Abstract
We previously reported the identification of a gene, rbf, involved in the regulation of biofilm formation by Staphylococcus aureus 8325-4. In an effort to study the mechanism of regulation, microarrays were used to compare the transcription profiles of the wild-type strain with an rbf mutant and an rbf overexpression strain of the clinical isolate UAMS-1. Among the genes affected by rbf overexpression are those of the intercellular adhesion (ica) locus; however, expression of these genes was not affected by an rbf deletion in the chromosome. The icaADBC genes are responsible for production of poly-N-acetylglucosamine (PNAG), a major constituent of biofilm. The icaR gene encodes a negative regulator of icaADBC. In UAMS-1 carrying an Rbf-encoding plasmid, Rbf was found to repress icaR transcription with a concomitant increase in icaADBC expression and increased PNAG and biofilm production relative to isogenic strains lacking the plasmid. Sequencing of the rbf gene from UAMS-1 showed that there was a 2-bp insertion affecting the 50th codon of the rbf open reading frame, suggesting that rbf is a pseudogene in UAMS-1. This finding explains why deletion of rbf had no effect on biofilm formation in UAMS-1. To further characterize the Rbf regulation on biofilm we compared biofilm formation, icaA and icaR transcription, and PNAG production in 8325-4 and its isogenic rbf and icaR single mutants and an rbf icaR double mutant. Our results are consistent with a model wherein rbf represses synthesis of icaR, which in turn results in derepression of icaADBC and increased PNAG production. Furthermore, purified rbf did not bind to the icaR or icaA promoter region, suggesting that rbf controls expression of an unknown factor(s) that represses icaR. The role of rbf in controlling the S. aureus biofilm phenotype was further demonstrated in a clinical strain, MW2.
Staphylococcus aureus is a major human pathogen capable of causing a broad range of diseases. Some S. aureus infections, such as endocarditis and osteomyelitis, are associated with biofilm formation (3, 4, 13, 29). Biofilms are composed of layers of bacteria within a glycocalyx composed of polysaccharides, DNA, and proteins. In addition to aiding bacterial colonization of surfaces, biofilms are believed to increase tolerance to antibiotics and immune defenses (3, 13, 17, 24, 36, 39, 51).
The biofilm-associated polysaccharide of S. aureus is referred to as the polysaccharide intercellular adhesin or PIA and has been well characterized (13, 30, 36). It is composed of polymeric N-acetylglucosamine (PNAG) that is synthesized by the products of four genes in the ica operon, icaADBC (10, 20). In some strains, genetic disruption of the icaADBC genes results in the loss of biofilm formation (10, 23, 33); however, ica-independent biofilm formation has also been described (2, 9, 37, 38, 47). Expression of icaADBC is, in part, regulated by icaR, a member of the TetR family of regulatory proteins (22). IcaR is encoded at the ica locus but is divergently transcribed from icaADBC. IcaR binds to the icaADBC promoter, 5′ to the icaA start codon, and negatively regulates ica expression (20). Transcription of icaADBC is also subject to positive regulation by the global regulatory factor SarA and in some strains by the stress-induced sigma factor SigB (2, 6, 20, 42). The teicoplanin-associated locus regulator, TcaR, is also a weak negative regulator of icaADBC (21).
A number of reports have shown that extracellular DNA is often an important component of biofilms (34, 40, 54), including S. aureus biofilms (14, 43, 50). The source of the extracellular DNA is bacterial, with DNA release occurring via lysis of bacterial cells. It has been proposed that DNA influences the early stages of biofilm development (43). Rice et al. (43) reported that inactivation of the cidA gene in S. aureus strain UAMS-1 inhibits the release of genomic DNA. CidA is a regulatory factor affecting cell lysis and antibiotic resistance. The cidA mutant was found to have a decreased capacity to form biofilms both in vitro and in vivo (43). Our laboratory has also found that DNase can disrupt UAMS-1 biofilms (unpublished results), suggesting that DNA is an important structural component.
Biofilm formation by staphylococci is subject to complex regulation that is influenced by a number of environmental factors, including osmolarity, glucose levels, anaerobiosis, temperature, and levels of iron, citrate, ethanol, and nitrites (11, 17, 23, 26, 46, 47). We (26) previously described a novel gene, rbf, which regulates biofilm production in response to glucose and NaCl. The rbf gene codes for a 716-amino-acid protein with homology to the AraC/XylS family of transcriptional activators. From this we proposed that rbf is involved in positive regulation of a protein or proteins that are important for biofilm formation. A mutant strain with an insertion in rbf was impaired in biofilm formation on polystyrene and glass. The mutant strain exhibited no defect in primary attachment to polystyrene, however, suggesting that inactivation of rbf affected the multicellular aggregation step rather than the primary attachment step in biofilm formation. In addition, disruption of rbf in S. aureus 8325-4 had no measurable effect on expression of a Pica::xylE reporter construct, suggesting that rbf may regulate an ica-independent pathway for biofilm formation in 8325-4 (26). More recently, we reported that rbf enhances bacterial survival in a murine model of foreign body infection (29).
In this report we utilized microarrays and genetic approaches to further investigate the function of rbf. Our results suggest that Rbf enhances biofilm formation by activating icaADBC expression and that activation is indirect, occurring by rbf repression of icaR. We also discovered that rbf was a pseudogene in UAMS-1. However, the rbf defect in UAMS-1 could be complemented with the rbf gene from 8325-4, suggesting that rbf controls biofilm formation in strains 8325-4 and UAMS-1 via a similar mechanism. The effect of rbf on biofilm was also observed in a recently isolated clinical strain, MW2.
MATERIALS AND METHODS
Bacterial strains, culture media, and growth conditions.
The bacterial strains used in this study are listed in Table 1. S. aureus UAMS-1 is a clinical isolate cultured from the bone of a patient with osteomyelitis (16). S. aureus MW2, obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus, was originally isolated from a child with fatal septicemia and septic arthritis (5). Staphylococci were cultured in tryptic soy broth (TSB; Difco Laboratories, Detroit, MI) or tryptic soy agar (Difco). In some experiments growth medium was supplemented with glucose and NaCl as described below. Antibiotics were added to culture media, as appropriate and unless otherwise specified, at final concentrations of 10 μg per ml chloramphenicol (Cm) and 100 μg per ml penicillin. Escherichia coli strains DH5α and XL1-Blue were used for plasmid construction and maintenance. E. coli was cultivated in Luria-Bertani broth or agar (Difco).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| S. aureus strains | ||
| 8325-4 | Prophage-free laboratory strain | J. Iandolo |
| UAMS-1 | Wild-type clinical isolate | 14 |
| MW2 | Wild-type clinical isolate | 5 |
| CYL6939 | UAMS-1(pLI50) | 29 |
| CYL6933 | UAMS-1(pYL8565) | 29 |
| CYL6970 | UAMS-1 rbf(pLI50) | 29 |
| CYL1112 | 8325-4(pLI50) | This study |
| CYL6968 | 8325-4 rbf(pLI50) | This study |
| CYL6973 | 8325-4(pYL8565) | This study |
| CYL6974 | 8325-4 rbf(pYL8565) | This study |
| CYL1097 | 8325-4 rbf::tet | 26 |
| CYL11688 | 8325-4 icaR(pLI50) | This study |
| CYL11689 | 8325-4 icaRrbf(pLI50) | This study |
| CYL11690 | 8325-4 icaRrbf(pYL8565) | This study |
| CYL11696 | 8325-4 icaADBC | This study |
| CYL11699 | 8325-4 icaR(pML3796) | This study |
| CYL11700 | 8325-4 icaRrbf(pML3796) | This study |
| CYL11683 | MW2(pLI50) | This study |
| CYL11712 | MW2 rbf(pLI50) | This study |
| CYL11713 | MW2 rbf (pYL8565) | This study |
| E. coli strains | ||
| DH5α | Host strain for plasmids | 45 |
| XL1-Blue | Host strain for plasmids | 45 |
| Plasmids | ||
| pLI50 | E. coli-S. aureus shuttle vector | 25 |
| pYL8565 | Rbf expression plasmid derived from pLI50 | 26 |
| pML3796 | IcaR expression plasmid derived from pLI50 | This study |
| pKOR1 | Vector for allele replacement | 1 |
Plasmid and strain construction.
