATCGG | 1483-1526 |
| 1603Cf | TTGGTTTAAACAGCAATGTTCCCGGCCAAAATACCGTTGCA | 1583-1623 |
| 1603Cr | TGCAACGGTATTTTGGCCGGGAACATTGCTGTTTAAACCAA | 1583-1623 |
| SmaI1603f | ACAGCAATGTTCCCGGGCAAAATACCGTTGCATT | 1592-1625 |
| SmaI1603r | AATGCAACGGTTATTTTGCCCGGGAACATTGCTGT | 1592-1625 |
| SmaI1831f | CGTCAAATTAAAAATGTGGCACCCGGGAATGTTGCGGCAAACTC | 1810-1853 |
| SmaI1831r | GAGTTTGCCGCAACATTCCCGGGTGCCACATTTTTAATTTGACG | 1810-1853 |
| SmaI1924f | CAAGGTTGGCAAATCACTCCCGGGGTAGAAAATGGTGG | 1906-1943 |
| SmaI1924r | CCACCATTTTCTACCCCGGGAGTGATTTGCCAACCTTG | 1906-1943 |
The generated SmaI sites are underlined and the mutated nucleotides are indicated in boldface.
The position of the indicated oligonucleotide in the emaA sequence downstream of the start codon (accession number AY344064).
Collagen binding assay.
Binding of A. actinomycetemcomitans strains to human type V collagen (Sigma Chemical Co., St. Louis, MO) was determined using an enzyme-linked immunoabsorbent assay as previously described (20, 29, 39). Each strain was tested in triplicate. The OD490 value of the background (no added bacteria) was subtracted from the mean OD490 value of the experimental sample and the data are represented as the percentage of binding activity of the complemented strain: [(OD490 target − OD490 background)/(OD490 complemented − OD490 background)] × 100%. The final results are presented as the means ± the standard deviations of three independent experiments. The data were statistically analyzed using Student's t test. A P value of <0.05 was considered significant.
Electron microscopy sample preparation and data acquisition.
Bacterial samples were prepared as described by Ruiz et al. (29). Briefly, mid-logarithmic bacteria were collected and resuspended in phosphate-buffered saline (10 mM sodium phosphate, 150 mM NaCl, pH 7.4) on ice to a final concentration of 5 × 109 CFU/ml. Five-microliter aliquots of bacterial suspensions were applied to copper grids coated with a thin layer of carbon, rinsed with phosphate-buffered saline, and stained using either 2% phosphotungstic acid (pH 7; Ted Pella, Redding, CA) or Nano W (Nanoprobes, Yaphank, NY). For electron tomography, the carbon-coated grids were pretreated with 5 μl polylysine (molecular weight, 2,500 to 4,000; 0.05 mg/ml; Sigma) for 1 min followed by 5 μl of colloidal gold (SPI, West Chester, PA) for 1 min before adding the bacterial suspension. Data collection was carried out using a Tecnai 12 electron microscope (FEI, Hillsboro, OR) equipped with a LaB6 cathode, a 14-μm 2,048- by 2048-pixel charge-coupled device (CCD) camera (TVIPS, Gauting, Germany) and a dual axis tilt tomography holder (Fischione, Export, PA), operating at 100 kV. 2D micrographs were recorded in the CCD camera at a 67,000× nominal magnification, which corresponds to a 0.196-nm pixel size on the specimen scale. Single-axis tomography tilt series were recorded in the CCD camera over a ±60° or ±70° angular range in 2° increments. The series of projections were collected at either 52,000× or 42,000× nominal magnification, which corresponds to either a 0.250-nm or 0.308-nm pixel size on the specimen scale, respectively.
2D image analysis.
Electron micrographs of each strain were visualized using ImageJ (10), and the distances of the bending positions from the apical end of the EmaA appendages were measured. The number of molecules showing a specific bend was represented as the percentage relative to the total number of EmaA appendages. The electron micrographs of each strain were divided, for statistical purposes, into groups containing at least 70 EmaA appendages per group. Averages and standard deviations for each of the bend distances were calculated for each group and analyzed for statistical significance. The final results were presented as the mean of the percentage ± the standard deviation. Statistical analysis used Student's t test, and a P value of <0.05 was considered significant.
Tomography data analysis.
The projections of each single-axis tomography tilt series were aligned to a common origin using IMOD (17). A preliminary alignment was obtained by cross-correlation and was subsequently refined by fiducial marker alignment. The tomograms were reconstructed using a weighted back-projection algorithm (25) in the SPIDER image processing system (version 5 with modifications [8]). EmaA appendages were identified in the projection of the whole tomogram. The z slices of the tomogram containing the appendages were visualized using WEB (8). The x and y coordinates defining the edges of a box encompassing the appendages were recorded, and subvolumes comprising the whole thickness of the tomogram were extracted. Projection onto convex sets (Pocs) on the extracted subvolumes was used to minimize the effect of the missing wedge (4). The subvolumes were rewindowed to the appropriate z dimension for each EmaA appendage, and the appendages in the subvolumes were segmented semiinteractively using the program JUST (31). The segmented information was used to create a mask for subsequent processing.
Alignment of 3D volumes of the EmaA appendages.
