Abstract
Bacterial cytoskeletal proteins participate in a variety of processes, including cell division and DNA segregation. Polymerization of one plasmid-encoded, actin-like protein, ParM, segregates DNA by pushing two plasmids in opposite directions and forms the current paradigm for understanding active plasmid segregation. An essential feature of ParM assembly is its dynamically instability, the stochastic switching between growth and disassembly. It is unclear whether dynamic instability is an essential feature of all actin-like protein-based segregation mechanisms or whether bacterial filaments can segregate plasmids by different mechanisms. We expressed and purified AlfA, a plasmid-segregating actin-like protein from Bacillus subtilis, and found that it forms filaments with a unique structure and biochemistry; AlfA nucleates rapidly, polymerizes in the presence of ATP or GTP, and forms highly twisted, ribbon-like, helical filaments with a left-handed pitch and protomer nucleotide binding pockets rotated away from the filament axis. Intriguingly, AlfA filaments spontaneously associate to form uniformly sized, mixed-polarity bundles. Most surprisingly, our biochemical characterization revealed that AlfA does not display dynamic instability and is relatively stable in the presence of diphosphate nucleotides. These results (i) show that there is remarkable structural diversity among bacterial actin filaments and (ii) indicate that AlfA filaments partition DNA by a novel mechanism.
Bacteria contain multiple filament-forming proteins related to eukaryotic actin (6). These actin-like proteins have multiple cellular roles, including determination of cell shape (18), arrangement of organelles (20), and segregation of DNA (36). Little is known about the assembly dynamics of most of these proteins or about the identities and activities of the factors that regulate them. The widely expressed actin-like protein MreB, for example, has been purified and studied in vitro, but its assembly appears to be strongly inhibited by physiological concentrations of monovalent cations, suggesting that its assembly in vivo is facilitated by as-yet-unknown factors (23). At present, the best-understood actin-like protein is ParM, a plasmid-encoded protein that constructs a bipolar spindle capable of pushing plasmids to opposite poles of rod-shaped cells (5, 25). In contrast to the eukaryotic actin cytoskeleton, whose assembly and architecture are regulated by a variety of accessory factors, ParM dynamics are regulated by a single factor, a complex composed of multiple copies of the repressor protein ParR bound to a DNA locus, parC (17). The ParR/parC complex binds the ends of ParM filaments and is pushed through the cytoplasm by filament elongation (5, 14, 25). The ability of ParM to function with such minimal regulation appears to be due to its unique assembly dynamics, which are dramatically different from those of eukaryotic actins. One of the most important differences is that ParM filaments are dynamically unstable (13). That is, similar to eukaryotic microtubules, they can exist in one of two states: stably growing or rapidly (catastrophically) shrinking. This property is required for the ability of ParM to segregate DNA in vivo and appears to solve several fundamental problems associated with DNA segregation. First, spontaneous disassembly of the polymer overcomes the need for an accessory factor to take filaments apart. Second, because filaments bound to ParR/parC complexes are selectively stabilized, the catastrophic disassembly of unattached filaments provides excess monomers that can preferentially elongate them. This is significant because, if the stabilities of attached and unattached filaments were similar, the concentration of free ParM monomers would equilibrate at a level not capable of promoting DNA segregation. And finally, pairs of plasmids appear to find each other via a search-and-capture mechanism (5, 14) that is dramatically enhanced by the continual growth and shortening of filaments attached to single plasmids (16).
Because we have little information on the dynamics of other actin-like proteins, it is unclear to what extent ParM's behavior reflects general properties of bacterial actins rather than specific adaptations to its role in DNA segregation. Furthermore, it is unclear whether all plasmid-segregating actins employ the same dynamic instability-based strategy to find and transport DNA molecules. To better understand the structural and functional diversity of bacterial actins, we studied a second, recently discovered plasmid-segregating actin-like protein, AlfA (1). The AlfA gene is part of an operon (alf) that is located close to the origin of replication of a ∼70-kb, low-copy-number plasmid, pLS32. This plasmid was initially isolated from a natto strain of Bacillus subtilis used in soybean fermentation (33), but a similar plasmid with an identical alf operon is also present in a colony-forming laboratory strain of B. subtilis, strain NCIB 3610 (8, 32). The function of these plasmids is cryptic. They are present at levels of only two or three copies per chromosome equivalent (33), and maintenance of their derivatives requires both AlfA and a downstream gene, alfB (1). Becker and coworkers (1) identified AlfA as a member of the actin superfamily based on the presence of a conserved nucleotide binding fold (4), although the sequence of AlfA is as different from the sequences of ParM and MreB as all three are from the sequence of conventional eukaryotic actin (∼20% identity). These authors also showed that fluorescent derivatives of AlfA form a single filamentous structure running along the long axis of the cell. Photobleached filaments recover from both ends in approximately 1 min, indicating that the structures are composed of multiple, dynamic filaments (1). By analogy with the ParR/parC complex, AlfB might be a DNA binding protein that couples AlfA assembly to plasmid movement. To date, no centromeric sequences involved in segregation have been identified in this plasmid.
We expressed and purified AlfA and characterized its assembly dynamics by using light scattering, high-speed pelleting, and fluorescence microscopy, and we determined the structure of AlfA polymers by high-resolution electron microscopy (EM). We found that in the presence of ATP and GTP, AlfA forms two-strand helical filaments and filament bundles. Like ParM filaments, AlfA filaments are left-handed two-start helices, but otherwise their filament architecture is quite different. AlfA filaments appear to be more tightly twisted and ribbon-like, and AlfA subunits have a significantly different orientation with respect to the filament axis. Unlike other actin-like proteins described thus far, AlfA spontaneously forms regularly sized, mixed-polarity filament bundles driven by electrostatic interactions between filaments, even in the absence of molecular crowding. Finally, AlfA shows no evidence of the dynamic instability crucial to the function of ParM. Thus, AlfA assembles into a unique structure with a unique set of biochemical and structural properties, suggesting a novel mechanism for DNA segregation.
MATERIALS AND METHODS
Expression and purification of AlfA.
We designed a codon-optimized gene encoding the amino acid sequence of AlfA (Gene Designer [DNA 2.0]). This gene was synthesized (Bio Basic) and cloned into pET20b with and without addition of a 3-amino-acid (KCK) tag at the C terminus for use in maleimide labeling reactions. Our cloning strategy also introduced an additional M residue at the N terminus of both constructs used in this study. However, we believe that this residue is irrelevant because AlfA expressed with a single M performed identically in a low-ATP light scattering assay (data not shown). Restriction sites were chosen to exclude the pelB leader peptide and His tag from the expressed open reading frame, which was confirmed by sequencing. C43 cells were transformed, grown at 37°C to an optical density at 600 nm of 0.8, and induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 14 h at 20°C. Cell pellets were frozen at −80°C, thawed, and resuspended in depolymerization buffer (100 mM KCl, 25 mM Tris HCl [pH 7.5], 1 mM dithiothreitol [DTT], 1 mM EDTA) before lysis with a microfluidizer and clarification by high-speed centrifugation. Ammonium sulfate cuts were taken from the high-speed supernatant, and the 30 to 50% pellet was resuspended in polymerization buffer (100 mM KCl, 25 mM Tris HCl [pH 7.5], 1 mM DTT, 1 mM MgCl2). After the resuspended ammonium sulfate pellet was precleared, ATP was added to a final concentration of 5 mM, and the polymer was pelleted by centrifugation at 80,000 rpm for 15 min. This polymerized pellet was then resuspended and dialyzed for 8 h in depolymerization buffer with 5 mM EDTA, after which it was gel filtered with a Superdex 75 column equilibrated in depolymerization buffer. Peak fractions were dialyzed into polymerization buffer containing 20 to 50% glycerol and frozen.
