Abstract
Recently, Nie and coworkers (L. Nie, Y. Ren, A. Janakiraman, S. Smith, and H. Schulz, Biochemistry 47:9618-9626, 2008) reported a new Escherichia coli thioesterase encoded by the ybaW gene that cleaves the thioester bonds of inhibitory acyl-coenzyme A (CoA) by-products generated during β-oxidation of certain unsaturated fatty acids. These authors suggested that ybaW expression might be regulated by FadR, the repressor of the fad (fatty acid degradation) regulon. We report mapping of the ybaW promoter and show that ybaW transcription responded to FadR in vivo. Moreover, purified FadR bound to a DNA sequence similar to the canonical FadR binding site located upstream of the ybaW coding sequence and was released from the promoter upon the addition of long-chain acyl-CoA thioesters. We therefore propose the designation fadM in place of ybaW. Although FadR regulation of fadM expression had the pattern typical of fad regulon genes, its modulation by the cyclic AMP (cAMP) receptor protein-cAMP complex (CRP-cAMP) global regulator was the opposite of that normally observed. CRP-cAMP generally acts as an activator of fad gene expression, consistent with the low status of fatty acids as carbon sources. However, glucose growth stimulated fadM expression relative to acetate growth, as did inactivation of CRP-cAMP, indicating that the complex acts as a negative regulator of this gene. The stimulation of fadM expression seen upon deletion of the gene encoding adenylate cyclase (Δcya) was reversed by supplementation of the growth medium with cAMP. Nie and coworkers also reported that growth on a conjugated linoleic acid isomer yields much higher levels of FadM thioesterase activity than does growth on oleic acid. In contrast, we found that the conjugated linoleic acid isomer was only a weak inducer of fadM expression. Although the gene is not essential for growth, the high basal level of fadM expression under diverse growth conditions suggests that the encoded thioesterase has functions in addition to β-oxidation.
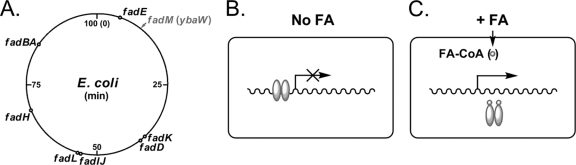
Most of our knowledge of the genetics and biochemistry of fatty acid degradation in bacteria has been derived from studies with Escherichia coli. The synthesis of at least five proteins involved in fatty acid β-oxidation is coordinately induced when long-chain fatty acids are present in the growth medium (Fig. 1). The genetic and enzymological studies of Overath and coworkers constitute the basis of the field (30, 37). The genes encoding the enzymes of aerobic β-oxidation (the fad regulon) are scattered around the E. coli chromosome, and fadBA is the only operon (11) (Fig. 1). The fad regulon is primarily responsible for the transport, activation, and β-oxidation of medium-chain fatty acids (C7 to C11) and long-chain fatty acids (C12 to C18) and is defined as those genes controlled by the fadR gene product under aerobic conditions.
FIG. 1.
The fad regulon and the current model of its regulation by fatty acids. (A) Physical location of the fad regulon loci in the E. coli chromosome. The ybaW (fadM) gene is in bold font. (B) Repression of a fad gene in the absence of exogenous fatty acids. (C) Induction of fad gene upon the addition of exogenous fatty acids. The ovals represent FadR, whereas the circles denote the acyl-CoA regulatory ligand. FA, fatty acids.
The levels of the enzymes of aerobic fatty acid degradation in E. coli depend on at least three regulatory systems, the global cyclic AMP (cAMP) receptor protein-cAMP complex (CRP-cAMP) and ArcAB system plus the fatty acid-specific FadR regulator (8, 18, 38). The CRP-cAMP system exerts its classical positive control of carbon utilization (16, 24, 31, 40) (putative CRP binding sites are found in the promoter regions of the fad genes [27]), whereas FadR negatively regulates both the fad regulon (4, 5) and the aceBA operon (23, 33). The ArcAB system has been reported to strongly (>20-fold) repress the expression of the 3-hydroxyacyl-coenzyme A (CoA) dehydrogenase encoded by the fadB gene and weakly repress acyl-CoA dehydrogenase activity (8). The mechanism(s) of repression of these genes by the ArcAB system have not yet been explored. The FadR protein has a dual role in fatty acid metabolism. It acts as a repressor of the β-oxidation pathway and an activator of the unsaturated fatty acid biosynthetic genes (3, 13, 25, 26). In the fad regulon (41), FadR acts in a manner similar to LacI in the lac system (1); it is a classical transcriptional repressor (see below). The first evidence that the fadR gene product regulates the expression of the fad regulon at the level of transcription was the lacZ transcriptional fusion studies of Clark (10). LacZ expression in strains carrying such fusions was inducible by long-chain fatty acids in wild-type strains and constitutive in fadR strains. Furthermore, the expression of β-galactosidase was repressed in these strains under catabolite-repressing growth conditions (as are the levels of the fatty acid degradative enzymes) and overexpression of FadR gave increased repression, presumably due to increased occupation of operator sites (17). Overath et al. (30, 37) originally postulated that long-chain acyl-CoA thioesters are the in vivo inducers of the fad regulon. This proposal has been confirmed by in vitro (25) and in vivo (12) studies that demonstrated that long-chain but not short-chain acyl-CoAs regulate DNA binding by FadR.
YbaW is a newly identified thioesterase, called thioesterase III, which is involved in fatty acid β-oxidation (35, 36). Nie and coworkers (36) proposed that the physiologically important reaction catalyzed by YbaW is hydrolysis of 3,5-tetradecadienoyl-CoA, a minor metabolite of β-oxidation of oleic acid, to 3,5-cis-tetradecadienoic acid which is released into the growth medium. The flux through the YbaW-dependent pathway was estimated to account for 10% of β-oxidation when oleic acid was the sole carbon source (35, 36). YbaW is proposed to hydrolyze β-oxidation metabolites that are resistant to further degradation and which therefore accumulate and inhibit the pathway. More recently, thioesterase III was shown to contribute to the degradation of 9-cis,11-trans-octadecadienoic acid, one of the major isomers of conjugated linoleic acid (CLA), by cleavage of the thioester bond of the terminal degradation product, 3,5-dodecadienoyl-CoA (35).