Volumes containing segmented EmaA appendages were aligned in three steps. First, the volumes were visualized using the program Chimera (22), and the appendages were coarsely aligned by hand to a common orientation. Second, the alignment of the whole EmaA appendages was refined using Radon transform algorithms (24). Finally, the outermost region of the EmaA appendage (30 nm from the apical end) was windowed out and used for subsequent alignment steps. For each round of refinement, three orthogonal projections of the volumes (0°, 0°; 0°, 90°; and 90°, 90°) were computed and rotationally/translationally aligned to a selected reference. Initially, the alignment was carried over an angular range of 180° in 5° angular increments. The averaged x, y, and z shifts were calculated from the 2D shifts of the three projections (2, 28, 30). The Euler angles for the volume rotation were determined from the angles obtained from the alignment of the (90°, 90°) projection. The volumes that showed high cross-correlation coefficients were averaged to create a volume with a higher signal/noise ratio. The averaged volume served as a new reference in the next round of refinement, which was carried out over a smaller angular range and with smaller angular increments.
RESULTS
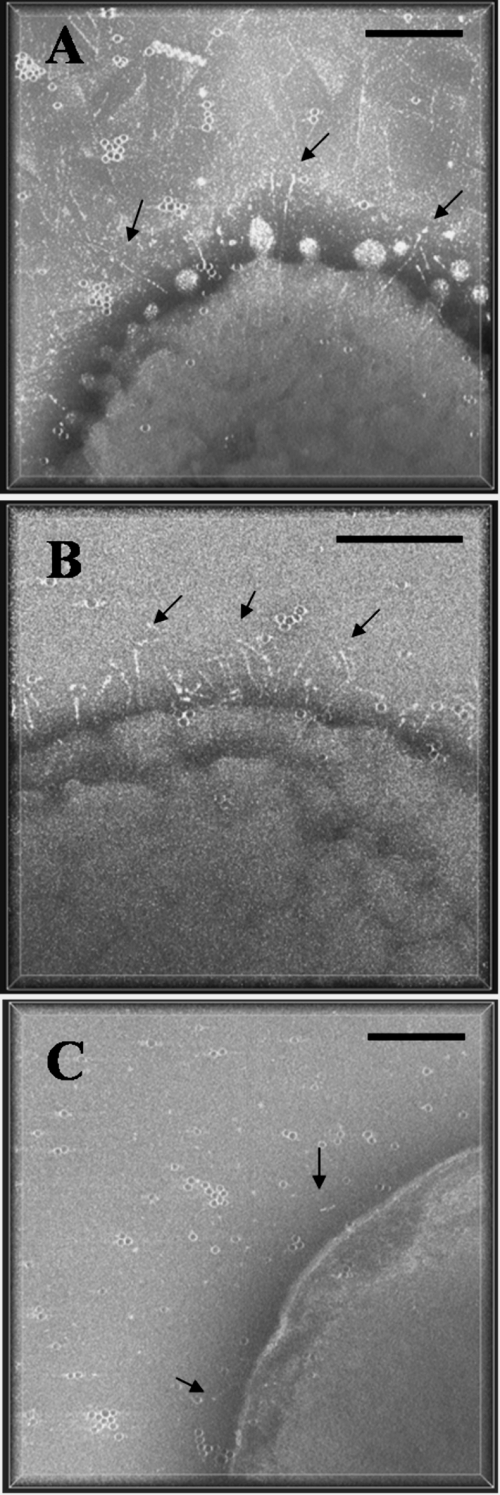
The surface of A. actinomycetemcomitans was investigated using negatively stained whole-mount preparations of the bacterial cells. Preliminary studies on three different strains were performed to determine the optimal strain for EmaA appendage visualization. The strains chosen for this study were (i) the wild type (WT), which contains a moderate number of EmaA molecules, (ii) a wild-type strain transformed with emaA (WT/emaA+), which overexpresses EmaA appendages, and (iii) a morC/emaA+ mutant strain, constructed from the parent morC mutant strain, that shows minimal convolutions (9) and may aid in the visualization of the entire EmaA appendage. The 3D tomograms presented in perspective clearly delineated the surface features of the three strains (Fig. 1). In all of the strains, the EmaA appendages were more abundant at the apical ends of the bacteria. Although in the morC/emaA+ strain the EmaA appendages were unobscured by membrane convolutions or secreted vesicles, the low aspect ratio of the bacterial dimensions made it difficult to distinguish the apical end of the bacteria to collect the data under the low-dose conditions used in this study (Fig. 1C). EmaA appendages were very abundant in the WT/emaA+ strain but were not well separated in 3D (Fig. 1B). Therefore, the WT strain was selected to study the structure of the EmaA appendages (Fig. 1A). A total of 23 tomograms were collected from this strain for further analysis.
FIG. 1.
Perspective view along the z axis of 3D tomograms of A. actinomycetemcomitans strains acquired from whole-mount preparations of the bacterium. The amount of perspective is indicated by the white bounding box framing the tomogram. (A) WT strain stained with Nano W; (B) WT/emaA+ strain stained with 2% phosphotungstic acid (pH 7); (C) morC/emaA+ strain, a morC mutant strain transformed with pKM9, stained with Nano W. Arrows indicate EmaA appendages on the bacterial surface. Dark points are colloidal gold particles. Bars, 100 nm.
Whole tomograms were segmented semiinteractively to separate the different structures on the surface of the bacterium, thus allowing the investigation of the relationship between these structures in situ. Densities representing the EmaA appendages and the bacterial membrane surface were labeled in blue and green, respectively (see Fig. S1 in the supplemental material). Although regions of the EmaA appendage were found at the level of the membrane, the loci where EmaA anchors to the membrane could not be resolved due to the high electron density of proteins and membrane components inside the bacterial cell. Therefore, the complete length of the EmaA appendage could not be determined under these experimental conditions. Since whole tomograms are very large to manipulate, subvolumes containing single EmaA appendages were extracted from the tomograms for a more detailed analysis. In addition, the EmaA appendages were segmented from the subvolumes (data not shown), and a 3D mask was created to aid in the visualization and subsequent alignment of the EmaA appendages.