EM.
We polymerized wild-type AlfA by adding 5 mM ATP in polymerization buffer and applied 4 μl of the reaction mixture to glow-discharged 200-mesh carbon-Formvar-coated copper grids at 25°C. After the grids were washed with 3 drops of polymerization buffer, they were negatively stained with 3 drops of 0.75% uranyl formate. Images of samples were obtained at 25°C with a Tecnai T12 microscope using an acceleration voltage of 120 kV and a magnification of ×52,000. Images were recorded with a Gatan 4k × 4k charge-coupled device camera.
Image processing.
Nonoverlapping segments of two-filament bundles were boxed out in 396-Å boxes using the program Boxer, which is part of the EMAN software suite (22). Reference-free class averages were calculated with the startnrclasses program from EMAN.
For single-filament reconstruction, the defocus of each micrograph was determined using CTFFIND (24), and the entire micrograph was corrected by phase flipping. Straight sections of single AlfA filaments were identified using Boxer; subsequent image processing was performed using SPIDER (12). Segments were boxed out along the filament in 265-Å boxes that overlapped by 90% along the helical axis. Each particle was binned twofold to a final pixel size of 4.4 Å. Iterative helical real-space reconstruction was used to refine the helical structure and symmetry parameters using a featureless cylinder as an initial model (9). Forty rounds of reconstruction with a final angular increment of 3° were carried out for each reconstruction.
Reconstructed volumes were viewed and manual placement of crystal structures was performed with Chimera (29).
Labeling of AlfA with fluorescent dyes and biotin.
Glycerol and DTT were removed by using PD10 or Nap5 salt exclusion columns equilibrated in polymerization buffer without DTT. We incubated AlfA-KCK with a maleimide label using a molar ratio of 1:1 to 1:2 for 15 min at 25°C. The reaction was quenched with 10 mM DTT, and free label was removed by gel filtration with G25 resin in the case of Cy3 and Alexa 488 labels. Protein was frozen in 0.2 M sucrose. The labeling efficiency was 60 to 100%.
High-speed pelleting assays.
We prepared serial dilutions of wild-type AlfA in polymerization buffer and determined the concentrations using absorbance at 280 nm. Immediately after addition of 5 mM ATP, samples were spun at 80,000 rpm in a TLA 100.4 rotor at 25°C for 15 min. Supernatants were recovered and separated by gel electrophoresis on 4 to 12% polyacrylamide gradient gels. The gels were stained with SYPRO red and scanned with a Typhoon variable-mode imager before quantitation with ImageQuant TL. The critical concentration was considered the x intercept in the plot of material missing from the supernatant.
Light scattering assays.
Right angle light scattering was conducted by mixing wild-type AlfA with ATP or GTP in polymerization buffer (with either 100 mM or 1.8 M KCl) in an SFA-20 rapid mixer (Hi-Tech) in a digital K2 fluorimeter (ISS, Champagne, IL) using an excitation wavelength of 320 nm. To calculate the critical concentrations, maximum-intensity values were collected from 5-s moving averages of background-subtracted 300-s traces. The maximum intensity was plotted against concentration as measured by absorbance at 280 nm, corrected for dilution. The critical concentration was considered the x intercept in the plot.
Phosphate release assays.
Phosphate release assays were performed using an EnzChek phosphate assay kit (Invitrogen) with an Ultrospec 2100 Pro spectrophotometer controlled with SWIFT II software (GE Healthcare Life Sciences). A360 values were converted to [Pi] by using a phosphate standard.
Total internal reflection fluorescence (TIRF) microscopy.
We used a Nikon Eclipse TE2000-E inverted microscope equipped with an Andor iXon+ EM digital camera and a CrystaLaser 100-mW 542-nm solid-state laser. Microscopy chambers were constructed from glass which was base washed in 0.5 M KOH and treated with 1% aminopropyltriethoxysilane. After assembly, the chambers were treated with 9 mg/ml polyethylene glycol-N-hydroxysuccinimide and 1 mg/ml biotin-polyethylene glycol-N-hydroxysuccinimide. Before use, chambers were treated with 2 μM streptavidin.
RESULTS
AlfA filaments spontaneously assemble in the presence of ATP and GTP and self-associate to form mixed-polarity bundles.
We initially attempted to express AlfA in Escherichia coli using an E. coli expression vector containing the native alfA gene. Despite systematic variation of the expression conditions (temperature and time of induction, cell density, E. coli strain, concentration of IPTG, etc.), we never observed significant expression of AlfA in E. coli. More careful analysis of the alfA sequence revealed a significant number of codons rarely found in E. coli genes. We therefore redesigned the gene (Gene Designer [DNA 2.0] and Bio Basic) by optimizing codon usage for E. coli and observed high levels of AlfA expression. To purify AlfA, we used a scheme similar to the protocol that we used for ParM, relying on ATP-dependent pelleting of the protein in an ultracentrifuge (13).
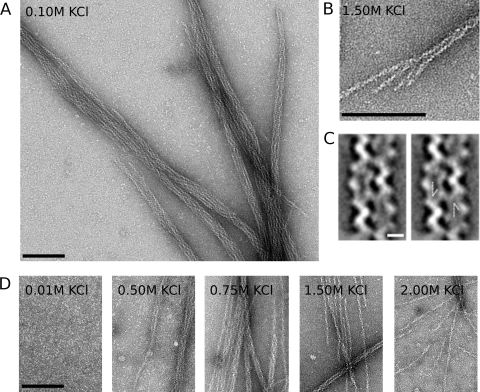
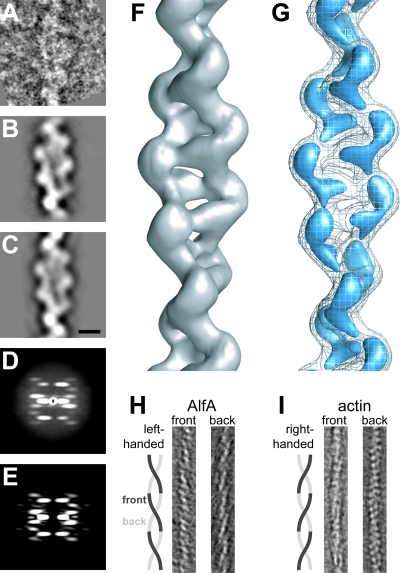
The success of our purification scheme suggested strongly that AlfA polymerizes upon addition of ATP. To see what sort of polymers AlfA forms, we mixed 5 μM AlfA with 5 mM ATP, negatively stained samples with uranyl formate, and obtained images of the samples using EM. We observed filamentous bundles that were approximately 20 nm wide and had an internal structural repeat every 100 nm (Fig. 1A). Many bundles have blunt ends, while others appear to be splayed, demonstrating that the bundles are composed of individual filaments. These single filaments are approximately 7 nm in diameter and appear to be double stranded, similar to actin, ParM, and MreB. Similar to assembly of the previously characterized plasmid actin and ParM, AlfA filaments and bundles can be assembled by addition of either ATP or GTP, and there are no appreciable differences in morphology. Unlike the dynamically unstable ParM molecules, however, AlfA filaments can also assemble in the presence of ADP and GDP (see Fig. S1 in the supplemental material). The small bundle shown in Fig. 1B is composed of four filaments, suggesting that the larger bundles shown in Fig. 1A contain at least twice this number of filaments. To better understand the relative orientations of filaments in the bundles, we generated reference-free averages for two-filament bundles; in one of these averages the filaments appear to run antiparallel to one another (Fig. 1C). The antiparallel orientation of the filaments was confirmed by reference-based matching with projections of the single-filament reconstruction (see below). This configuration suggests that the larger bundles contain filaments with a mixture of polarities.
