Abstract
Streptococcus pyogenes, in addition to causing fulminant disease, can be carried asymptomatically and may survive in the host without causing disease. Long-term stationary-phase cultures were used to characterize the metabolism of cultures surviving after glucose depletion. Survival of stationary-phase cultures in glucose-depleted rich medium was truncated by switching the cells to phosphate-buffered saline or by the addition of antibiotics, suggesting that survival depended on the presence of nutrients and metabolic activity. The metabolites of the pyruvate-to-acetate (PA) pathway (acetate and formate) and amino acid catabolic pathways (ammonia) accumulated throughout long-term stationary phase (12 weeks). Acid and ammonia production was balanced so that the culture pH was maintained above pH 5.6. Strains isolated from long-term stationary-phase cultures accumulated mutations that resulted in unique exponential-phase metabolisms, with some strains expressing the PA pathway, some strains producing ammonia, and some strains expressing both in the presence of glucose. Strains expressing high levels of PA pathway activity during exponential growth were unable to survive when regrown in pure culture due to the production of excess acid. These data suggest that S. pyogenes diversifies during survival in stationary phase into distinct strains with different metabolisms and that complementary metabolism is required to control the pH in stationary-phase cultures. One of three survivor strains isolated from tonsillar discard material from patients expressed high levels of the PA pathway during exponential growth. Sequencing of multiple group A streptococcus regulators revealed two different mutations in two different strains, suggesting that random mutation occurs during survival.
The human pathogen Streptococcus pyogenes (group A streptococcus [GAS]) is the causative agent of mild infections (e.g., impetigo and pharyngitis), severe disease (e.g., toxic shock syndrome and necrotizing fasciitis), and secondary sequelae (e.g., rheumatic fever and glomerulonephritis) (reviewed in references 17 and 32). Asymptomatic carriage and antibiotic treatment failure suggest that S. pyogenes may be able to persist in the host after the initial infection (5, 7). S. pyogenes asymptomatic carriage rates of 2.5% to 32% have been observed with large cohorts of school-age children worldwide (2, 16, 23, 46, 56, 57, 63, 64). Carriage in adults is less frequent (1.3% to 4.9%) but has been observed with adult health care workers and military recruits (9, 34). Although not all cases of recurrent tonsillitis are clonal, carriage of S. pyogenes has been implicated as one of the causes of recurrent tonsillitis (5, 7, 8, 46, 52, 53, 56, 59). In addition to recurrent tonsillitis, carriers have been associated with outbreaks of invasive disease (9, 15).
Since the carried bacteria do not cause fulminant infection during carriage, it is likely that they survive in an altered growth state. Survival of bacteria has been characterized for both gram-negative and gram-positive bacteria grown in vitro and in animal models (reviewed in reference 49). Bacteria survive in states ranging from completely dormant to slowly growing. These states include sporulation (e.g., Bacillus subtilis) (reviewed in reference 58), viable but nonculturable bacteria (e.g., Micrococcus luteus) (reviewed in reference 50), and survival in a nonreplicating state (e.g., Mycobacterium tuberculosis) (6, 33, 36). Another mechanism of bacterial survival is that they can grow slowly during survival without an obvious increase in cell numbers, suggesting a balance of dividing and dying cells, as is the case for Escherichia coli and Staphylococcus aureus. During long-term stationary-phase survival of E. coli, there is a succession of strains that arise by mutation, and each strain is more fit to survive in stationary phase than the previous strains (growth advantage in stationary phase) (reviewed in references 27 and 86). S. aureus survives within eukaryotic cells in which the bacteria form small colonies when regrown on agar plates (small-colony variants) because the mutations reduce the ability of the bacteria to produce energy by respiration, and they rely solely on fermentation for the production of energy (reviewed in references 61 and 62). Small-colony variants are less susceptible to antibiotics and are associated with recurrent and latent infections (reviewed in reference 62).
How S. pyogenes survives in long-term stationary phase is relatively uncharacterized. Studies of virulence factor regulation and transcriptomes clearly demonstrate that virulence factor and metabolic gene expression varies significantly depending on growth state and available nutrients (reviewed in references 4, 12, and 38). These patterns are complex and can vary between M-protein serotypes; however, some general trends of early-stationary-phase behavior can be observed. S. pyogenes does not appear to have a stationary-phase global regulator RpoS homolog. Only one alternate sigma factor, SigX, has been described for S. pyogenes (51). SigX is not important for exponential growth, regulation of most known virulence factors, stress tolerance, biofilm formation, or stationary-phase survival (51, 83, 84). SigX directly influences transcription of putative competence genes and the putative cell wall cross-linking enzyme gene femB (84). Instead of alternate sigma factors, regulation of stationary-phase responses in S. pyogenes depends on global regulators, such as RelA, Rgg/RopB, CsrR/CovR, CcpA, and CodY, which affect expression of metabolic genes and virulence factors (11, 13, 21, 43, 44, 73, 75, 77, 78). In addition, the stability of some mRNAs is altered upon entrance into stationary phase (1).
Bacteria carried in the oropharynx are likely to be in a glucose-limited environment, so we are interested in studying the unique changes in metabolism that occur during long-term stationary-phase survival, in particular shifts in metabolic expression upon glucose depletion. Amino acid catabolism, pyruvate metabolism, and the use of alternate sugar sources have been shown to be important in early stationary phase, in sugar-depleted conditions, in saliva, and in the oropharynx (4, 12, 73, 75). For the utilization of amino acids, increases in proteases, peptide transporters, and arginine deiminase in early stationary phase have been observed (4, 12). Under some conditions, RelA and CodY are responsible for changes in transcription of genes encoding proteins involved in amino acid uptake, di- and oligopeptide permeases and proteases (43, 44). S. pyogenes with mutations in rgg catabolize serine and arginine in the presence of glucose (11, 13). Serine dehydratase converts serine to pyruvate and the arginine deiminase pathway converts arginine to ornithine and citrulline with the production of ATP (11, 13). Enzymes and sugar transporters that allow for the use of alternate sugars and pyruvate are also increased in early stationary phase (4, 12, 73, 75). Pyruvate metabolism and NADH-oxidase activity under conditions of sugar starvation have been observed (71).
Carriage of S. pyogenes may require longer periods of survival in stationary phase. We have found that S. pyogenes can survive up to 1.5 years in complex medium (Todd-Hewitt [TH] broth) (83). Long-term survival is dependent on the maintenance of culture pH above ∼5.6, and any decrease of the pH below this threshold significantly truncates survival (83). Individual strains isolated from long-term stationary-phase cultures accumulate mutations that lead to altered colony morphologies and unique exponential-phase proteomes (40, 83). In the present studies, metabolism during long-term survival of S. pyogenes was further characterized. Surviving S. pyogenes M49 CS101 cultures were metabolically active, and metabolites of the pyruvate-to-acetate (PA) pathway and amino acid catabolism accumulated throughout survival. The surviving population was a mixture of mutants that had stable, and often unique, changes in the expression of their exponential-phase metabolic pathways. Metabolic diversity within the culture maintained a pH above the critical threshold of ∼5.6 by a combination of strains expressing high PA-pathway activity, high amino acid catabolism, or both. Survival after regrowth of individual survivor strains was truncated for strains producing high levels of acid. One of three intracellular survivor strains isolated from patients showed changes in its exponential-phase metabolism. In addition, sequencing of regulator genes showed unique mutations in two strains, suggesting that random mutation may play a role in the generation of diversity during survival.
MATERIALS AND METHODS
Bacterial strains and incubation conditions.
S. pyogenes serotype M49 strain CS101 was used for this study and was provided by P. P. Cleary. JRS4 was kindly provided by J. Scott. Strain Alt. 1 was isolated from one independent 14-week stationary-phase CS101 culture. Alt. 2 was isolated from a second independent 14-week stationary phase CS101 culture. Alt. 4A, Alt. 4B, and Alt. 4D were isolated from a third independent 14-week-old culture. Alt. 5A, Alt. 5B, Alt. 5C, and Alt. 5D were isolated from a fourth independent 14-week-old culture. Each independent culture was grown in a different batch of TH broth and inoculated on a different day. All Alt. strains were identified as S. pyogenes by sequencing a 16S rRNA fragment generated by PCR with the primers EubA and EubB as previously described (83). The tonsillar survivor isolates 221, Sfr321, and MK322 were obtained from a previous study (59). All S. pyogenes strains were frozen in 30% glycerol stocks at −80°C prior to use. All cultures were inoculated from single colonies on plates streaked from the glycerol stocks, making each culture a third passage of each strain. Cultures were grown in TH broth or on TH agar (DIFCO, Sparks, MD). All S. pyogenes cultures were maintained static at 37°C under a 5% CO2 atmosphere. It should be noted that lots of TH media occasionally would not support survival due to a drop in culture pH for unclear reasons.
mRNA isolation, detection, and comparison.
