Abstract
We have generated a set of plasmids, based on the mobilizable shuttle vector pMIDG100, which can be used as tools for genetic manipulation of Actinobacillus pleuropneumoniae and other members of the Pasteurellaceae. A tandem reporter plasmid, pMC-Tandem, carrying promoterless xylE and gfpmut3 genes downstream of a multiple-cloning site (MCS), can be used for identification of transcriptional regulators and conditions which favor gene expression from different cloned promoters. The ability to detect transcriptional regulators using the tandem reporter system was validated in A. pleuropneumoniae using the cloned rpoE (σE) promoter (P). The resulting plasmid, pMCrpoEP, was used to identify a mutant defective in production of RseA, the negative regulator of σE, among a bank of random transposon mutants, as well as to detect induction of σE following exposure of A. pleuropneumoniae to ethanol or heat shock. pMCsodCP, carrying the cloned sodC promoter of A. pleuropneumoniae, was functional in A. pleuropneumoniae, Haemophilus influenzae, Haemophilus parasuis, Mannheimia haemolytica, and Pasteurella multocida. Two general expression vectors, pMK-Express and pMC-Express, which differ in their antibiotic resistance markers (kanamycin and chloramphenicol, respectively), were constructed for the Pasteurellaceae. Both plasmids have the A. pleuropneumoniae sodC promoter upstream of the gfpmut3 gene and an extended MCS. Replacement of gfpmut3 with a gene of interest allows complementation and heterologous gene expression, as evidenced by expression of the Haemophilus ducreyi nadV gene in A. pleuropneumoniae, rendering the latter NAD independent.
Actinobacillus pleuropneumoniae is the etiological agent of pleuropneumonia, an economically significant disease responsible for substantial morbidity and mortality in the worldwide pig industry (47). Understanding the molecular basis of pathogenicity is important in the design and implementation of vaccine and treatment strategies. Established virulence factors include surface polysaccharides (5, 24), Apx toxins (19, 29), iron uptake systems (24), components of anaerobic metabolism (3, 4, 10, 11, 23), and outer membrane proteins (12). In general, these have been discovered through hypothesis-driven research (9). As with other bacterial pathogens, further advances will be facilitated by the availability of microarrays and genetic tools such as transposons and reporter gene plasmids.
Whole-genome sequences are available for A. pleuropneumoniae serovars 3 (accession no. NC_010278), 5b (accession no. NC_009053), and 7 (accession no. NC_010942), with others in progress. Microarrays, developed from the whole-genome sequences, have been used for genotyping (22) as well as to identify genes important for iron uptake (15), anaerobicity (10, 11), interaction with host cells (2), and the role of the global regulators H-NS and RpoE in biofilm formation (J. T. Bossé, S. Sinha, C. A. O'Dwyer, J. H. E. N. Nash, A. N. Rycroft, J. S. Kroll, and P. R. Langford, submitted for publication). It is envisaged that microarray analysis will provide further insights into pathogenicity in the future. Random transposon mutagenesis is a valuable technique for the discovery of new virulence factors in bacteria. For A. pleuropneumoniae, the transposon Tn10 has been the most widely used (8, 20, 38, 39, 44, 46). We (44) and others (20), have modified the Tn10 delivery vehicle described by Tascon et al. (46) to allow high-throughput screening for virulence factors using signature-tagged mutagenesis. The results suggested that genes involved in the stress response and anaerobicity are important for survival of A. pleuropneumoniae in the lungs during acute infection. Microarrays and transposon mutagenesis are valuable tools for identifying potential virulence factors, although they are not without limitations. Microarrays are not always readily available and can be expensive due to the number of replicates required for statistical power. This can be cost prohibitive when screening under multiple growth conditions is being done. Tn10 transposition is far from ideal since insertion is dependent on the DNA sequence recognized by the transposase, resulting in hot spots rather than random insertion (6). Thus, many virulence factors of A. pleuropneumoniae may have been missed through the use of Tn10-based analysis (20, 44). Furthermore, potential virulence-associated genes identified by transposon mutagenesis and/or microarrays require validation and characterization using conventional genetic tools.
Plasmids have been widely used in A. pleuropneumoniae for complementation of mutants with defined genetic defects. Such plasmids include those based on pJFF224 (18), pIG112 (51), pYG10 (26), pSL88 (17), and pGZRS (50). In contrast, in comparison to other gram-negative bacteria, reporter plasmids have been underused in A. pleuropneumoniae for the discovery of virulence factors or to monitor promoter activity of specific genes. One reason is that A. pleuropneumoniae harbors a lacZ homologue encoding a β-galactosidase (1), a widely used reporter gene in other bacteria. Luciferase activity has been used, though not extensively, as a reporter for identification of promoter activity and to monitor gene expression in A. pleuropneumoniae (21, 23) (constrained by the need for specialized equipment for quantitative analysis). The promoter-trap vector pTF86, containing the promoterless Vibrio harveyi luxAB genes, was used for in vivo expression technology to identify A. pleuropneumoniae genes upregulated in vivo (21). Similarly, the luxAB genes from Photorhabdus luminescens have been used to generate a chromosomal fusion to the A. pleuropneumoniae aspA gene in order to monitor induction of this gene in response to anaerobic culture or exposure to bronchoalveolar lavage fluid (23). The xylE gene (encoding catechol 2,3-dioxygenase) from the Pseudomonas putida TOL plasmid has been shown to be functionally active in A. pleuropneumoniae (18) but has not been used as a reporter gene. XylE has great potential for the rapid screening of bacteria on agar plates, as catechol 2,3-dioxygenase production results in development of a yellow color when colonies are sprayed with catechol. To our knowledge, there is no description of the use of green fluorescent protein (GFP) as a reporter system in A. pleuropneumoniae, a surprising omission given its usefulness in virulence factor discovery through, for example, differential fluorescence induction (48).