FIG. 1.
AlfA polymerizes into filaments that spontaneously associate into mixed-polarity bundles. (A) AlfA filament bundles formed in the presence of Mg2+ and ATP and in the absence of crowding agents. AlfA (5 μM) was polymerized with 5 mM ATP, stained with 0.75% uranyl formate, and visualized by transmission EM. Similar bundles were observed with GTP. The conditions used were as follows: 100 mM KCl, 25 mM Tris HCl (pH 7.5), 1 mM MgCl, 1 mM DTT, and 25°C. (B) Splayed ends show that bundles are composed of individual filaments. (C) One of four reference-free class averages from a 102-segment data set for filament pairs, showing that filaments can associate with one another in an antiparallel fashion. The arrows in the panel on the right indicate the antiparallel features. Scale bar, 5 nm. (D) Bundles can be disassociated in the presence of high salt concentrations, and single filaments predominate in the presence of 2.00 M KCl. Scale bar, 100 nm.
AlfA bundles form spontaneously due to electrostatic interactions between filaments. Bundling was not due to molecular crowding or depletion interactions since we obtained EM images in the absence of crowding agents, such as methylcellulose. Unlike actin paracrystals (19), AlfA bundles appear even at low concentrations of MgCl2 (concentrations as low as 100 μM) (data not shown). High concentrations of KCl, however, support polymerization but efficiently suppress formation of AlfA filament bundles. In the presence of 0.1 M KCl virtually all AlfA is in bundles. In the presence of 1.0 M KCl, about one-half of the structures are single filaments and the bundles appear to be smaller, and in the presence of 2.0 M KCl, almost all AlfA bundles are disassociated into single filaments. Even in the presence of 2.0 M KCl, however, a few small bundles are observed (Fig. 1D). Concentrations of KCl greater than 0.1 M are required for efficient polymerization. With KCl concentrations less than 0.1 M, fewer bundles are observed on a grid, and in the presence of 0.01 M KCl only monomers are present (Fig. 1D).
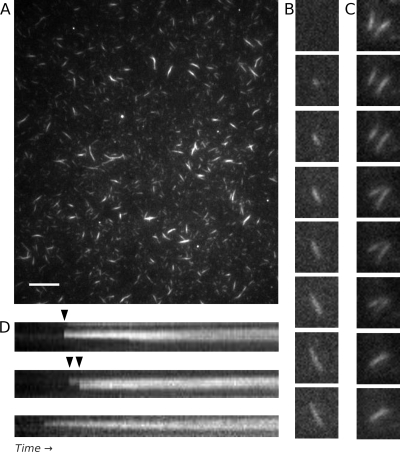
We next observed assembly of AlfA bundles using TIRF microscopy. For these experiments we tethered AlfA filaments to biotin-polyethylene glycol-streptavidin-coated coverslips by including 10% biotin-labeled AlfA in the reaction mixture. We visualized the filaments by including 20% Cy3-labeled AlfA. All experiments were performed in the absence of crowding agents. At steady state, 5 min after addition of ATP under conditions under which few single filaments are observed by EM (0.1 M KCl), AlfA forms filamentous structures that are various sizes and have various fluorescence intensities (Fig. 2A). The variation in the fluorescence intensities of filamentous structures in the same field suggests that the structures are composed of various numbers of filaments. Although the bundles seen by EM appear to be fairly uniform in size, they often appear to be grouped laterally alongside other bundles (Fig. 1A; see Fig. S1 in the supplemental material), and the structures observed by TIRF microscopy may represent such groups of many bundles that are parallel to one another.
FIG. 2.
AlfA filament and bundle assembly observed by TIRF microscopy, with bundles showing no evidence of dynamic instability. (A) TIRF microscopy of a field containing Cy3- and biotin-labeled 3 μM AlfA 5 min after polymerization revealed stable filamentous structures with various fluorescence intensities. The variation in intensity is consistent with filament bundles. Scale bar, 10 μm. The conditions used were as follows: 20% Cy3-labeled AlfA and 5% biotin-labeled AlfA (total concentration, 3 μM) with 5 mM ATP in the buffer described in the legend to Fig. 1A. (B) Bundles appear to elongate bidirectionaly, but the polarity of the filaments (and the directionality of their elongation) cannot be determined. The time interval was 40 s. The conditions were the same as those described above, except that 0.5% bovine serum albumin was added. (C) Annealing between two bundles adhering to a coverslip. The time interval was 7 s. The conditions were the same as those described above for panel A. (D) Kymographs collected immediately after addition of nucleotide, showing that bundles can grow by lateral annealing events (arrowheads). The time scale was 2.5 min. The conditions were the same as those described above for panel A.
Consistent with AlfA's ability to polymerize in the presence of ADP or GDP, time lapse imaging of AlfA at steady state revealed that bundles do not undergo cycles of growth and shrinkage, which are characteristic of dynamic instability. Instead, they persist for many minutes without changing in length, although they do exhibit stepwise changes in intensity that may be due to bundles recruiting filaments from solution, disassociating from the coverslip, or photobleaching.
To understand how AlfA bundles grow to reach their steady-state size, we observed the onset of filament assembly by flowing AlfA into microscopy cells immediately after addition of ATP. Bundles were observed to grow over a period of several minutes by apparent bidirectional elongation (Fig. 2B). However, bundles were also frequently observed to anneal to one another (Fig. 2C), and most growing structures increased in size predominantly by annealing to filaments from the solution. In kymographs, these events appear as stepwise increases in bundle length and intensity (Fig. 2D). The majority of the filaments that we observed grew by lateral annealing, and adhesion of filaments from the solution to existing seeds occurred much more frequently than we expected for random landing on a coverslip based on the rate of filament adhesion to unoccupied places on the coverslip. Since annealing events were responsible for the majority of the growth under our assay conditions, it was difficult to determine elongation rates for single AlfA filaments.
Spontaneous AlfA assembly is rapid under bundling and nonbundling conditions and shows no evidence of dynamic instability.