RNA was prepared by growing cells overnight in TH broth. The following day, the cultures (<15 h old) were diluted 1:10 into fresh prewarmed TH broth. Incubation was continued until cultures reached an optical density at 600 nm (OD600) between 0.50 and 0.65. These cells represent mid-exponential-phase cells, and total RNA was isolated from them using a modified hot-phenol extraction method of Shaw and Clewell (72). Cells from 40 ml of culture were resuspended in 2 ml of lysis buffer and added to 1.5 g of zirconia silica beads. The bacteria were lysed by bead beating three times for 1 min at 4,800 rpm (Biospec Products, Bartlesville, OK). The lysate was processed by hot-phenol extraction (72). The isolated RNA was quantitated at OD260. Denaturing agarose gel electrophoresis and Northern blotting were performed on dilutions of the RNA representing 5, 2.5, and 1.25 μg of total cellular RNA (60). Even loading of total RNA was confirmed by visualization of ethidium bromide-stained gels. Detection of gyrA served as a loading control (42). Digoxigenin (DIG)-dUTP-labeled probes for the genes encoding lactate oxidase (lctO); pyruvate dehydrogenase, alpha chain, subunit E1 (acoA); pyruvate formate lyase (pfl); phosphotransacetylase (pta); acetate kinase (ackA); and gyrase A (gyrA) were generated by PCR using the primer pairs listed in Table 1. Hybridization and visualization using disodium-3-(4-methoxy- spiro[1,2-dioxetane-3′2′-(5-chloro)tricyclo(3.3.1.33,7)decan]-4-yl)phenylphosphate (CSPD) (Roche, Indianapolis, IN) were done as previously described (60). The Northern blots represent the result of at least three independent experiments using freshly isolated RNA for each blot.
TABLE 1.
Oligonucleotides used in this study
| Name | Sequence (5′-3′) | Name | Sequence (5′-3′) |
|---|---|---|---|
| lctO FWD | AGCACCTGTAGCGGCTCATAAACT | ccpA REV | AGATGGTGCTCATAATTCAC |
| lctO REV | ATACCTGATGCTCCTGCGTCCAAT | srv FWD 1 | TACTATCAAAGGGCATTAGC |
| acoA FWD | GCTTGCTGGTAAAGCAACTGGTGT | srv FWD 2 | TGTATGTTAACTAACGGCCG |
| acoa REV | ATAGCTGGACCATTTCCACCACGA | srv REV | TGCAGATCCAGATCAAAGCC |
| pfl FWD | TCGTCTTGCTCTTTACGGTGCTGA | SPy1548 FWD | GGAACGTATGAGTTGGTGAC |
| pfl REV | TCTGGCAGTTGGTCAGTCCAAAGA | SPy1548 REV | TTCTTTACCGGGTCTGTGGAGC |
| pta FWD | TGACACTGTTCGTCCAGCTCTTCA | relA FWD 1 | ACTAAGCGCTTTCTTAGCAG |
| pta REV | GCCATCAAGTGCCAAATCAGGGTT | relA FWD 2 | TCGCATCAAATGGGAACTAG |
| ackA FWD | CGTGTTGTTGCTGGTGGTGAACTT | relA REV | TTGACCCGTTGCAAGACAAG |
| ackA REV | AAGTGGTGTAAAGCCCATCGAGGT | SPy0145 FWD 1 | TCAAGAGCAAAGGTGGTGAGAGGA |
| gyrA FWD | ACAGGTCGGGAACGTATTGTGGTT | SPy0145 FWD 2 | CCAGTGACAGGTCAATTGTC |
| gyrA REV | GTTCCAAACCAGTCAAACGACGCA | SPy0145 REV | GAGGCACAAGCTGCCGAAGC |
| codY FWD 1 | ACAAGCTAGTGCTTATCTCC | SPy1630 FWD | AGCTGAGCGGGTTAAGCGTATCAT |
| codY FWD 2 | TATCCAGGAGGTCTAACGAC | SPy1630 Rev | TGGCTTGTCCGTTTGCATCAATGG |
| codY REV | AAACAGGGAAACCTCTCCCC | CovRS FWD 1 | GATAGATTAAGAGGATAAGGGTTGGT |
| ropB FWD 1 | AAGCGACTATCATCCGAAAC | CovRS FWD 2 | TGTTTGGAAATATGATGAAGCCGT |
| ropB FWD 2 | ACTTGGAGTCACTATGAGAC | CovRS FWD3 | TGGTCCTATCGGTCGTGTGTATCA |
| ropB REV | AAGCTAACACCATAAGAGCG | CovRS FWD 4 | TGAGGCTGACCGTATGGCAATCAT |
| ccpA FWD 1 | AAAGTGGTTACAAATCATGC | CovRS REV | CATCAGCTTCTAACCAGTTGTGGC |
| ccpA FWD 2 | ACGCTCTCGTACTCCAGTTG |
Truncation of survival by PBS and antibiotics.
To determine the role of nutrients in survival, surviving S. pyogenes CS101 cultures were harvested by centrifugation at 6,000 rpm (3,300 × g). Cells were resuspended in the same volume of phosphate buffered saline (PBS) or spent medium from the original culture, as a centrifugation control. Antibiotics were added to the cultures at the following final concentrations: rifampin (rifampicin), 5 μg ml−1; gentamicin, 10 μg ml−1; penicillin, 0.75 μg ml−1; and vancomycin, 1 μg ml−1. After treatment of the cultures, incubation was continued at 37°C in 5% CO2. Samples (50 μl) were removed at various time points and spotted on TH plates. Any colony formation after incubation of the plates at 37°C in 5% CO2 was scored as positive for survival, with a lower limit of detection of 20 CFU ml−1.
Metabolite analysis.
Culture supernatants used for metabolite analysis were collected from stationary-phase or exponential-phase cultures. For exponential-phase cultures, an overnight (<15-h) culture of S. pyogenes was diluted 1:100 in fresh medium. Growth of newly inoculated cultures was monitored by spectrometry at OD600, and samples were removed from each tube at the indicated time points. Culture samples were filter sterilized and subsequently heat inactivated at 90°C for 5 minutes before being stored at −20°C, except for the samples for ethanol readings which were used immediately after filtration to minimize evaporation. l-Lactate, formate, acetate, ethanol, and ammonia concentrations were determined from culture supernatants using R-Biopharm test kits (Marshall, MI), as described by the manufacturer. Each data point represents the metabolite concentrations from at least three independent cultures. Statistical analysis for lactate, formate, acetate, and ammonia was done by J. Gaughn at the Temple University School of Medicine, Department of Biostatistics. The dependent variables lactate, acetate, formate, and ammonia were treated as continuous variables. Means, standard deviations, and number of observations were presented. The experimental unit was each individual culture. The experiment used a randomized block design (strain and culture) with three to eight cultures per strain. The null hypothesis was that there would be no difference between strains. Prior to analysis, the data were tested for normality using the Shapiro-Wilk test. The data had a nonnormal distribution. In order to apply analysis of variance (ANOVA) methods, a “normalized-rank” transformation was applied. The rank-transformed data were analyzed using a generalized linear-model ANOVA and then multiply compared to detect significant mean differences between strains. Differences between means (rejection of the null hypothesis) were considered significant if the probability of chance occurrence was ≤0.05 using two-tailed tests. Due to the exploratory nature of the experiments, no multiple-comparison adjustments were made. Statistical differences in ethanol concentrations were determined using the SPSS statistics program (SPSS Inc., Chicago, IL). Equal variances were assumed for the samples, and they were analyzed by one-way ANOVA using Tukey's posthoc test. Differences between means were considered significant at P values of ≤0.05.
Ammonia supplementation assays.
S. pyogenes CS101, Alt. 1, and Alt. 2 were grown in sterile TH broth. Approximately 24 h after entry into stationary phase, exogenous ammonia was added to cultures to achieve a final concentration of 5, 10, or 20 mM ammonia. Culture pH was determined after the addition of exogenous ammonia. Survival was assayed by spotting 50-μl culture samples on TH agar plates in 24-h intervals postentry into stationary phase. These plates were incubated at 37°C under a 5% CO2 atmosphere, and formation of any colonies in these culture spots was scored as positive for survival (lower limit of detection of 20 CFU ml−1).
Supernatant switch assays.
S. pyogenes CS101, Alt. 1, and Alt. 2 were each grown individually in TH broth. Approximately 12 h after entry into stationary phase, cultures were pelleted by centrifugation, culture supernatant for each strain was filter sterilized, and supernatant pH was determined. CS101 cell pellets were resuspended in either Alt. 1- or Alt. 2-conditioned supernatants. Alt. 1 and Alt. 2 cell pellets were resuspended in CS101-conditioned supernatant. Survival was assayed by spotting 50 μl of culture samples on TH agar plates in 24-h intervals postentry into stationary phase. These plates were incubated at 37°C, under a 5% CO2 atmosphere, and formation of any colonies in these culture spots were scored as positive for survival.
Regulator gene sequencing.
S. pyogenes strains were grown overnight in TH broth, and DNA was isolated using a phenol extraction. Briefly, cells from 10 ml of culture were resuspended in 2 ml of lysis buffer and added to 0.8 g of zirconia silica beads with 50 μl 10% sodium dodecyl sulfate. The bacteria were lysed by bead beating four times for 1 min at 4,800 rpm (Biospec Products, Bartlesville, OK). The lysate was extracted with phenol. The isolated DNA was quantitated at an OD260. The genes of interest were PCR amplified using oligonucleotides external to the open reading frame of each gene and using Platinum Pfx polymerase (Invitrogen, Carlsbad, CA). The regulatory genes codY, ccpA, srv, SPY1548, relA, ropB, SPY1630, SPY0145, and covRS were PCR amplified using primer pairs based on the genome sequence of the M1 serotype strain SF370, and they are listed in Table 1. The PCR products were cleaned up using a Qiaquick PCR cleanup kit (Qiagen, Valencia, CA), and PCR products were quantitated at an OD260. The PCR products were sent in for sequencing at the Kimmel Cancer Center (Thomas Jefferson University School of Medicine, Philadelphia, PA). The FWD primers used for sequencing are listed in Table 1. The resulting sequences were aligned using the ClustalW2 program (http://www.ebi.ac.uk/Tools/clustalw2/index.html). Sequencing was performed once for most strains, except for those that showed a mutation compared to CS101. DNA was isolated, amplified, and sequenced two additional times for any strain that showed a mutation.