Although the biochemical roles of thioesterase III have been investigated, the mechanisms that regulate ybaW expression are unknown. A putative FadR binding site is found upstream of ybaW, but its role in regulation was untested. Moreover, it was reported that the activity of thioesterase III is 20-fold higher in cells grown on the major CLA isomer, 9-cis,11-trans-octadecadienoic acid, than on cells grown on oleate, suggesting a second level of regulation. We report that ybaW is indeed expressed from a FadR-responsive promoter but find that induction of ybaW expression by the CLA isomer is weak. We also show that ybaW expression is negatively controlled by CRP-cAMP, in contrast to the other fad regulon genes, which are positively regulated by this transcription complex. The ybaW gene is clearly a member of the fad regulon, and we propose that it should be named fadM.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are derivatives of E. coli K-12 (Table 1). The media were LB medium (Luria-Bertani medium containing 10 g of tryptone, 5 g of yeast extract, and 10 g of NaCl per liter), Rich broth (RB medium containing 10 g of tryptone, 1 g of yeast extract, and 5 g of NaCl per liter), or minimal medium M9 (28) supplemented with 0.4% glucose, 0.1% vitamin-free Casamino Acids, 1 mM MgSO4, 0.1 mM CaCl2, and 0.001% thiamine. Fatty acids (e.g., oleic acid) (Sigma) were neutralized with KOH, solubilized with Tergitol NP-40, and used at a 5 mM final concentration for induction assays. Antibiotics were used at the following concentrations (in mg/liter): ampicillin, 100; kanamycin, 50; tetracycline, 15; and chloramphenicol, 20.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| E. coli strains | ||
| MG1655 | Wild-type K-12 strain | Laboratory stock |
| UB1005 | metB1 spoT1 relA1 spoT1 gyrA216 | 25 |
| MFH8 | UB1005 fadR::Tn10 | 25 |
| MFH9 | UB1005 fadR::Tn5 | 25 |
| SI91 | UB1005 fabR::Cm Cmr | Laboratory stock |
| MC1061 | Δ(codB-lacI)3 | 6 |
| Topo10 | lacX74 | Invitrogen |
| BL21(DE3) | Expression host for protein production | Laboratory stock |
| MC4100 | araD139 Δ(argF-lac)169 | Laboratory stock |
| RH74 | MC4100 Δcya-851 | Laboratory stock |
| RH77 | MC4100 Δcya-851 Δcrp | Laboratory stock |
| SI203 | MC1061 fadBA-lacZ fusion | Laboratory stock |
| LE392 | Wild-type strain | 9 |
| HC70 | LE392 tesA::Km tesB::Cm | 9 |
| FYJ14 | MC1061 ybaW::Km | This work |
| FYJ15 | MC1061 ΔybaW | This work |
| FYJ16 | FYJ15 carrying pCP20 | This work |
| FYJ17 | FYJ15 ybaW-lacZ fusion | This work |
| FYJ18 | FYJ17 fadR::Tn10 | This work |
| FYJ19 | FYJ17 fabR::Cm | This work |
| FYJ20 | MC4100 ybaW-lacZ fusion | This work |
| FYJ21 | RH74 ybaW-lacZ fusion Δcya-851 | This work |
| FYJ22 | RH77 ybaW-lacZ fusion Δcya-851 Δcrp | This work |
| FYJ23 | FYJ20 fadR::Tn10 | This work |
| Plasmids | ||
| pCR2.1-TOPO | Topo cloning vector | Invitrogen |
| pKD46 | λred recombinase plasmid, temperature sensitive | 15 |
| pKD4 | bla FRT ahp FRT oriR6K | 15 |
| pCP20 | bla cat cI857 λPRflp oriTS | 7, 19 |
| pCE70 | ahp FRT lacZY+ thisoriR6K | 19 |
| pET28-fadRec | Expression plasmid encoding E. coli fadR | This work |
| pET28-fabRec | Expression plasmid encoding E. coli fabR | This work |
Plasmids and DNA manipulations.
The chromosomal ybaW-lacZ transcriptional fusion strain FYJ17 (Table 1) was constructed in two steps. First, a ybaW::Km strain was constructed using the λ Red system of Datsenko and Wanner (15). Primers ybaW_fusion-F and ybaW_fusion-R, containing ybaW sequences at their 5′ ends (Table 2), were designed to amplify the kanamycin resistance cassette using plasmid pKD4 as the template. The PCR products were purified using a Qiagen gel purification kit and then electroporated into competent cells of strain MC1061 carrying the temperature-sensitive λred plasmid, pKD46 (15). Kanamycin-resistant colonies were selected, one of which became the ybaW::Km strain FYJ14 (Table 1). The construct was verified by PCR analyses and direct DNA sequencing of PCR products. Subsequently, the kanamycin resistance cassette in strain FYJ14 was removed by the expression of FLP recombinase from plasmid pCP20 to give strain FYJ15, which retained a single FLP recombinase target (FRT) site. In the second step, the FRT site was used for site-specific integration of the lacZ fusion plasmid, pCE70, which contains an FRT site upstream of the lacZY genes that lack a promoter, a kanamycin resistance gene, and the R6K origin of replication (19). Finally, the transformants were plated on LB agar plates containing kanamycin and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside at 37°C to give the ybaW-lacZ fusion strain FYJ17 (Table 1), which carries a lacZ fusion plasmid integrated at the ybaW chromosomal location. The fusion is stable due to lack of function of the R6K origin in the host strain and the loss of the temperature-sensitive pCP20 plasmid (19).
TABLE 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| ybaW-F | 5′-CAT CTC GAC GTT TAC CAG CAC G-3′ |
| ybaW-R | 5′-GTT AAC AGG TCA CTT AAT ACC GCT G-3′ |
| ybaW_P-F | 5′-TCT GGC GGT ATT AAC CCT GT-3′ |
| ybaW_P-R | 5′-GTG TTT GCA TAG CGC AAT AAC-3′ |
| 16S-F | 5′-GCA TAA CGT CGC AAG ACC AAA G-3′ |
| 16S-R | 5′-TTC TTC ATA CAC GCG GCA TGG-3′ |
| ybaW-F2 | 5′-AGC TGG AGC AGA TGG TTA AG-3′ |
| ybaW-GSP | 5′-TTA CTT AAC CAT CTG CTC CAG-3′ |
| AAPa | 5′-GGC CAC GCG TCG ACT AGT ACG GGI IGG GII GGG IIG-3′ |
| AUAP | 5′-GGC CAC GCG TCG ACT AGT AC-3′ |
| RLM-RACE Adaptor | 5′-GCU GAU GGC GAU GAA UGA ACA CUG CGU UUG CUG GCU UUG AUG AAA-3′ |
| RLM-RACE Outer | 5′-GCT GAT GGC GAT GAA TGA ACA CTG-3′ |
| RLM-RACE Inner | 5′-CGC GGA TCC GAA CAC TGC GTT TGC TGG CTT TGA TG-3′ |
| ybaW-check1 | 5′-CAA CAA CGT AAG GTT ATT GCG C-3′ |
| ybaW-check2 | 5′-AG CAT CCG GCA CCA CAA AAC-3′ |
| ybaV-F | 5′-AGC ACT GCT CAT TAC CCT GTC-3′ (18-38) |
| ybaV-R | 5′-CAG TTT TAA ACG GAC CGT ACT C-3′ (277-298) |
| queC-F | 5′-GGG CAT GGC GAA AGA TAT TCG-3′ (444-464) |
| queC-R | 5′-CTC AAC CCG GTT TTC TGC TTC-3′ (678-693) |
| ybaW_fusion-F | 5′-TAC CAG TTA TGA CCT CTG TAC TTA TAA CAA CAA CGT AAG GTT ATT GCG CT TGT GTA GGC TGG AGC TGC TT-3′ |
| ybaW_fusion-R | 5′-TAT TCC GGG TGT CGC CGG ATG CGG CTT GAG CAT CCG GCA CCA CAA AAC GT CAT ATG AAT ATC CTC CTT AG-3′ |
The letter “I” in the AAP primer denotes deoxyinosine, which is included in an improved anchor primer to eliminate the need for the mixture of anchor and adaptor primers used in the previous 5′-RACE version. Numbers in parentheses are the locations of the primers within the coding sequence of the gene of interest.