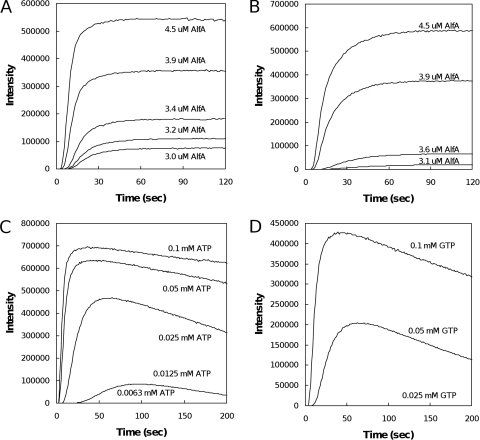
We next examined the kinetics of AlfA polymerization by light scattering. The spontaneous assembly of AlfA is much more rapid than that of conventional eukaryotic actin and more similar to that of plasmid-encoded ParM. In the presence of a saturating concentration of ATP (5 mM), AlfA at concentrations greater than 3 μM assembles rapidly, reaching equilibrium approximately 1 min after addition of the nucleotide (Fig. 3A). The lag in polymerization kinetics at early time points suggests that, like actin and ParM, AlfA assembles via nucleation-condensation. Data collapse of the curves after normalization of the intensity (100%) and time (half time) indicated that assembly occurs via a single mechanism at all concentrations (Fig. 4D) (11). The intensity of the signal generated by 4 μM AlfA in the presence of low concentrations of ATP (Fig. 3C) did not decrease rapidly after an initial burst of polymerization, as has been observed for ParM (13). Rather, the signal intensity decreased slowly, and only in the presence of an extremely low concentration of ATP, 12.5 μM (approximately 3:1 molar excess over AlfA in this experiment), did we observe complete depolymerization in a 5-min time course. With one-half this concentration of ATP (6.25 μM; approximately 1.5:1 molar excess), we observed no polymerization.
FIG. 3.
Light scattering. (A and B) Similar assembly of filaments in the presence of saturating levels (5 mM) of ATP (A) and GTP (B). The conditions were the same as those described in the legend to Fig. 1A. (C and D) AlfA at a concentration of 4.7 μM does not rapidly depolymerize in the presence of low levels of either ATP (C) or GTP (D), providing evidence of a lack of dynamic instability. AlfA appears to be less stable in the presence of GTP. The conditions were the same as those described above for panels A and B.
FIG. 4.
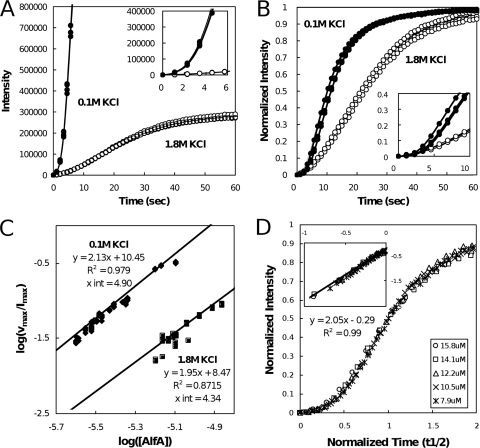
Light scattering in high-salt conditions. (A and B) Assembly of filaments in the presence of 5 mM ATP and 0.1 M KCl (filled circles) is more rapid than assembly of filaments in the presence of 1.8 M KCl (open circles). (A) At identical instrument settings, assembly of 3.8 μM AlfA in the presence of 0.1 M KCl quickly saturates the detector, while the signal for 7.4 μM AlfA in the presence of 1.8 M KCl remains modest. Both concentrations of AlfA are approximately 1 μM greater than the critical concentrations for the relevant KCl concentrations. (Inset) Early time points. Otherwise, the conditions were the same as those described in the legend to Fig. 3A. (B) Intensity-normalized traces obtained with similar instrument settings, showing that polymerization is slower in the presence of 1.8 M KCl. The conditions were the same as those described above for panel A, except that 0.1 M KCl was used with 3.1 μM AlfA. (C) Determination of nucleus size and relative nucleation rates for AlfA in high- and low-salt conditions. To generate a data point, the maximal velocity of a light scattering trace (vmax) was divided by the maximal intensity of the signal (Imax), and the log of the resulting value was plotted against the log of the AlfA concentration. The slopes of the regression lines are proportional to n/2, where n is the number of monomers required to form a nucleus. This analysis suggests that AlfA assembles with a nucleus size of four monomers in the presence of both 0.1 M KCl and 1.8 M KCl but that the nucleation is approximately 10 times faster in low-salt conditions. (D) Data collapse for light scattering traces obtained with 1.8 M KCl using various concentrations of AlfA after normalization of maximum intensities and half times (t1/2). (Inset) Log-log plot of the same data, showing that the slope of a regression line is 2. The conditions were the same as those described above for panel A.
AlfA assembles with similar kinetics in the presence of a saturating concentration of GTP (Fig. 3B), but it appears to be less stable in the presence of low concentrations of GTP than in the presence of low concentrations of ATP. We say this because the signal intensity decreases more rapidly in the presence of low concentrations of GTP and because the threshold GTP concentration for observing AlfA polymerization is higher. No AlfA, for example, polymerizes in the presence of 25 μM GTP (Fig. 3D), while we observed a significant signal with the same concentration of AlfA in the presence of 25 μM ATP (Fig. 3C).
We used light scattering (see Fig. S2B in the supplemental material) and high-speed pelleting (Table 1) to estimate critical concentrations of AlfA for assembly in the presence of ATP, ADP, adenylyl-imidodiphosphate (AMP-PNP), GTP, and GDP. Both methods indicated that the critical concentrations of AlfA are similar in the presence of ATP and GTP. Furthermore, high-speed pelleting demonstrated that the critical concentrations in the presence of ADP and GDP are within an order of magnitude of those in the presence of ATP and GTP. The ratios of the critical concentrations in the presence of di- and triphosphate nucleotides are comparable to the ratio in the presence of ADP and ATP for conventional actin but strikingly different from the very high ratio for ParM filaments (13). The relative stability in the presence of diphosphate nucleotides is consistent with the lack of dynamic instability observed by TIRF microscopy.
TABLE 1.
Critical concentrations of AlfA in high- and low-salt conditions
| KCl concn (M) | Compound | Critical concn (μM) bya:
|
|
|---|---|---|---|
| Pelleting | Light scattering | ||
| 0.1 | ATP | 2.4 ± 0.6 (4) | 2.7 ± 0.2 (3) |
| ADP | 10.1 ± 3.1 (6) | ||
| AMP-PNP | 1.8 ± 0.02 (2) | ||
| GTP | 2.0 ± 0.8 (4) | 3.4 ± 0.3 (3) | |
| GDP | 19.5 ± 2.2 (5) | ||
| 1.8 | ATP | 4.3 ± 0.6 (3) | 6.1 ± 1.3 (4) |
| ADP | 33.8 (1) | ||
| GTP | 7.9 ± 4.1 (2) | >8 (1) | |
| GDP | 47.1 (1) | ||
The critical concentrations were determined as described in Materials and Methods. The numbers in parentheses are numbers of experiments.
To remove potential effects of bundling on filament dynamics, we compared the behavior of AlfA in low-salt buffers that permit bundling to the behavior of AlfA in high-salt buffers that favor single filaments (Fig. 4). In buffers containing 1.8 M KCl, AlfA pellets in the presence of ATP, ADP, GTP, and GDP with critical concentrations that are two- to fourfold higher than the critical concentrations in low-salt buffers (Table 1), suggesting that AlfA does not exhibit dynamic instability under conditions that dissociate bundles. The intensity of light scattering is much lower in a high-salt buffer (1.8 M KCl), likely because the bundles scatter light more effectively than single filaments, making direct comparison between the two conditions difficult. We compared the polymerization of 3.8 μM AlfA in the presence of 0.1 M KCl to the polymerization of 7.4 μM AlfA in the presence of 1.8 M KCl (Fig. 4A). Based on the critical concentrations measured by light scattering (Table 1), the two reactions produced equivalent quantities (∼1 μM) of polymer at steady state. However, it is apparent that the low-salt reactions produced a far stronger signal.