RESULTS
Stationary-phase-surviving cultures are metabolically active.
To determine if long-term stationary-phase survival of S. pyogenes required one or more nutrients present in the TH broth supernatant, cultures of S. pyogenes CS101 were inoculated and incubated for 1 week. The supernatant of the surviving cultures was then removed, the bacterial cells were washed with PBS, and the supernatant was replaced with PBS. Control samples did not have their supernatants replaced. Survival was measured by plating samples from the cultures at various time points after supernatant replacement. Cultures that had their supernatants replaced with PBS survived less than 48 h in stationary phase compared to >12 weeks for control cultures.
To determine if surviving populations of S. pyogenes engaged in the basic cellular processes during survival, antibiotics that inhibited transcription (rifampin), translation (gentamicin), and cell wall synthesis (penicillin or vancomycin) were added to week-old stationary-phase cultures. Bacterial survival was monitored by plating. Addition of gentamicin caused surviving populations to become nonculturable in less than 72 h, while addition of the remaining antibiotics caused loss of culturability in less than 120 h. Taken together, these data suggest that the long-term stationary-phase cultures remain metabolically active.
PA pathway and amino acid catabolic pathway metabolites accumulate in long-term stationary-phase cultures.
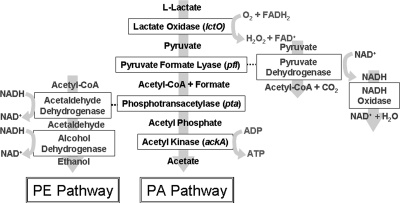
Seki and coworkers reported activity of NADH oxidase and the production of acetate in glucose-starved cultures, suggesting a role for pyruvate metabolism under conditions of glucose starvation (71). Since all of the glucose is depleted from surviving cultures upon entrance into stationary phase (83), KEGG (http://www.genome.jp/kegg/pathway.html) and the M1 genome sequence (26) were used to identify the complete PA pathway that could convert lactate to acetate with the production of ATP as well as the pyruvate-to-ethanol (PE) pathway (Fig. 1).
FIG. 1.
Pyruvate metabolic pathway. The PA pathway for the catabolism of lactate generates a single molecule of ATP for each molecule of lactate consumed. The PE pathway recycles NAD+ but does not directly produce ATP. The conversion of pyruvate to acetyl-CoA can be achieved through either pyruvate formate lyase or pyruvate dehydrogenase. The enzymes are boxed, and the shaded arrow represents the direction of the pathway.
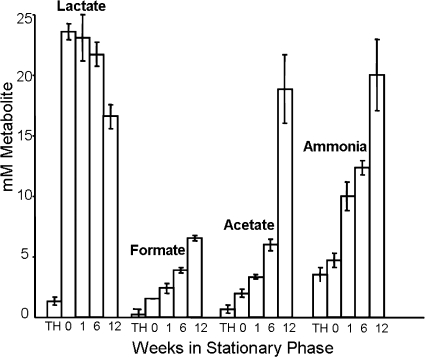
To determine if the PA pathway activity could be detected in late-stationary-phase cultures, the levels of lactate, formate, and acetate were measured during survival (Fig. 2). Cultures of S. pyogenes CS101 contained 24 mM lactate and low levels of formate and acetate at entry into stationary phase. The lactate levels slowly decreased, and after 12 weeks the concentration of lactate had decreased by 8 mM (Fig. 2). Decrease in lactate was accompanied by an increase in formate to 7 mM at 12 weeks in stationary phase (Fig. 2). An acetate concentration of 19 mM was observed at 12 weeks with surviving cultures (Fig. 2). Since only 1/1,000 cells survive late into stationary phase (83), isolation of quality mRNA for transcriptional analysis was precluded by the large background of dead cells present in the surviving cultures.
FIG. 2.
PA pathway metabolites accumulated in surviving S. pyogenes CS101 cultures. S. pyogenes strain CS101 was grown in static TH broth cultures under a 5% CO2 atmosphere. Culture samples were removed at various points after entrance into stationary phase. The samples were filter sterilized, and the concentrations of lactate, formate, acetate, and ammonia were determined. Metabolite concentrations present in sterile TH broth are represented by “TH”. Time is given in weeks under each bar, and T0 (“0”) is entry into stationary phase. Each bar represents the mean value from at least four independent cultures. Error bars are the standard deviations of the means.
Despite the accumulation of formate and acetate, the pH of the cultures did not change throughout stationary-phase survival (data not shown). Two amino acid degradation pathways, the arginine deiminase and serine dehydratase pathways, have been shown to be active in S. pyogenes before the depletion of sugar in rgg mutants (11, 13). The arginine deiminase pathway produces ATP and ammonia from arginine, whereas the serine dehydratase pathway produces pyruvate and ammonia (11, 13). In both cases, the production of ammonia would help to neutralize the acid being produced by the PA pathway. In addition to ammonia, serine dehydratase would also produce pyruvate that could then enter into the PA pathway. Consistent with amino acid catabolism during survival, cultures of S. pyogenes CS101 contained 10 mM ammonia after 1 week in stationary phase and 20 mM ammonia after 12 weeks in stationary phase (Fig. 2).
Metabolic heterogeneity of survivor strains.
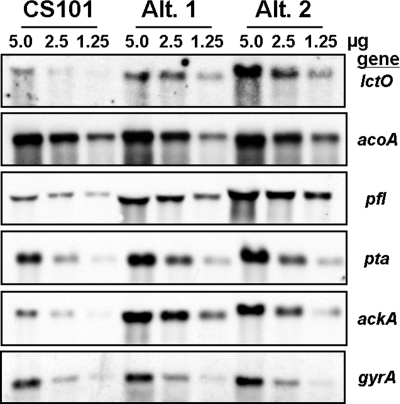
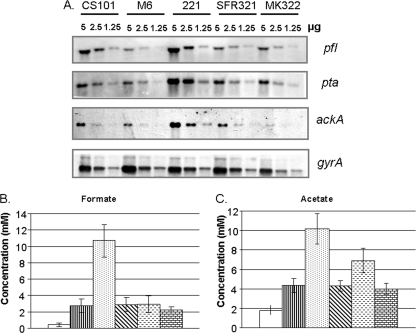
In previous studies, two S. pyogenes survivor strains isolated from 14-week stationary-phase cultures (Alt. 1 and Alt. 2) were found to accumulate mutations such that they regrew with altered exponential-phase proteomes compared to the parental strain CS101 (83). Proteomic analysis of Alt. 1 and Alt. 2 revealed increases in NADH oxidase during exponential growth (83). The other PA pathway enzymes were not identified in the two-dimensional gels (83). To determine if the entire PA pathway (Fig. 1) was transcriptionally upregulated in Alt. 1 and Alt. 2 during exponential growth, transcription of the PA pathway was compared between mid-exponential cultures of CS101, Alt. 1, and Alt. 2 by Northern blotting (Fig. 3). The genes encoding lactate oxidase (lctO), pyruvate formate lyase (pfl), phosphotransacetylase (pta), and acetyl kinase (ackA) were all found to be transcriptionally upregulated in Alt. 1 and Alt. 2 during exponential growth (Fig. 3). The alpha chain of the E1 subunit of the pyruvate dehydrogenase complex (acoA) was not transcriptionally upregulated. DNA gyrase (gyrA) was used as a loading control (42). The transcript sizes corresponded well to those of predicted monocistronic or polycistronic transcripts identified in the M1 sequence. Consistent with the changes in PA pathway transcription, exponential-phase cultures of Alt. 1 and Alt. 2 showed decreased production of lactate and increased production of formate and acetate (Fig. 4A).
FIG. 3.
Transcription of PA pathway genes was increased in Alt. 1 and Alt. 2 during exponential growth. Total RNA was isolated from mid-exponential cells (OD600 of 0.50 to 0.65) grown in TH broth. RNA concentrations were determined by spectrophotometric absorbance at 260 nm. Total RNA (5.0, 2.5, and 1.25 μg) was separated on a denaturing agarose gel. RNA gels underwent Northern blotting, and the binding of DIG-labeled DNA probes was detected by CSPD development and then by autoradiography. Equal loading was confirmed by ethidium bromide staining of the gel and by probing for DNA gyrase (gyrA). RNA concentrations in μg are noted above the lanes in each image. Fresh RNA was isolated for each experiment, and the results presented here are representative of three independent preparations.
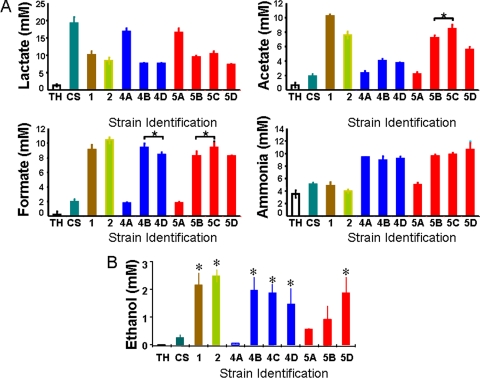
FIG. 4.