P1vir phage transduction.
Phage P1vir transductions were carried out as described by Miller (34), with minor modifications. Strains FYJ18 and FYJ19 were generated by P1vir transduction of strain FYJ17, using independent lysates grown on strains MFH8 (fadR::Tn10) and SI91 (fabR::Cm). Three strains, FYJ20, FYJ21, and FYJ22, were obtained by P1vir transduction of strains MC4100, RH74, and RH77, respectively, with a lysate grown on strain FYJ17 and selection for kanamycin resistance (Table 1). Strain FYJ23 was generated by transduction with a P1vir lysate grown on strain MFH8 (fadR::Tn10) of strain MC4100 with selection for tetracycline resistance.
β-Galactosidase and thioesterase assays.
Overnight cultures grown in either RB or minimal medium (with or without various fatty acids as sole carbon sources) were diluted 100-fold into the same medium and shaken at 37°C. When the cultures reached the mid-log phase, the collected cells were washed twice and assayed for β-galactosidase activity after sodium dodecyl sulfate-chloroform lysis (25, 28, 34). The data were collected in triplicate in three independent experiments. For thioesterase III assays, strain HC70, a tesA tesB double mutant lacking both thioesterase I (TesA) and thioesterase II (TesB) (Table 1), was used to compare enzymatic activities following growth on glucose, oleic acid (Sigma), or 9-cis,11-trans-octadecadienoic acid [9(Z),11(E)-octadecadienoic acid from Matreya LLC], a CLA isomer. The cells of 20-ml cultures were collected by centrifugation, washed three times with potassium phosphate buffer (0.1 M, pH 7.4), and then concentrated into 2 ml. After three rounds of passage through a French pressure apparatus, the extract was centrifuged (21,000 × g for 30 min) to remove debris. The concentration of cell-free crude extract was determined by using a Bio-Rad protein assay kit.
Assays for bacterial thioesterase III activity were performed as described by Nie et al. (36). The 500-μl-total-volume enzymatic reaction system was composed of crude extract, 6.8 μg/ml; acyl-CoA, 20 μM; 5,5′-dithiobis-2-nitrobenzoic acid, 0.4 mM; and potassium phosphate buffer (pH 7.4), 100 mM. The assay measures the free thiol of the CoA resulting from thioesterase action on the acyl-CoA substrate. The background due to thiols in the extracts was subtracted using samples lacking acyl-CoA. The reactions were run in triplicate at 30°C for 10 min. The data were recorded at 412 mM, and the molar extinction coefficient used was 13,600 M−1 cm−1 (9, 36).
RNA isolation and reverse transcription (RT)-PCR.
Bacterial RNA extraction was carried out as recommended by Qiagen. Briefly, the cells of 0.5-ml cultures were collected by a brief centrifugation and the pellets were suspended in 100 μl of Tris-EDTA buffer containing 400 μg/ml of lysozyme for 10 min at room temperature. RNeasy lysis buffer (350 μl, containing guanadine thiocyanate to inactivate nucleases) and ethanol (250 μl) were added successively, and the resulting mixtures (∼700 μl) were loaded on RNeasy mini columns (Qiagen). After the columns were washed, the total RNA samples were eluted in RNase-free water and treated with RNase-free DNase I (at room temperature for ∼1 h). The RNA samples were analyzed by 0.8% agarose gel electrophoresis to assess the quality of the rRNA profiles. To rule out the possibility of residual DNA contamination, general PCR-based detection was performed using total RNA samples as template and a pair of specific primers (16S-F/16S-R) that hybridize to chromosomal sequences that lie outside the sequence of the mature 16S rRNA such that a negative result indicated the absence of detectable contamination with genomic DNA.
The RNA samples that survived validation were subjected to RT-PCR analyses as follows: 1 μg of RNA was mixed with 0.5 μg of random primers (22 μl total volume), denatured (70°C for 5 min), and then chilled on ice (5 min). Reverse transcription was then done with a reaction mixture of denatured RNA template, 20 μl; random primers, 2 μl; ImProm-II 5× reaction buffer, 8 μl; MgCl2, 5 μl; deoxynucleoside triphosphate mix, 2 μl; recombinant RNasin RNase inhibitor, 1 μl; ImProm-II reverse transcriptase, 2 μl using a program consisting of equilibration at 25°C for 5 min, extension at 42°C for 60 min, and inactivation of enzyme at 70°C for 15 min. Finally, 1 μl of cDNA served as template for PCR amplification of the genes of interest using specific primers (Table 2) on an Eppendorf thermal cycler.
Real-time qPCR.
Assay of in vivo levels of ybaW transcription by real-time quantitative PCR (qPCR) used the SYBR green method (20). The qPCR reaction system (20 μl) consisted of the following components: 12.5 μl of iQ SYBR green Supermix, 1 μl each of the primers, 1 μl of the diluted cDNA sample, and 4.5 μl of sterile water. All analyses were performed in triplicate on a Mastercycler ep realplex instrument (Eppendorf) using a program consisting of a denaturing cycle at 95°C for 15 min; 45 cycles comprising 94°C for 20 s, 60°C for 20 s, and 72°C for 20 s; and a final step in which temperature was elevated on a gradient from 60°C to 90°C to dissociate double-stranded DNA products. The internal reference was the16S rRNA gene (Table 2), and water functioned as blank sample to monitor cross-contamination of cDNA samples. The relative transcription levels were measured by the method of Livak and Schmittgen (32).
5′-RACE and RLM-RACE.
For determination of transcriptional start sites of the ybaW gene, RNA samples were isolated from strains MG1655 (wild type) and MFH9 (fadR) plus strain MG1655 grown on minimal medium with oleic acid as the sole carbon source. Random amplification of cDNA ends (5′-RACE) was performed using two complementary kits, 5′-RACE (Invitrogen) and Ambion RNA ligase-mediated (RLM)-RACE (Applied Biosystems).
For 5′-RACE analyses, total RNA samples from overnight cultures were prepared for the first-strand cDNA synthesis with a specific ybaW reverse primer (ybaW-GSP) (Table 2). Following purification of the cDNA on a small nucleic acid purification column (S.N.A.P. column from Invitrogen), oligo(dC) tails were added to the 5′ end of the first strand of cDNA via a terminal transferase-mediated reaction at 65°C. The oligo(dC)-tailed cDNA was amplified by PCR using an abridged anchor primer (primer AAP) plus the nested ybaW primer, ybaW-R. Subsequently, nested PCR was performed using the 20-fold-diluted PCR products obtained as described above with primers ybaW-R and AUAP (Table 2). The PCR amplification cycling protocol was a predenaturing step at 94°C for 5 min and 35 cycles consisting of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, followed by a final extension step at 72°C for 8 min. The resulting PCR products were directly cloned into pCR2.1-TOPO vectors (Invitrogen) for further DNA sequencing. The first nucleotide after the oligo(dG) sequence on the complementary strand is taken as the transcriptional start site (43).