Here we describe a series of conjugative reporter plasmids that can be used for detection of promoter activity via rapid screening of colonies (through XylE activity) and/or GFP expression (via fluorescence-activated cell sorter [FACS] analysis), as well as for complementation of defined mutations. They are based on the RK6 plasmid, pMIDG100, that we used to express meningococcal outer membrane proteins in commensal Neisseria strains (35, 49). The plasmids are designed to facilitate cloning of either promoters or genes of interest and provide a choice of antibiotic resistance cassettes (kanamycin or chloramphenicol) for selection. They are of broad host range and permit heterologous gene expression in A. pleuropneumoniae, Haemophilus influenzae, Haemophilus parasuis, Mannheimia haemolytica, Pasteurella multocida, and Escherichia coli.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli TOP10 (Invitrogen) and S17.1λpir (33) were cultured in Luria-Bertani (LB) (Difco) broth or on LB agar supplemented, when required, with either kanamycin (100 μg/ml) or chloramphenicol (20 μg/ml). The various Pasteurellaceae strains (A. pleuropneumoniae S4074T; H. influenzae Rd; H. parasuis serotype 3 [clinical isolate; MIDG3176]; M. haemolytica [clinical isolate; strain MIDG1579]; and P. multocida [clinical isolate; strain MIDG1570]) were grown on brain heart infusion (BHI) (Difco) agar supplemented with Levinthal's base (BHI-Lev) or in BHI broth supplemented with 0.01% NAD and, for Haemophilus strains, hemin (10 μg/ml). When required, the BHI was supplemented with kanamycin (50 μg/ml), chloramphenicol (1 μg/ml), or nalidixic acid (20 μg/ml). Spontaneous nalidixic acid-resistant derivatives of the various Pasteurellaceae species were selected by exposure of high-density broth cultures to increasing concentrations of nalidixic acid up to 20 μg/ml, followed by overnight growth on agar plates supplemented with 20 μg/ml nalidixic acid (44).
General molecular biology techniques.
Genomic DNA was prepared from bacterial strains using a QIAamp minikit or DNA maxikit, and plasmid extractions were performed using Qiaprep spin columns (Qiagen). DNA concentrations were measured using a NanoDrop ND-1000 UV-visible spectrophotometer (NanoDrop Technologies). Unless otherwise stated, restriction enzymes were obtained from Roche and used according to the manufacturer's protocol. PCR was performed according to standard procedures (42) using HotStarTaq DNA polymerase (Qiagen). The ABI Prism BigDye Terminator cycle sequencing kit and an ABI3700 DNA sequencer were used for sequencing.
Conjugation.
Conjugal transfer of plasmids from E. coli S17.1λpir into recipient strains was carried out using a modification of the method of Dehio and Meyer (14). Briefly, donor and recipient strains were grown in broth cultures overnight at 37°C. Bacteria were harvested by centrifugation for 5 min at 6,000 × g, washed twice, and resuspended to the original volume in cold 10 mM MgSO4. Cells were mixed (100 μl donor cells plus 200 μl recipient cells) and concentrated to 100 μl, and 20 μl was spread onto 0.22-μm-pore-size filters (Millipore). Filters were placed onto BHI-Lev plates and incubated at 37°C with 5% CO2 for 5 h. Thereafter, the bacteria were removed into 1 ml of sterile phosphate-buffered saline (PBS), and dilutions were plated either onto BHI-Lev supplemented with nalidixic acid (20 μg/ml) to determine CFU/ml of recipient or onto plates with nalidixic acid and either kanamycin (100 μg/ml) or chloramphenicol (1 μg/ml), as appropriate, for selection of transconjugants.
Construction of plasmids.
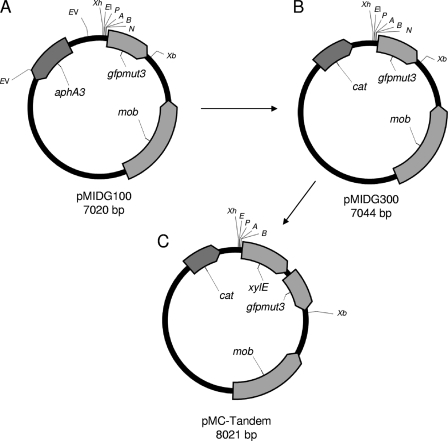
During cloning, plasmids were initially transformed into E. coli TOP10 cells using the heat shock protocol supplied by the manufacturer (Invitrogen). All of the vectors constructed in this study were derived from the broad-host-range shuttle vector pMIDG100 (49) (Fig. 1A). To generate pMIDG300 (Fig. 1B), the aphA3 gene was replaced with a chloramphenicol acetyltransferase gene (cat) originally from the Staphylococcus aureus plasmid pC194 (30). To create the tandem reporter plasmid pMC-Tandem, a second promoterless reporter gene, xylE, encoding catechol 2,3-dioxygenase, was PCR amplified from pJFF224-NX:xylE (18) using the primers XylEFBamHI and XylERXbaI (Table 1) and was inserted upstream of the gfpmut3 gene in pMIDG300 (Fig. 1C).
FIG. 1.
Construction of the reporter vector, pMC-Tandem. (A) pMIDG100 (49) containing aphA3 (kanamycin resistance), gfpmut3 (GFP), and mob (mobilization protein). (B) pMIDG300 was derived from pMIDG100 by cloning the cat gene (chloramphenicol resistance) from S. aureus pC194 (30) into the two EcoRV sites, replacing the aphA3 gene. (C) pMC-Tandem was derived from pMIDG300 by insertion of the xylE gene (catechol 2,3-dioxygenase) from pJFF224-NX:xylE (18) into the unique BamHI and NheI sites upstream of gfpmut3. Relevant restriction sites are labeled as follows: A, ApaI; B, BamHI; EI, EcoRI; EV, EcoRV; K, KpnI; N, NheI; P, PstI; Xb, XbaI; Xh, XhoI. All labeled restriction sites are unique except for EcoRV, which cuts twice in pMIDG100.
TABLE 1.
Primers used in this study
| Primer name | Sequence, 5′ to 3′ |
|---|---|
| XylEFBamHI | GCGCGGATCCATGAACTATGAAGAGGTGAC |
| XylERXbaI | TGCTCTAGAGGTACCTCTCTGCAATAAGTCGTA |
| Actinosod 6 | CGCAAGCTTCGTGAATCATTAAAGAATGAC |
| SodCGSP1 | TGCGTTATCAGACCACGGAT |
| RseAInvI | GTCCATAAAAGCGGAAAGGGT |
| RpoEGSP1 | GCATCTTCCGCCAAAATATC |
| SodCFEcoRI | GCGCGAATTCCTTCGTTCGTGTAGTCACCG |
| SodCRBamHI | GCGCGGATCCCGACAAGTTTTTCCACTGAG |
| RpoEFEcoRI | GCGCGAATTCGTAGCATCCTTAGTCTT |
| RpoERBamHI | GCGCGGATCCCACCGCAATACGATAAAGC |
| SodCPFXhoI | GCTCGAGCCGCGCCAACCGATA |
| SodCPREcoRI | GGAATCCTCCTTTTATTTTGGTT |
| M13FXbaI | GTCTAGAGAGTAAAACGACGGCCA |
| M13R | CAGGAAACAGCTATGACC |
| NadVF | CTGTATGAGATTTAAGGAAAGAAATTATTATGGATAACC |
| NadVR | GCGTATTAAGTACAAATATCATAGCGTAGTGC |
Validation of pMC-Tandem.