We next compared the rates of polymerization under high- and low-salt conditions. Based on the different critical concentrations, we chose starting concentrations of AlfA that would produce similar concentrations of polymer at steady state under the two conditions (3.1 μM AlfA with 0.1 M KCl and 7.4 μM AlfA with 1.8 M KCl). We collected complete light scattering curves at instrument settings appropriate for each condition and normalized the resultant traces to their maximal intensities. This comparison demonstrates that AlfA polymerization is slightly slower in the presence of 1.8 M KCl than in the presence of 0.1 M KCl (Fig. 4B).
We used kinetics to investigate the mechanism of AlfA filament assembly. We first estimated the size of the AlfA nucleus, defined as the smallest oligomer to which monomers add with approximately the same affinity as to stable filaments, under bundling and nonbundling conditions. We did this in several ways. First, we plotted the concentration dependence of the maximal velocity of spontaneous polymerization as described by Nishida and Sakai (26). The maximal velocity of each light scattering curve was normalized by using its maximal intensity. We plotted the log of the resulting value against the log of the AlfA concentration (Fig. 4C) and fit the data to a straight line. In the Nishida-Sakai formalism, the slope of this line is equal to the nucleus size divided by two (see the supplemental material for derivation). Regardless of whether the analysis was performed with low-salt (0.1 M KCl) or high-salt (1.8 M KCl) data, the slope of the line generated was approximately 2, suggesting that AlfA polymerized from tetrameric nuclei in both cases. Thus, AlfA assembles by the same mechanism in both buffer conditions, although nucleation appears to be somewhat slower in high-salt conditions.
We also estimated the nucleus size from the kinetics of assembly at early time points. Using a method described by Flyvberg et al. (11), we normalized light scattering data by using the maximum intensity and half times and observed a collapse of all the data onto a single curve, regardless of the concentration of AlfA (Fig. 4D). This type of “phenomenological scaling” indicates that the same mechanism governs polymer assembly at different protein concentrations. On a log-log scale the slope of the early time plots of the normalized data is related to the number of steps in the nucleation mechanism. In this case, we estimated a slope of 2, consistent with a trimeric nucleus. The slight nonlinearity of the light scattering signal at the earliest time points when the average polymer is short means that the Nishida-Sakai formalism may provide the more accurate estimate. Nevertheless, the two independent methods indicate that the size of the AlfA filament nucleus is three or four subunits.
Finally, we numerically simulated AlfA assembly using the Berkeley Madonna software package. For this simulation, we assumed a nucleus size of four, a filament elongation rate similar to those of actin and ParM, and a disassembly rate consistent with measured critical concentrations. We varied the rate constants for formation of AlfA dimers, trimers, and tetramers to obtain a best fit to light scattering data sets collected using four different AlfA concentrations (from 7.9 to 14.1 μM). All data used for fitting were collected in high-salt conditions (1.25 M KCl) to minimize nonlinearities associated with strong light scattering from bundles. Global, nonlinear, least-squares fitting yielded a single set of rate constants that fit the data at all concentrations (see Fig. S3 in the supplemental material). From this analysis we estimated an affinity for AlfA dimerization of ∼10 mM. This affinity is approximately 500-fold higher than the estimated affinity of a conventional actin dimer (31).
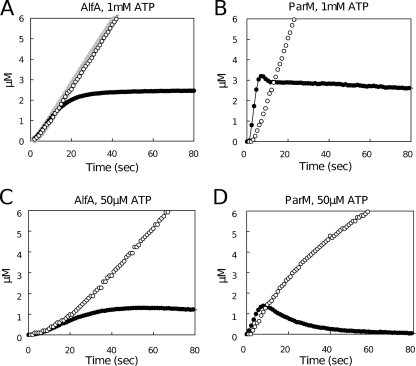
We used a coupled enzyme assay to monitor ATP hydrolysis and phosphate release from 5 μM AlfA in the presence of both limiting (50 μM) and saturating (5 mM) concentrations of ATP (Fig. 5). At early time points the two ATP concentrations produced similar curves, while at steady state, when the light scattering signal was relatively constant, phosphate release was faster with the saturating ATP concentration (Fig. 5A and C). At early time points, in the presence of both low and high concentrations of ATP, phosphate release kept pace with polymerization, and we observed almost no lag between the two curves. Using these data it is not possible to obtain accurate values for ATP hydrolysis and phosphate release, but we placed lower boundaries on both processes of around 0.4 s−1. This rate is significantly higher than the rate of phosphate dissociation from conventional actin or previously characterized actin-related proteins (3, 7, 21).
FIG. 5.
Phosphate release assays. (A) Phosphate release ([Pi], determined by a colorimetric assay) (open circles) and amount of polymer formed ([polymeric AlfA], determined by light scattering) (filled circles) with 8.6 μM AlfA in the presence of 1 mM ATP and 1.8 M KCl. Otherwise, the conditions were the same as those described in the legend to Fig. 1A. The gray line indicates the [Pi] release predicted by our model. (B) Phosphate release and amount of polymer formed with 4.9 μM ParM in the presence of 0.1 M KCl and 1 mM ATP. Otherwise, the conditions were the same as those described above for panel A. (C) Phosphate release and amount of polymer formed with 8.6 μM AlfA in the presence of 50 μM ATP. Otherwise, the conditions were the same as those described above for panel A. (D) Phosphate release and amount of polymer formed with 4.9 μM ParM in the presence of 50 μM ATP. Otherwise, the conditions were the same as those described above for panel B.
We compared ATP hydrolysis and phosphate release of AlfA with ATP hydrolysis and phosphate release of 5 μM ParM using the same ATP concentrations. With both concentrations of ATP, the initial burst of phosphate release by ParM lagged significantly behind polymerization (Fig. 5A and 5D). With the saturating concentration of ATP, the steady-state rate of phosphate release by ParM was higher than the steady-state rate of phosphate release by AlfA (Fig. 5A and B), indicating that in the steady-state plateau phase, when total polymer levels are constant, the flux of ParM subunits through filaments is faster than that of AlfA subunits.
Reconstruction of the AlfA filament structure: AlfA forms left-handed filaments with an architecture distinct from that of ParM.
We polymerized AlfA in the presence of 1.25 M KCl to produce EM grids rich in single filaments. We first collected electron micrographs at multiple tilt angles and generated three-dimensional tomograms of individual AlfA filaments to establish the handedness of the long-pitch helix (Fig. 6H). These tomograms revealed that, like ParM filaments, AlfA filaments are two-strand helical polymers with a left-handed pitch. As a control, the right-handedness of the actin helix was confirmed in parallel experiments (Fig. 6I) (15).
FIG. 6.