In the RLM-RACE method, the full-length mRNA samples were obtained as described in the manufacturer's protocol. After removal of the 5′-phosphate by calf intestinal phosphatase from any rRNA, tRNA, or degraded mRNA species present and inactivation of the phosphatase, the mRNA 5′-triphosphates of the intact mRNAs were converted to 5′-monophosphates by tobacco acid pyrophosphatase treatment. The RNA molecules that carried a 5′-monophosphate were then ligated to the 5′-RACE adaptor using RNA ligase. Reverse transcription was then done using random primers as recommended by the manufacturer. The resulting PCR products were used as a template for nested PCR using an outer 5′-RLM-RACE PCR (with primers 5′-RLM-RACE Outer and ybaW-GSP) followed by an inner 5′-RLM-RACE PCR (with primers 5′-RLM-RACE Inner and fabW-R) (Table 2). The final PCR products were cloned into vector pCR2.1 TOPO for direct DNA sequencing. The first nucleotide after the 5′-RACE adaptor was taken as the transcriptional start site.
In vitro translation of FabR protein.
Our preliminary studies have shown that it is difficult to obtain E. coli FabR protein in a soluble and functional form using expression in E. coli. Thus, we utilized a PURExpress in vitro protein synthesis kit (New England Biolabs), a cell-free transcription/translation system in which all components are well defined. The reaction system (total volume, 25 μl), in which the gel-purified pET28-fabRec plasmid (∼1 μg) was mixed with 12.5 μl of solution A and 5.0 μl of solution B, was kept at 37°C for 1.5 h. The protein synthesized was used immediately for functional analyses.
Electrophoretic mobility shift assays.
Gel shift assays were used to test whether the ybaW promoter region interacted with the two regulatory proteins FadR and FabR. A DNA fragment covering the ybaW promoter region was obtained by PCR using primers ybaW_P-F and ybaW_P-R (Table 2) and labeled by terminal transferase treatment (37°C for 15 min) with digoxigenin-ddUTP (Roche) as substrate. The digoxigenin-labeled DNA (∼ 3.8 pmol) was incubated with serial dilutions of either purified FadR protein or in vitro-translated FabR protein in 5× binding buffer [which has the composition 100 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.6, 5 mM EDTA, 50 mM (NH4)2SO4, 5 mM dithiothreitol, 150 mM KCl, and 1% (vol/wt) Tween 20] at room temperature for 15 min. The DNA-protein mixtures were separated by electrophoresis of a 6% native polyacrylamide gel electrophoresis gel in 0.5× Tris-borate-EDTA buffer for 3.5 h (90 V) after the gel had been prerun for ∼1.5 h. The DNA samples were transferred from the gel to an equilibrated, positively charged nylon membrane (Roche) by contact blotting followed by UV cross-linking (120 mJ for 180 s). The membrane was rinsed in washing buffer for 5 min followed by 30 min of incubation with blocking buffer. The membrane was then incubated with an antidigoxigenin antibody solution (diluted 1:10,000) at room temperature for ∼1 h, washed five times using 50 ml of washing buffer (15 min each), and equilibrated with 30 ml of detection buffer for 5 min before development of the luminescent reaction in 1 ml of CSPD working solution (Roche) at 37°C for 10 min. Finally, the signals were exposed to high-performance chemiluminescence film (Amersham Hyperfilm ECL).
RESULTS
Genetic organization and transcriptional analyses of ybaW.
The E. coli genome sequence shows that ybaW is located between ybaV and queC (Fig. 2). The upstream gene, ybaV, is predicted to encode a ComEA competence protein homolog (2), whereas the downstream queC gene (encoded on the other DNA strand) plays a role in an initial step of the biosynthesis of the modified ribonucleoside, queuosine (22). The ybaW gene encodes a small (132 residue) protein recently demonstrated to be a novel thioesterase (thioesterase III) that plays an editing role in β-oxidation (35, 36) and has homologues in species closely related to E. coli.
FIG. 2.
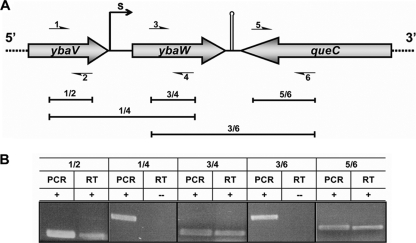
Transcriptional analyses of the ybaW gene and neighboring loci. (A) Genetic organization of the ybaW gene and the neighboring loci. The gray arrows represent open reading frames, and the small, numbered arrows correspond to the primers. 1, ybaV-F; 2, ybaV-R; 3, ybaW-F; 4, ybaW-R; 5, queC-F; 6, queC-R (Table 2). The primer combinations 1/2, 3/4, 5/6, 1/4, and 3/6 are shown. S denotes the transcriptional start site. The hairpin indicates a predicted terminator. The primer pairs used are shown above the brackets. (B) Results of PCR and RT-PCR (RT) analyses of the ybaW gene and its two neighboring genes.
To address the possibility that ybaW may be cotranscribed with ybaV, PCR and RT-PCR assays were performed using three primer pairs (Table 2 and Fig. 2A). Positive amplifications using the three primer sets 1/2, 3/4, and 5/6 were obtained by both PCR and RT-PCR, which showed that all three genes are transcribed (Fig. 2B). However, the 1/4- and 3/6-primed amplicons were observed only with PCR and not with RT-PCR (Fig. 2B), indicating that ybaW is a monocistronic transcriptional unit.
The ybaW transcriptional initiation site.
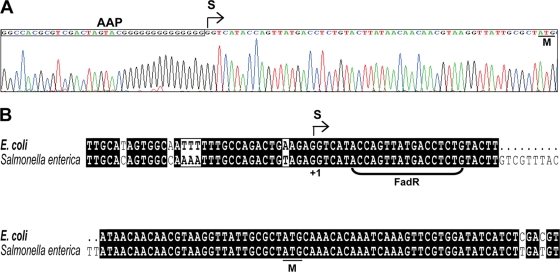
Total RNA samples from an E. coli wild-type strain (grown on either LB medium or on minimal medium with oleic acid as sole carbon source) and a fadR null mutant strain were used to map the ybaW transcriptional start site using two techniques, 5′-RACE and RLM-RACE, the second of which requires that the RNAs detected originally carried a 5′-triphosphate (and thus were primary transcription products rather than processed species). Although several truncated ybaW transcripts were observed (data not shown), the same start site 55 nucleotides upstream of the ybaW translation initiation codon was found by both methods in all strains tested (Fig. 3A), a result in complete agreement with that predicted by prokaryotic promoter analysis with the Neutral Network Program of Promoter Prediction (http://www.fruitfly.org/seq_tools/promoter.html). A putative FadR binding site (5′-ACCAGTTATGACCTCTG-3′, where the underlined bases were found in at least 80% of the in vitro-selected oligonucleotides that define the consensus binding site [see reference 23]), was found downstream of the determined transcriptional start site in a position where FadR binding could impede RNA polymerase action (Fig. 3B). Moreover, the ybaW promoter and putative FadR binding sequences of E. coli and Salmonella enterica were highly similar (Fig. 3B). However, a caveat is that FadR binding sites show less sequence conservation than is generally seen with other sequences that specifically bind a given regulatory protein (3, 23, 42) (Fig. 4A).