Survivor strains showed metabolic heterogeneity in exponential phase. S. pyogenes strain CS101 and nine strains which were isolated from 14-week-old stationary-phase cultures of strain CS101 were grown statically in TH broth under a 5% CO2 atmosphere, and exponential cell density was monitored by culture absorbance at 600 nm. Late-exponential-phase (OD600 of 0.80 to 0.90) culture samples were removed and filter sterilized, and lactate, acetate, formate, ammonia, and ethanol concentrations were determined. Metabolite concentrations present in sterile TH broth are represented in each graph by the bar at the left. Each bar represents the mean value from at least three independent cultures. (A) The survivor strains were compared against each other and CS101 for lactate, formate, and ammonia production. The standard deviation of the mean is represented by the error bars. “*” denotes a P value of <0.5. Unmarked bars that appear similar are not significantly different. Unmarked bars that appear different are statistically significantly different (P value of <0.05). (B) Ethanol production in each survivor strain was compared to CS101. The standard deviation of the mean is represented by the error bars. “*” denotes a P value of <0.5. Strain identification is as follows: 1, strain Alt. 1; 2, strain Alt. 2; 4A, strain Alt. 4A; 4B, strain Alt. 4B; 4D, strain Alt. 4D; 5A, strain Alt. 5A; 5B, strain Alt. 5B; 5C, strain Alt. 5C; 5D, strain Alt. 5D. CS, CS101.
To determine if all survivor strains had accumulated mutations such that they expressed the PA pathway during exponential phase, the exponential-phase metabolic profile of multiple survivor strains, some isolated from the same surviving culture, were characterized (Fig. 4A). To obtain survivor strains, a sample of a surviving culture was plated on TH broth. Individual colonies were picked and grown in TH broth, and freezer stocks were prepared in glycerol. For each experiment, the freezer stock was plated on TH agar, and an isolated colony was used as inoculum. Alt. 4A, Alt. 4B, and Alt. 4D were isolated from a 14-week-old culture. Alt. 5A, Alt. 5B, Alt. 5C, and Alt. 5D were isolated from a second independent 14-week-old culture. Alt. 1, Alt. 2, Alt. 4B, Alt. 5C, and Alt. 5D grew as small colonies, and Alt. 4A, Alt. 4D, Alt. 5A, and Alt. 5B grew as atypical large colonies. The identity of each strain as S. pyogenes was confirmed by 16S rRNA gene sequencing. Concentrations of glucose, lactate, formate, acetate, and ammonia were compared between CS101 and CS101-derived survivor strains during exponential growth. Statistics were done as described in Materials and Methods to determine if the strains produced significantly different patterns of metabolites. A P value of less than 0.05 was used to denote statistical difference between concentrations. All of the strains had statistically different metabolisms, suggesting that expression of the PA pathway was diverse between the strains (Fig. 4A). While most of the strains had high levels of PA pathway activity, Alts. 4A and 5A had only modest increases in PA pathway activity. One strain (strain 5A) produced levels of ammonia comparable to those of CS101, Alt. 1, and Alt. 2, while others produced significantly higher levels of ammonia (strains 4A, 4B, 4D, 5B, 5C, and 5D). There was no correlation between PA pathway activity and ammonia concentrations. There was also no correlation between colony size and metabolism. In addition to the by-products of the PA pathway and amino acid catabolism, ethanol production during exponential growth, which could result from PE pathway activity, was measured (Fig. 4B). Ethanol can be produced from acetyl coenzyme A (acetyl-CoA) through the enzymes acetaldehyde dehydrogenase and alcohol dehydrogenase (Fig. 1). Conversion of acetyl-CoA to ethanol can restore the NAD+ levels in the cells. Some, but not all, strains increased production of ethanol, indicating that these strains were heterofermentative during exponential phase, even with glucose present in the culture.
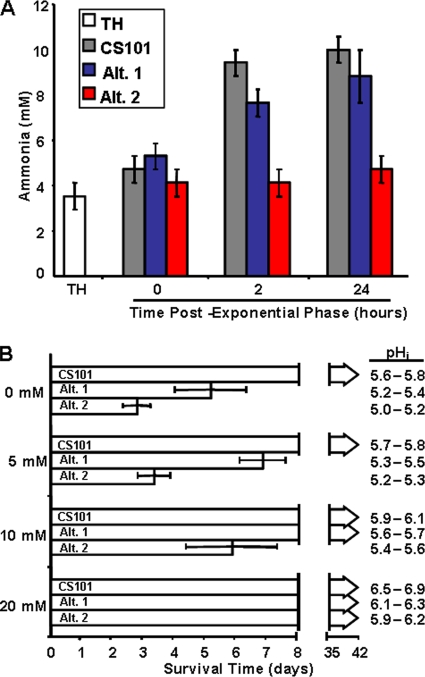
Alt. 1 and Alt. 2 cannot survive in pure culture due to high levels of acid production.
Alt. 1 and Alt. 2 had increased PA pathway activity but did not show increased ammonia production during exponential growth. In fact, Alt. 2 did not appear to produce a measurable increase in ammonia even in early stationary phase (Fig. 5A). Since the pH of a surviving culture must remain above pH 5.6 (83), strains like Alt. 5C that had increased expression of both the PA pathway and amino acid catabolism still had a pH of 5.6 to 5.8 upon entry into stationary phase and survive, like the parental strain, for longer than 6 weeks. However, Alt. 1 and Alt. 2 had a significantly lower pH upon entry into stationary phase (Fig. 5B). The decreased pH significantly reduced survival of the strains, with Alt.1 surviving for approximately 5 days in stationary phase, with a pH ranging from 5.2 to 5.4 upon entry into stationary phase (Fig. 5B). Alt. 2 survived for approximately 3 days in stationary phase, with a pH ranging from 5.0 to 5.2 (Fig. 5B).
FIG. 5.
Ammonia production and its effect upon survival in Alt. 1 and Alt. 2 cultures. (A) S. pyogenes strains CS101, Alt. 1, and Alt. 2 were grown to stationary phase in static TH broth cultures under a 5% CO2 atmosphere. Early-stationary-phase culture samples were removed and filter sterilized, and ammonia concentrations were determined. For this graph, T0 is entry into stationary phase (OD600 of ∼1.0). The concentration of ammonia present in sterile TH broth is represented by the bar at the left marked “TH.” Each bar represents the mean value from at least three independent cultures. The error bars are the standard deviations from the means. (B) S. pyogenes strains CS101, Alt. 1, and Alt. 2 were grown to stationary phase in static TH broth cultures under a 5% CO2 atmosphere. Approximately 24 h after entry into stationary phase, exogenous ammonia was added from a 2 M stock to yield a final culture concentration of 20.0 mM, 10.0 mM, or 5.0 mM. Survival was assayed by spotting 50-μl culture samples on TH agar plates at 24-h time intervals, postentry into stationary phase. The formation of any S. pyogenes colonies within the culture spot was scored positively for survival. The lower limit of detection was 20 CFU per ml. Bars represent the mean duration of survival for each culture condition. For this graph, T0 corresponds with entry into stationary phase. The pH range for each culture condition (pHi), measured after the addition of exogenous ammonia, is noted next to each survival bar. Each data set is the mean of at least four independent cultures. Error bars represent the standard deviations between these experiments.
To determine if pH was the major factor governing the decreased survival of Alt.1 and Alt. 2, ammonia was used to neutralize the excess acid and restore the ability of Alt. 1 and Alt. 2 to survive in stationary phase. Exogenous ammonia was added to early-stationary-phase cultures of Alt. 1 and Alt. 2 (Fig. 5B). The addition of exogenous ammonia at a final concentration of 10 mM and 20 mM to cultures of Alt. 1 and Alt. 2 raised culture pH above 5.6 and resulted in culture survival in excess of 6 weeks (Fig. 5B). The difference in Alt. 1 and Alt.2 to survive at 5 mM ammonia (6 versus 3 days) may be due to the lack of production of endogenous ammonia in exponential growth in Alt. 2 that results in a slight decrease in culture pH (Fig. 5A). It was not possible to buffer the pH of Alt. 1 or Alt. 2 cultures because trying to buffer TH broth above pH 5.6 led to salt toxicity of the media before reaching the buffering point (data not shown). However, in further support of pH being the driving force for truncated survival, lowering the pH with the addition of 12.5 mM lactic acid or HCl truncated survival of the parental strain CS101 to 2 days (data not shown).
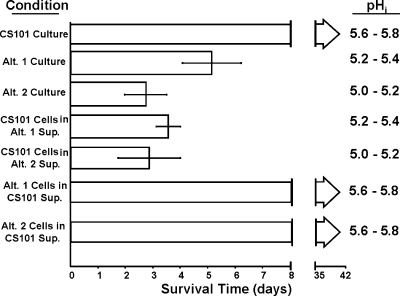
To further verify that the differences in survival were due to the supernatant and not due to differences in the other defects in Alt. 1 and Alt. 2 cells, supernatant switching experiments were done. TH broth cultures of all three strains were grown to early stationary phase, cells were pelleted by centrifugation, and supernatants were removed. The cells were washed and resuspended in the supernatant of a different culture (Fig. 6). Resuspension of CS101 cells in an equivalent volume of Alt. 1-conditioned supernatant (pH 5.2 to 5.4) truncated CS101 survival to under 4 days, while an equivalent volume of Alt. 2-conditioned supernatant (5.0 to 5.2) truncated CS101 survival to under 3 days (Fig. 6). Resuspension of Alt. 1 and Alt. 2 cells in CS101-conditioned supernatant (pH 5.6 to 5.8) allowed for survival in excess of 6 weeks, the last time point taken for this experiment (Fig. 6).