Three-dimensional reconstruction of single AlfA filaments. (A) Representative segment of an AlfA filament in a negatively stained micrograph. The conditions used were as follows: 1.25 M KCl, 25 mM Tris HCl (pH 7.5), 1 mM MgCl, 1 mM DTT, 25°C, 5 mM AMP-PNP, and 7 μm AlfA. (B) Average of 757 AlfA segments after iterative helical real-space reconstruction. (C) Reprojection of the final AlfA filament model in the same orientation as the average shown in panel B. Scale bar, 5 nm. (D) Average power spectrum for 2,000 nonoverlapping segments. (E) The two-dimensional power spectrum of the final AlfA filament model is very similar to the average power spectrum shown in panel D. (F) Three-dimensional reconstruction of an AlfA filament, contoured to enclose the expected mass of the AlfA subunits. (G) AlfA filament at the same contour as the contour shown in panel F (mesh) and at a higher contour level (blue), clearly showing the separation between the two strands of the filament and between AlfA subunits in each strand. (H) Approximately 3.5-nm slices from a tomographic reconstruction of a single AlfA filament, demonstrating that the long pitch of the helix is left-handed. (I) Tomogram of actin, prepared like the tomogram for AlfA in panel H as a control, demonstrating the well-established right-handed pitch of the long helix. In panels H and I the width of the image is 25 nm.
We determined the structure of single, unbundled AlfA filaments by iterative helical real-space reconstruction, a single-particle reconstruction approach for helical structure determination (9, 10). We first collected a set of 50 micrographs of negatively stained AlfA filaments at defocus values from 0.8 to 1.6 μm (average, 1.1 μm), as determined with CTFfind, and corrected for the contrast transfer function by flipping the phases of the whole micrographs. Filaments were extracted from corrected micrographs in 265-Å segments (Fig. 6A), with 90% overlap of boxes along the filament length. The high degree of overlap enabled us to make full use of the helical symmetry during refinement of the structure. The approximate orientation of each filament was recorded and used as a constraint on the segment rotation during the alignment procedure. Altogether, we extracted and used 42,547 segments for the reconstruction.
As an initial model for projection matching, we used a featureless cylinder. Reference projections were generated at 3° intervals perpendicular to the cylinder axis. We carried out 40 rounds of iterative projection matching alignment and reconstruction, and in each round a search was performed for the optimal helical symmetry, defined as the rotation around and translation parallel to the helical axis between adjacent subunits, as described by Egelman (9). This approach requires an initial estimate of the helical symmetry parameters, and it is an indication of the robustness of the reconstruction that the refinement of the symmetry parameters converges when different initial estimates are used (see Fig. S4 in the supplemental material). After rejection of segments with excessive shifts parallel to the helix axis or with rotations deviating from the initial estimates of the helical axis orientation, 36,158 segments were included in the final reconstruction.
In addition to the convergence of symmetry parameters for different starting values, there are several other indicators of the reliability of the reconstruction. Average images of particles with the same refined orientations are nearly identical to reprojections of the final model in the same orientation (Fig. 6B and C), showing that our model is self-consistent. More convincingly, the average power spectrum of 2,000 nonoverlapping segments has the same features as the power spectrum of the reconstruction (Fig. 6D and E). While the resolution indicated by the FSC 0.5 criterion is 15 Å (see Fig. S5 in the supplemental material), the lack of surface detail suggests that the effective resolution may be somewhat lower; this is likely due to the use of a negative stain as a contrast agent. Finally, parallel reconstruction of an Acanthamoeba actin filament using the same sample preparation and reconstruction techniques yielded a structure with clearly identifiable features of conventional eukaryotic actin.
The architecture of the AlfA filament is significantly different from that of previously described actins. The subunits in the AlfA filament are related by a rotation of 157° and by a translation of 25 Å along the helical axis. The filament is two stranded, with a rotation of 46° and a translation of 50 Å between subunits in each strand (Fig. 6F). While the translation between subunits is similar to that of ParM, the rotation is 18° greater in AlfA than in ParM (27). At higher contour levels, the map clearly shows the separation between strands and suggests boundaries between AlfA subunits (Fig. 6G). The helix is relatively open, with minimal buried surface between the two strands. The overall shape of the helix is more ribbon-like than the helix of either conventional actin or ParM, with a maximum width of ∼100 Å and minimum width of ∼45 Å.
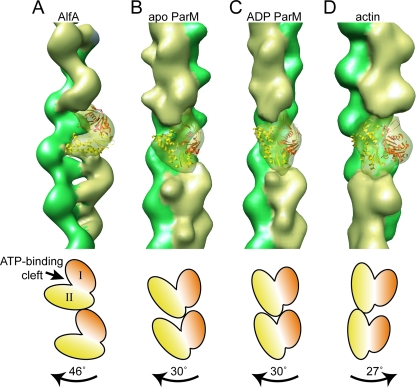
We made several attempts to fit the crystal structures of AlfA homologs into the EM reconstruction. The crystal structures of actin (PDB 2BTF) (30) and nucleotide-bound ParM (PDB 1MWM) (36) did not fit well into our AlfA filament structure. However, a reasonable fit was obtained with the apo conformation of ParM, in which the ATP binding cleft is opened by about 25° relative to the ADP state (PDB 1MWM) (36). In the best fit, the ATP binding cleft of the crystal structure is tilted roughly 45° relative to the direction of the helix axis (Fig. 7A). Of course, the reliability of this fit is limited both by the distant relationship between ParM and AlfA and by the resolution limitations inherent in negative-stain reconstructions, and the assignment of the AlfA orientation within the filament should be considered preliminary. A more detailed understanding of the orientation of the individual AlfA subunits will require a higher-resolution cryo-EM reconstruction of the filament.
FIG. 7.
The architecture of the AlfA filament is distinct from the architecture of ParM and the architecture of actin. (A) Reconstruction of the AlfA filament, with the strands indicated by different colors. A single subunit of the filament is transparent, with the crystal structure of apo ParM manually fitted into the density. The crystal structure is orange in domain I and yellow in domain II. (B to D) For comparison, the simulated EM densities calculated from models of the ParM filament in the apo and ADP-bound conformations (E. H. Egelman, personal communication) and of actin filaments (28) are shown using the same color scheme. The cartoons at the bottom indicate the orientations of subunits in the filaments; the magnitude and direction of the rotation between subunits moving up each strand are indicated by labeled arrows.
The overall symmetry of the AlfA filament is distinct from that of both the ParM and actin filaments. Recent reconstructions of ParM filaments in both the apo and ADP conformations have a rotation of 30° between subunits along each strand (Fig. 7B and C) (E. H. Egelman, personal communication). The actin filament has a rotation of 27° between subunits but in the opposite direction, producing a right-handed helix (Fig. 7D) (28). By contrast, AlfA has a twist of 46° between subunits, producing a more twisted helix (Fig. 7A). Furthermore, the AlfA filament appears to make fewer contacts between the two strands, producing a filament with a more open architecture than that of ParM or actin. These differences in the overall architecture may be explained in part by the apparently unique orientation of the AlfA protomer within the filament; in the AlfA filament the ATP binding cleft seems to be tilted by about 45° relative to the helical axis, whereas in both ParM and actin the ATP binding cleft is nearly parallel to the helical axis.