FIG. 3.
Mapping of the ybaW transcriptional start site. (A) Result of 5′-RACE-based mapping of the transcription start site of the ybaW gene. The sequencing profile of the product is shown. The AAP sequence is within the rectangle. The same start site was found in the wild-type strain in the presence or absence of oleate induction and in a ΔfadR strain. RLM-RACE analyses (data not shown) also gave the same start site. The site identified was identical to that predicted by the Neutral Program of Promoter Prediction (http://www.fruitfly.org/seq_tools/promoter.html). (B) Sequence alignments of ybaW promoter regions in E. coli and Salmonella enterica. The bracketed sequence is the putative FadR binding site. S denotes the transcriptional start site, and M denotes the ybaW initiation codon. Black and gray shading shows identity and similarity, respectively. Boxes highlight differences, and dots show deletions relative to the other sequence.
FIG. 4.
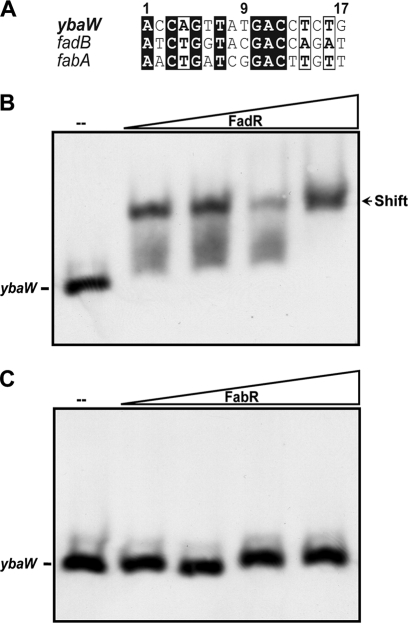
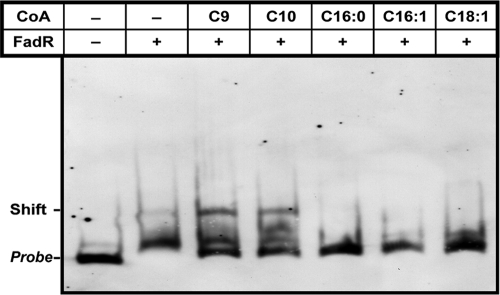
FadR binds the ybaW promoter region. (A) Comparison of FadR binding sites in ybaW, fadB, and fabA. The sequence is highlighted as described for Fig. 3B. (B) Gel shift assay of the interaction of the ybaW promoter region with FadR. (C) Gel shift assay of the ybaW promoter region with FabR. Both the FadR and FabR proteins were used at concentrations of 0 (−), 0.1, 0.5, 1.0, and 2.0 μM, denoted by the triangles. The DNA probe is a ybaW promoter region of 125 bp obtained by amplification with primers ybaW_P-F and ybaW_P-R (Table 2) and then end labeled. The arrow indicates the shifted DNA-protein complex. A representative result from three independent experiments is shown.
The ybaW promoter region binds FadR, and this binding is specifically reversed by long-chain acyl-CoAs.
The function of the putative FadR binding site of the ybaW promoter region (Fig. 3B) was tested in electrophoretic mobility shift assays using purified FadR (Fig. 4). We amplified and digoxigenin end labeled a promoter region DNA fragment of ∼120 bp that overlapped the candidate FadR binding site. Using this probe, we found that the DNA fragment bound FadR regulator in a dose-dependent manner (Fig. 4B). The interaction was specific in that the DNA fragment failed to bind FabR protein (Fig. 4C) and FadR-DNA interaction was reversed by acyl-CoAs having long acyl chains (Fig. 5). As discussed above, long-chain but not short-chain acyl-CoAs regulate DNA binding by FadR (12, 25), and hence, we tested five acyl-CoA species of differing acyl chain lengths. The results of gel shift experiments showed that the short-chain acyl-CoAs (C9:0 and C10:0) did not interfere with FadR binding to the ybaW promoter region, whereas the long-chain acyl-CoA species (C16:0, C16:1, and C18:1) strongly impaired DNA binding (Fig. 5). Therefore, these in vitro data indicate that long-chain acyl-CoAs regulate ybaW transcription through their interaction with FadR.
FIG. 5.
Functional impairment of interaction between FadR and the ybaW promoter by long-chain fatty acyl-CoA species. The assay mixtures contained labeled probe, 1 μM; FadR protein, 3 μM; and acyl-CoA, 10 μM. +, present; −, absent.
YbaW expression is directly repressed by FadR.
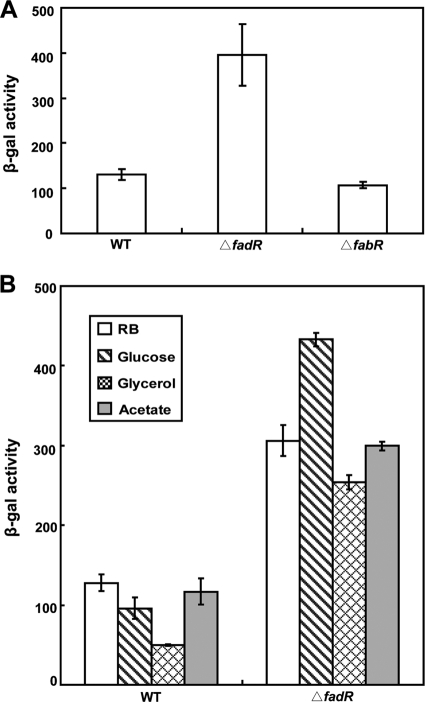
Nie and coworkers (35), working with an E. coli strain lacking the major thioesterases (TesA and TesB), reported that growth of the strain on oleate gave a modest (3.6-fold) increase in thioesterase activity, assayed by tetradecanoyl-CoA hydrolysis, that was attributed to YbaW. However, ion exchange fractionation showed the presence of two additional peaks of thioesterase activity present in extracts of both the tesA tesB strain and a tesA tesB ybaW strain that could have contributed a portion of the activity detected (35). Moreover, the control strain was grown on glucose, which generally decreases fad gene expression by lowering cAMP levels (10, 38). Hence, the levels of induction of ybaW expression by long-chain fatty acids and FadR were unclear. We used a strain carrying a chromosomal ybaW-lacZY transcriptional fusion to assay ybaW expression. Derivatives of this strain with an otherwise intact fad regulon and isogenic strains carrying null mutations in either fadR or fabR (which encodes a regulator of fatty acid biosynthesis [42]) served as controls. The level of ybaW transcription in the fadR strain was about threefold higher than the levels in the wild-type and fabR strains (Fig. 6A), indicating that ybaW is a member of the fad regulon. We propose that the gene should be named fadM and will use this designation for the remainder of this paper except when referring to the prior literature.