In order to validate the usefulness of the tandem reporter system for identification of transcriptional regulators and conditions for promoter induction, we cloned the sodC and rpoE promoters from A. pleuropneumoniae. The sodC promoter was chosen, as previous studies showed that the A. pleuropneumoniae gene was expressed in all growth conditions tested (28), thus providing a positive control for reporter gene expression. The rpoE promoter was chosen because rpoE encodes an alternative sigma factor, σE, for which there is a putative negative regulator (RseA) and because we had previously isolated A. pleuropneumoniae mutants with Tn10 insertions in both rpoE and rseA, with both mutants being attenuated for virulence in pigs (44).
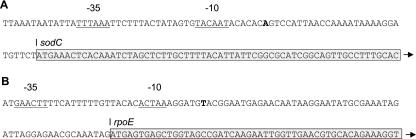
A 5′ rapid amplification of cDNA ends kit (Invitrogen) was used to confirm or identify the transcriptional start sites of the sodC and rpoE genes (Fig. 2), respectively. In addition to the primers provided in the kit, gene-specific primers for sodC (Actinosod 6 and SodCGSP1) and rpoE (RseAInvI and RpoEGSP1) were used for production of cDNA and a subsequent nested PCR (Table 1). The resulting PCR products were sequenced to determine the transcriptional start sites.
FIG. 2.
Identification of transcriptional start sites for sodC and rpoE in A. pleuropneumoniae by 5′ rapid amplification of cDNA ends. (A) Sequence showing the transcriptional start site for sodC (bold) and the predicted −10 and −35 promoter regions (underlined). The −10 sequence agrees with that previously predicted by Langford et al. (28) using primer extension. In that study, the −35 region was predicted to be TTATT, although an alternative (TTTAAA), which shows closer homology to the consensus for the −35 region of E. coli σ70 promoters (TTGACA), is present. (B) Sequence showing the transcriptional start site for rpoE (bold) and the predicted −10 and −35 promoter regions (underlined). The predicted −35 region is a perfect match for the −35 region of the E. coli σE promoter (GAACTT), whereas the −10 region (ACTAA) differs slightly from that in E. coli (TCAAA). For both sodC and rpoE, the boxed regions show the beginning of the translated genes.
Once the promoter regions had been identified, PCR primers were designed to amplify regions of DNA containing the sodC and rpoE promoters. These primer pairs, SodCFEcoRI/SodCRBamHI and RpoEFEcoRI/RpoERBamHI (Table 1), incorporated EcoRI and BamHI sites to allow directional cloning of the promoters upstream of the reporter genes. The resulting plasmids, pMCsodCP and pMCrpoEP, as well as pMC-Tandem, were purified from E. coli TOP10 and transformed by heat shock into chemically competent E. coli S17.1λpir, the donor strain used for conjugation as described above. For pMC-Tandem and pMCsodCP, recipient strains were nalidixic acid-resistant derivatives of A. pleuropneumoniae, H. influenzae, H. parasuis, M. haemolytica, and P. multocida. For the pMCrpoEP plasmid, recipient strains were nalidixic acid-resistant derivatives of A. pleuropneumoniae S4074, as well as a pool of 48 Tn10 mutants of S4074, including the rseA::Tn10 mutant 19B10 (44).
Induction of PrpoE by heat shock and exposure to ethanol.
The ability to detect induction of promoters using the tandem reporter system was tested with the pMCrpoEP-containing strains exposed to two conditions known to induce expression of rpoE in other bacteria, namely, heat shock and exposure to ethanol (25, 32, 43). For induction by heat shock, A. pleuropneumoniae S4074 containing the reporter plasmids pMC-Tandem (promoterless), pMCsodCP, and pMCrpoEP were grown overnight at 30°C. The following day, the plates were shifted to various temperatures (from 30°C to 50°C) for one hour. The bacteria were harvested from the plates, washed once in PBS, and resuspended to a concentration of approximately 1 × 106 CFU/ml in PBS containing 2% formaldehyde. The fluorescent signal from formaldehyde-fixed strains was quantified using a FACSCalibur flow cytometer (Becton Dickinson) as previously described (35). For induction by exposure to ethanol, the same strains were grown overnight on BHI-Lev agar plates and resuspended in fresh BHI broth to a concentration of 106 CFU per ml. After 2 hours of growth at 37°C, 3% ethanol was added, and thereafter aliquots were removed every hour for 5 hours in order to (i) determine the levels of fluorescence per cell by FACS and (ii) determine viability by plating dilutions onto BHI-Lev plates. For FACS analysis, samples were collected by centrifugation, washed once with PBS, and resuspended in PBS containing 2% formaldehyde to approximately 1 × 106 bacteria/ml prior to being analyzed.
Identification of regulators of rpoE in A. pleuropneumoniae.
In order to determine whether RseA is a negative regulator of σE in A. pleuropneumoniae as it is in other bacteria (13, 34), we compared xylE expression in A. pleuropneumoniae S4074 and 19B10 (rseA::Tn10) strains containing pMC-Tandem, pMCsodCP, or pMCrpoEP. Once the role of RseA as a negative regulator was confirmed, we tested the feasibility of using the catechol screening system to identify a regulatory mutant (rseA::Tn10) within a mixed population of A. pleuropneumoniae Tn10 mutants containing pMCrpoE. The transconjugants were plated to give well-isolated colonies, which were then sprayed with a fine mist of freshly prepared catechol (50 mg/ml in water). Yellow colonies were isolated, and the location of the Tn10 insertion was confirmed by inverse PCR and sequencing as previously described (44).
Construction of the expression vectors, pMK-Express and pMC-Express.