FIG. 6.
Stationary-phase survival of S. pyogenes CS101, Alt. 1, and Alt. 2 after supernatant switch. S. pyogenes CS101, Alt. 1, and Alt. 2 were grown to stationary phase in static TH broth cultures under a 5% CO2 atmosphere. Approximately 12 h after entry into stationary phase, cultures were pelleted, and culture supernatants for each strain were pooled. CS101 cells were resuspended in Alt. 1 or Alt. 2 supernatants. Alt. 1 and Alt. 2 cells were resuspended in CS101 supernatant (Sup.). Survival was assayed by spotting 50-μl culture samples on TH agar plates at 24-h time intervals, postentry into stationary phase. The formation of any S. pyogenes colonies within the culture spot was scored positively for survival. The lower limit of detection was 20 CFU per ml. Bars represent the mean duration of survival for each culture condition. For this graph, T0 corresponds with entry of cultures into stationary phase. The initial pH range for each culture condition (pHi), measured upon supernatant switch, is noted next to each survival bar. Each data set is the mean of at least four independent cultures. Error bars represent the standard deviations between these experiments.
Taken together, these data suggest that in order to survive the culture pH must remain above 5.6. A culture of metabolically diverse strains can survive as long as acid production through the PA pathway is balanced with ammonia production to maintain this pH threshold.
Metabolic diversity may exist in clinical isolates from tonsils.
Stationary-phase cultures are a closed system; therefore, it is possible that metabolic diversity is a result of a selective pressure unique to in vitro cultures. It has been proposed that S. pyogenes can survive in tonsillar cells, since they are in a location protected from penicillin and the immune response (52-54, 59). Using increased PA pathway activity during exponential growth as a marker of metabolic diversity, three intracellular clinical isolates were examined (59). The strains were isolated previously from tonsillar material from three males 20, 21, and 22 years of age that reported suffering from recurrent tonsillitis (59). S. pyogenes cells were cultured from the tonsil swab and surgical specimen from one patient (isolate 221) and exclusively from the surgically removed tonsil material of two patients (isolates Sfr321 and MK322) (59). In all tonsillar samples, intracellular S. pyogenes cells could be visualized by immunohistochemistry and light microscopy with antibodies directed against the streptococcal cell wall (59). Like CS101, strains 221 and Sfr321 were M serotype 49 strains; MK322 was M serotype 6. One complication of these studies is that only the survivor strains and not the parental strain of the clinical isolates were available; therefore, matched serotype controls were used for the clinical isolates (CS101 for M49 and JRS4 for M6). Northern blotting for PA pathway gene transcription and the production of PA pathway by-products, formate and acetate, during exponential growth were used to screen the strains for increased PA pathway activity. Strain 221 significantly increased PA pathway transcription and formate and acetate production (Fig. 7). MK322 had no increases in acetate or formate production or in PA pathway transcription. Sfr321 did not have detectable increases in PA pathway activity. These data suggest that it is possible to isolate intracellular survivors, such as strain 221, that have increased PA pathway expression during exponential growth. In the studies by Podbielski et al., only a single strain from each tonsillar sample was saved (59). Since metabolic diversity may be a hallmark of stationary-phase survival, it would be interesting to determine if metabolic diversity would be observed with multiple strains isolated from a single patient.
FIG. 7.
PA pathway activity is increased during exponential growth in one of three clinical strains. (A) Total RNA was isolated from mid-exponential cells (OD600 nm of 0.50 to 0.65) grown in TH broth. RNA concentrations were determined by spectrophotometric absorbance at 260 nm. Total RNA (5.0, 2.5, and 1.25 μg) was separated on a denaturing agarose gel. RNA gels were subjected to Northern blotting, and the binding of DIG-labeled DNA probes was detected by CSPD development and by autoradiography. Equal loading was confirmed by ethidium bromide staining of the gel and by probing for DNA gyrase (gyrA). RNA concentrations in μg are noted above the lanes in each image. Fresh RNA was isolated for each experiment, and the results presented here are representative of three independent preparations. (B and C) S. pyogenes strains were grown statically in TH broth under a 5% CO2 atmosphere, and exponential cell density was monitored by culture absorbance at 600 nm. Late-exponential-phase (OD600 of 0.80 to 0.90) culture samples were removed and filter sterilized, and lactate, acetate, and formate concentrations were determined. Metabolite concentrations present in sterile TH broth are represented in each graph by the bar at the left. Each bar represents the mean value from at least three independent cultures. The standard deviation of the mean is represented by the error bars. Each strain is represented by the following bars: □ TH, ▥, CS101; ░⃞, 221; ▧, Sfr321;  , JRS4; and
, JRS4; and  , MK322.
, MK322.
The generation of diversity in survivor strains may be due to random mutation.
Strains isolated from long-term stationary-phase cultures are metabolically diverse compared to each other, even between strains isolated from the same culture. These phenotypes are stable even after multiple passages, suggesting that the changes may be genetic. Accumulation of mutations in long-term stationary-phase cultures has been well documented with E. coli (reviewed in reference 86). To determine if mutations in global regulators of S. pyogenes could be detected in strains isolated from surviving cultures, the genes of nine known regulators were amplified by PCR and sequenced in each of the survivor strains. The sequences were then compared to the parental CS101 strain to look for mutations in these genes or their promoter regions. Two strains showed two different types of mutations (Table 2). Alt. 1 had a point mutation in codY. This mutation codes for an amino acid change at position 128 of glycine to glutamic acid. CodY is a repressor that responds to levels of branched-chain amino acids. Its structure has been determined (41), and position 128 lies within the N-terminal cofactor binding domain. It is possible that a mutation in this area could affect its regulatory activity. A different mutation was found in Alt. 2. A 12-bp insertion, which is a direct repeat of the sequence directly preceding it, occurred in the SPY1548 gene. SPY1548 is a hypothetical protein in the GAS genome that has homology to an Fnr (fumarate nitrate reductase) family protein. Streptococcus gordonii produces Flp (Fnr-like protein) that activates the arginine deiminase operon (22). Fnr proteins are involved in the acetate switch in bacteria such as E. coli, in which the cells switch from producing acetate during exponential phase to catabolizing acetate during stationary phase (39). These mutations were found only in single strains and were not present in other strains.
TABLE 2.
Mutations in global regulators of survivor strainsa
| Gene | Sequencing result compared to sequence of parental strain CS101 | Function of gene product | Reference |
|---|---|---|---|
| codY | Point mutation of G to A in Alt. 1. Causes aa 128 to change from G to E. | Controls expression of stationary genes by repressing exponential phase genes. | 44 |
| ccpA | No mutations | Repressor that plays a role in catabolite repression. | 20 |
| srv | No mutations | Homologous to a member of Crp/Fnr family (involved in the acetate switch). Regulates virulence factors in S. pyogenes. | 68 |
| SPY1548 | Alt 2 showed an insertion of 12 bases. Adds Ile-Val-Val-Ala. Insertion is a repeat of sequence preceding it. | FNR-like protein, which is involved in the acetate switch. FNR is an activator of the Arc operon in S. gordonii. | 22 |
| relA | No mutations | Converts GTP to pppGpp during the stringent response. | 44 |
| ropB | No mutations | A mutation in rgg causes utilization of serine and arginine in the presence of carbon. Rgg affects growth phase proteins associated with amino acid utilization. | 11,13 |
| SPY1630 | No mutations | Omega subunit of RNA polymerase. Has a role in stringent response in E. coli. | 82 |
| SPY0145 | No mutations | Homology to AldR, which is a negative regulator of transcription for genes involved in amino acid metabolism in L. lactis. | 31 |
| covRS | No mutations | Two-component response regulator that acts as a repressor of ∼15% of the GAS genome. Has a role in stress response and regulation of multiple virulence factors. | 14 |
aa, amino acid.
DISCUSSION
In the present study we found that long-term stationary-phase S. pyogenes cultures were metabolically active. Addition of the transcriptional inhibitor rifampin and the protein synthesis inhibitor gentamicin truncated survival. Penicillin and vancomycin truncated survival, indicating that the survivors required continued production of the cell wall. It is not possible to conclude whether the bacteria were still dividing or surviving in a nonreplicating state, since the production of new peptidoglycan has been shown to occur in E. coli for the purpose of cell wall repair during stationary phase (55). Consistent with metabolic activity during survival, depletion of all nutrients, except phosphate, by resuspending cells in PBS truncated survival. The initial 3-log decrease in cell numbers during the first week was consistent with the observations of Trainor et al. (81), and the remaining S. pyogenes cells survived for about 1.5 years in Todd-Hewitt broth (83). Regardless of whether death after 1.5 years results from a final depletion of nutrients, build-up of toxins, or the eventual senescence of the cultures, these data suggested that S. pyogenes bacteria survive in a metabolically active state.