FIG. 6.
ybaW transcription is repressed by FadR. (A) The effects of the two regulatory proteins FadR and FabR on ybaW expression. (B) Levels of expression of fabM in a fadR strain grown on various sole carbon sources are shown. Expression was monitored using the fabM-lacZ transcriptional fusion strain and β-galactosidase (β-Gal) assays, which were performed in triplicate. Levels of activity are shown in Miller units, and error bars show standard deviations. WT, wild type.
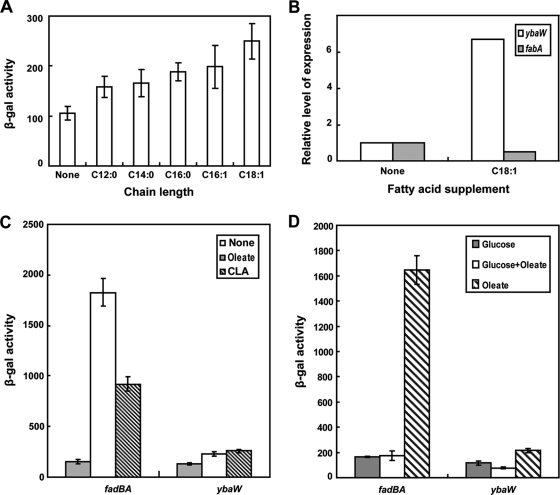
The levels of fadM induction of the fadM-lacZY fusion strain were largely independent of the carbon source (Fig. 6B) except that growth on fatty acids as the sole carbon source induced fadM expression and growth on glycerol resulted in decreased expression (Fig. 7). Five fatty acids of various chain lengths and levels of saturation (dodecanoic acid, C12:0; tetradecanoic acid, C14:0; hexadecanoic [palmitic] acid, C16:0; palmitoleic acid, C16:1; and oleic acid, C18:1) were used as sole carbon sources, and fadM expression was measured using the fadM-lacZ transcriptional fusion strain. β-Galactosidase assays showed that, relative to the level of induction in cells grown on glucose as the carbon source, C18:1 was the best inducer, C16:0 and C16:1 were less effective and equivalent to eachother, and C12:0 and C14:0 were relatively poor inducers (Fig. 7A). Induction of fadM expression by oleate was confirmed by qPCR, which also showed the expected decrease in transcription of the fabA fatty acid biosynthesis gene (25, 26) (Fig. 7B). Taken together, the levels of fadM induction were in good agreement with the enzymatic activity measurements of Nie et al. (36). However, Nie et al. (36) also reported that growth on 9-cis,11-trans-octadecadienoic acid (one of the major CLA isomers) gave 10-fold higher levels of YbaW/FadM thioesterase activity than did oleic acid. This result was surprising since other fad operon genes are fully derepressed in oleate-grown cells, and thus, we tested the effects of growth on the conjugated acid on the expression of fadBA, which are the most highly regulated genes of the regulon, and on fadM. In contrast to the report of Nie (36), we found that the levels of fadM induction by the CLA isomer were similar to those seen in oleic acid-grown cells, whereas the conjugated acid induced fadBA only half as well as oleate (Fig. 7C). We also compared thioesterase levels in a tesA tesB strain (which lacks the two major thioesterases of E. coli) grown on either oleate or the CLA isomer and found that the levels of induction were essentially identical when assayed using three different acyl-CoAs as substrates (Table 3). These results are consistent with those obtained using the fadM-lacZY fusion strain.
FIG. 7.
Induction of fadM transcription. The cultures grown without a fatty acid carbon source were grown on glucose. (A) β-Galactosidase activities of a chromosomal fabM-lacZ transcriptional fusion strain grown on different fatty acids. (B) RT-qPCR analyses of fabM expression induced by oleate. The level of expression of fabA, which is known to be repressed upon the addition of oleate (25), serves as a control. Each sample was assayed in triplicate. (C) Effects of CLA on fad gene expression. (D) Effects of glucose on fad gene expression. The data are the means of the results of three independent assays. Levels of β-galactosidase activity are shown in Miller units, and error bars show standard deviations.
TABLE 3.
Thioesterase III activities of crude extracts of cultures of the tesA tesB strain HC70 grown on different carbon sourcesa
| Substrate | Sp act (mU/mg protein) with indicated carbon source
|
||
|---|---|---|---|
| Glucose | Oleic acid | CLA | |
| Palmitoyl-CoA | 11.4 ± 0.7 | 50.3 ± 2.8 | 59.1 ± 2.4 |
| Palmitoleoyl-CoA | 17.0 ± 1.5 | 91.1 ± 2.8 | 97.4 ± 4.2 |
| Oleoyl-CoA | 19.9 ± 3.6 | 67.0 ± 5.4 | 64.6 ± 3.1 |
The thioesterase assays were performed in triplicate, and the data are expressed as the means ± standard deviations of the results.
Inhibition of fadM transcription by CRP-cAMP.
The fadM expression levels of a ΔfadR strain were similar in glucose-grown and acetate-grown cells (Fig. 6B), which is atypical of fad gene regulation. Clark (10) showed that the levels of expression of both fadBA and fadE were decreased 8- to10-fold when glucose rather than acetate was the sole carbon source. Pauli et al. (38) reported similar data for FadD enzyme activity. However, in common with findings for other fad regulon genes (10, 38), induction of fadM expression by oleate was blocked by the addition of glucose (Fig. 7D). Thus, the induction of fadM expression was inhibited by glucose, although glucose does not lower the basal level of expression (taken as the level in the acetate-grown cells, since fatty acids are metabolized as acetate units). Glucose inhibition of the expression of other fad regulon genes has been shown to be exerted through CRP-cAMP (10, 38), and thus, we examined the role of this complex in fadM expression by using both the fadM-lacZY fusion construct and real-time qPCR assays of transcription. In vivo transcription analyses were carried out using two mutant strains, strain RH74, which lacks cya, the gene that encodes the adenylate cyclase responsible for conversion of AMP into cAMP, and strain RH77, which lacks both cya and crp, the gene encoding CRP. Analyses using the fadM-lacZ fusion introduced into the strains indicated that, relative to the levels in the wild-type parent, the levels of fadM expression were elevated in the cya strain by three- to fourfold, whereas the levels in the cya crp strain were elevated about twofold (Table 4). The results of real-time qPCR assays were in general agreement with the lacZ fusion results (Table 4). Therefore, unlike the expression of the typical fad regulon genes that are activated by CRP-cAMP, the basal expression of fadM was repressed by the complex. The intact complex was required for repression; the addition of 1 mM cAMP to the medium of the cya strain resulted in repression, but only when CRP was available (Table 5). The addition of cAMP gave a slight but reproducible decrease in fadM expression in the wild-type and ΔfadR strains. Moreover, glucose metabolism was required. Substitution of the nonmetabolizable glucose analogue α-methyl-d-glucoside for glucose had no effect on expression in the wild-type or fadR strain or the strains lacking CRP-cAMP (data not shown).