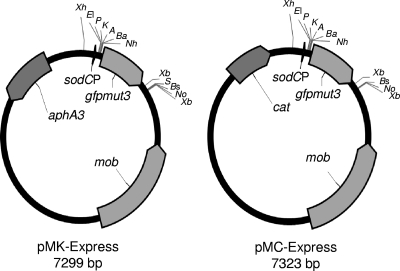
We exploited the fact that the A. pleuropneumoniae sodC promoter can be used to express heterologous genes in various Pasteurellaceae in order to construct a pair of expression vectors, pMK-Express and pMC-Express, differing in their selective markers (aphA3 and cat, respectively) (Fig. 3). In order to preserve more of the multiple-cloning site (MCS) upstream of the gfpmut3 gene in pMIDG100, we recloned the A. pleuropneumoniae sodC promoter using primers SodCPFXhoI and SodCPREcoRI (Table 1) to allow directional cloning into the unique XhoI and EcoRI sites. Additionally, since pMIDG100 has limited restriction sites downstream of the reporter gene, an 80-bp fragment from the MCS of pBluescript IIKS (Stratagene) was amplified using the primers M13FXbaI and M13R (Table 1), cloned into pGEM-T (Invitrogen), and then subcloned (as an XbaI fragment) downstream of gfpmut3 to generate the plasmid pMK-Express (Fig. 3), which contains a kanamycin cassette. Subsequently, the kanamycin cassette was replaced with the S. aureus pC194 chloramphenicol cassette (as described above) to generate pMC-Express. The presence of the gfpmut3 gene in the two shuttle vectors allows easy demonstration of restriction site cleavage (i.e., removal of an approximately 800-bp fragment) prior to insertion of genes of interest.
FIG. 3.
Maps of the expression vectors pMK-Express and pMC-Express. Genes aphA3 (kanamycin resistance), cat (chloramphenicol resistance), gfpmut3 (GFP), and mob (mobilization protein) are indicated by solid arrows. The A. pleuropneumoniae sodC promoter (sodCP) is indicated by the arrowhead upstream of gfpmut3. Relevant restriction sites are labeled as follows: A, ApaI; Ba, BamHI; Bs, BstXI; EI, EcoRI; EV, EcoRV; K, KpnI; Nh, NheI; No, NotI; P, PstI; S, SacI; Xb, XbaI; Xh, XhoI. All labeled restriction sites are unique except for XbaI, which cuts twice in both pMK-Express and pMC-Express.
Expression of nadV from Haemophilus ducreyi in A. pleuropneumoniae.
A 1.5-kb fragment of genomic DNA from H. ducreyi strain 35000HP (27) containing the nadV gene was amplified by PCR using primers NadVF and NadVR (Table 1). The resulting PCR product was cloned into pGEM-T (Invitrogen). The nadV gene was then subcloned (as an ApaI/SacI fragment) into pMK-Express and transformed into E. coli TOP10. The resulting kanamycin-resistant clones were screened by colony PCR to confirm the presence of the nadV gene. Plasmid was purified from a selected clone, transformed into E. coli S17.1λpir, and then transferred by conjugation into A. pleuropneumoniae S4074 as described above. Following selection of transconjugants on BHI-NAD plates containing kanamycin and nalidixic acid, colonies were subcultured onto BHI plates without NAD to demonstrate expression of NadV.
Nucleotide sequence accession numbers.
The complete sequences of pMC-Tandem, pMC-Express, and pMK-Express have been deposited in the GenBank database under accession numbers GQ334691, GQ334689, and GQ334690, respectively.
RESULTS
Evaluation of the tandem reporter system.
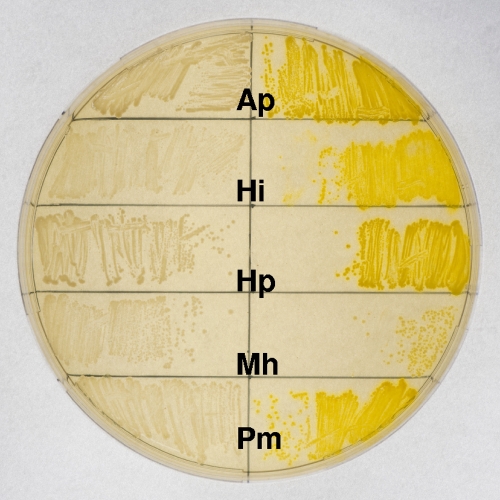
The plasmids pMC-Tandem and pMCsodCP were successfully conjugated into A. pleuropneumoniae, H. influenzae, H. parasuis, M. haemolytica, and P. multocida with frequencies of between 10−5 and 10−4 per recipient for each strain. As an initial nonquantitative screen for promoter activity, plate-grown bacteria were assayed for catechol 2,3-dioxygenase activity. Each of the Pasteurellaceae strains containing the pMCsodCP plasmid produced bright yellow colonies when sprayed with catechol, whereas there was no detectable color change for any of the strains containing pMC-Tandem (Fig. 4).
FIG. 4.
Assay for catechol 2,3-dioxygenase activity. Each of the Pasteurellaceae strains containing pMCsodCP (with the cloned A. pleuropneumoniae sodC promoter; right side of plate) produced bright yellow colonies (including those that are single and isolated) when sprayed with catechol, whereas there was no detectable color change for any of the strains containing pMC-Tandem (promoterless; left side of plate). Bacteria: Ap, A. pleuropneumoniae; Hi, H. influenzae; Hp, H. parasuis; Mh, M. haemolytica; Pm, P. multocida.
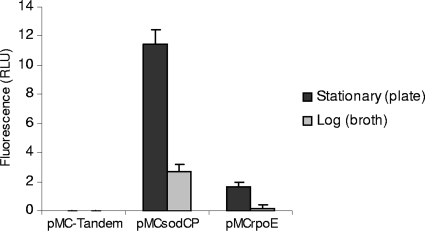
Individual cells of all the Pasteurellaceae were green when viewed under a fluorescence microscope at a wavelength of 495 nm (data not shown). Quantitative analysis of GFP expression was done by measuring the fluorescence of A. pleuropneumoniae S4074 cells containing pMC-Tandem, pMCsodCP, or pMCrpoE. The negative control containing pMC-Tandem showed a low background level of fluorescence, which was used as a baseline. Cells grown in broth to mid-log phase showed lower levels of fluorescence than plate-grown bacteria in stationary phase (Fig. 5). The strain containing pMCsodCP showed detectable levels of fluorescence for both broth- and plate-grown bacteria, whereas fluorescence was barely detectable for the strain containing pMCrpoEP grown in broth at 37°C. Low-level fluorescence was detected in the pMCrpoEP-containing strain grown to stationary phase on agar plates at 37°C, indicating a low level of transcription from the rpoE promoter under these growth conditions.
FIG. 5.