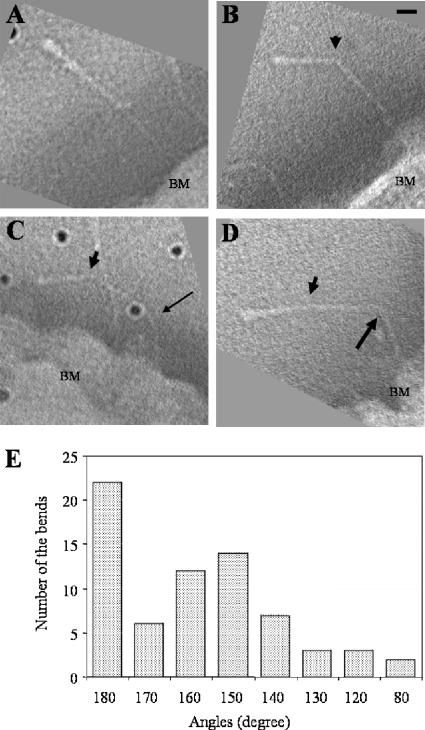
The morphologies of 114 EmaA 3D structures were analyzed. Twenty-two appendages displayed straight stalks (Fig. 2A), while each of the remaining 92 appendages contained either one (Fig. 2B) or multiple (Fig. 2C and D) bends at different positions in the structure. The positions of the bends were calculated as the distance from the bend to the distal end of the appendage. The distances for all the bending positions were recorded, analyzed for statistical significance, and divided into 14 groups (Table 3). The data suggest that the most common bend was found at a distance of 29.4 ± 1.8 nm from the distal end of the appendage (Fig. 2B). Two other bends were less frequently observed within the first 29.4 nm of the EmaA appendage at 10.3 nm and 23.5 nm (Table 3). The angles formed by the bend at 29.4 nm were mainly distributed between 140° and 170° (Fig. 2E), with only a few angles smaller than 140°. The angles of the bends at other positions in the structure were similar (data not shown).
FIG. 2.
Projections through EmaA subvolumes extracted from the tomograms showing different morphologies of the EmaA appendages. Subvolumes have been rotated to visualize the EmaA appendage parallel to the imaging plane. (A) Straight EmaA appendage; (B) EmaA appendage with a bend at 29.4 nm (arrowhead) from the apical ending of the structure; (C) EmaA appendage with bends at 29.4 nm (arrowhead) and 65.6 nm (thin arrow); (D) EmaA appendage with bends at 29.4 nm (arrowhead) and 68.8 nm (thick arrow). BM, bacterial membrane. Bar, 10 nm. Dark points are gold particles. (E) Angular distribution of the bends at 29.4 nm.
TABLE 3.
Specific bending positions measured from the distal end the EmaA appendage and the number of EmaA appendages bent at that location
| Distance (nm) of bend from distal enda | No. of EmaA appendagesb |
|---|---|
| 10.3 ± 0.5 | 4 |
| 23.5 ± 0.6 | 4 |
| 29.4 ± 1.8 | 47 |
| 34.0 ± 0.6 | 7 |
| 38.2 ± 1.6 | 17 |
| 44.6 ± 1.3 | 14 |
| 51.2 ± 1.2 | 6 |
| 59.8 ± 0.7 | 3 |
| 62.4 ± 0.9 | 3 |
| 65.6 ± 0.9 | 8 |
| 68.8 ± 0.8 | 5 |
| 71.9 ± 1.2 | 5 |
| 76.6 ± 1.1 | 3 |
| 86.5 ± 2.6 | 4 |
Each group of bending positions is significantly different from the neighboring groups (P < 0.05).
The total number of EmaA appendages is 114. A total of 130 bends were found in these appendages, while 22 appendages were straight.
The protein sequence corresponding to the bend at 29.4 nm was broadly mapped to the region between amino acids 502 and 641, which is localized between the first two neck sequences. Three in-frame deletion mutants (Δ502-534, Δ535-610, and Δ611-641) were constructed to refine the mapping of this sequence. Plasmids containing these constructs were transformed into the emaA mutant strain, 2D electron micrographs were collected, and the surface appendages formed by the truncated EmaA proteins were analyzed. The bend localized at ≤10.3 nm was excluded, since it was unrelated to these sequences. The emaA mutant strain complemented with the full-length emaA displayed 53% of the appendages with a bend at 29.4 nm (Table 4). The strains containing the Δ502-534 and Δ535-610 constructs had similar percentages of bends at 29.4 nm as the complemented strain (Table 4). In contrast, the strain containing the Δ611-641 construct displayed only 13% of the EmaA appendages with a bend at 29.4 nm compared to the complemented strain (Table 4). It was surprising that this value did not approach zero, if these amino acids are truly correlated with the bend at 29.4 nm. In this regard, it is useful to compare the results from the 2D and 3D analysis of the WT strain. Concomitant with the reduction of the percentage of bends at 29.4 nm, the percentage of the straight EmaA appendages of the strain containing the Δ611-641 construct increased compared with the other strains (Table 4). There was no significant difference between the percentages of the bends at other positions in the structures of these strains (Table 4). The apparent discrepancy can be accounted for by the foreshortening of distances (by cos[α]) in the 2D projections, which affects the correct assignment of the bend location. Based on this fact, the 13% of EmaA appendages displaying a bend at 29.4 nm is within the discrepancy margin and is essentially equivalent to zero.