Although the glucose present in TH broth was consumed by entry into stationary phase, cultures survived for ∼1.5 years (83). Previous studies of survival in chemically defined medium (CDM) show that CDM does not support long-term stationary-phase survival for S. pyogenes, even under low-glucose conditions, suggesting that some component(s) from TH broth are necessary for survival (83). TH broth is rich in nutrients, such as amino acids, proteins, glycoproteins and pentoses, which could potentially be used to generate ATP. Even though many pathogens prefer glucose for growth in the laboratory, the use of alternative energy sources in stationary phase and during survival in the host is well documented. For example, M. tuberculosis generates energy during survival by using the glyoxylate shunt of the Krebs cycle, which uses fatty acids to generate energy (reviewed in reference 48). In other instances of survival, pyruvate metabolism has been shown to provide energy after the depletion of glucose in lactic acid bacteria (29, 30). Consistent with previous observations, S. pyogenes shows evidence of pyruvate metabolism after the depletion of glucose (71). In the current studies, the slow decrease in lactate and the increase in formate and acetate over 12 weeks suggests PA pathway activity throughout stationary-phase survival. Caution must be used in interpreting the exact ratios of PA pathway intermediates due to possible further metabolism of formate and the diverse metabolism of individual strains in the surviving culture. However, the presence of formate suggested pyruvate formate lyase was responsible for at least some of the conversion of pyruvate to acetyl-CoA. There was significantly more acetate produced than lactate consumed. One possible explanation for this observation is that there were other intermediates present that could enter the PA pathway. For example, serine dehydratase converts serine to pyruvate, which could enter then into the PA pathway. Activity of amino acid catabolic pathways is suggested by the accumulation of ammonia in surviving cultures.
Stationary-phase cultures of E. coli accumulate mutations that confer growth advantages in stationary phase (recently reviewed in reference 27). When the survivors isolated from 1-week-old stationary-phase cultures of E. coli are competed against unaged cells, the aged cells are better able to survive (28). The mutations map to several genes, including the stationary-phase sigma factor gene rpoS and genes responsible for amino acid catabolism (85, 87, 88). Therefore, the surviving E. coli cultures have a succession of strains, each one more fit to survive in stationary phase. In contrast, S. pyogenes cultures appear to diversify during survival. For S. pyogenes to survive in vitro, it is essential to maintain the pH above ∼5.6 (83), and the addition of HCl to a pH below 5.6 will truncate survival. The individual strains in the culture have mutated such that they now express increased levels of the PA pathway, PE pathway, or amino acid catabolism, even in the presence of glucose, suggesting that the pathways are no longer subject to normal regulation. Metabolites from these pathways accumulate throughout stationary survival, suggesting that these pathways are active in the surviving culture. Maintenance of the pH of late-stationary-phase cultures likely requires metabolic activity of multiple strains to maintain the culture pH. Single strains, such as Alt. 1 and Alt. 2, that produce high levels of acid lower the culture pH during survival and are unable to survive in pure culture beyond 5 days. The production of ammonia from amino acid catabolism has been shown to protect other bacteria, such as oral bacteria and Lactobacillus sakei, from the effects of metabolic acids (reviewed in references 10 and 45). The addition of ammonia to Alt. 1 and Alt. 2 prolongs survival of those cultures, and high ammonia producers can be isolated from surviving cultures. Therefore, stationary-phase cultures probably survive as a metabolically mixed population, which maintains the culture pH above 5.6.
The generation of metabolic diversity in S. pyogenes CS101 may not be an artifact of a closed in vitro system. One of three clinical isolates from the tonsillar tissue of patients with recurrent tonsillitis (59) showed increased PA pathway activity during exponential growth, suggesting that in vivo survival during carriage may result in metabolic diversity. It is currently unknown how common these strains might be during infection. It is possible that metabolic diversity is a property of survival, and it may be selected against or out-competed in an active infection.
Stationary-phase survivor strains, such as Alt. 1 and Alt. 2, have not only changes in metabolism, but also proteome-wide changes (83), suggesting that survival in stationary phase may generate diversity beyond metabolism. S. pyogenes clinical isolates are very diverse, even within the same M type (18, 19, 35), and considerable effort is being made to differentiate the core genome from variable regions of the chromosome (3, 25, 79). S. pyogenes appears to have a core genome that is relatively conserved between M types (3). This conservation has been observed on the level of genome sequencing and multilocus sequence typing (25). There can be significant divergence of strains outside this core region. The divergence is partially due to chromosomal rearrangements induced by insertion sequences and phages and partially due to allelic mutation (3, 18, 37, 74). Both genic gain and loss and allelic mutation can contribute to the processes of S. pyogenes diversification and virulence (47). This genetic flexibility may be important for the ability of S. pyogenes to survive in so many host niches. Mutation during survival may contribute to the allelic diversification of S. pyogenes strains. In the present study, sequencing of nine regulator genes of survivor strains revealed two unique mutations, a point mutation and an insertion, in two different survivor strains. Since the mutations observed in our studies occurred in late stationary phase, it is possible that the bacteria entered a hypermutable state due to stress-induced mutation or as a result of other mechanisms, such as phage inactivation of mutS (70). Diversification by random allelic mutation would result in increased fitness for the population as a whole but not necessarily for each strain generated, which is consistent with the observations that some strains, such as Alt. 1 and Alt. 2, do not survive better than the parental strain in pure culture. Selective pressures after survival may determine the phenotypic characteristics of the infectious strains. For example, adaptation of S. pyogenes to the host niche has been observed during passage of S. pyogenes in animal models and during passage in human blood (65-67, 69, 76). In these studies, differences in virulence factor expression levels have been suggested to be the result of the mutation of response regulators (24). Recent studies suggest that mutations in the global virulence regulator genes covR and covS may be responsible for differences between pharyngeal transcriptome profiles and invasive transcriptome profiles of human disease isolates (80). Therefore, intracellular survival during carriage may provide an environment in which S. pyogenes diversifies. Upon reemergence, selective pressures determine the properties of the infectious strains. Analysis of diversity between strains isolated from eukaryotic cell cocultures, colonized animals, and tonsillar tissue from a single asymptomatic carrier could yield further insight into the generation of diversity during S. pyogenes carriage.
Acknowledgments
We thank Patrick J. Piggot, Shannon Morgan, Vasant Chary, Javier Izquierdo, and Bryan Utter for helpful discussions.
This work was supported in part by grant 01606430U from the American Heart Association (to B.A.B). This project is funded, in part, under a grant from the Pennsylvania Department of Health. The Department specifically disclaims responsibility for any analyses, interpretations, or conclusions. The work of B.K. was supported by a grant from the German BMBF in the framework of the SysMO program (FKZ0313978B).
Footnotes
Published ahead of print on 7 August 2009.