Expression of GFP in A. pleuropneumoniae S4074 bacteria containing different reporter plasmids. The relative fluorescence of the bacteria, grown either to stationary phase on BHI-Lev plates or to mid-log phase in BHI broth, was measured on gated populations of bacterial cells by flow cytometry, producing data in relative light units (RLU) per cell. In each case, the background fluorescence from A. pleuropneumoniae S4074 containing the pMC-Tandem (promoterless) has been subtracted. Experiments were performed in triplicate, and the error bars indicate the standard deviations of the means.
In order to confirm that plasmids containing genomic sequences from A. pleuropneumoniae are stably maintained as extrachromosomal elements, rather than integrating into the genome of the host A. pleuropneumoniae S4074 strain, the stability of pMCsodCP was tested. The plasmid pMCsodCP was used due to the large size of the promoter region, which would make recombination more likely. After five successive days of subculture of A. pleuropneumoniae S4074 containing pMCsodCP on BHI-Lev agar plates, similar numbers of bacteria were recovered on plates with and without chloramphenicol, all of which were positive for catechol 2,3-dioxygenase activity. Plasmids recovered from six independent colonies all showed identical restriction maps (data not shown), confirming that the plasmid had been stably maintained.
Analysis of promoter induction using the tandem reporter system.
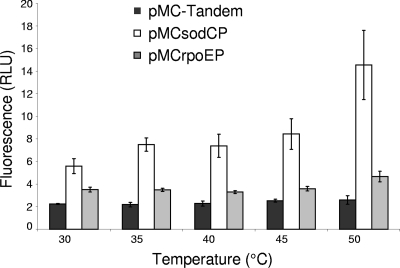
We tested two conditions known to induce expression of σE-regulated genes in other bacteria: heat shock and exposure to 3% ethanol. The levels of fluorescence in A. pleuropneumoniae S4074 containing pMC-Tandem, pMCsodCP, and pMCrpoEP were measured following a shift from overnight growth on agar plates at 30°C to incubation for 1 hour at various temperatures up to 50°C (Fig. 6). At each temperature, the level of fluorescence from pMC-Tandem (promoterless) was constant. In contrast, as the temperature increased, the level of fluorescence per cell containing pMCsodCP increased. The level of fluorescence of cells containing pMCrpoEP showed only a slight but significant increase following incubation at 50°C (P = 0.03).
FIG. 6.
Induction of the A. pleuropneumoniae sodC and rpoE promoters following heat shock. The level of fluorescence per cell containing pMC-Tandem (promoterless) did not change following a shift from growth at 30°C to temperatures ranging from 35°C to 50°C. In contrast, the level of fluorescence per cell containing pMCsodCP or containing pMCrpoEP increased significantly after a temperature increase from 30°C to 50°C (P = <0.01 and P = 0.03, respectively), as determined by Student's t test. Experiments were performed in triplicate, and the error bars indicate the standard deviations of the means.
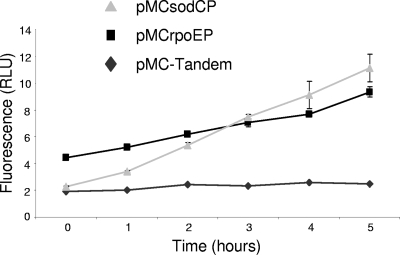
Following exposure of broth cultures to ethanol, the background level of fluorescence, as determined with S4074 containing pMC-Tandem, remained constant throughout the time course, while the levels of fluorescence in cells containing either pMCsodCP or pMCrpoEP increased over time, indicating induction of these promoters in the presence of ethanol (Fig. 7). Following addition of ethanol, viable cell counts did not increase, and by 5 h, there was a 10-fold reduction in viable cell counts.
FIG. 7.
Induction of the A. pleuropneumoniae sodC and rpoE promoters following exposure to ethanol. The level of fluorescence per cell was measured following addition of 3% (vol/vol) ethanol to mid-log phase cultures (at 0 h). In A. pleuropneumoniae S4074 containing pMC-Tandem (promoterless), the levels of fluorescence remained constant throughout the time course, while the levels of fluorescence in cells containing pMCsodCP or pMCrpoE increased over time. Following addition of ethanol, viable cell counts did not increase, and by 5 h there was a 10-fold reduction in viable cell counts. Experiments were performed in triplicate, and the error bars indicate the standard deviations of the means.
Detection of transcriptional regulators using the tandem reporter system.
The reporter plasmid, pMCrpoEP, was further used to validate the ability of the tandem reporter system to screen for transcription factors that repress expression from the cloned promoter. In order to test the ability of the catechol screening system to identify a single mutant within a mixed population, pMCrpoEP was conjugated into a mixed pool of 48 tagged Tn10 mutants of A. pleuropneumoniae S4074, one of which was 19B10 (rseA::Tn10). The transconjugants were plated to give well-isolated colonies, which were then sprayed with catechol. Yellow colonies were isolated only in the case of 19B10 and were confirmed to be rseA knockouts by inverse PCR and sequencing.
Validation of pMK-Express as an expression vector.
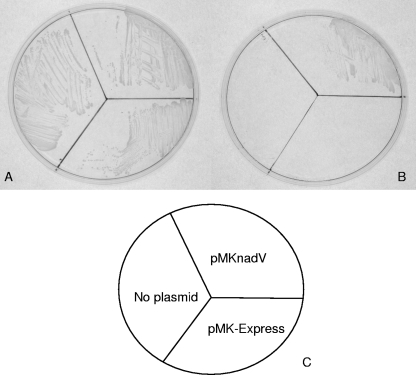
pMK-Express and pMC-Express were designed to allow expression of cloned genes under the control of the A. pleuropneumoniae sodC promoter, which we have shown to be active under all conditions investigated so far and in all of the Pasteurellaceae strains tested. As a test of this series of vectors, we cloned the nadV gene from H. ducreyi (31, 40) into pMK-Express and conjugated it into A. pleuropneumoniae S4074. The resulting kanamycin-resistant transconjugants were screened by PCR to confirm the presence of the nadV gene. Colonies patched onto BHI agar without NAD confirmed expression of functional NadV (Fig. 8).
FIG. 8.
Complementation of the requirement for NAD in A. pleuropneumoniae S4074 by H. ducreyi nadV. Bacterial strains were plated on BHI plus NAD (A) or BHI with no supplement (B). (C) Cartoon showing the relative positions of A. pleuropneumoniae containing no plasmid, empty vector (pMK-Express), or pMKnadV (expressing the cloned H. ducreyi nadV gene). Only the strain containing pMKnadV was able to grow on BHI media in the absence of added NAD.