TABLE 4.
EmaA appendages bending at different positionsa and straight EmaA appendages based on both 2D and 3Db analyses
| Analysis | Strain | % of EmaA appendages
|
||
|---|---|---|---|---|
| With bends at 29.4 nmc | With bends at other positions | That were straight | ||
| 2D | Complemented | 53 ± 3.5 | 40 ± 4.6 | 25 ± 3.7 |
| 2D | Δ502-534 | 53 ± 1.6 | 51 ± 8.0 | 25 ± 3.2 |
| 2D | Δ535-610 | 48 ± 9.0 | 56 ± 5.6 | 29 ± 6.3 |
| 2D | Δ611-641 | 13 ± 1.7d | 53 ± 8.3 | 49 ± 3.7 |
| 2D | Wild type | 69 ± 0.3 | 58 ± 8.8 | 16 ± 3.2 |
| 3D | Wild type | 45e | 69 | 19 |
An EmaA appendage may contain multiple bending positions; therefore, the sums of the percentages for a strain may be more than 100%.
The 3D analysis used the 114 EmaA 3D structures and the bending data listed in Table 3.
The bending positions that had a distance between 10.3 nm and 29.4 nm were all considered as the bend at 29.4 nm.
The percentage of the appendages bending at 29.4 nm formed by the deletion construct Δ611-641 was significantly different from those of other strains (P < 0.05).
The percentage includes bends at both 29.4 nm and 23.5 nm (see Table 3).
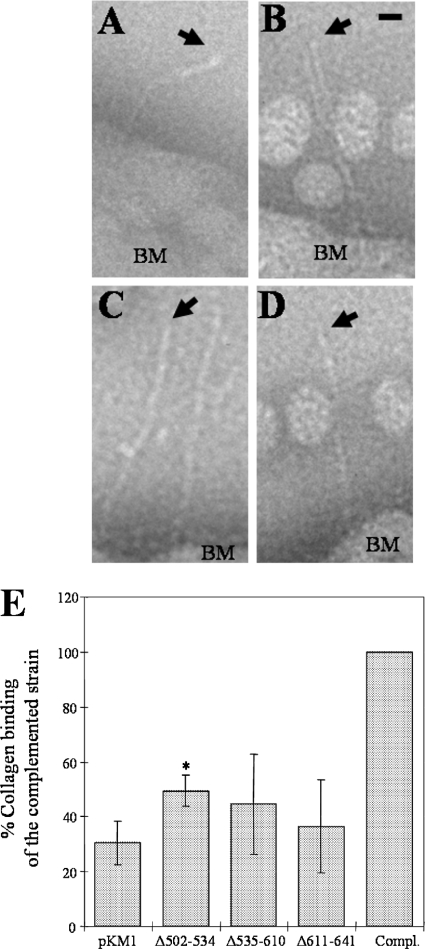
Gain-of-function studies of the strains containing the deletion constructs were performed to determine the role of these sequences in collagen binding. All the mutant strains showed a decrease in collagen binding activity compared to the complemented strain (Fig. 3E). The strain containing Δ502-534 restored 49.5% of the collagen binding activity compared with the complemented strain. This level of binding was slightly higher than that of the negative control (emaA mutant transformed with the empty shuttle plasmid pKM1). The strains containing the Δ535-610 and Δ611-641 constructs restored 44.6% and 36.5% of the collagen binding activity, respectively, compared with the activity of the complemented strain. The percentages of binding of these two strains were not significantly different from that of the negative control (Fig. 3E). The distal end of the EmaA appendages of the three deletion constructs did not form the prototypic ellipsoidal ending domain when observed by transmission electron microscopy (Fig. 3; see also Fig. S2 in the supplemental material). In some cases, a small enlargement of the stalk domain was observed (Fig. 3B). Nonetheless, the ending was still quite distinct from the prototypic ending domain of the WT strain. Therefore, the lack of these sequences appears to affect the structure of the binding domain and the collagen binding ability concurrently.
FIG. 3.
EmaA appendages windowed from electron micrographs of A. actinomycetemcomitans strains acquired from whole-mount preparations of the bacteria stained with 2% phosphotungstic acid (pH 7). (A) WT strain; (B) emaA mutant strain transformed with pKMΔ502-534; (C) emaA mutant strain transformed with pKMΔ535-610; (D) emaA mutant strain transformed with pKMΔ611-641. Arrows point to the apical end of the EmaA appendages. Spherical particles in the images are extracellular vesicles. BM, bacterial membrane. Bar, 10 nm. (E) Collagen binding activities of the emaA mutant strains in panels A to D and pKM9 (plasmid containing the full-length emaA gene and the upstream putative promoter region [Compl.]) as measured by enzyme-linked immunosorbent assay. The data are represented as the percentage of the binding of the full-length complemented strain (Compl.). *, statistically significantly different from the emaA mutant/pKM1 strain.