REFERENCES
- 1.Barnett, T. C., J. V. Bugrysheva, and J. R. Scott. 2007. Role of mRNA stability in growth phase regulation of gene expression in the group A streptococcus. J. Bacteriol. 189:1866-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Begovac, J., E. Bobinac, B. Benic, B. Desnica, T. Maretic, A. Basnec, and N. Kuzmanovic. 1993. Asymptomatic pharyngeal carriage of beta-haemolytic streptococci and streptococcal pharyngitis among patients at an urban hospital in Croatia. Eur. J. Epidemiol. 9:405-410. [DOI] [PubMed] [Google Scholar]
- 3.Beres, S. B., and J. M. Musser. 2007. Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PLoS ONE 2:e800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beyer-Sehlmeyer, G., B. Kreikemeyer, A. Horster, and A. Podbielski. 2005. Analysis of the growth phase-associated transcriptome of Streptococcus pyogenes. Int. J. Med. Microbiol. 295:161-177. [DOI] [PubMed] [Google Scholar]
- 5.Bingen, E., E. Denamur, N. Lambert-Zechovsky, N. Braimi, M. el Lakany, and J. Elion. 1992. DNA restriction fragment length polymorphism differentiates recurrence from relapse in treatment failures of Streptococcus pyogenes pharyngitis. J. Med. Microbiol. 37:162-164. [DOI] [PubMed] [Google Scholar]
- 6.Boshoff, H. I., and C. E. Barry. 2005. A low-carb diet for a high-octane pathogen. Nat. Med. 11:599-600. [DOI] [PubMed] [Google Scholar]
- 7.Brandt, C. M., B. Spellerberg, M. Honscha, N. D. Truong, B. Hoevener, and R. Lutticken. 2001. Typing of Streptococcus pyogenes strains isolated from throat infections in the region of Aachen, Germany. Infection 29:163-165. [DOI] [PubMed] [Google Scholar]
- 8.Brook, I., P. Yocum, and K. Shah. 1980. Surface vs core-tonsillar aerobic and anaerobic flora in recurrent tonsillitis. JAMA 244:1696-1698. [PubMed] [Google Scholar]
- 9.Campbell, J. R., C. A. Arango, J. A. Garcia-Prats, and C. J. Baker. 1996. An outbreak of M serotype 1 group A Streptococcus in a neonatal intensive care unit. J. Pediatr. 129:396-402. [PubMed] [Google Scholar]
- 10.Champomier Verges, M. C., M. Zuniga, F. Morel-Deville, G. Perez-Martinez, M. Zagorec, and S. D. Ehrlich. 1999. Relationships between arginine degradation, pH and survival in Lactobacillus sakei. FEMS Microbiol. Lett. 180:297-304. [DOI] [PubMed] [Google Scholar]
- 11.Chaussee, M. A., E. A. Callegari, and M. S. Chaussee. 2004. Rgg regulates growth phase-dependent expression of proteins associated with secondary metabolism and stress in Streptococcus pyogenes. J. Bacteriol. 186:7091-7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaussee, M. A., A. V. Dmitriev, E. A. Callegari, and M. S. Chaussee. 2008. Growth phase-associated changes in the transcriptome and proteome of Streptococcus pyogenes. Arch. Microbiol. 189:27-41. [DOI] [PubMed] [Google Scholar]
- 13.Chaussee, M. S., G. A. Somerville, L. Reitzer, and J. M. Musser. 2003. Rgg coordinates virulence factor synthesis and metabolism in Streptococcus pyogenes. J. Bacteriol. 185:6016-6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Churchward, G., C. Bates, A. A. Gusa, V. Stringer, and J. R. Scott. 2009. Regulation of streptokinase expression by CovR/S in Streptococcus pyogenes: CovR acts through a single high-affinity binding site. Microbiology 155:566-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cockerill, F. R., III, K. L. MacDonald, R. L. Thompson, F. Roberson, P. C. Kohner, J. Besser-Wiek, J. M. Manahan, J. M. Musser, P. M. Schlievert, J. Talbot, B. Frankfort, J. M. Steckelberg, W. R. Wilson, and M. T. Osterholm. 1997. An outbreak of invasive group A streptococcal disease associated with high carriage rates of the invasive clone among school-aged children. JAMA 277:38-43. [PubMed] [Google Scholar]
- 16.Cornfeld, D., and J. P. Hubbard. 1961. A four-year study of the occurrence of beta-hemolytic streptococci in 64 school children. N. Engl. J. Med. 264:211-215. [DOI] [PubMed] [Google Scholar]
- 17.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai, M., A. Tanna, A. Efstratiou, R. George, J. Clewley, and J. Stanley. 1998. Extensive genetic diversity among clinical isolates of Streptococcus pyogenes serotype M5. Microbiology 144:629-637. [DOI] [PubMed] [Google Scholar]
- 19.Desai, M., A. Tanna, R. Wall, A. Efstratiou, R. George, and J. Stanley. 1998. Fluorescent amplified-fragment length polymorphism analysis of an outbreak of group A streptococcal invasive disease. J. Clin. Microbiol. 36:3133-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deutscher, J., R. Herro, A. Bourand, I. Mijakovic, and S. Poncet. 2005. P-Ser-HPr-a link between carbon metabolism and the virulence of some pathogenic bacteria. Biochim. Biophys. Acta 1754:118-125. [DOI] [PubMed] [Google Scholar]
- 21.Dmitriev, A. V., E. J. McDowell, K. V. Kappeler, M. A. Chaussee, L. D. Rieck, and M. S. Chaussee. 2006. The Rgg regulator of Streptococcus pyogenes influences utilization of nonglucose carbohydrates, prophage induction, and expression of the NAD-glycohydrolase virulence operon. J. Bacteriol. 188:7230-7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong, Y., Y. Y. Chen, and R. A. Burne. 2004. Control of expression of the arginine deiminase operon of Streptococcus gordonii by CcpA and Flp. J. Bacteriol. 186:2511-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durmaz, R., B. Durmaz, M. Bayraktar, I. H. Ozerol, M. T. Kalcioglu, E. Aktas, and Z. Cizmeci. 2003. Prevalence of group A streptococcal carriers in asymptomatic children and clonal relatedness among isolates in Malatya, Turkey. J. Clin. Microbiol. 41:5285-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eberhard, T. H., D. D. Sledjeski, and M. D. Boyle. 2001. Mouse skin passage of a Streptococcus pyogenes Tn917 mutant of sagA/pel restores virulence, beta-hemolysis and sagA/pel expression without altering the position or sequence of the transposon. BMC Microbiol. 1:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enright, M. C., B. G. Spratt, A. Kalia, J. H. Cross, and D. E. Bessen. 2001. Multilocus sequence typing of Streptococcus pyogenes and the relationships between emm type and clone. Infect. Immun. 69:2416-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finkel, S. E. 2006. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat. Rev. Microbiol. 4:113-120. [DOI] [PubMed] [Google Scholar]
- 28.Finkel, S. E., and R. Kolter. 1999. Evolution of microbial diversity during prolonged starvation. Proc. Natl. Acad. Sci. USA 96:4023-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fordyce, A. M., V. L. Crow, and T. D. Thomas. 1984. Regulation of product formation during glucose or lactose limitation in nongrowing cells of Streptococcus lactis. Appl. Environ. Microbiol. 48:332-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibello, A., M. D. Collins, L. Dominguez, J. F. Fernandez-Garayzabal, and P. T. Richardson. 1999. Cloning and analysis of the l-lactate utilization genes from Streptococcus iniae. Appl. Environ. Microbiol. 65:4346-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goupil-Feuillerat, N., M. Cocaign-Bousquet, J. J. Godon, S. D. Ehrlich, and P. Renault. 1997. Dual role of alpha-acetolactate decarboxylase in Lactococcus lactis subsp. lactis. J. Bacteriol. 179:6285-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hahn, R. G., L. M. Knox, and T. A. Forman. 2005. Evaluation of poststreptococcal illness. Am. Fam. Physician 71:1949-1954. [PubMed] [Google Scholar]
- 33.Hingley-Wilson, S. M., V. K. Sambandamurthy, and W. R. Jacobs, Jr. 2003. Survival perspectives from the world's most successful pathogen, Mycobacterium tuberculosis. Nat. Immunol. 4:949-955. [DOI] [PubMed] [Google Scholar]
- 34.Hoe, N. P., K. E. Fullerton, M. Liu, J. E. Peters, G. D. Gackstetter, G. J. Adams, and J. M. Musser. 2003. Molecular genetic analysis of 675 group A streptococcus isolates collected in a carrier study at Lackland Air Force Base, San Antonio, Texas. J. Infect. Dis. 188:818-827. [DOI] [PubMed] [Google Scholar]
- 35.Holden, M. T., A. Scott, I. Cherevach, T. Chillingworth, C. Churcher, A. Cronin, L. Dowd, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Moule, K. Mungall, M. A. Quail, C. Price, E. Rabbinowitsch, S. Sharp, J. Skelton, S. Whitehead, B. G. Barrell, M. Kehoe, and J. Parkhill. 2007. Complete genome of acute rheumatic fever-associated serotype M5 Streptococcus pyogenes strain Manfredo. J. Bacteriol. 189:1473-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karakousis, P. C., T. Yoshimatsu, G. Lamichhane, S. C. Woolwine, E. L. Nuermberger, J. Grosset, and W. R. Bishai. 2004. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J. Exp. Med. 200:647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kratovac, Z., A. Manoharan, F. Luo, S. Lizano, and D. E. Bessen. 2007. Population genetics and linkage analysis of loci within the FCT region of Streptococcus pyogenes. J. Bacteriol. 189:1299-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kreikemeyer, B., K. S. McIver, and A. Podbielski. 2003. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 11:224-232. [DOI] [PubMed] [Google Scholar]
- 39.Kumari, S., C. M. Beatty, D. F. Browning, S. J. Busby, E. J. Simel, G. Hovel-Miner, and A. J. Wolfe. 2000. Regulation of acetyl coenzyme A synthetase in Escherichia coli. J. Bacteriol. 182:4173-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leonard, B. A., M. Woischnik, and A. Podbielski. 1998. Production of stabilized virulence factor-negative variants by group A streptococci during stationary phase. Infect. Immun. 66:3841-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levdikov, V. M., E. Blagova, P. Joseph, A. L. Sonenshein, and A. J. Wilkinson. 2006. The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in gram-positive bacteria. J. Biol. Chem. 281:11366-11373. [DOI] [PubMed] [Google Scholar]
- 42.Loughman, J. A., and M. Caparon. 2006. Regulation of SpeB in Streptococcus pyogenes by pH and NaCl: a model for in vivo gene expression. J. Bacteriol. 188:399-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malke, H., and J. J. Ferretti. 2007. CodY-affected transcriptional gene expression of Streptococcus pyogenes during growth in human blood. J. Med. Microbiol. 56:707-714. [DOI] [PubMed] [Google Scholar]
- 44.Malke, H., K. Steiner, W. M. McShan, and J. J. Ferretti. 2006. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: the action of CodY and RelA. Int. J. Med. Microbiol. 296:259-275. [DOI] [PubMed] [Google Scholar]
- 45.Marquis, R. E., G. R. Bender, D. R. Murray, and A. Wong. 1987. Arginine deiminase system and bacterial adaptation to acid environments. Appl. Environ. Microbiol. 53:198-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin, J. M., M. Green, K. A. Barbadora, and E. R. Wald. 2004. Group A streptococci among school-aged children: clinical characteristics and the carrier state. Pediatrics 114:1212-1219. [DOI] [PubMed] [Google Scholar]
- 47.McMillan, D. J., K. S. Sriprakash, and G. S. Chhatwal. 2007. Genetic variation in group A streptococci. Int. J. Med. Microbiol. 297:525-532. [DOI] [PubMed] [Google Scholar]
- 48.Munoz-Elias, E. J., and J. D. McKinney. 2006. Carbon metabolism of intracellular bacteria. Cell Microbiol. 8:10-22. [DOI] [PubMed] [Google Scholar]
- 49.Nataro, J. P., M. J. Blaser, and S. Cunningham-Rundles. 2000. Persistent bacterial infections. ASM Press, Washington, DC.