DISCUSSION
We have designed a versatile set of mobilizable plasmids (pMC-Tandem, pMC-Express, and pMK-Express), based on the broad-host-range shuttle vector pMIDG100 (49), that can be used in members of the Pasteurellaceae for monitoring of gene expression via XylE or GFP, for identification of transcriptional regulators, and for complementation of gene defects and/or heterologous expression. The XylE approach neatly complements inadequacies in the use of LacZ in the Pasteurellaceae. While β-galactosidase activity has been used for both qualitative and quantitative analysis of promoter activity through creation of lacZ fusions in H. influenzae (16, 45), the presence of lacZ homologues in A. pleuropneumoniae, H. parasuis, and M. haemolytica makes it unsuitable for general use as a reporter gene in the Pasteurellaceae. Catechol 2,3-dioxygenase activity provides an alternative qualitative colorimetric reporter assay, through fusions with xylE, a gene that is not present in any of the sequenced Pasteurellaceae genomes. The presence of the promoterless xylE gene in pMC-Tandem allows rapid nonquantitative screening of cloned promoter activity. We have demonstrated that the xylE gene, under the control of the A. pleuropneumoniae sodC promoter, is functional in A. pleuropneumoniae, H. influenzae, H. parasuis, P. multocida, and M. haemolytica. Thus, pMC-Tandem can be exploited for the analysis of endogenous promoters in each of these species. Furthermore, the presence of a promoterless gfpmut3 gene, downstream of xylE in pMC-Tandem, allows for quantitative analysis, by FACS, of cloned promoter activity, as shown in the validation studies described.
We have validated the usefulness of pMC-Tandem for identification of transcriptional regulators and conditions which favor gene expression from cloned promoters using the A. pleuropneumoniae rpoE and sodC promoters as examples. The pMCrpoEP plasmid was used to confirm that the A. pleuropneumoniae RseA protein is a negative regulator of σE, as it is in other bacteria (13, 34). Furthermore, we have demonstrated that the tandem reporter construct can be used to screen for regulatory mutants. Using catechol 2,3-dioxygenase screening, we could detect and isolate an rseA::Tn10 mutant from a mixed pool of 48 random Tn10 mutants into which pMCrpoEP had been conjugated. In this case, we have shown detection of a mutation in a negative regulator resulting in increased expression from the normally inactive promoter. It should also be possible to screen for mutations in transcriptional activators resulting in decreased expression from cloned positively regulated promoters.
The tandem reporter construct was also used to study environmental regulation of cloned promoters. Increased fluorescence was detected following exposure of A. pleuropneumoniae cells containing pMCrpoEP to ethanol and, to a lesser extent, following exposure to extreme heat shock, two conditions shown to upregulate σE expression in other bacteria (36, 41, 43). Similarly, exposure to both ethanol and elevated temperature resulted in increased fluorescence of A. pleuropneumoniae cells containing pMCsodCP but not of cells containing the promoterless control, pMC-Tandem. Previous work showed no change in levels of SodC activity under any of the conditions tested (different growth phases, aerobic versus anaerobic growth, and presence or absence of iron), suggesting possible constitutive expression (28). This is the first report of regulation of sodC expression in A. pleuropneumoniae. Because promoter activity is determined as a result of translation of functional XylE and/or GFP, the reporter construct may not be as rapid and/or sensitive as detection of increased levels of transcripts, which can be detected within 10 min following exposure to the inducing stimulus (36). Therefore, this method can be used as a simple screen for identification of inducing stimuli, which can then be monitored more accurately using other methods, such as quantitative reverse transcription-PCR.
In addition to the promoter-probe vector, we have also constructed a pair of shuttle vectors, pMK-Express and pMC-Express, that can be used in complementation experiments or to express heterologous genes. We have exploited the facts that the A. pleuropneumoniae sodC promoter shows detectable, though variable, activity under all of the conditions tested so far and is also active in other members of the Pasteurellaceae in order to place cloned genes under a generally active promoter. The pMIDG100 vector was again used as the backbone to construct the plasmids. In order to provide a greater selection of restriction sites for excision of the gfpmut3 gene and insertion of cloned genes of interest, we ligated the A. pleuropneumoniae sodC promoter into the unique XhoI and EcoRI sites upstream and cloned a portion of the pBluescriptIIKS MCS into the unique XbaI site. As these plasmids are designed to facilitate complementation of mutated genes, we have allowed for the use of either kanamycin (pMK-Express) or chloramphenicol (pMC-Express) for the selection of plasmid-containing strains. These plasmids can also be used to express heterologous genes. In this study, we cloned the nadV gene from H. ducreyi into pMK-Express and expressed it in A. pleuropneumoniae S4074, rendering it NAD independent.
The advantages of these plasmids include conjugative mobilization to facilitate use in bacterial strains difficult to transform by electroporation, stable maintenance with or without antibiotic pressure, versatile cloning sites and choice of antibiotic selection for pMC-Express and pMK-Express, and choice of qualitative or quantitative assays for promoter activity using pMC-Tandem. It was possible to conjugate pMC-Tandem into a range of Pasteurellaceae. While all of the tested strains were amenable to conjugation, with frequencies of transconjugants of between 10−5 and 10−4 per recipient, it is likely that there will be strain variation among the different species. In the case of A. pleuropneumoniae, conjugation into the serotype 3 (S1421) and serotype 15 (HS143) reference strains was also achieved (data not shown). Should it not be possible to conjugate into strains of interest, then alternative methods may be possible, including electroporation (18) or, in those strains that are naturally transformable, the addition of either the Hin or Apl uptake signal sequences (37) into the plasmid backbone. In the case of H. parasuis, the latter approach has been successful (7). The plasmids constructed in this study will greatly add to the tools available for genetic analyses in the Pasteurellaceae.
Acknowledgments
This work was supported by grants from the UK Biotechnology and Biological Sciences Research Council (to P.R.L., J.S.K., A.N.R., and J.T.B.) and a studentship from the States of Guernsey (to A.L.D.).
We are grateful to Joachim Frey (University of Berne) for the gift of plasmid pJFF224-NX:xylE, to Dan Tucker (University of Cambridge) for the serotype 3 H. parasuis strain, and to Charles Gervais for photographic expertise.
Footnotes
Published ahead of print on 7 August 2009.