Fine detailed structural information was difficult to obtain from individual EmaA subvolumes extracted from the tomograms. The low electron dose used to minimize radiation damage during data collection and the reduced number of projections, 71, contributing to each volume resulted in structures with high noise levels. However, a higher signal to noise (S/N) ratio structure can be obtained by averaging volumes that have been previously well aligned. Since 41% of the subvolumes contained a bend at 29.4 nm, the alignment steps were carried out using an additional mask of 30 nm from the apical end of the EmaA appendages, thus increasing the number of subvolumes that could be included in the alignment. Alignment procedures showed good performance when applied to volumes containing isolated structures and comprising no additional features (e.g., vesicles or gold particles). The 114 subvolumes containing EmaA appendages extracted from the tomograms were analyzed to determine good candidates for 3D alignment. Many of the original EmaA subvolumes contained additional features (e.g., gold particles, vesicles, EmaA appendages) very close to the EmaA appendage of interest. Although these features do not prevent a reliable manual study, they pose a hindrance for the alignment procedures. Thus, only 25 EmaA subvolumes were selected for further processing. Further reductions in the number of subvolumes available for averaging arose from bends located at distances closer than 29.4 nm from the apical end and from deficient alignment due to the low S/N ratios of single subvolumes. Therefore, only 9 EmaA subvolumes could be averaged.
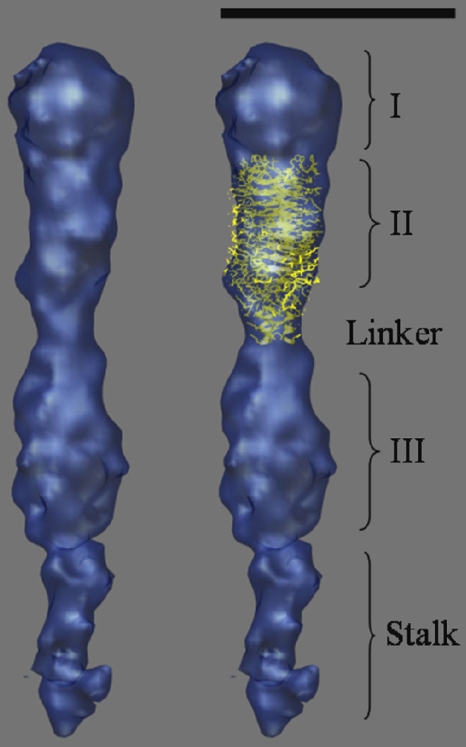
An average of 9 EmaA subvolumes that showed the highest cross-correlation coefficients, after several rounds of alignment and refinement, was calculated. The averaging resulted in a 3D structure of the apical end of the EmaA structure with increased contrast, higher S/N ratio, and well-defined densities. The structure can be clearly divided into three main subdomains, which show strong densities and have been labeled as subdomains I, II, and III (Fig. 4). The amino-terminal subdomain I has a globular shape with a diameter of 5 nm and corresponds to the ellipsoidal ending structure observed in 2D micrographs and reported in the previous 2D analysis of EmaA (29). Adjacent to this globular domain is subdomain II, which has a more cylindrical shape with dimensions of 4.4 nm by 5.8 nm (Fig. 4). Subdomain III, with dimensions of 4.6 nm by 6.6 nm, has a similar shape as subdomain II. Between subdomains II and III, at 10.8 nm from the apical end of the appendage, a narrow density of 3 nm in diameter and 3 nm long is observed (labeled as the linker). Subdomain III is followed by a narrow density (∼3 nm in diameter), which represents the beginning of the stalk domain of EmaA. All the subdomains show finer detailed features represented as mounds and valleys on the surface of the structure. A more detailed interpretation of these features, however, awaits the availability of a higher-resolution structure.
FIG. 4.
(Left) Surface representation of the 3D structure of the apical 30 nm of the EmaA appendage obtained by averaging 9 EmaA subvolumes extracted from the tomograms. (Right) Manual docking of the atomic model of the YadA head domain into subdomain II. Three subdomains, the linker region, and the stalk domain are labeled. Bar, 10 nm.
DISCUSSION
A. actinomycetemcomitans is considered to be the major pathogen in the development of an acute form of periodontal disease that infects young individuals (12, 32). Localized aggressive periodontitis is characterized by rapid periodontal attachment loss and leads to bone destruction (16). Genetic and biochemical studies have revealed a number of determinants involved in the initiation of oral and nonoral infections associated with this bacterium (7). These virulence factors are either membrane associated or require the membrane for secretion. Even though there is a wealth of information at the genetic and biochemical levels, there is a paucity of structural information describing these known or putative virulence factors that are involved in establishing disease. Our work attempts to characterize, for the first time, the 3D architecture of the associated surface structures of this periodontal pathogen.
The outer membrane surface of afimbriated A. actinomycetemcomitans strains is decorated with antenna-like appendages which are composed of a multimer of the collagen binding adhesin EmaA (19, 29). EmaA is also present on the surface of fimbriated strains obtained from clinical isolates (data not shown). Sequence analysis has shown that EmaA possesses all the characteristics of the Oca family of adhesins (19). The monomer molecular mass of 202 kDa makes EmaA one of the larger members of this family of proteins, compared with YadA (42 kDa), UspA1 (83 kDa), UspA2 (60 kDa), Hia (114 kDa), and BadA (340 kDa) (13, 33, 38). In contrast with YadA, UspA1/UspA2 and BadA, which are expressed at high relative densities on the bacterial surface (13, 33), EmaA is sparsely distributed on the surface and can be more easily found at the apical end of the bacterium (29, 36, 39). The low number of EmaA adhesins on the bacterial surface may appear to put A. actinomycetemcomitans at a disadvantage to interact with extracellular matrix components. However, the long length and the inherent flexibility of the appendages, expressed as multiple bending points along the stalk region, may overcome the relatively low surface coverage. The angular distribution of the bends of the EmaA appendages with the stalk domain was found to range between 40° and 10° (Fig. 2). These experimentally derived parameters support the hypothesis, based on the crystal structure of YadA, that a 30° angle of the head domain with the stalk domain is required for optimal positioning of the functional domain to interact with collagen fibers (21). Therefore, the expression of a long, flexible EmaA adhesin should mitigate the low level of surface expression and achieve a physiologically relevant level of binding for A. actinomycetemcomitans.