TABLE 4.
Inhibition of fadM transcription by the cAMP-CRP complexa
| Strain | Transcriptional level of fadM in indicated assay
|
|
|---|---|---|
| LacZ activity | RT-qPCR | |
| Wild type | 1.00 ± 0.07 | 1.00 ± 0.05 |
| ΔcyaA strain | 3.65 ± 0.31 | 5.68 ± 0.66 |
| ΔcyaA Δcrp strain | 2.07 ± 0.27 | 3.65 ± 0.38 |
Data are shown relative to the results for the wild type. LacZ activity was measured as β-galactosidase activity; the activity of the wild-type strain was 138.5 ± 10 Miller units. The β-galactosidase activities are the means ± standard deviations of the results of three independent assays, whereas each sample of the RT-qPCR experiments was analyzed in triplicate. The strains used for LacZ activity measurements were FYJ20, FYJ21, and FYJ22, whereas those used for RT-qPCR analyses were MC4100, RH74, and RH77.
TABLE 5.
Effect of exogenous cAMP on fadM expression measured using fadM-lacZ fusion strainsa
| Medium | cAMP | β-Galactosidase activity (Miller units) in:
|
|||
|---|---|---|---|---|---|
| Wild type | ΔcyaA strain | ΔcyaA Δcrp strain | ΔfadR strain | ||
| RB | − | 131 ± 11 | 387 ± 61 | 240 ± 40 | 351 ± 43 |
| M9 | − | 130 ± 12 | 363 ± 35 | 256 ± 14 | 364 ± 38 |
| M9 | + | 97 ± 5 | 117 ± 7 | 239 ± 14 | 277 ± 21 |
The cultures were grown in either RB medium or glucose minimal medium (M9). Growth in the latter medium was either in the absence (−) or presence (+) of 1 mM cAMP. The β-galactosidase activities are the means ± standard deviations of the results of three independent assays. The strains used for LacZ activity measurements were FYJ20, FYJ21, FYJ22, and FYJ23.
DISCUSSION
Our data indicate that ybaW, which we now propose be called fadM, is a new member of the fad regulon. The expression of fadM has characteristics in common with the expression of the other members of the regulon but differs markedly in other parameters. FadM is only weakly induced by oleate, as shown by both transcriptional fusion strain and enzymatic assays (Fig. 7 and Table 3). The FadM induction levels are similar to those of fadL and fadD, two genes required for exogenous fatty acids to enter the β-oxidation pathway, and are markedly lower than that of fadBA. The weak FadR regulation of fadL and fadD induction is readily rationalized by the fact that the induction of the fad regulon is triggered by the accumulation of long-chain acyl-CoAs. Therefore, if fadL and fadD expression was tightly regulated by FadR, induction would fail or be very sluggish because of a lack of the acyl-CoAs needed to neutralize the repressor (Fig. 1). In contrast, fadBA induction by oleate is very robust (10- to 20-fold). The locations and number of the FadR binding sites in the fad regulon promoter regions do not correlate with induction levels in any straightforward manner. Like the highly regulated fadBA promoter, the FadR binding site of fadM overlaps the transcriptional initiation site and, also, DNA sequences downstream of that site. In contrast, the weakly regulated fadD and fadL genes each have two FadR binding sites (18).
The fact that its induction levels resemble those of FadL and FadD suggests that FadM has a nonessential cellular role in addition to its detoxification functions during β-oxidation of unsaturated fatty acids. One possibility is that FadM may cleave inappropriate acyl-acyl carrier protein species. However, activity on acyl-acyl carrier protein substrates has not yet been tested. Another indication that FadM may have a cellular role other than in β-oxidation is that its basal expression is not markedly decreased by growth in the presence of glucose, although glucose does block induction by oleate. This is in contrast to the other fad regulon genes, where glucose addition both decreases the basal levels of expression and blocks induction by oleate. Moreover, CRP-cAMP negatively regulates fadM, whereas the complex acts as a positive regulator of the other genes of the regulon. Since we failed to find a CRP-cAMP consensus binding site region (21, 29, 39) in the fadM promoter (or within 500 bp upstream of the coding sequence), we believe that regulation of fadM expression by CRP-cAMP is indirect. This is unusual, but not without precedent. Some of the genes of the E. coli citric acid cycle show clear regulation by CRP-cAMP but lack a recognizable canonical CRP binding site (14), and in a class of promoters, CRP-cAMP activates transcription by binding to noncanonical DNA sequences with the aid of the Sxy protein (2). Perhaps fadM regulation also requires a protein to partner CRP-cAMP.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grant AI15650.
Footnotes
Published ahead of print on 14 August 2009.
REFERENCES
- 1.Beckwith, J. R. 1967. Regulation of the lac operon. Recent studies on the regulation of lactose metabolism in Escherichia coli support the operon model. Science 156:597-604. [DOI] [PubMed] [Google Scholar]
- 2.Cameron, A. D., and R. J. Redfield. 2006. Non-canonical CRP sites control competence regulons in Escherichia coli and many other gamma-proteobacteria. Nucleic Acids Res. 34:6001-6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell, J. W., and J. E. Cronan, Jr. 2001. Escherichia coli FadR positively regulates transcription of the fabB fatty acid biosynthetic gene. J. Bacteriol. 183:5982-5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell, J. W., and J. E. Cronan, Jr. 2002. The enigmatic Escherichia coli fadE gene is yafH. J. Bacteriol. 184:3759-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, J. W., R. M. Morgan-Kiss, and J. E. Cronan, Jr. 2003. A new Escherichia coli metabolic competency: growth on fatty acids by a novel anaerobic beta-oxidation pathway. Mol. Microbiol. 47:793-805. [DOI] [PubMed] [Google Scholar]
- 6.Casadaban, M. J., J. Chou, and S. N. Cohen. 1980. In vitro gene fusions that join an enzymatically active β-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J. Bacteriol. 143:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 8.Cho, B. K., E. M. Knight, and B. O. Palsson. 2006. Transcriptional regulation of the fad regulon genes of Escherichia coli by ArcA. Microbiology 152:2207-2219. [DOI] [PubMed] [Google Scholar]
- 9.Cho, H., and J. E. Cronan, Jr. 1993. Escherichia coli thioesterase I, molecular cloning and sequencing of the structural gene and identification as a periplasmic enzyme. J. Biol. Chem. 268:9238-9245. [PubMed] [Google Scholar]
- 10.Clark, D. 1981. Regulation of fatty acid degradation in Escherichia coli: analysis by operon fusion. J. Bacteriol. 148:521-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark, D. P., and J. Cronan. October 2005, posting date. Chapter 3.4.4. Two-carbon compounds and fatty acids as carbon sources. In A. Böck, R. Curtiss III, J. Kaper, P. Karp, F. Neidhardt, T. Nyström, J. Slauch, C. Squires, and D. Ussery (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org/.