REFERENCES
- 1.Anderson, T. J., and J. I. MacInnes. 1997. Expression and phylogenetic relationships of a novel lacZ homologue from Actinobacillus pleuropneumoniae. FEMS Microbiol. Lett. 152:117-123. [DOI] [PubMed] [Google Scholar]
- 2.Auger, E., V. Deslandes, M. Ramjeet, I. Contreras, J. H. Nash, J. Harel, M. Gottschalk, M. Olivier, and M. Jacques. 2009. Host-pathogen interactions of Actinobacillus pleuropneumoniae with porcine lung and tracheal epithelial cells. Infect. Immun. 77:1426-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baltes, N., I. Hennig-Pauka, I. Jacobsen, A. D. Gruber, and G. F. Gerlach. 2003. Identification of dimethyl sulfoxide reductase in Actinobacillus pleuropneumoniae and its role in infection. Infect. Immun. 71:6784-6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baltes, N., M. N′Diaye, I. D. Jacobsen, A. Maas, F. F. Buettner, and G. F. Gerlach. 2005. Deletion of the anaerobic regulator HlyX causes reduced colonization and persistence of Actinobacillus pleuropneumoniae in the porcine respiratory tract. Infect. Immun. 73:4614-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bandara, A. B., M. L. Lawrence, H. P. Veit, and T. J. Inzana. 2003. Association of Actinobacillus pleuropneumoniae capsular polysaccharide with virulence in pigs. Infect. Immun. 71:3320-3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender, J., and N. Kleckner. 1992. Tn10 insertion specificity is strongly dependent upon sequences immediately adjacent to the target-site consensus sequence. Proc. Natl. Acad. Sci. USA 89:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bigas, A., M. E. Garrido, A. M. de Rozas, I. Badiola, J. Barbe, and M. Llagostera. 2005. Development of a genetic manipulation system for Haemophilus parasuis. Vet. Microbiol. 105:223-228. [DOI] [PubMed] [Google Scholar]
- 8.Bossé, J. T., H. D. Gilmour, and J. I. MacInnes. 2001. Novel genes affecting urease acivity in Actinobacillus pleuropneumoniae. J. Bacteriol. 183:1242-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bossé, J. T., H. Janson, B. J. Sheehan, A. J. Beddek, A. N. Rycroft, J. S. Kroll, and P. R. Langford. 2002. Actinobacillus pleuropneumoniae: pathobiology and pathogenesis of infection. Microbes Infect. 4:225-235. [DOI] [PubMed] [Google Scholar]
- 10.Buettner, F. F., I. M. Bendallah, J. T. Bosse, K. Dreckmann, J. H. Nash, P. R. Langford, and G. F. Gerlach. 2008. Analysis of the Actinobacillus pleuropneumoniae ArcA regulon identifies fumarate reductase as a determinant of virulence. Infect. Immun. 76:2284-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buettner, F. F., J. T. Bossé, J. H. Nash, E. Hartig, P. R. Langford, and G. F. Gerlach. 2009. Analysis of the Actinobacillus pleuropneumoniae HlyX (FNR) regulon and identification of iron-regulated protein B as an essential virulence factor. Proteomics 9:2383-2398. [DOI] [PubMed] [Google Scholar]
- 12.Chung, J. W., C. Ng-Thow-Hing, L. I. Budman, B. F. Gibbs, J. H. Nash, M. Jacques, and J. W. Coulton. 2007. Outer membrane proteome of Actinobacillus pleuropneumoniae: LC-MS/MS analyses validate in silico predictions. Proteomics 7:1854-1865. [DOI] [PubMed] [Google Scholar]
- 13.da Silva Neto, J. F., T. Koide, S. L. Gomes, and M. V. Marques. 2007. The single extracytoplasmic-function sigma factor of Xylella fastidiosa is involved in the heat shock response and presents an unusual regulatory mechanism. J. Bacteriol. 189:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dehio, C., and M. Meyer. 1997. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J. Bacteriol. 179:538-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deslandes, V., J. H. Nash, J. Harel, J. W. Coulton, and M. Jacques. 2007. Transcriptional profiling of Actinobacillus pleuropneumoniae under iron-restricted conditions. BMC Genomics 8:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dixon, K., C. D. Bayliss, K. Makepeace, E. R. Moxon, and D. W. Hood. 2007. Identification of the functional initiation codons of a phase-variable gene of Haemophilus influenzae, lic2A, with the potential for differential expression. J. Bacteriol. 189:511-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dixon, L. G., W. L. Albritton, and P. J. Willson. 1994. An analysis of the complete nucleotide sequence of the Haemophilus ducreyi broad-host-range plasmid pLS88. Plasmid 32:228-232. [DOI] [PubMed] [Google Scholar]
- 18.Frey, J. 1992. Construction of a broad host range shuttle vector for gene cloning and expression in Actinobacillus pleuropneumoniae and other Pasteurellaceae. Res. Microbiol. 143:263-269. [DOI] [PubMed] [Google Scholar]
- 19.Frey, J. 1995. Virulence in Actinobacillus pleuropneumoniae and RTX toxins. Trends Microbiol. 3:257-261. [DOI] [PubMed] [Google Scholar]
- 20.Fuller, T. E., S. Martin, J. F. Teel, G. R. Alaniz, M. J. Kennedy, and D. E. Lowery. 2000. Identification of Actinobacillus pleuropneumoniae virulence genes using signature-tagged mutagenesis in a swine infection model. Microb. Pathog. 29:39-51. [DOI] [PubMed] [Google Scholar]
- 21.Fuller, T. E., R. J. Shea, B. J. Thacker, and M. H. Mulks. 1999. Identification of in vivo induced genes in Actinobacillus pleuropneumoniae. Microb. Pathog. 27:311-327. [DOI] [PubMed] [Google Scholar]
- 22.Goure, J., W. A. Findlay, V. Deslandes, A. Bouevitch, S. J. Foote, J. I. MacInnes, J. W. Coulton, J. H. Nash, and M. Jacques. 2009. Microarray-based comparative genomic profiling of reference strains and selected Canadian field isolates of Actinobacillus pleuropneumoniae. BMC Genomics 10:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobsen, I., I. Hennig-Pauka, N. Baltes, M. Trost, and G. F. Gerlach. 2005. Enzymes involved in anaerobic respiration appear to play a role in Actinobacillus pleuropneumoniae virulence. Infect. Immun. 73:226-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacques, M. 2004. Surface polysaccharides and iron-uptake systems of Actinobacillus pleuropneumoniae. Can. J. Vet. Res. 68:81-85. [PMC free article] [PubMed] [Google Scholar]