The EmaA bending sites are not randomly distributed throughout the structure but appear to be localized at specific positions. The neck sequences act as transitional elements between two distinct structural domains of the adhesin (21). Our results have revealed the localization of two of the neck sequences in the 3D structure of the EmaA appendages. The residues just after the first neck sequence (amino acids 415 to 433) correspond with the bend at 10.3 nm (Fig. 4), while the bend located at 29.4 nm from the distal end of the molecule correlates well with the bend observed in 2D micrographs (29). Collectively, all the data presented here suggest that the bend at 29.4 nm is associated with the amino acids following the second neck sequence (619 to 628). Assuming that the stalk domain, after the second neck sequence, forms an α-helical structure for at least 42 nm, the bending positions can be correlated with the amino acid sequence. Based on this assumption, the sequences of seven bending positions, which include the majority of the bends, contain the amino acids NGS/T or NGGS/T (Table 5). Glycine and serine residues are predicted to have a poor helix-forming probability (3). Hence, the disruption of the helix may allow for the increased flexibility observed in this adhesin. Under these assumptions, the bends in the EmaA appendage would correlate with the consensus sequence NG(G)S/T.
TABLE 5.
Correlation between bending position and EmaA protein sequencea
| Bending position (nm) | Amino acid position | Protein sequenceb |
|---|---|---|
| 29.4 ± 1.8 | 619 ± 12 | 607KNVAAGNVAANSTDAVNGSQLFAVA631 |
| 34.0 ± 0.6 | 649 ± 4 | 645ENGGTQNGA653 |
| 38.2 ± 1.6 | 678 ± 10 | 668LAGKNLAVKQNGTNFTFSTQE688 |
| 44.6 ± 1.3 | 720 ± 8 | 712SVQNGGTINMGNNRITG728 |
| 62.4 ± 0.9 | 839 ± 6 | 833EAAKSIPLTYKANGS847 |
| 65.6 ± 0.9 | 860 ± 6 | 854LDKGLNFTNGMMT866 |
| 71.9 ± 1.2 | 902 ± 8 | 894VAGTPGTNGANGTDGKD910 |
Values are means ± standard deviations.
The consensus sequences are shown in bold. The underlined sequence is the second conserved neck sequence (amino acids 619 to 628).
The EmaA structure inferred from the 2D electron microscopy analysis shows a simple geometry containing an ellipsoidal head domain (with the longest axis of ∼5 nm) and a long stalk (29). The 3D structure, obtained using electron tomography and subvolume averaging, revealed a more complex architecture for the distal end of the appendages, which is composed of defined subdomains (Fig. 4). Together subdomain I, which corresponds to the head domain described in the previous 2D study (29), and subdomain II, which includes the first neck sequence at the proximal end, comprise the region of the EmaA sequence consisting of amino acids 70 to 450. A calculation of the volumes encompassed by each subdomain suggests that subdomain II contains ∼1.35 times the molecular mass of subdomain I. Revisiting the electron micrographs with this knowledge permits the recognition of these features on the 2D images of the EmaA appendages (see Fig. S3A in the supplemental material; see also Fig. 8B of reference 39). In addition, a broadening of the distal end of the stalk domain is observed in images of the Δ70-206 mutant strain (see Fig. S3B in the supplemental material), which may correspond to the presence of subdomain II in these mutant EmaA appendages.
Additional insights into the EmaA structure were gained by submitting the sequence to the daTAA server (http://toolkit.tuebingen.mpg.de/dataa), a trimeric autotransporter adhesin domain prediction program (34). Based on these predictions, YadA-like domains are predicted in subdomains I, II and III, and a HIN2 domain (defined by an FxG motif) is predicted in subdomain III (See Fig. S4 in the supplemental material). The YadA structure results from the periodicity of specific SVAIG degenerate motifs. Since the periodicity of these motifs in the region of the EmaA protein (residues 323 to 415) is similar to that found in YadA, we have manually fitted the YadA head domain (Protein Data Bank (PDB) code 1p9h, (21) into the 3D electron tomography structure of subdomain II of EmaA (Fig. 4). The fitted YadA head domain accounts for a large percentage of the electron density of subdomain II, although small external densities still remain unfilled. Moreover, this analysis allowed us to define the location of the first EmaA neck sequence, which is found just above the linker region (Fig. 4).
The 3D electron microscopy data in combination with the docking of the YadA head trimer into the subdomain II and the linker region strongly suggest that EmaA is a trimeric adhesin. Fitting of the YadA head domain into the subdomain I of EmaA was not satisfactory (data not shown). The suboptimal fit might be due to the variable periodicity of the SVAIG motifs found in this region of EmaA (39). In some instances, the SVAIG motifs have a much larger separation than that found in the YadA sequence. The additional amino acids between the SVAIG motifs could be present as large loops between the β-rolls. This hypothesis is supported by the extreme functional and structural effect of a single amino acid mutation (G162S) in subdomain I (39). It is possible that the presence of glycine at this position is required to stabilize the secondary structure of the regions between 134GIALG138 and 162GIAIG168 and 162GIAIG168 and 190STAIG194. Similar effects were not observed when comparable amino acid substitutions were carried out in other motifs (data not shown). Thus, it is expected that the architecture of subdomain I is not identical to YadA.