DISCUSSION
Two major results of our study—(i) that AlfA filaments are not dynamically unstable and (ii) that these filaments spontaneously form mixed-polarity bundles—provide a molecular basis for previous in vivo observations made by Becker and coworkers (1). These authors found that, unlike ParM, which forms short-lived filaments in E. coli (5), AlfA forms stable structures that run from pole to pole in B. subtilis and display no evidence of catastrophic disassembly. These data are consistent with our discovery that, like eukaryotic actin, AlfA can polymerize in the presence of ADP with a critical concentration only fourfold higher than that in the presence of ATP. Furthermore, as determined by TIRF microscopy, fluorescent AlfA filaments are stable and do not undergo obvious growth and shortening phases. Together, these data argue strongly that, unlike ParM filaments, AlfA filaments are not dynamically unstable. When Becker and coworkers photobleached fluorescent AlfA structures in vivo, they found that the bleached region recovered symmetrically from both sides, suggesting that in vivo AlfA forms bundles of multiple (possibly treadmilling) filaments. The fact that the new filaments follow exactly the track of the original bleached structure suggests that there is close lateral association between the filaments. This fits with our EM analysis of purified AlfA, which revealed that in the presence of ATP or GTP, AlfA has a strong tendency to form tight, mixed-polarity filament bundles. Furthermore, using TIRF microscopy, we directly observed labeled AlfA structures zippering together to form larger bundles. Together, our results and those of Becker et al. demonstrate that the in vivo and in vitro assembly dynamics of AlfA are unique, and they strongly suggest that the mechanism of AlfA-dependent DNA segregation is significantly different from the previously characterized ParM-dependent process.
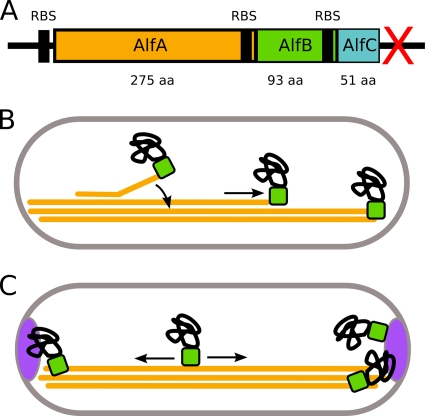
Dynamic instability of ParM filaments provides a mechanism for balanced plasmid movement. Because the filament is unstable at both ends, only bivalently attached filaments are competent to elongate, and the movement of one plasmid to a pole is always coupled to the movement of a second plasmid to the opposite pole. Our finding that AlfA is not dynamically unstable raises important questions about AlfA-dependent DNA segregation. It is easy to imagine elongating AlfA filaments aligning with the long axis of a cell and forming a track running from pole to pole. Here is the mystery: if plasmids are not required for filament assembly or stability, how does such a track segregate plasmids? First, plasmids must somehow interact with AlfA filaments. This interaction might be mediated by accessory factors, possibly located in nearby open reading frames in the pLS32 plasmid. One potential regulator, alfB, is a gene directly downstream of alfA that is also required for plasmid maintenance (1). Based on homology to transcription regulators, alfB is predicted to encode a DNA binding protein. The role of AlfB in DNA segregation may be similar to that of the type II segregation factor ParR, which attaches plasmid DNA to the growing ends of ParM filaments (5, 25). Another potential regulator might be encoded by a previously undescribed open reading frame downstream of the AlfB gene that we call “alfC.” This open reading frame is very short and has no obvious homology to known protein-encoding genes. It is not known whether AlfC interacts with AlfA, and further studies are required to determine whether AlfC plays a role in plasmid segregation.
The two most general mechanisms by which a plasmid could interact with an AlfA filament are (i) end binding and (ii) side binding. If plasmids (perhaps via AlfB) bind to the end of AlfA filaments, they may be pushed poleward by assembly of filaments growing along the existing bundle (Fig. 8B). Alternately, plasmids may bind to the side of the filaments and be carried in one direction or the other by filament treadmilling. Both of these models explain how one plasmid might find a pole, but neither model explains how two plasmids could be partitioned to opposite poles. We know from in vivo experiments that AlfA does, in fact, partition plasmids rather than simply place them at the poles (1). One potential solution to this problem is a mechanism for ejecting supernumerary plasmids from an occupied pole and encouraging them to travel to the other pole. Such a mechanism could involve strictly limiting the number of polar plasmid binding sites. These sites could be either on the ends of AlfA filaments or provided by host cell factors. The existence of host cell anchors is plausible since pLS32 is found only in Bacillus species and one Bacillus gene, racA, is already known to help maintain pLS32 during sporulation. Normally, RacA helps anchor the bacterial chromosome in the forespore (2, 37), but deletion of the racA gene also reduces plasmid retention (1). Because RacA is expressed only during sporulation, however, another factor would have to be responsible for maintaining plasmids during vegetative growth.
FIG. 8.
Hypothetical mechanisms for plasmid segregation by a bundling polymer lacking dynamic instability. (A) Diagram of the alf operon, showing the relative sizes and positions of open reading frames and their ribosome binding sites. The red X indicates a transcriptional terminator. RBS, ribosome binding site; aa, amino acids. (B) “End interaction.” Plasmids, through an adapter protein (perhaps AlfB), can bind to the ends of individual filaments. The ends are either encountered randomly by diffusion or created through nucleation of a new filament, which soon anneals to the existing bundle (curved arrow). Plasmids track with the growing end (straight arrow) until they reach one of the cell poles, where they are likely to remain because of the high concentration of filament ends. With some frequency, plasmids are transmitted to the other pole by tracking with the end of a new filament. (C) “Lateral interaction.” Plasmids bind filaments laterally and can travel with treadmilling or sliding filaments. If a plasmid reaches a cell pole, it is maintained there by another factor (such as RacA).
AlfA forms bundles in vivo and in vitro, and this behavior likely plays a role in plasmid segregation. Although the interaction appears to be driven by electrostatics, we observed AlfA bundles in the presence of salt concentrations as high as 2 M KCl (Fig. 1D), suggesting that the bundling observed in vivo occurs by the same mechanism as the spontaneous lateral association that we observed in vitro. We do not know the precise role of bundling in plasmid segregation, but there are several attractive possibilities. First, by providing high local concentrations of filaments, bundles can stabilize weak interactions with accessory proteins. In addition, spontaneous bundling may gather all AlfA filaments in the cytoplasm into a single, DNA-segregating structure. By restricting movement of filaments, such a structure could provide a positional “memory” that persists beyond the lifetime of an individual filament. In other words, a filament bundle could act as a stable template, guiding the treadmilling or sliding of individual plasmid-attached filaments.
The unique architecture of the AlfA filament demonstrates the remarkable diversity in the actin family. While we think of filament formation as the “function” of an actin-like protein, the residues that mediate filament formation are not highly conserved. The most highly conserved sequences in the actin family line the nucleotide binding pocket. The residues that mediate subunit-subunit contacts in the filament are quite divergent. This divergence leads to remarkable differences in filament architecture. To date, structures have been determined for four actin family filaments: conventional actin (34), MreB (35), ParM (27), and now AlfA. While all of these filaments can be described as two-stranded, the relationship between the strands is different in each case. In one filament (actin) the strands form a right-handed helix; in two (ParM and AlfA) the helix is left-handed; and one (MreB) lacks a helical twist altogether. Furthermore, even though ParM and AlfA filaments both form left-handed helices, the symmetries of the two types of filaments are quite different, as are the orientations of subunits with respect to the filament axis, and the ATP binding cleft of AlfA filaments is rotated roughly 45° relative to its orientation in the other types of filaments (Fig. 7). In this regard, a similar orientation of the ATP binding cleft was observed in a “ribbon” model based on crystal contacts from the structure of the β-actin-profilin complex (30). The Schutt model suggested that actin (and actin-like) monomers can be assembled into polymers with significantly different architectures, and subsequent studies of bacterial actins have borne this out.