- 50.Oliver, J. D. 2005. The viable but nonculturable state in bacteria. J. Microbiol. 43:93-100. [PubMed] [Google Scholar]
- 51.Opdyke, J. A., J. R. Scott, and C. P. Moran, Jr. 2001. A secondary RNA polymerase sigma factor from Streptococcus pyogenes. Mol. Microbiol. 42:495-502. [DOI] [PubMed] [Google Scholar]
- 52.Osterlund, A., and L. Engstrand. 1997. An intracellular sanctuary for Streptococcus pyogenes in human tonsillar epithelium-studies of asymptomatic carriers and in vitro cultured biopsies. Acta Otolaryngol. 117:883-888. [DOI] [PubMed] [Google Scholar]
- 53.Osterlund, A., and L. Engstrand. 1995. Intracellular penetration and survival of Streptococcus pyogenes in respiratory epithelial cells in vitro. Acta Otolaryngol. 115:685-688. [DOI] [PubMed] [Google Scholar]
- 54.Osterlund, A., R. Popa, T. Nikkila, A. Scheynius, and L. Engstrand. 1997. Intracellular reservoir of Streptococcus pyogenes in vivo: a possible explanation for recurrent pharyngotonsillitis. Laryngoscope 107:640-647. [DOI] [PubMed] [Google Scholar]
- 55.Park, J. T. 1995. Why does Escherichia coli recycle its cell wall peptides? Mol. Microbiol. 17:421-426. [DOI] [PubMed] [Google Scholar]
- 56.Pichichero, M. E., J. L. Green, A. B. Francis, S. M. Marsocci, A. M. Murphy, W. Hoeger, C. Noriega, A. Sorrento, and J. Gootnick. 1998. Recurrent group A streptococcal tonsillopharyngitis. Pediatr. Infect. Dis. J. 17:809-815. [DOI] [PubMed] [Google Scholar]
- 57.Pichichero, M. E., S. M. Marsocci, M. L. Murphy, W. Hoeger, J. L. Green, and A. Sorrento. 1999. Incidence of streptococcal carriers in private pediatric practice. Arch. Pediatr. Adolesc. Med. 153:624-628. [DOI] [PubMed] [Google Scholar]
- 58.Piggot, P. J., and D. W. Hilbert. 2004. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 7:579-586. [DOI] [PubMed] [Google Scholar]
- 59.Podbielski, A., S. Beckert, R. Schattke, F. Leithauser, F. Lestin, B. Gossler, and B. Kreikemeyer. 2003. Epidemiology and virulence gene expression of intracellular group A streptococci in tonsils of recurrently infected adults. Int. J. Med. Microbiol. 293:179-190. [DOI] [PubMed] [Google Scholar]
- 60.Podbielski, A., A. Flosdorff, and J. Weber-Heynemann. 1995. The group A streptococcal virR49 gene controls expression of four structural vir regulon genes. Infect. Immun. 63:9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Proctor, R. A., P. van Langevelde, M. Kristjansson, J. N. Maslow, and R. D. Arbeit. 1995. Persistent and relapsing infections associated with small-colony variants of Staphylococcus aureus. Clin. Infect. Dis. 20:95-102. [DOI] [PubMed] [Google Scholar]
- 62.Proctor, R. A., C. von Eiff, B. C. Kahl, K. Becker, P. McNamara, M. Herrmann, and G. Peters. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4:295-305. [DOI] [PubMed] [Google Scholar]
- 63.Quinn, R. W. 1980. Hemolytic streptococci in Nashville school children. South Med. J. 73:288-296. [DOI] [PubMed] [Google Scholar]
- 64.Quinn, R. W., and C. F. Federspiel. 1973. The occurrence of hemolytic streptococci in school children in Nashville, Tennessee, 1961-1967. Am. J. Epidemiol. 97:22-33. [DOI] [PubMed] [Google Scholar]
- 65.Raeder, R., and M. D. Boyle. 1993. Association between expression of immunoglobulin G-binding proteins by group A streptococci and virulence in a mouse skin infection model. Infect. Immun. 61:1378-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raeder, R., and M. D. Boyle. 1993. Association of type II immunoglobulin G-binding protein expression and survival of group A streptococci in human blood. Infect. Immun. 61:3696-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raeder, R., E. Harokopakis, S. Hollingshead, and M. D. Boyle. 2000. Absence of SpeB production in virulent large capsular forms of group A streptococcal strain 64. Infect. Immun. 68:744-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reid, S. D., A. G. Montgomery, and J. M. Musser. 2004. Identification of srv, a PrfA-like regulator of group A Streptococcus that influences virulence. Infect. Immun. 72:1799-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rezcallah, M. S., M. D. Boyle, and D. D. Sledjeski. 2004. Mouse skin passage of Streptococcus pyogenes results in increased streptokinase expression and activity. Microbiology 150:365-371. [DOI] [PubMed] [Google Scholar]
- 70.Scott, J., P. Thompson-Mayberry, S. Lahmamsi, C. J. King, and W. M. McShan. 2008. Phage-associated mutator phenotype in group A streptococcus. J. Bacteriol. 190:6290-6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seki, M., K. Iida, M. Saito, H. Nakayama, and S. Yoshida. 2004. Hydrogen peroxide production in Streptococcus pyogenes: involvement of lactate oxidase and coupling with aerobic utilization of lactate. J. Bacteriol. 186:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shaw, J. H., and D. B. Clewell. 1985. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J. Bacteriol. 164:782-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shelburne, S. A., III, D. Keith, N. Horstmann, P. Sumby, M. T. Davenport, E. A. Graviss, R. G. Brennan, and J. M. Musser. 2008. A direct link between carbohydrate utilization and virulence in the major human pathogen group A Streptococcus. Proc. Natl. Acad. Sci. USA 105:1698-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shelburne, S. A., III, P. Sumby, I. Sitkiewicz, C. Granville, F. R. DeLeo, and J. M. Musser. 2005. Central role of a bacterial two-component gene regulatory system of previously unknown function in pathogen persistence in human saliva. Proc. Natl. Acad. Sci. USA 102:16037-16042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shelburne, S. A., III, P. Sumby, I. Sitkiewicz, N. Okorafor, C. Granville, P. Patel, J. Voyich, R. Hull, F. R. DeLeo, and J. M. Musser. 2006. Maltodextrin utilization plays a key role in the ability of group A Streptococcus to colonize the oropharynx. Infect. Immun. 74:4605-4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith, T. C., D. D. Sledjeski, and M. D. Boyle. 2003. Regulation of protein H expression in M1 serotype isolates of Streptococcus pyogenes. FEMS Microbiol. Lett. 219:9-15. [DOI] [PubMed] [Google Scholar]
- 77.Steiner, K., and H. Malke. 2000. Life in protein-rich environments: the relA-independent response of Streptococcus pyogenes to amino acid starvation. Mol. Microbiol. 38:1004-1016. [DOI] [PubMed] [Google Scholar]
- 78.Steiner, K., and H. Malke. 2001. relA-independent amino acid starvation response network of Streptococcus pyogenes. J. Bacteriol. 183:7354-7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sumby, P., S. F. Porcella, A. G. Madrigal, K. D. Barbian, K. Virtaneva, S. M. Ricklefs, D. E. Sturdevant, M. R. Graham, J. Vuopio-Varkila, N. P. Hoe, and J. M. Musser. 2005. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J. Infect. Dis. 192:771-782. [DOI] [PubMed] [Google Scholar]
- 80.Sumby, P., A. R. Whitney, E. A. Graviss, F. R. DeLeo, and J. M. Musser. 2006. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog 2:e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Trainor, V. C., R. K. Udy, P. J. Bremer, and G. M. Cook. 1999. Survival of Streptococcus pyogenes under stress and starvation. FEMS Microbiol. Lett. 176:421-428. [DOI] [PubMed] [Google Scholar]
- 82.Vrentas, C. E., T. Gaal, W. Ross, R. H. Ebright, and R. L. Gourse. 2005. Response of RNA polymerase to ppGpp: requirement for the omega subunit and relief of this requirement by DksA. 19:2378-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wood, D. N., M. A. Chaussee, M. S. Chaussee, and B. A. Buttaro. 2005. Persistence of Streptococcus pyogenes in stationary-phase cultures. J. Bacteriol. 187:3319-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Woodbury, R. L., X. Wang, and C. P. Moran, Jr. 2006. Sigma X induces competence gene expression in Streptococcus pyogenes. Res. Microbiol. 157:851-856. [DOI] [PubMed] [Google Scholar]
- 85.Zambrano, M. M., D. A. Siegele, M. Almiron, A. Tormo, and R. Kolter. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259:1757-1760. [DOI] [PubMed] [Google Scholar]
- 86.Zinser, E. R., and R. Kolter. 2004. Escherichia coli evolution during stationary phase. Res. Microbiol. 155:328-336. [DOI] [PubMed] [Google Scholar]
- 87.Zinser, E. R., and R. Kolter. 1999. Mutations enhancing amino acid catabolism confer a growth advantage in stationary phase. J. Bacteriol. 181:5800-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zinser, E. R., and R. Kolter. 2000. Prolonged stationary-phase incubation selects for lrp mutations in Escherichia coli K-12. J. Bacteriol. 182:4361-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]