Allele replacement of rbf in strains 8325-4 and MW2 was performed using the same primers and methods as previously described for UAMS-1 Δrbf (29). Allele replacement was confirmed by PCR. Similarly, the icaADBC and icaR deletion mutants of 8325-4 were constructed by allele replacement using pKOR1 (1). DNA inserts with target region deletions were constructed by overlapping PCR. PCR primers attB1-icaKO1, comp-icaKO2, comp-icaKO3, and attB2-icaKO4 were used in construction of the icaADBC mutant (Table 2). Primers attB1-icaRKO1, comp-icaRKO2, comp-icaRKO3, and attB2-icaRKO4 (Table 2) were used in construction of the icaR mutant.
TABLE 2.
Oligonucleotide primers used for plasmid and strain construction
| Primer | Sequence |
|---|---|
| adh14 | CACTCATAAAAGCTTCTTC |
| adh15 | GGGCCCAAGCGACTTAAATTCGATTCGT |
| rbf20 | ATACCGCGGCGCGTTGTCGCATATTCATT |
| rbf62 | CTTAAATATAGAAAGAGGTA |
| attB1-icaKO1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTCCACTGCTCCAAATTTTTGCG |
| attB2-icaKO4 | GGGGACCACTTTGTACAAGAAAGCTGGGTCGATCTGACGCGTGAGGGTGC |
| attB1-icaRKO1 | GGGGACAAGTTTGTACAAAAAAGCAGGCTGAGCCCATCTCACGCGTTGC |
| attB2-icaRKO4 | GGGGACCACTTTGTACAAGAAAGCTGGGTCACTTACTACAAGACATATTGCCG |
| comp-icaKO2 | TATACATAATCCGCGGCCGCGAGTGCAAGAACATTAGACAACG |
| comp-icaKO3 | GTTCTTGCACTCGCGGCCGCGGATTATGTATAGGTGTCGGC |
| comp-icaRKO2 | TTCCACTGCTCCGCGGCCGCCATCAAGTGTTGTACCGTC |
| comp-icaRKO3 | CAACACTTGATGGCGGCCGCGGAGCAGTGGAAGAAAGTA |
| icaR-1 | GGATCCGAACCGACAATCCAGTAAATAGAC |
| icaR-3 | GAATTCGTAAGTTAATTATTACAAACTAGTAAC |
The icaR gene was amplified by PCR using primers icaR-1 and icaR-3 (Table 2) and cloned into pLI50 at BamHI and EcoRI sites to generate expression plasmid pML3796. The construct was verified by DNA sequencing. Plasmids pLI50, pYL8565, and pML3796 were transduced into the S. aureus strains listed in Table 1 by phage 52A.
Biofilm and PNAG assays.
Flow cell biofilm assays were performed in flow cells purchased from Stovall Life Science, Greensboro, NC, as previously described (29). UAMS-1 derivatives were cultured in TSB containing 3.5% NaCl, 0.75% glucose, and 10 μg per ml Cm. MW2 derivatives were cultured similarly except that the NaCl concentration was reduced to 1.5% and Cm was used at 5 μg/ml. Bacterial cells were harvested for RNA isolation at the peak biofilm formation. Microtiter biofilm assays were performed in 96-well microtiter plates as described by Lim et al. (26) with minor modifications (29). Assays for PNAG were performed as previously described (10, 55).
RNA methods.
RNA was isolated as described by Luong et al. (29). For Northern analysis of the rbf transcript in UAMS-1, RNA was isolated from flow cells and separated on a 1% agarose-formaldehyde gel and transferred to a nylon membrane (Roche Diagnostics, Mannheim, Germany) as described previously (7). For hybridization, a 1-kb fragment of rbf, amplified using rbf62 and adh14 primers (Table 2), was gel purified and labeled with [32P]dATP by using a High Prime DNA labeling kit from Roche Diagnostics. The membrane was hybridized with the rbf-specific probe at 42°C for 22 h in Ultrahyb hybridization buffer (Ambion Inc., Austin, TX). RNA size standards were from Roche. Microarray profiling was performed essentially as described previously (2). Briefly, two RNA samples from each strain were prepared from two separate flow cells and were independently labeled following the manufacturer's recommendations for prokaryotic antisense arrays (Affymetrix, Santa Clara, CA). A 1.5-μg aliquot of a labeled sample was hybridized to a commercially available Affymetrix S. aureus GeneChip. Scanned intensity values for each detected RNA species were normalized to the median GeneChip signal value, and biological replicates were averaged using GeneSpring GX 7.3.1 analysis platform software (Agilent Technologies, Redwood City, CA). Genes that were found to be differentially expressed exhibited a ≥2-fold change in expression under the indicated conditions were determined to be above background signal intensity values and considered “present” by Affymetrix algorithms under the induced condition and were considered statistically differentially expressed as determined by Student's t test (P ≤ 0.05). Quantitative real-time reverse transcription-PCR (RT-PCR) was performed as described previously (27).
For isolation of RNA from S. aureus strain 8325-4, cultures were grown overnight in TSB containing 3.5% NaCl and 0.75% glucose. Cm was added to cultures of plasmid-bearing strains. The cultures were adjusted to an optical density at 660 nm of 3 and then diluted 1:250 into fresh TSB-glucose-NaCl and cultured for 6 h, with aeration, at 37°C.
RESULTS
Characterization of rbf transcription in wild-type, rbf, and rbf overexpression strains.
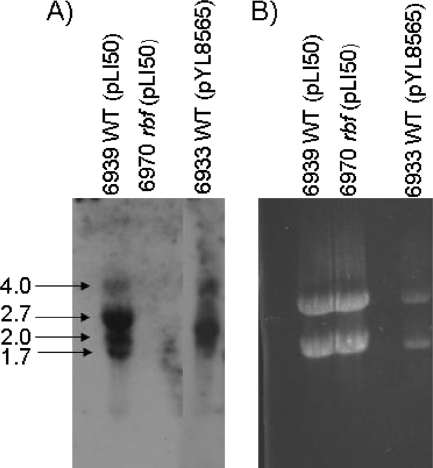
To verify expression of rbf in UAMS-1, RNA was isolated from the wild-type strain UAMS-1 (strain 6939), an isogenic rbf mutant (strain 6970), and an Rbf overexpression strain (strain 6933). The RNA was subjected to Northern blot analysis using an rbf-specific probe. As shown in Fig. 1, the probe hybridized to multiple transcripts in wild-type RNA ranging in size from 1.7 to 4.0 kb. Since rbf is 2.2 kb in length, the results suggest that there may be multiple transcriptional start sites and/or processing/degradation of the primary transcript. No transcript was detected in RNA from the rbf mutant. As anticipated, a much higher level of hybridizing RNA was detected in the rbf overexpression strain. These results were confirmed using real time RT-PCR to measure rbf transcription (not shown). These results suggested that our strains and experimental conditions were appropriate to characterize the cellular role of rbf.