- 12.Cronan, J. E., Jr. 1997. In vivo evidence that acyl coenzyme A regulates DNA binding by the Escherichia coli FadR global transcription factor. J. Bacteriol. 179:1819-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cronan, J. E., Jr., and S. Subrahmanyam. 1998. FadR, transcriptional co-ordination of metabolic expediency. Mol. Microbiol. 29:937-943. [DOI] [PubMed] [Google Scholar]
- 14.Cronan, J. E., and D. C. LaPorte. November 2006, posting date. Chapter 3.5.2. Tricarboxylic acid cycle and glyoxylate bypass. In A. Böck, R. I. Curtiss, J. B. Kaper, P. D. Karp, F. C. Neidhardt, T. Nyström, J. M. Slauch, and C. L. Squires (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org/.
- 15.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deutscher, J. 2008. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 11:87-93. [DOI] [PubMed] [Google Scholar]
- 17.DiRusso, C. C., and W. D. Nunn. 1985. Cloning and characterization of a gene (fadR) involved in regulation of fatty acid metabolism in Escherichia coli. J. Bacteriol. 161:583-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiRusso, C. C., and T. Nystrom. 1998. The fats of Escherichia coli during infancy and old age: regulation by global regulators, alarmones and lipid intermediates. Mol. Microbiol. 27:1-8. [DOI] [PubMed] [Google Scholar]
- 19.Ellermeier, C. D., A. Janakiraman, and J. M. Slauch. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153-161. [DOI] [PubMed] [Google Scholar]
- 20.Feng, Y., M. Li, H. Zhang, B. Zheng, H. Han, C. Wang, J. Yan, J. Tang, and G. F. Gao. 2008. Functional definition and global regulation of Zur, a zinc uptake regulator in a Streptococcus suis serotype 2 strain causing streptococcal toxic shock syndrome. J. Bacteriol. 190:7567-7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaston, K., B. Chan, A. Kolb, J. Fox, and S. Busby. 1988. Alterations in the binding site of the cyclic AMP receptor protein at the Escherichia coli galactose operon regulatory region. Biochem. J. 253:809-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaur, R., and U. Varshney. 2005. Genetic analysis identifies a function for the queC (ybaX) gene product at an initial step in the queuosine biosynthetic pathway in Escherichia coli. J. Bacteriol. 187:6893-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gui, L., A. Sunnarborg, and D. C. LaPorte. 1996. Regulated expression of a repressor protein: FadR activates iclR. J. Bacteriol. 178:4704-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harman, J. G. 2001. Allosteric regulation of the cAMP receptor protein. Biochim. Biophys. Acta 1547:1-17. [DOI] [PubMed] [Google Scholar]
- 25.Henry, M. F., and J. E. Cronan, Jr. 1992. A new mechanism of transcriptional regulation: release of an activator triggered by small molecule binding. Cell 70:671-679. [DOI] [PubMed] [Google Scholar]
- 26.Henry, M. F., and J. E. Cronan, Jr. 1991. Escherichia coli transcription factor that both activates fatty acid synthesis and represses fatty acid degradation. J. Mol. Biol. 222:843-849. [DOI] [PubMed] [Google Scholar]
- 27.Higashitani, A., Y. Nishimura, H. Hara, H. Aiba, T. Mizuno, and K. Horiuchi. 1993. Osmoregulation of the fatty acid receptor gene fadL in Escherichia coli. Mol. Gen. Genet. 240:339-347. [DOI] [PubMed] [Google Scholar]
- 28.Iram, S. H., and J. E. Cronan. 2006. The β-oxidation systems of Escherichia coli and Salmonella enterica are not functionally equivalent. J. Bacteriol. 188:599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimata, K., H. Takahashi, T. Inada, P. Postma, and H. Aiba. 1997. cAMP receptor protein-cAMP plays a crucial role in glucose-lactose diauxie by activating the major glucose transporter gene in Escherichia coli. Proc. Natl. Acad. Sci. USA 94:12914-12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein, K., R. Steinberg, B. Fiethen, and P. Overath. 1971. Fatty acid degradation in Escherichia coli. An inducible system for the uptake of fatty acids and further characterization of old mutants. Eur. J. Biochem. 19:442-450. [DOI] [PubMed] [Google Scholar]
- 31.Li, L., V. N. Uversky, A. K. Dunker, and S. O. Meroueh. 2007. A computational investigation of allostery in the catabolite activator protein. J. Am. Chem. Soc. 129:15668-15676. [DOI] [PubMed] [Google Scholar]
- 32.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 33.Maloy, S. R., and W. D. Nunn. 1981. Role of gene fadR in Escherichia coli acetate metabolism. J. Bacteriol. 148:83-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, J. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 35.Nie, L., Y. Ren, A. Janakiraman, S. Smith, and H. Schulz. 2008. A novel paradigm of fatty acid beta-oxidation exemplified by the thioesterase-dependent partial degradation of conjugated linoleic acid that fully supports growth of Escherichia coli. Biochemistry 47:9618-9626. [DOI] [PubMed] [Google Scholar]
- 36.Nie, L., Y. Ren, and H. Schulz. 2008. Identification and characterization of Escherichia coli thioesterase III that functions in fatty acid beta-oxidation. Biochemistry 47:7744-7751. [DOI] [PubMed] [Google Scholar]
- 37.Overath, P., G. Pauli, and H. U. Schairer. 1969. Fatty acid degradation in Escherichia coli. An inducible acyl-CoA synthetase, the mapping of old-mutations, and the isolation of regulatory mutants. Eur. J. Biochem. 7:559-574. [PubMed] [Google Scholar]
- 38.Pauli, G., R. Ehring, and P. Overath. 1974. Fatty acid degradation in Escherichia coli: requirement of cyclic adenosine monophosphate and cyclic adenosine monophosphate receptor protein for enzyme synthesis. J. Bacteriol. 117:1178-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramseier, T. M., and M. H. Saier, Jr. 1995. cAMP-cAMP receptor protein complex: five binding sites in the control region of the Escherichia coli mannitol operon. Microbiology 141:1901-1907. [DOI] [PubMed] [Google Scholar]
- 40.Stulke, J., and W. Hillen. 1999. Carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 41.Vanden Boom, T., and J. E. Cronan, Jr. 1989. Genetics and regulation of bacterial lipid metabolism. Annu. Rev. Microbiol. 43:317-343. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, Y. M., H. Marrakchi, and C. O. Rock. 2002. The FabR (YijC) transcription factor regulates unsaturated fatty acid biosynthesis in Escherichia coli. J. Biol. Chem. 277:15558-15565. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, Y. M., K. Zhu, M. W. Frank, and C. O. Rock. 2007. A Pseudomonas aeruginosa transcription factor that senses fatty acid structure. Mol. Microbiol. 66:622-632. [DOI] [PubMed] [Google Scholar]