The divergence in the subunit-subunit contact sites in the family of actin filaments indicates one of three things: (i) the architectural constraints of filament formation are loose enough to permit significant genetic drift; (ii) there is positive evolutionary pressure on the bacterial actin-like proteins to produce filaments with divergent architecture; or (iii) filament formation evolved multiple times in the actin family.
Supplementary Material
Acknowledgments
We thank members of the Mullins and Vale labs for discussions and support, J. Pogliano (University of California at San Diego) for providing plasmids, E. H. Egelman (University of Virginia) for discussions on the structure of ParM, and M. Braunfeld, S. Zheng, and E. Branlund (University of California at San Francisco) for technical assistance.
This work was supported by grants to R.D.M. from the Sandler Family Supporting Foundation and the National Institutes of Health (grant R01-GM079556). J.P. was supported by an NSF predoctoral fellowship.
Footnotes
Published ahead of print on 7 August 2009.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Becker, E., N. C. Herrera, F. Q. Gunderson, A. I. Derman, A. L. Dance, J. Sims, R. A. Larsen, and J. Pogliano. 2006. DNA segregation by the bacterial actin AlfA during Bacillus subtilis growth and development. EMBO J. 25:5919-5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Yehuda, S., D. Z. Rudner, and R. Losick. 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299:532-536. [DOI] [PubMed] [Google Scholar]
- 3.Blanchoin, L., and T. D. Pollard. 2002. Hydrolysis of ATP by polymerized actin depends on the bound divalent cation but not profilin. Biochemistry 41:597-602. [DOI] [PubMed] [Google Scholar]
- 4.Bork, P., C. Sander, and A. Valencia. 1992. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA 89:7290-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, C. S., and R. D. Mullins. 2007. In vivo visualization of type II plasmid segregation: bacterial actin filaments pushing plasmids. J. Cell Biol. 179:1059-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carballido-López, R. 2006. The bacterial actin-like cytoskeleton. Microbiol. Mol. Biol. Rev. 70:888-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dayel, M. J., E. A. Holleran, and R. D. Mullins. 2001. Arp2/3 complex requires hydrolyzable ATP for nucleation of new actin filaments. Proc. Natl. Acad. Sci. USA 98:14871-14876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Earl, A. M., R. Losick, and R. Kolter. 2007. Bacillus subtilis genome diversity. J. Bacteriol. 189:1163-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egelman, E. H. 2000. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 85:225-234. [DOI] [PubMed] [Google Scholar]
- 10.Egelman, E. H. 2007. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J. Struct. Biol. 157:83-94. [DOI] [PubMed] [Google Scholar]
- 11.Flyvbjerg, H., E. Jobs, and S. Leibler. 1996. Kinetics of self-assembling microtubules: an “inverse problem” in biochemistry. Proc. Natl. Acad. Sci. USA 93:5975-5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190-199. [DOI] [PubMed] [Google Scholar]
- 13.Garner, E. C., C. S. Campbell, and R. D. Mullins. 2004. Dynamic instability in a DNA-segregating prokaryotic actin homolog. Science 306:1021-1025. [DOI] [PubMed] [Google Scholar]
- 14.Garner, E. C., C. S. Campbell, D. B. Weibel, and R. D. Mullins. 2007. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science 315:1270-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes, K. C., D. Popp, W. Gebhard, and W. Kabsch. 1990. Atomic model of the actin filament. Nature 347:44-49. [DOI] [PubMed] [Google Scholar]
- 16.Holy, T. E., and S. Leibler. 1994. Dynamic instability of microtubules as an efficient way to search in space. Proc. Natl. Acad. Sci. USA 91:5682-5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen, R. B., and K. Gerdes. 1997. Partitioning of plasmid R1. The ParM protein exhibits ATPase activity and interacts with the centromere-like ParR-parC complex. J. Mol. Biol. 269:505-513. [DOI] [PubMed] [Google Scholar]
- 18.Jones, L. J., R. Carballido-López, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 19.Kawamura, M., and K. Maruyama. 1970. Polymorphism of F-actin. I. Three forms of paracrystals. J. Biochem. 68:885-899. [DOI] [PubMed] [Google Scholar]
- 20.Komeili, A., Z. Li, D. K. Newman, and G. J. Jensen. 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311:242-245. [DOI] [PubMed] [Google Scholar]
- 21.Korn, E. D., M. F. Carlier, and D. Pantaloni. 1987. Actin polymerization and ATP hydrolysis. Science 238:638-644. [DOI] [PubMed] [Google Scholar]
- 22.Ludtke, S. J., P. R. Baldwin, and W. Chiu. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82-97. [DOI] [PubMed] [Google Scholar]
- 23.Mayer, J. A., and K. J. Amann. 2009. Assembly properties of the Bacillus subtilis actin, MreB. Cell Motil. Cytoskelet. 66:109-118. [DOI] [PubMed] [Google Scholar]
- 24.Mindell, J. A., and N. Grigorieff. 2003. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142:334-347. [DOI] [PubMed] [Google Scholar]
- 25.Møller-Jensen, J., R. B. Jensen, J. Löwe, and K. Gerdes. 2002. Prokaryotic DNA segregation by an actin-like filament. EMBO J. 21:3119-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishida, E., and H. Sakai. 1983. Kinetic analysis of actin polymerization. J. Biochem. 93:1011-1020. [DOI] [PubMed] [Google Scholar]
- 27.Orlova, A., E. C. Garner, V. E. Galkin, J. Heuser, R. D. Mullins, and E. H. Egelman. 2007. The structure of bacterial ParM filaments. Nat. Struct. Mol. Biol. 14:921-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orlova, A., A. Shvetsov, V. E. Galkin, D. S. Kudryashov, P. A. Rubenstein, E. H. Egelman, and E. Reisler. 2004. Actin-destabilizing factors disrupt filaments by means of a time reversal of polymerization. Proc. Natl. Acad. Sci. USA 101:17664-17668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 30.Schutt, C. E., J. C. Myslik, M. D. Rozycki, N. C. Goonesekere, and U. Lindberg. 1993. The structure of crystalline profilin-beta-actin. Nature 365:810-816. [DOI] [PubMed] [Google Scholar]
- 31.Sept, D., and J. A. McCammon. 2001. Thermodynamics and kinetics of actin filament nucleation. Biophys. J. 81:667-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srivatsan, A., Y. Han, J. Peng, A. K. Tehranchi, R. Gibbs, J. D. Wang, and R. Chen. 2008. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 4:e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka, T., and M. Ogura. 1998. A novel Bacillus natto plasmid pLS32 capable of replication in Bacillus subtilis. FEBS Lett. 422:243-246. [DOI] [PubMed] [Google Scholar]
- 34.Trinick, J., J. Cooper, J. Seymour, and E. H. Egelman. 1986. Cryo-electron microscopy and three-dimensional reconstruction of actin filaments. J. Microsc. 141:349-360. [DOI] [PubMed] [Google Scholar]
- 35.van den Ent, F., L. A. Amos, and J. Löwe. 2001. Prokaryotic origin of the actin cytoskeleton. Nature 413:39-44. [DOI] [PubMed] [Google Scholar]
- 36.van den Ent, F., J. Møller-Jensen, L. A. Amos, K. Gerdes, and J. Löwe. 2002. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 21:6935-6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, L. J., and J. Errington. 2003. RacA and the Soj-Spo0J system combine to effect polar chromosome segregation in sporulating Bacillus subtilis. Mol. Microbiol. 49:1463-1475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.