FIG. 1.
Northern analysis of rbf transcription in UAMS-1. (A) Northern analysis with an rbf-specific probe. RNA was isolated from wild-type UAMS-1 (strain 6939), an rbf mutant (strain 6970), and an rbf overexpression strain (6933). Numbers to the left of the figure are the sizes of the indicated RNA bands (in kb). (B) Ethidium bromide-stained gel of RNAs, showing the relative level of rRNA in each preparation.
Identification of rbf-regulated genes by microarray.
In order to identify what genes are regulated by rbf, microarray experiments were performed with RNA isolated from wild-type UAMS-1 (strain 6939) and the rbf mutant (strain 6970). In addition, RNA was isolated from an Rbf overexpression strain (strain 6933), as we had previously observed that overexpression of rbf enhanced biofilm formation. In these experiments, bacteria were cultured in commercially acquired flow cells. The microarray experiments were performed with RNA from two independent cultures of each strain. Comparing the wild type and the rbf mutant, we identified 16 genes upregulated by rbf by at least twofold. In the Rbf overexpression strain, 6 genes were upregulated while 35 genes were downregulated compared to the wild-type strain (Tables 4 and 5). Notably, overexpression of rbf in strain 6933 resulted in increased expression of the icaADBC operon, the genes required for PNAG synthesis, by five- to sixfold. In addition, two genes known to repress icaADBC expression, icaR and tcaR, were repressed in the Rbf-overproducing strain. These findings could account, at least in part, for the role of rbf in biofilm formation.
TABLE 4.
Genes upregulated by rbfa
| ORF | Fold change in:
|
Gene | Description | |
|---|---|---|---|---|
| rbfΔ vs wt | rbf++ vs wt | |||
| SACOL2641 | 3.7 | gpxA | Glutathione peroxidase | |
| SACOL2689 | 6.1 | icaA | Intercellular adhesion protein A | |
| SACOL2691 | 4.5 | icaB | Intercellular adhesion protein B | |
| SACOL2690 | 5.9 | icaD | Intercellular adhesion protein D | |
| SACOL0725 | 5.4 | 14.0 | rbf | Transcriptional regulator, AraC family |
| SACOL0726 | 11.5 | sarX | Transcription factor | |
| 7.1 | Intergenic region upstream of rbf | |||
| SACOL2198 | 2.1 | aldC | α-Acetolactate decarboxylase | |
| SACOL1351 | 2.2 | cls1 | Cardiolipin synthetase | |
| SACOL0357 | 2.2 | dut | Prophage L54a, deoxyuridine 5-triphosphate nucleotidohydrolase | |
| SACOL0600 | 2.1 | ilvE | Branched-chain amino acid aminotransferase | |
| SACOL2070 | 2.1 | kdpD | Sensor histidine kinase | |
| SACOL2397 | 2.3 | nirD | Small subunit nitrite reductase | |
| SACOL0696 | 2.1 | tagB | Teichoic acid biosynthesis protein B | |
| SACOL0569 | 2.1 | ATP:guanido phosphotransferase family protein | ||
| SACOL1591 | 2.0 | Lipoate-protein ligase A family protein | ||
| SACOL2138 | 2.4 | Cation efflux family protein | ||
| SACOL2357 | 2.0 | ABC transporter, permease protein | ||
| SACOL2386 | 2.0 | narT/nirK | Nitrite extrusion protein | |
| SACOL2396 | 2.0 | Uroporphyrinogen III methylase, SirB, putative | ||
| N315-A1617 | 2.1 | Fragment, conserved hypothetical protein | ||
| SACOL0568 | 2.1 | Conserved hypothetical protein | ||
| SACOL1790 | 2.1 | Conserved hypothetical protein | ||
rbfΔ/wt data indicate the increased expression level in the wild type compared to the rbf deletion mutant. rbf++/wt data indicate the increased expression level in the Rbf-overproducing strain compared to the wild type.
TABLE 5.
Genes downregulated by rbfa
| ORF | Fold change in rbf++ vs wt | Gene | Description |
|---|---|---|---|
| SACOL2627 | −2.3 | betA | Choline dehydrogenase |
| SACOL2628 | −2.5 | betB | Betaine aldehyde dehydrogenase |
| SACOL0302 | −2.1 | brnQ | Branched-chain amino acid transport system II carrier protein |
| MSRA252-SAR2036 | −3.9 | chp | Chemotaxis-inhibiting protein |
| SACOL2652 | −2.7 | clfB | Clumping factor B |
| SACOL1168 | −5.1 | efb/fbp | Fibrinogen binding protein (efb) |
| SACOL2511 | −4.3 | fnbA | Fibronectin binding protein A |
| SACOL2003 | −3.7 | hlb | Phospholipase C |
| SACOL2688 | −2.5 | icaR | Intercellular adhesion regulator |
| SACOL0600 | −2.2 | ilvE | Branched-chain amino acid aminotransferase |
| SACOL0247 | −3.3 | lrgA | Holin-like protein LrgA |
| SACOL0248 | −4.6 | lrgB | LrgB protein |
| SACOL0245 | −2.1 | lytS | Sensor histidine kinase |
| SACOL2002 | −3.3 | map | Map protein, authentic frameshift |
| N315-SA0387 | −4.0 | set11 | Exotoxin 11 |
| SACOL2353 | −2.3 | tcaR | Transcriptional regulator TcaR |
| SACOL1364 | −2.2 | thrB | Homoserine kinase |
| SACOL2004 | −4.0 | Leukocidin F subunit, putative | |
| SACOL2418 | −2.1 | Immunoglobulin G binding protein SBI | |
| SACOL0468 | −2.6 | Exotoxin 3, putative | |
| SACOL0857 | −2.0 | Staphylocoagulase precursor, putative | |
| SACOL1164 | −2.8 | Fibrinogen-binding-related protein | |
| SACOL1751 | −3.5 | Truncated cell surface protein map | |
| SACOL1892 | −2.3 | Membrane protein, putative | |
| SACOL1904 | −2.3 | Transcriptional regulator, putative | |
| SACOL2499 | −2.1 | Helicase, putative | |
| MU50-SAV1941 | −4.6 | Putative membrane protein | |
| SACOL0199 | −3.6 | Conserved hypothetical protein | |
| SACOL0480 | −2.4 | Hypothetical protein | |
| SACOL0767 | −3.0 | Conserved hypothetical protein | |
| SACOL0768 | −2.7 | Conserved hypothetical protein | |
| SACOL1777 | −4.2 | Conserved hypothetical protein | |
| SACOL2492 | −2.2 | Hypothetical protein | |
| SACOLR1913 | −2.1 | Hypothetical protein | |
| N315-SAS058 | −2.1 | Conserved hypothetical protein |
The rbf++ versus wt data indicate the decreased expression level in the rbf-overproducing strain compared to the wild type.
Overexpression of rbf also affected the transcription of at least four other genes likely to impact on biofilm formation. These included tagB, encoding teichoic acid biosynthesis protein B, which is upregulated by rbf. It has been argued that teichoic acid synthesis is important for biofilm formation (17, 36). Three genes involved in the regulation of cell lysis, lytS and lrgAB, are repressed in the overproducing strain. It has been shown that cell lysis is the source of extracellular genomic DNA, which is an important constituent of S. aureus biofilms (43, 44, 50).