Based on the current genetic, biochemical, and structural data, the functional domain identified by Yu et al. (39) has to be reassigned. The collagen binding domain should include at least subdomains I and II until the linker region, which corresponds to the amino acids following the first neck sequence and might be correlated with the bend observed at 10.3 nm. It is possible that subdomain III might also play a functional role. However, at the present time the data do not show strong support for this possibility. A more detailed structure of these subdomains awaits higher-resolution studies.
In summary, we have initiated the investigation of the 3D architecture of the EmaA adhesin on the surface of A. actinomycetemcomitans by using electron tomography of whole-mount bacterial preparations. Our structural, functional, and mutagenesis studies have shown that the EmaA appendages, composed of three identical monomers, contain specific amino acid sequences that are proposed to confer flexibility to the adhesins for optimal collagen binding. The first 3D structure of the functional collagen binding domain of EmaA was calculated using electron tomography and subvolume averaging. The functional domain of the EmaA molecule is suggested to be composed of two subdomains, or probably three. Additional studies are under way to optimize this technology and obtain more detailed information on the structure of this adhesin.
Supplementary Material
Acknowledgments
We thank Michael Radermacher, Grace Tang, Montserrat Bárcena, and Todd Clason for helpful discussions and critical reading of the manuscript.
This work was supported by NIH-NIDCR grants RO1-DE13824 (to K.P.M.) and RO1-DE017474 (to T.R.) and DOE EPSCoR grant DE-F602-00ER45828 (to K.M.) and has benefited from developments supported by NIH grant RO1-GM069551 (to T.R.).
Footnotes
Published ahead of print on 28 August 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Asakawa, R., H. Komatsuzawa, T. Kawai, S. Yamada, R. B. Goncalves, S. Izumi, T. Fujiwara, Y. Nakano, N. Suzuki, Y. Uchida, K. Ouhara, H. Shiba, M. A. Taubman, H. Kurihara, and M. Sugai. 2003. Outer membrane protein 100, a versatile virulence factor of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 50:1125-1139. [DOI] [PubMed] [Google Scholar]
- 2.Barcena, M., M. Radermacher, J. Bar, G. Kopperschlager, and T. Ruiz. 2007. The structure of the ATP-bound state of S. cerevisiae phosphofructokinase determined by cryo-electron microscopy. J. Struct. Biol. 159:135-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaber, M., X. J. Zhang, and B. W. Matthews. 1993. Structural basis of amino acid alpha helix propensity. Science 260:1637-1640. [DOI] [PubMed] [Google Scholar]
- 4.Carazo, J. M., and J. L. Carrascosa. 1987. Restoration of direct Fourier three-dimensional reconstructions of crystalline specimens by the method of convex projections. J. Microsc. 145:159-177. [PubMed] [Google Scholar]
- 5.Fine, D. H., D. Furgang, H. C. Schreiner, P. Goncharoff, J. Charlesworth, G. Ghazwan, P. Fitzgerald-Bocarsly, and D. H. Figurski. 1999. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology 145:1335-1347. [DOI] [PubMed] [Google Scholar]
- 6.Fine, D. H., K. Velliyagounder, D. Furgang, and J. B. Kaplan. 2005. The Actinobacillus actinomycetemcomitans autotransporter adhesin Aae exhibits specificity for buccal epithelial cells from humans and Old World primates. Infect. Immun. 73:1947-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fives-Taylor, P. M., D. H. Meyer, K. P. Mintz, and C. Brissette. 1999. Virulence factors of Actinobacillus actinomycetemcomitans. Periodontol. 2000 20:136-167. [DOI] [PubMed] [Google Scholar]
- 8.Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190-199. [DOI] [PubMed] [Google Scholar]
- 9.Gallant, C. V., M. Sedic, E. A. Chicoine, T. Ruiz, and K. P. Mintz. 2008. Membrane morphology and leukotoxin secretion are associated with a novel membrane protein of Aggregatibacter actinomycetemcomitans. J. Bacteriol. 190:5972-5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Girish, V., and A. Vijayalakshmi. 2004. Affordable image analysis using NIH Image/ImageJ. Indian J. Cancer 41:47. [PubMed] [Google Scholar]
- 11.Grant, S. G., J. Jessee, F. R. Bloom, and D. Hanahan. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 87:4645-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haubek, D., O. K. Ennibi, L. Abdellaoui, N. Benzarti, and S. Poulsen. 2002. Attachment loss in Moroccan early onset periodontitis patients and infection with the JP2-type of Actinobacillus actinomycetemcomitans. J. Clin. Periodontol. 29:657-660. [DOI] [PubMed] [Google Scholar]
- 13.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 19:5989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inouye, T., H. Ohta, S. Kokeguchi, K. Fukui, and K. Kato. 1990. Colonial variation and fimbriation of Actinobacillus actinomycetemcomitans. FEMS Microbiol. Lett. 57:13-17. [DOI] [PubMed] [Google Scholar]
- 15.Kachlany, S. C., P. J. Planet, R. Desalle, D. H. Fine, D. H. Figurski, and J. B. Kaplan. 2001. flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 40:542-554. [DOI] [PubMed] [Google Scholar]
- 16.Kinane, D. F., and P. J. Hodge. 2001. Periodontal disease in children and adolescents: introduction and classification. Periodontol. 2000 26:7-15. [DOI] [PubMed] [Google Scholar]