- 25.Kovacikova, G., and K. Skorupski. 2002. The alternative sigma factor sigma(E) plays an important role in intestinal survival and virulence in Vibrio cholerae. Infect. Immun. 70:5355-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lalonde, G., and P. D. O'Hanley. 1989. Development of a shuttle vector and a conjugative transfer system for Actinobacillus pleuropneumoniae. Gene 85:243-246. [DOI] [PubMed] [Google Scholar]
- 27.Langford, P. R., and J. S. Kroll. 1997. Distribution, cloning, characterisation and mutagenesis of sodC, the gene encoding copper/zinc superoxide dismutase, a potential determinant of virulence, in Haemophilus ducreyi. FEMS Immunol. Med. Microbiol. 17:235-242. [DOI] [PubMed] [Google Scholar]
- 28.Langford, P. R., B. M. Loynds, and J. S. Kroll. 1996. Cloning and molecular characterization of Cu,Zn superoxide dismutase from Actinobacillus pleuropneumoniae. Infect. Immun. 64:5035-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, J., X. Chen, C. Tan, Y. Guo, Y. Chen, S. Fu, W. Bei, and H. Chen. 2009. In vivo induced RTX toxin ApxIVA is essential for the full virulence of Actinobacillus pleuropneumoniae. Vet. Microbiol. 137:282-289. [DOI] [PubMed] [Google Scholar]
- 30.Lofdahl, S., J. E. Sjostrom, and L. Philipson. 1978. Characterization of small plasmids from Staphylococcus aureus. Gene 3:145-159. [PubMed] [Google Scholar]
- 31.Martin, P. R., R. J. Shea, and M. H. Mulks. 2001. Identification of a plasmid-encoded gene from Haemophilus ducreyi which confers NAD independence. J. Bacteriol. 183:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mecsas, J., P. E. Rouviere, J. W. Erickson, T. J. Donohue, and C. A. Gross. 1993. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 7:2618-2628. [DOI] [PubMed] [Google Scholar]
- 33.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Missiakas, D., M. P. Mayer, M. Lemaire, C. Georgopoulos, and S. Raina. 1997. Modulation of the Escherichia coli sigmaE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol. 24:355-371. [DOI] [PubMed] [Google Scholar]
- 35.O'Dwyer, C. A., K. Reddin, D. Martin, S. C. Taylor, A. R. Gorringe, M. J. Hudson, B. R. Brodeur, P. R. Langford, and J. S. Kroll. 2004. Expression of heterologous antigens in commensal Neisseria spp.: preservation of conformational epitopes with vaccine potential. Infect. Immun. 72:6511-6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raina, S., D. Missiakas, and C. Georgopoulos. 1995. The rpoE gene encoding the sigma E (sigma 24) heat shock sigma factor of Escherichia coli. EMBO J. 14:1043-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Redfield, R. J., W. A. Findlay, J. Bossé, J. S. Kroll, A. D. Cameron, and J. H. Nash. 2006. Evolution of competence and DNA uptake specificity in the Pasteurellaceae. BMC Evol. Biol. 6:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rioux, S., C. Begin, J. D. Dubreuil, and M. Jacques. 1997. Isolation and characterization of LPS mutants of Actinobacillus pleuropneumoniae serotype 1. Curr. Microbiol. 35:139-144. [DOI] [PubMed] [Google Scholar]
- 39.Rioux, S., C. Galarneau, J. Harel, M. Kobisch, J. Frey, M. Gottschalk, and M. Jacques. 2000. Isolation and characterization of a capsule-deficient mutant of Actinobacillus pleuropneumoniae serotype 1. Microb. Pathog. 28:279-289. [DOI] [PubMed] [Google Scholar]
- 40.Rongvaux, A., R. J. Shea, M. H. Mulks, D. Gigot, J. Urbain, O. Leo, and F. Andris. 2002. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur. J. Immunol. 32:3225-3234. [DOI] [PubMed] [Google Scholar]
- 41.Rouviere, P. E., P. A. De Las, J. Mecsas, C. Z. Lu, K. E. Rudd, and C. A. Gross. 1995. rpoE, the gene encoding the second heat-shock sigma factor, sigma E, in Escherichia coli. EMBO J. 14:1032-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Schurr, M. J., H. Yu, J. C. Boucher, N. S. Hibler, and V. Deretic. 1995. Multiple promoters and induction by heat shock of the gene encoding the alternative sigma factor AlgU (sigma E) which controls mucoidy in cystic fibrosis isolates of Pseudomonas aeruginosa. J. Bacteriol. 177:5670-5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheehan, B. J., J. T. Bossé, A. J. Beddek, A. N. Rycroft, J. S. Kroll, and P. R. Langford. 2003. Identification of Actinobacillus pleuropneumoniae genes important for survival during infection in its natural host. Infect. Immun. 71:3960-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szabo, M., D. Maskell, P. Butler, J. Love, and R. Moxon. 1992. Use of chromosomal gene fusions to investigate the role of repetitive DNA in regulation of genes involved in lipopolysaccharide biosynthesis in Haemophilus influenzae. J. Bacteriol. 174:7245-7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tascon, R. I., E. F. Rodriguez-Ferri, C. B. Gutierrez-Martin, I. Rodriguez-Barbosa, P. Berche, and J. A. Vazquez-Boland. 1993. Transposon mutagenesis in Actinobacillus pleuropneumoniae with a Tn10 derivative. J. Bacteriol. 175:5717-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor, D. J. 1999. Actinobacillus pleuropneumoniae, p. 343-354. In B. E. Shaw, S. D'Allaire, W. L. Mengeling, and D. J. Taylor (ed.), Diseases of swine. Blackwell Science, Oxford, United Kingdom.
- 48.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 49.Webb, S. A. R., P. R. Langford, and J. S. Kroll. 2001. A promoter probe plasmid based on green fluorescent protein. Methods Mol. Med. 67:663-677. [DOI] [PubMed] [Google Scholar]
- 50.West, S. E., M. J. Romero, L. B. Regassa, N. A. Zielinski, and R. A. Welch. 1995. Construction of Actinobacillus pleuropneumoniae-Escherichia coli shuttle vectors: expression of antibiotic-resistance genes. Gene 160:81-86. [DOI] [PubMed] [Google Scholar]
- 51.Wright, C. L., R. A. Strugnell, and A. L. Hodgson. 1997. Characterization of a Pasteurella multocida plasmid and its use to express recombinant proteins in P. multocida. Plasmid 37:65-79. [DOI] [PubMed] [Google Scholar]