Overexpression of rbf has the general effect of decreasing transcription of secreted and surface-associated proteins, including many of which are known or suspected virulence factors (Table 5). These include chp (chemotaxis-inhibiting protein), clfB (clumping factor B), fbp (fibrinogen binding protein), fnbA (fibronectin binding protein), hlb (β-hemolysin), map (major histocompatibility complex class II analogue protein), set11 (exotoxin 11), SACOL2004 (leukocidin F), SACOL2418 (immunoglobulin G binding protein), and SACOL0468 and SACOL0857, which are a phage-encoded exotoxin and coagulase, respectively. Transcription of sarX, which encodes a transcriptional regulator located immediately downstream of the rbf gene, was upregulated over 10-fold in the Rbf overexpression strain (strain 6933). Other potential regulatory factors affected by rbf are lytS and kdpD, both encoding sensor histidine kinases, and SACOL1904 (a putative transcription regulator).
Confirmation of microarray results using real-time PCR.
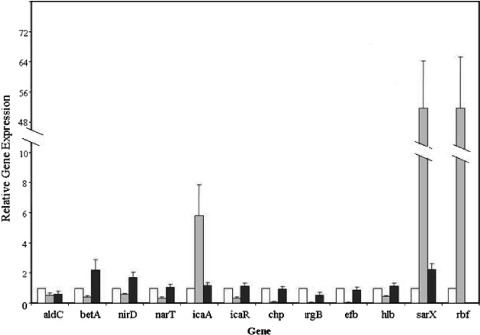
In order to verify our microarray results we used real-time RT-PCR to compare expression levels of 12 select UAMS-1 genes (Fig. 2). Bacteria for these experiments were cultured in the same manner as cultures used in the microarray studies. RNA from two independent cultures of each strain was analyzed. Relative expression levels were determined using gyrB expression for normalization of data. With some exceptions, the real-time PCR results correlated well with the microarray results. Two exceptions were sarX and rbf when comparing expression levels between the wild type (strain 6939) and the Rbf overexpression strain (6933) (Table 4). For the Rbf overexpression strain, expression levels of sarX and rbf were 11.5- and 14-fold higher, respectively, in the microarray experiments (Table 4), whereas both genes were expressed approximately 52-fold higher in the real-time PCR experiments (Fig. 2). The underestimation of transcript levels in microarray studies has been reported previously by our laboratory and others (3, 27) and may simply reflect the increased sensitivity of real-time PCR over microarrays. The other exception was nirD, encoding the small subunit of nitrite reductase, for which we had anticipated twofold-higher expression in the rbf mutant, strain 6970, compared to the wild-type strain, 6939. This result was intriguing, as nitrite has been shown to inhibit biofilm formation by S. aureus (46). The real-time PCR results, however, showed an approximate 64% reduction in nirD RNA in strain 6933.
FIG. 2.
Confirmation of microarray results using real-time RT-PCR. RNA was isolated from the wild-type strain, 6939 (white bars), an Rbf overexpression strain, 6933 (gray bars), and an rbf mutant strain, 6970 (black bars) in flow cell cultures and subjected to real-time RT-PCR using primers specific for the indicated genes (Table 3). Expression levels are expressed relative to that of strain 6939, which was arbitrarily assigned a value of 1. The expression of gyrB was used to correct for differences in RNA quantities added to each reaction mixture. Data represent the means ± standard errors from two independent experiments.
Evidence that rbf may not be expressed in UAMS-1.
The results presented above demonstrate that high-level expression of rbf has a profound effect on gene expression by UAMS-1. Moreover, rbf overexpression had a dramatic effect on biofilm formation and PNAG synthesis (Fig. 3). In contrast, the rbf mutation had only small effects on gene expression, suggesting that rbf may be poorly expressed in UAMS-1. Sequencing of the UAMS-1 rbf gene revealed the presence of a 2-bp insertion (relative to several sequenced S. aureus strains, including NCTC 8325) affecting the 50th codon of the predicted rbf open reading frame (ORF). Thus, UAMS-1 appears not to carry an intact rbf gene and therefore may not produce a functional protein. It is important to note here that the rbf gene carried on pYL8565 was cloned from S. aureus 8325-4 (26).
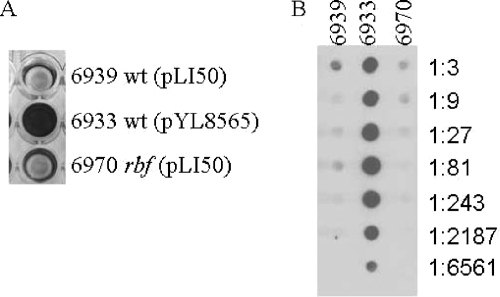
FIG. 3.
Rbf enhances PNAG synthesis and biofilm formation in UAMS-1. (A) Biofilm formation by wild-type UAMS-1 (6939), an rbf mutant (6970), and a UAMS-1 derivative that overexpresses rbf (6933). The 96-well plates were inoculated with each of the indicated strains. Following a 24-h incubation, wells were washed and biofilms were stained with crystal violet. (B) PNAG production. PNAG was extracted from each UAMS-1 derivative, serially diluted, and applied to a membrane. PNAG was detected by incubating the membrane, successively, with rabbit anti-PNAG serum, goat anti-rabbit-horseradish peroxidase, and a chemiluminescent substrate. Numbers to the right of the figure indicate dilutions.
To test whether the Rbf protein is produced by UAMS-1, we used anti-Rbf antibody in Western analyses (data not shown). No detectable Rbf protein was found in UAMS-1, whereas an ∼80-kDa band, matching the predicted size of Rbf, was readily detected in 8325-4. This result indicates that Rbf is not produced in UAMS-1, apparently due to the 2-bp insertion in the rbf ORF.
rbf regulation of ica operon expression in S. aureus 8325-4.
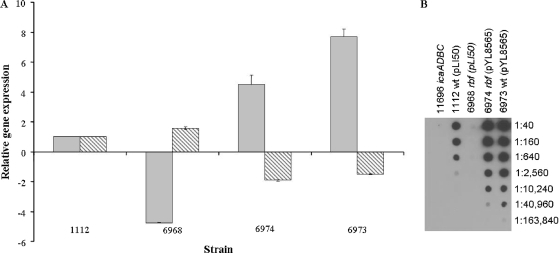
The microarray and the real-time RT-PCR experiments using UAMS-1 and its derivative strains suggest that rbf may regulate biofilm formation by affecting ica gene expression. Because we found that the rbf gene was functional in 8325-4 but not in UAMS-1, we chose strain 8325-4 for further studies. However, our previous finding using a Pica::xylE reporter construct showed that an rbf mutation in S. aureus strain 8325-4 was not associated with altered ica expression (26). This inconsistency may have been due to the relative insensitivity of the xylE reporter used in the former study. To investigate this apparent discrepancy we reassessed the role of the strain 8325-4 rbf gene in the ica operon and PNAG regulation. For these experiments, a stable, internal deletion of rbf was constructed in strain 8325-4 by allele replacement. As shown in Fig. 4A, real-time RT-PCR experiments revealed a significant decrease in icaA transcript levels in the rbf deletion strain (strain 6968) and expression of rbf from plasmid pYL8565 effectively complemented biofilm formation in the rbf mutant (strain 6974).
FIG. 4.