- 17.Kremer, J. R., D. N. Mastronarde, and J. R. McIntosh. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71-76. [DOI] [PubMed] [Google Scholar]
- 18.Meng, G., N. K. Surana, J. W. St. Geme III, and G. Waksman. 2006. Structure of the outer membrane translocator domain of the Haemophilus influenzae Hia trimeric autotransporter. EMBO J. 25:2297-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mintz, K. P. 2004. Identification of an extracellular matrix protein adhesin, EmaA, which mediates the adhesion of Actinobacillus actinomycetemcomitans to collagen. Microbiology 150:2677-2688. [DOI] [PubMed] [Google Scholar]
- 20.Mintz, K. P., and P. M. Fives-Taylor. 1999. Binding of the periodontal pathogen Actinobacillus actinomycetemcomitans to extracellular matrix proteins. Oral Microbiol. Immunol. 14:109-116. [DOI] [PubMed] [Google Scholar]
- 21.Nummelin, H., M. C. Merckel, J. C. Leo, H. Lankinen, M. Skurnik, and A. Goldman. 2004. The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel beta-roll. EMBO J. 23:701-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 23.Pierce, C. S., W. R. Bartholomew, D. Amsterdam, E. Neter, and J. J. Zambon. 1984. Endocarditis due to Actinobacillus actinomycetemcomitans serotype C and patient immune response. J. Infect. Dis. 149:479. [DOI] [PubMed] [Google Scholar]
- 24.Radermacher, M. 1994. Three-dimensional reconstruction from random projections: orientational alignment via Radon transforms. Ultramicroscopy 53:121-136. [DOI] [PubMed] [Google Scholar]
- 25.Radermacher, M., T. Wagenknecht, A. Verschoor, and J. Frank. 1987. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J. Microsc. 146:113-136. [DOI] [PubMed] [Google Scholar]
- 26.Roggenkamp, A., N. Ackermann, C. A. Jacobi, K. Truelzsch, H. Hoffmann, and J. Heesemann. 2003. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 185:3735-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rose, J. E., D. H. Meyer, and P. M. Fives-Taylor. 2003. Aae, an autotransporter involved in adhesion of Actinobacillus actinomycetemcomitans to epithelial cells. Infect. Immun. 71:2384-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruiz, T., G. Kopperschlager, and M. Radermacher. 2001. The first three-dimensional structure of phosphofructokinase from Saccharomyces cerevisiae determined by electron microscopy of single particles. J. Struct. Biol. 136:167-180. [DOI] [PubMed] [Google Scholar]
- 29.Ruiz, T., C. Lenox, M. Radermacher, and K. P. Mintz. 2006. Novel surface structures are associated with the adhesion of Actinobacillus actinomycetemcomitans to collagen. Infect. Immun. 74:6163-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruiz, T., I. Mechin, J. Bar, W. Rypniewski, G. Kopperschlager, and M. Radermacher. 2003. The 10.8-Å structure of Saccharomyces cerevisiae phosphofructokinase determined by cryoelectron microscopy: localization of the putative fructose 6-phosphate binding sites. J. Struct. Biol. 143:124-134. [DOI] [PubMed] [Google Scholar]
- 31.Salvi, E., F. Cantele, L. Zampighi, N. Fain, G. Pigino, G. Zampighi, and S. Lanzavecchia. 2008. JUST (Java User Segmentation Tool) for semi-automatic segmentation of tomographic maps. J. Struct. Biol. 161:287-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slots, J., H. S. Reynolds, and R. J. Genco. 1980. Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation. Infect. Immun. 29:1013-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szczesny, P., D. Linke, A. Ursinus, K. Bar, H. Schwarz, T. M. Riess, V. A. Kempf, A. N. Lupas, J. Martin, and K. Zeth. 2008. Structure of the head of the Bartonella adhesin BadA. PLoS Pathog. 4:e1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szczesny, P., and A. Lupas. 2008. Domain annotation of trimeric autotransporter adhesins: daTAA. Bioinformatics 24:1251-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang, G., T. Kitten, C. L. Munro, G. C. Wellman, and K. P. Mintz. 2008. EmaA, a potential virulence determinant of Aggregatibacter actinomycetemcomitans in infective endocarditis. Infect. Immun. 76:2316-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tang, G., T. Ruiz, R. Barrantes-Reynolds, and K. P. Mintz. 2007. Molecular heterogeneity of EmaA, an oligomeric autotransporter adhesin of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Microbiology 153:2447-2457. [DOI] [PubMed] [Google Scholar]
- 37.Wollmann, P., K. Zeth, A. N. Lupas, and D. Linke. 2006. Purification of the YadA membrane anchor for secondary structure analysis and crystallization. Int. J. Biol. Macromol. 39:3-9. [DOI] [PubMed] [Google Scholar]
- 38.Yeo, H. J., S. E. Cotter, S. Laarmann, T. Juehne, J. W. St. Geme III, and G. Waksman. 2004. Structural basis for host recognition by the Haemophilus influenzae Hia autotransporter. EMBO J. 23:1245-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, C., T. Ruiz, C. Lenox, and K. P. Mintz. 2008. Functional mapping of an oligomeric autotransporter adhesin of Aggregatibacter actinomycetemcomitans. J. Bacteriol. 190:3098-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zambon, J. J. 1985. Actinobacillus actinomycetemcomitans in human periodontal disease. J. Clin. Periodontol. 12:1-20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.