Effects of Rbf on icaADBC and icaR expression in strain 8325-4. (A) Comparative measurements of icaA (gray bars) and icaR (hatched bars) transcription by real-time RT-PCR in the following S. aureus strains: 1112, wild type (pLI50); 6968, rbf(pLI50); 6974, rbf(pYL8565); 6973, wild type (pYL8565). Total RNA was prepared from cultures grown for 6 h at 37°C in TSB containing 3.5% NaCl, 0.75% glucose. Real-time RT-PCR was used to measure the relative expression of icaA and icaR compared to the gyrB gene. Transcript levels in all strains were compared to transcript levels in the wild-type strain, 1112, which was assigned a value of 1. The data presented are the averages of two separate experiments and standard errors are indicated. Plasmid pYL8565 carries the wild-type rbf gene, and pLI50 is the plasmid vector. (B) Regulation of PNAG synthesis by rbf in 8325-4 and derivatives. Bacterial extracts were prepared from overnight cultures grown at 37°C in TSB containing 3.5% NaCl, 0.75% glucose. PNAG was extracted from cells, serially diluted, and applied to a membrane. PNAG was detected by incubating the membrane, successively, with rabbit anti-PNAG serum, goat anti-rabbit-horseradish peroxidase, and a chemiluminescent substrate. Dilutions are indicated to the right of the figure. Strain numbers and genotypes are listed at the top of the figure.
As was observed for UAMS-1, overexpression of wild-type rbf resulted in repression of icaR transcription (Fig. 4A). Carriage of pYL8565 reduced icaR expression by approximately twofold in both the rbf mutant (strain 6974) and the wild-type strain (strain 6973). These results support the argument that Rbf may promote transcription of icaADBC by repression of icaR.
To further examine the impact of rbf on biofilm regulation in 8325-4, we measured PNAG production by the wild-type strain and its derivatives (Fig. 4B). Consistent with the activation of ica operon expression, mutation of rbf was associated with decreased production of PNAG and overexpression of rbf was associated with increased production (Fig. 4B). Taken together, these data strongly indicate that, similar to our observations in UAMS-1, rbf controls biofilm formation in strain 8325-4, at least in part, by controlling expression of the ica genes.
Regulation of biofilm formation and icaADBC expression by icaR and rbf in S. aureus strain 8325-4.
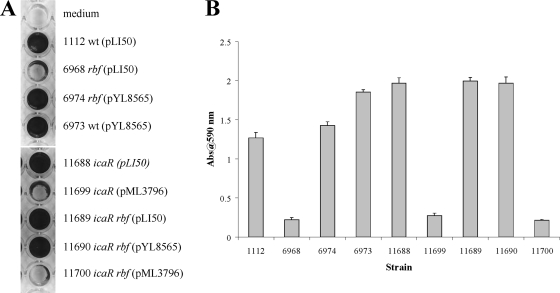
Our results thus far suggest that rbf may enhance biofilm formation, at least in part, by promoting transcription of icaADBC. The data further suggest that rbf activation of the ica operon could be indirect, being accomplished by rbf repression of icaR and/or tcaR, both of which are repressors of icaADBC (21). It should be noted here that strain 8325-4 contains a mutation in the tcaR gene (32); therefore, TcaR may not have an effect on biofilm in the 8325-4 background. To characterize rbf regulation of the ica locus and its potential interaction with icaR further, we compared the effects of single and double mutants of rbf and icaR in strain 8325-4. As shown in Fig. 5, inactivation of icaR (strain 11688), in contrast to the rbf mutant, promoted biofilm formation which could be complemented with an icaR-bearing plasmid, pML3796 (strain 11699). In the icaR rbf double mutant, biofilm formation was similar to that of the icaR mutant. These results indicate that the effect of icaR is epistatic to that of rbf, arguing that icaR acts downstream of rbf in the pathway for biofilm regulation. To further test this possibility, we complemented the icaR rbf double mutant with either the rbf-bearing plasmid pYL8565 (strain 11690) or the icaR-bearing plasmid pML3796 (strain 11700). We found that complementation of the double mutant with the icaR-bearing plasmid repressed biofilm formation to less than the wild-type level (Fig. 5). In contrast, the presence of the rbf-bearing plasmid (strain 11690) did not affect biofilm formation compared to the double mutant strain. These results support the proposal that icaR functions downstream of rbf.
FIG. 5.
Contributions of rbf and icaR to biofilm formation by 8325-4. (A) Biofilm formation under static incubation conditions. Microtiter plate wells were inoculated with each of the indicated 8325-4 derivatives. Following a 24-h incubation, wells were washed and biofilms were stained with crystal violet. Each assay was performed a minimum of two times. (B) Quantitation of biofilms. Crystal violet was extracted from wells by using ethanol-acetic acid and diluted 10-fold, and the absorbance of each extract was measured. Error bars indicate standard errors.
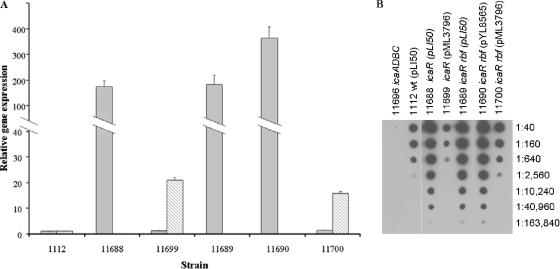
To confirm that increased biofilm formation was associated with increased icaADBC transcription, we compared icaA and icaR RNA levels in these strains using real-time RT-PCR (Fig. 6A). The results indicated that the icaR mutation in both the wild-type background (strain 11688) and the rbf mutant (strain 11689) increased icaA transcription by nearly 200-fold relative to the wild-type strain, 1112. Carriage of plasmid pYL8565 (strain 6973) increased icaA transcription by approximately eightfold with a concomitant reduction in icaR expression (Fig. 4A). Thus, measurements of icaA and icaR mRNA levels in these strains correlated well with the biofilm data, i.e., increased icaA expression (Fig. 6) correlated with decreased icaR expression and increased biofilm formation (Fig. 5).
FIG. 6.
Regulation of icaA expression and PNAG production by rbf and icaR in 8325-4. (A) Transcription of icaA (gray bars) and icaR (hatched bars). Experiments were performed as described in the legend to Fig. 4A. (B) PNAG production. PNAG was measured as described in the legend to Fig. 4B. Numbers to the right of the figure indicate dilutions. Strain numbers and genotypes are listed at the top of the figure.
Derepression of icaADBC was anticipated to result in increased production of PNAG and, in turn, increased biofilm formation. To confirm that PNAG levels correlated with biofilm formation, immunoassays for PNAG were performed (Fig. 6B). The icaR mutation increased PNAG production by roughly 64-fold, consistent with robust biofilm formation and increased icaA expression. Transformation of icaR mutants with pML3796 (strains 11699 and 11700) decreased PNAG synthesis to the wild-type level. Thus, for most strains there is a strong correlation between rbf expression, icaA transcription, PNAG production, and biofilm formation. The exceptions were the icaR single and icaR rbf double mutant strains carrying the icaR-encoding plasmid pML3796 (strains 11699 and 11700). These strains produced wild-type levels of icaA transcript and PNAG, yet neither strain was able to form a stable biofilm. These results suggest that icaR may regulate genes other than icaADBC, at least when the repressor is overexpressed.
rbf regulation of biofilm formation in S. aureus MW2.
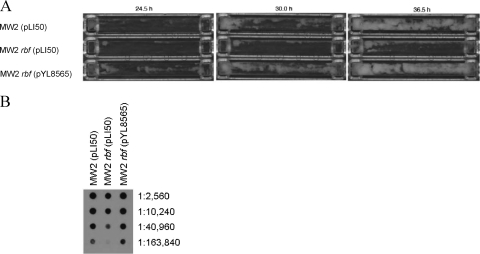
The MW2 strain was isolated from a child with fatal septicemia and septic arthritis (5). To determine whether rbf affected biofilm formation in this clinical isolate, we compared the growth of MW2 (pLI50), an rbf mutant [MW2 rbf(pLI50)], and a complemented mutant [MW2 rbf(pLY8565)] in flow cells. We found, based on three separate experiments, that the time required for initiation of visible biofilm formation was 10.50 ± 1.76 h, 13.33 ± 0.33 h, and 10.33 ± 0.17 h (means ± standard errors) for the wild type, rbf mutant, and the complemented mutant, respectively. The results from one of the experiments are shown in Fig. 7A. The average times to peak biofilm formation were 30.50 ± 1.76 h, 39.17 ± 2.68 h, and 31.5 ± 0.29 h for the same three strains. The differences in time to initiation were statistically significant (P = 0.0076 for MW2 compared to MW2 rbf and P = 0.0151 for MW2 rbf compared with the complemented strain). The differences for peak biofilm formation were not statistically significant (P > 0.05). We also compared PNAG production in flow cell cultures. The results, shown in Fig. 7B, revealed that PNAG production was approximately fourfold lower for MW2 rbf than it was for either the wild-type or complemented strains. These results indicate that rbf contributes to the regulation of biofilm formation in MW2. It should be noted here that MW2 forms strong biofilms in microtiter plates precoated with human serum but forms very weak biofilms without precoating. In either case, rbf had no effect on the biofilm.
FIG. 7.
Regulation of biofilm formation by rbf in MW2. (A) Biofilm formation in flow cells after 24.5, 30.0, and 36.5 h of incubation. The results are representative of three separate experiments. Strains grown in each flow cell are indicated to the left of the figure. (B) PNAG production by MW2 and derivatives. Cells were harvested from flow cells after 48 h of incubation. PNAG was extracted and then detected as described in the legend to Fig. 4B. Numbers to the right of the figure indicate dilutions. Strain numbers and genotypes are listed at the top of the figure.
DISCUSSION
Many staphylococcal infections are associated with biofilm formation, most notably, infections associated with indwelling medical devices. Therefore, studies of biofilm constituents and the regulatory mechanisms governing their synthesis is an important area of research that could ultimately lead to therapies for prevention of device-related infections. However, regulation of biofilm formation is complex, being affected by environmental factors such as osmolarity, anaerobiosis, temperature, and levels of glucose, iron, ethanol, citrate, and nitrites (11, 18, 23, 26, 46, 47, 55). Numerous staphylococcal regulatory factors have been implicated in biofilm formation as well, including agr, SarA, SigB, IcaR, TcaR, ArlRS, SrrAB, MgrA, and Rbf (2, 26, 42, 48, 50-52). In this study we attempted to define the role of one regulatory factor, Rbf, in biofilm formation by S. aureus.
Our microarray studies demonstrated a profound effect of overexpression of rbf on gene expression in the clinical isolate UAMS-1, an observation supported by our real-time PCR experiments. Overall, we determined that 6 genes were upregulated and 35 genes were downregulated in an Rbf overexpression strain in comparison to the wild-type parent strain. Several of these genes, including icaR, icaADBC, lytS, and lrgAB, have the potential to impact biofilm formation. These results indicate that Rbf could potentially regulate biofilm formation at multiple levels. It was anticipated that genes responsive to overexpression of Rbf would also respond to genetic inactivation of rbf. This was generally not the case, however, as comparatively few genes were affected in our UAMS-1 rbf deletion mutant and the measured effects were all quite small (typically around twofold). The reason for this is that UAMS-1 is unlikely to produce an active Rbf protein. While we found that rbf was actively transcribed, sequencing of the UAMS-1 rbf gene revealed a 2-bp insertion within the predicted Rbf coding region. The mutation, which affects codon 50 of Rbf, would result in the production of a truncated protein. Additionally, we were unable to detect Rbf in extracts of UAMS-1 by using Rbf antiserum, whereas we were able to detect Rbf in extracts of strain 8325-4. We concluded that UAMS-1 has acquired a mutation that inactivates rbf; therefore, subsequent experiments were performed with S. aureus 8325-4. We gathered evidence that rbf is functionally expressed in another clinical isolate, MW2. Mutation of rbf in this strain reduced both biofilm formation and PNAG production. In a previous study (26) we found that rbf was present in 22 of 27 clinical isolates.
Rbf is a member of the AraC/XylS family of transcriptional regulators (26). Although the members of this family are diverse, they all possess a 99-amino-acid conserved region containing a dual helix-turn-helix DNA binding motif (15). The functions of the nonconserved domains of AraC/XylS family members are unknown in most cases, but it is presumed that the nonconserved regions may bind small molecules that could enhance or inhibit transcriptional activation by the protein (15). It is unknown at present whether Rbf binds an inducer or cofactor.
It is also unknown whether Rbf can function as a transcriptional activator, but nearly all other AraC/XylS family members act in this way (15). In this study, we found that Rbf contributed to biofilm formation by activating icaADBC. Our genetic analyses further showed that the activation was indirect, occurring via repression of icaR. The ica operon encodes four genes, icaA, -D, -B, and -C, responsible for the synthesis and possible transport of the intercellular adhesin PNAG (18). Numerous studies have demonstrated that icaADBC is required for biofilm formation by some strains of S. aureus (23, 33, 55); however, recent studies suggested that some S. aureus strains can form biofilms independently of icaADBC (2, 37, 47) and many strains seem to possess both ica-dependent and ica-independent biofilm pathways (36).
Several factors that regulate icaADBC expression have been described, including SarA, agr, TcaR, and IcaR (6, 8, 13, 20-22, 49). IcaR regulation is probably the most studied and best understood. IcaR binds specifically to the icaADBC promoter just upstream of the icaA start site to block transcription of the ica genes (13, 20). When rbf is overexpressed in 8325-4, transcription of icaR is decreased by 33 to 48%, with a concomitant rise in icaA transcription of four- to eightfold. The rbf mutation increases icaR transcription, while icaA expression is decreased. The tcaR gene has been reported to encode a weak repressor of icaADBC (21). Our microarray results showed that like icaR, tcaR is repressed by overproduction of Rbf in UAMS-1. Thus, it is possible that activation of icaADBC by rbf could be attributed to rbf repression of tcaR. However, a deletion of the tcaR gene in UAMS-1 had no measurable impact on biofilm formation (unpublished data), and 8325-4 harbors a mutation in tcaR (32). Therefore, it appears that tcaR may not contribute significantly to regulation of icaADBC in the strains studied here.
Rbf does not seem to directly repress icaR transcription. In fact, our in vitro DNA binding assays revealed that recombinant Rbf protein did not bind directly to a DNA fragment encompassing the icaR-icaADBC regulatory region. The same protein did bind to the S. aureus sdrC promoter as well as the Staphylococcus epidermidis sdrF promoter region, suggesting that it is functional (results not shown). One possible alternative mechanism for icaR repression is that Rbf upregulates expression of a gene encoding a repressor of icaR. Our experiments did show that rbf activated expression of at least one regulatory factor, SarX, along with a putative transcription factor, SACOL1904. SarX is a member of the SarA protein family of gene regulators (31). In our microarray experiments sarX transcription was increased 11.5-fold in an Rbf-overproducing strain of UAMS-1. This was confirmed by real-time RT-PCR, wherein we found that rbf enhanced sarX expression by an average of 52-fold in UAMS-1. SarX has been shown to repress expression of agr and agr-dependent genes, so it seems possible that SarX is involved in repression of icaR. The chromosomal context of sarX may also be important, given its location just downstream of rbf. Manna and Cheung (31) identified a minor transcript containing sequences transcribed from both rbf and sarX; thus, it is possible that at least part of sarX activation could be due to readthrough transcription of rbf. SarX is under positive regulation by the global regulatory protein MgrA (31). It is unlikely that Rbf activates sarX via activation of mgrA transcription, as MgrA affects the expression of at least 255 genes (28), whereas rbf affects only 57 genes.
It remains a possibility that Rbf does bind to the icaR regulatory region, but not under the conditions of our in vitro DNA binding assay. DNA binding by some members of the AraC/XylS family, including AraC (15), Rns (35), and MelR (53), involves protein binding to sites up to several hundred base pairs upstream or downstream of a regulated promoter. Thus, it is possible that Rbf binds to DNA sites distal to the icaR promoter or requires binding to multiple sites to form a stable complex with DNA. It is also possible that Rbf requires a cofactor protein for regulation of ica. At least two AraC/XylS family members, MxiE from Shigella flexneri (9) and InvF from Salmonella enterica (12, 41), appear to require interaction with a cofactor protein to act as transcriptional activators of virulence genes.
Rbf represses at least three genes, lytS, lrgA, and lrgB, involved in the regulation of cell lysis. This finding is intriguing, as lysis of S. aureus would release DNA into biofilms (43). LytS is the sensor component of a two-component regulatory system, LytSR, which positively regulates expression of the lrgAB operon. The LrgA protein is a putative antiholin that inhibits cell lysis (19, 43); thus, rbf repression of lytSR would be predicted to decrease expression of LrgA, thereby increasing cell lysis and DNA release. Extracellular DNA has been proposed to be an important structural component of S. aureus biofilms (43, 50).
Although many questions remain, our data suggest a simple working model for how Rbf affects icaADBC transcription. Rbf may promote expression of an unknown factor that binds to the regulatory region of icaR, thus repressing synthesis of the IcaR repressor. Repression of icaR would, in turn, result in derepression of icaADBC and PNAG production. Current studies are directed toward testing this model.
TABLE 3.
Oligonucleotide primers used in real-time RT-PCR
| Primer | Gene | Sequence |
|---|---|---|
| SGaldC1 | aldC | CGTATGATGCCGGCTCAAGAACCACCTTAT |
| SGaldC2 | aldC | CGCAAAGTGTACATGAAATCCTGCTGATCCGA |
| SGbetA1 | betA | TGCCTGCTGCGTTAATGTTCCCTTCA |
| SGbetA2 | betA | ACGACCGCCCATATGTGGTTCTTCAT |
| SGchp1 | chp | CAGGAATCAGTACACACCATCATTCAGCGAAAGC |
| SGchp2 | chp | AATTTCCTAGCGTTGTAGGAAGACCACTATTT |
| SGefb1 | efb | ATGCGAGCGAAGGATACGGTCCAAGAGAAA |
| SGefb2 | efb | TGTGGACGTGCACCATATTCGAATGTACCA |
| SGgyrB3 | gyrB | GGAATCGGTGGCGACTTTGATCTAGCGAAA |
| SGgyrB4 | gyrB | CGCTCCATCCACATCGGCATCAGTCATAAT |
| SGhlb1 | hlb | AAACACCTGTACTCGGCCGTTCTCAATCAG |
| SGhlb2 | hlb | ACTTACAATCGCTACGCCACCATCTTCTGC |
| SGicaA1 | icaA | CTGGCGCAGTCAATACTATTTCGGGTGTCT |
| SGicaA2 | icaA | GACCTCCCAATGTTTCTGGAACCAACATCC |
| icaR for | icaR | TACGTTCAATTATCTAATACGCCTGAGGAATTTTCTGGAA |
| icaR rev | icaR | AGGATGCTTTCAAATACCAACTTTCAAGAAACAGCAAATATT |
| SGlrgB1 | lrgB | CTCAAGCAGCAACTACAGCGATTGCGTTAC |
| SGlrgB2 | lrgB | TTCCAAGTGCTAATCCTCGGGCAATAGGGT |
| SGnirD1 | nirD | AAGGACCATTGTCTGAAGGGACAGTGAGTG |
| SGnirD2 | nirD | ATATACGTTCCCGTCTGTAACTTCTACCTCA |
| SGnirK1 | nirK | TGAAGCGGGATCCGCAAATGGTATCGTATC |
| SGnirK2 | nirK | GTGGGAAGAATCCTCCTAAACCACCCATCA |
| Δrbf for | rbf | ACGCGTTGCCAAGATGGCATAGTCTT |
| Δ rbf rev | rbf | AGCCTAATTCCGCAAACCAATCGCTA |
| SGsarX1 | sarX | TGAATACTGAGAAATTAGAAACATTGCTTGGCTTCTATAAACA |
| SGsarX2 | sarX | TGTCCTACTTAAATCTAGCTCATCCATTGCAGTT |
Acknowledgments
We thank T. Maira-Litran and G. B. Pier for anti-PNAG antisera.
This work was supported by grant AI067857 from the National Institute of Allergy and Infectious Diseases (C.Y.L.) and the Irish Research Council for Science, Engineering and Technology (J.P.O.).
Footnotes
Published ahead of print on 14 August 2009.
REFERENCES
- 1.Bae, T., and O. Schneewind. 2006. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55:58-63. [DOI] [PubMed] [Google Scholar]
- 2.Beenken, K. E., J. S. Blevins, and M. S. Smeltzer. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beenken, K. E., P. M. Dunman, F. McAleese, D. Macapagal, E. Murphy, S. J. Projan, J. S. Blevins, and M. S. Smeltzer. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186:4665-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brady, R. A., J. G. Leid, J. H. Calhoun, J. W. Costerton, and M. E. Shirtliff. 2008. Osteomyelitis and the role of biofilms in chronic infection. FEMS Immunol. Med. Microbiol. 52:13-22. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus: Minnesota and North Dakota, 1997-1999. MMWR Morb. Mortal. Wkly. Rep. 48:707-710. [PubMed] [Google Scholar]
- 6.Cerca, N., J. L. Brooks, and K. K. Jefferson. 2008. Regulation of the intercellular adhesin locus regulator (icaR) by SarA, σB, and IcaR in Staphylococcus aureus. J. Bacteriol. 190:6530-6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Z., T. T. Luong, and C. Y. Lee. 2007. The sbcDC locus mediates repression of type 5 capsule production as part of the SOS response in Staphylococcus aureus. J. Bacteriol. 189:7343-7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. Regulation of icaR gene expression in Staphylococcus epidermidis. FEMS Microbiol. Lett. 216:171-177. [DOI] [PubMed] [Google Scholar]
- 9.Corrigan, R. M., D. Rigby, P. Handley, and T. J. Foster. 2007. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 153:2435-2446. [DOI] [PubMed] [Google Scholar]
- 10.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Gotz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cramton, S. E., M. Ulrich, F. Gotz, and G. Doring. 2001. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69:4079-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darwin, K. H., and V. L. Miller. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 20:1850-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzpatrick, F., H. Humphreys, and J. P. O'Gara. 2005. The genetics of staphylococcal biofilm formation—will a greater understanding of pathogenesis lead to better management of device-related infection? Clin. Microbiol. Infect. 11:967-973. [DOI] [PubMed] [Google Scholar]
- 14.Fournier, B., and D. C. Hooper. 2000. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillaspy, A. F., S. G. Hickmon, R. A. Skinner, J. R. Thomas, C. L. Nelson, and M. S. Smeltzer. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63:3373-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goerke, C., S. Campana, M. G. Bayer, G. Doring, K. Botzenhart, and C. Wolz. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68:1304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gotz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 19.Groicher, K. H., B. A. Firek, D. F. Fujimoto, and K. W. Bayles. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182:1794-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jefferson, K. K., S. E. Cramton, F. Gotz, and G. B. Pier. 2003. Identification of a 5-nucleotide sequence that controls expression of the ica locus in Staphylococcus aureus and characterization of the DNA-binding properties of IcaR. Mol. Microbiol. 48:889-899. [DOI] [PubMed] [Google Scholar]
- 21.Jefferson, K. K., D. B. Pier, D. A. Goldmann, and G. B. Pier. 2004. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 186:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeng, W. Y., T. P. Ko, C. I. Liu, R. T. Guo, C. L. Liu, H. L. Shr, and A. H. Wang. 2008. Crystal structure of IcaR, a repressor of the TetR family implicated in biofilm formation in Staphylococcus epidermidis. Nucleic Acids Res. 36:1567-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, M., A. Cockayne, and J. A. Morrissey. 2008. Iron-regulated biofilm formation in Staphylococcus aureus Newman requires ica and the secreted protein Emp. Infect. Immun. 76:1756-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kristian, S. A., T. A. Birkenstock, U. Sauder, D. Mack, F. Gotz, and R. Landmann. 2008. Biofilm formation induces C3a release and protects Staphylococcus epidermidis from IgG and complement deposition and from neutrophil-dependent killing. J. Infect. Dis. 197:1028-1035. [DOI] [PubMed] [Google Scholar]
- 25.Lee, C. Y., S. L. Buranen, and Z. H. Ye. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101-105. [DOI] [PubMed] [Google Scholar]
- 26.Lim, Y., M. Jana, T. T. Luong, and C. Y. Lee. 2004. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J. Bacteriol. 186:722-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luong, T. T., P. M. Dunman, E. Murphy, S. J. Projan, and C. Y. Lee. 2006. Transcription profiling of the mgrA regulon in Staphylococcus aureus. J. Bacteriol. 188:1899-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luong, T. T., and C. Y. Lee. 2006. The arl locus positively regulates Staphylococcus aureus type 5 capsule via an mgrA-dependent pathway. Microbiology 152:3123-3131. [DOI] [PubMed] [Google Scholar]
- 29.Luong, T. T., M. G. Lei, and C. Y. Lee. 2009. Staphylococcus aureus Rbf activates biofilm formation in vitro and promotes virulence in a murine foreign-body infection model. Infect. Immun. 77:335-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maira-Litran, T., A. Kropec, C. Abeygunawardana, J. Joyce, G. Mark, 3rd, D. A. Goldmann, and G. B. Pier. 2002. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 70:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manna, A. C., and A. L. Cheung. 2006. Expression of SarX, a negative regulator of agr and exoprotein synthesis, is activated by MgrA in Staphylococcus aureus. J. Bacteriol. 188:4288-4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCallum, N., M. Bischoff, H. Maki, A. Wada, and B. Berger-Bachi. 2004. TcaR, a putative MarR-like regulator of sarS expression. J. Bacteriol. 186:2966-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenney, D., J. Hubner, E. Muller, Y. Wang, D. A. Goldmann, and G. B. Pier. 1998. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect. Immun. 66:4711-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moscoso, M., E. Garcia, and R. Lopez. 2006. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J. Bacteriol. 188:7785-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munson, G. P., and J. R. Scott. 2000. Rns, a virulence regulator within the AraC family, requires binding sites upstream and downstream of its own promoter to function as an activator. Mol. Microbiol. 36:1391-1402. [DOI] [PubMed] [Google Scholar]
- 36.O'Gara, J. P. 2007. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270:179-188. [DOI] [PubMed] [Google Scholar]
- 37.O'Neill, E., C. Pozzi, P. Houston, H. Humphreys, D. A. Robinson, A. Loughman, T. J. Foster, and J. P. O'Gara. 2008. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 190:3835-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Neill, E., C. Pozzi, P. Houston, D. Smyth, H. Humphreys, D. A. Robinson, and J. P. O'Gara. 2007. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J. Clin. Microbiol. 45:1379-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Otto, M. 2008. Staphylococcal biofilms. Curr. Top. Microbiol. Immunol. 322:207-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petersen, F. C., L. Tao, and A. A. Scheie. 2005. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J. Bacteriol. 187:4392-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pilonieta, M. C., and G. P. Munson. 2008. The chaperone IpgC copurifies with the virulence regulator MxiE. J. Bacteriol. 190:2249-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rachid, S., K. Ohlsen, U. Wallner, J. Hacker, M. Hecker, and W. Ziebuhr. 2000. Alternative transcription factor σB is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 182:6824-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rice, K. C., E. E. Mann, J. L. Endres, E. C. Weiss, J. E. Cassat, M. S. Smeltzer, and K. W. Bayles. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 104:8113-8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rice, K. C., T. Patton, S. J. Yang, A. Dumoulin, M. Bischoff, and K. W. Bayles. 2004. Transcription of the Staphylococcus aureus cid and lrg murein hydrolase regulators is affected by sigma factor B. J. Bacteriol. 186:3029-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 46.Schlag, S., C. Nerz, T. A. Birkenstock, F. Altenberend, and F. Gotz. 2007. Inhibition of staphylococcal biofilm formation by nitrite. J. Bacteriol. 189:7911-7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shanks, R. M., M. A. Meehl, K. M. Brothers, R. M. Martinez, N. P. Donegan, M. L. Graber, A. L. Cheung, and G. A. O'Toole. 2008. Genetic evidence for an alternative citrate-dependent biofilm formation pathway in Staphylococcus aureus that is dependent on fibronectin binding proteins and the GraRS two-component regulatory system. Infect. Immun. 76:2469-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toledo-Arana, A., N. Merino, M. Vergara-Irigaray, M. Debarbouille, J. R. Penades, and I. Lasa. 2005. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J. Bacteriol. 187:5318-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trotonda, M. P., A. C. Manna, A. L. Cheung, I. Lasa, and J. R. Penades. 2005. SarA positively controls bap-dependent biofilm formation in Staphylococcus aureus. J. Bacteriol. 187:5790-5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trotonda, M. P., S. Tamber, G. Memmi, and A. L. Cheung. 2008. MgrA represses biofilm formation in Staphylococcus aureus. Infect. Immun. 76:5645-5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ulrich, M., M. Bastian, S. E. Cramton, K. Ziegler, A. A. Pragman, A. Bragonzi, G. Memmi, C. Wolz, P. M. Schlievert, A. Cheung, and G. Doring. 2007. The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol. Microbiol. 65:1276-1287. [DOI] [PubMed] [Google Scholar]
- 52.Valle, J., A. Toledo-Arana, C. Berasain, J. M. Ghigo, B. Amorena, J. R. Penades, and I. Lasa. 2003. SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48:1075-1087. [DOI] [PubMed] [Google Scholar]
- 53.Wade, J. T., T. A. Belyaeva, E. I. Hyde, and S. J. Busby. 2000. Repression of the Escherichia coli melR promoter by MelR: evidence that efficient repression requires the formation of a repression loop. Mol. Microbiol. 36:223-229. [DOI] [PubMed] [Google Scholar]
- 54.Whitchurch, C. B., T. Tolker-Nielsen, P. C. Ragas, and J. S. Mattick. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. [DOI] [PubMed] [Google Scholar]
- 55.Zhu, Y., E. C. Weiss, M. Otto, P. D. Fey, M. S. Smeltzer, and G. A. Somerville. 2007. Staphylococcus aureus biofilm metabolism and the influence of arginine on polysaccharide intercellular adhesin synthesis, biofilm formation, and pathogenesis. Infect. Immun. 75:4219-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]