Abstract
Male-specific RNA coliphages (F-RNA coliphages) have been proposed as a potential viral indicator of fecal contamination in water and foods because they are easy to culture and are a normal component of the mammalian gut flora. F-RNA coliphage plaque numbers are typically obtained by directly plating a 10-fold dilution of 1 g of fecal material, but the numbers of F-RNA coliphages shed by animals and humans may be too low for direct enumeration. Therefore, the sensitivity of detecting F-RNA coliphages in fecal material was improved by extracting and precipitating F-RNA coliphage from a 10-g fecal sample by use of polyethylene glycol (PEG). The highest recovery of F-RNA coliphage with 10% beef extract, pH 7.2, was obtained in the presence of 1 M NaCl and 10% PEG after 16 h of precipitation, but a pellet was not obtained after a short precipitation time of 2 h. There was no significant difference between eluant-to-fecal-material ratios of 4:1 and 9:1 or homogenization with a stomacher or pulsifier. F-RNA coliphage were detected in 64% (16 of 25 samples) of fecal samples from various sources when the sample size was 10 g but in 36% (9 of 25 samples) of samples when the sample size was 1 g. When F-RNA coliphage were detected in 1-g samples, they were also detected in 10-g samples. When F-RNA coliphage were detected in 10-g samples but not in 1-g samples, the levels were <100 PFU/g.
There are increasing concerns about zoonotic transmission of some animal enteric viruses which are closely related to human-pathogenic strains. Most enteric viruses are detected by molecular techniques because they cannot be cultured. Nonetheless, the presence of viral nucleic acid does not necessarily represent an infectious virus, particularly when microbial inactivation treatments are employed (25). Therefore, a culturable surrogate enteric virus of animal origin that is naturally present should be used to identify likely relationships between the detection of viral nucleic acid and the potential infectivity of the virus particle in foods of animal origin.
Male-specific RNA coliphages (F-RNA coliphages) have been proposed as a potential viral indicator of fecal contamination in water and foods because they do not readily multiply in the environment, are easy to culture, have a similar size and survival characteristics to those of human enteric viruses, and are a normal component of the mammalian gut flora (4, 8, 11, 28, 29). The frequencies of F-RNA coliphages recovered from feces of pigs, cattle, and poultry range from 4 to 70%, at levels of <1 to 6.8 log PFU/g (2, 8, 9, 21, 31). In general, F-RNA coliphages are recovered at higher frequencies and levels from pigs and poultry than from cattle. However, Calci et al. (2) reported that 53% of poultry shed <1 log PFU/g of F-RNA coliphage. Therefore, the amount of sample and the manner in which it is processed for enumeration may require closer examination to increase the probability of recovery, as in many cases the numbers of F-RNA coliphages in fecal samples may be too low for direct enumeration by culture and/or molecular techniques.
Coliphages and other enteric viruses can be unattached or absorbed onto or occluded within fecal particulates, where the fraction of viruses associated with solids varies from <1 to 100% (6, 10, 26). However, most coliphages associated with fecal solids are absorbed rather than embedded and are easily eluted (26). In spite of this, it is common practice to suspend 1 g of fecal material in phosphate-buffered saline (PBS) or saline-peptone and to use the supernatant after settling or clarification for detection and enumeration of F-RNA coliphages by plaque assay (8, 12, 21). Similarly, nucleic acids are often extracted from a portion of the clarified supernatant for the detection of enteric and hepatic viruses by reverse transcription-PCR (RT-PCR) (14, 16, 18, 19). Guzmán et al. (7) optimized the extraction of somatic coliphages from whole suspensions of sludge, soil, and treated biowaste from naturally contaminated samples because somatic coliphage numbers recovered from the supernatant from naturally contaminated sewage sludge, with and without elution, were about 1 log unit lower than those recovered from entire sewage sludge and the eluted pellet. The extraction procedure was developed for routine testing in minimally equipped laboratories and is proposed to be suitable for F-RNA coliphages because the recommended pH of the elution buffer is 7.2, in contrast to pH 9, which is often used for virus extraction but which may inactivate F-RNA coliphages. The objective of this study was to improve the sensitivity of detecting F-RNA coliphages in fecal samples containing <100 PFU/g by applying and modifying the extraction protocol of Guzmán et al. (7), using standard or modern laboratory equipment such as a pulsifier, and to combine the extraction with an optimized concentration step using polyethylene glycol (PEG) precipitation.
MATERIALS AND METHODS
F-RNA coliphage MS2 stock production and enumeration.
F-RNA coliphage prototype MS2 (ATCC 15597-B1), obtained from the American Type Culture Collection (Manassas, VA), was propagated, stored, and enumerated by plaque assay according to the ISO 10705-1 protocol (12), using Salmonella enterica WG49 as the host strain. Each dilution was plated in triplicate.
Optimization of MS2 precipitation.
Eluant solutions composed of 10% beef extract (Difco, Fisher Scientific, Edmonton, Alberta, Canada), pH 7.2, containing 0.2, 0.3, or 1.0 M NaCl were inoculated with MS2 to a final concentration of 1 × 104 PFU/ml. PEG 8000 (Sigma-Aldrich Canada Ltd., Oakville, Ontario, Canada) was added to 40-ml portions of inoculated eluant in 50-ml centrifuge tubes to obtain final concentrations (wt/vol) of 8, 10, 12, and 14% PEG. The centrifuge tubes were shaken by hand until the PEG was dissolved, incubated at 4°C for 2 h or overnight, and centrifuged at 10,000 × g for 30 min at 4°C. The pellets were suspended in sterile PBS to a final volume of 3 ml and used for enumeration of MS2.
Naturally contaminated fecal material.
Portions of fecal material that were previously tested for the presence or absence of F-RNA coliphages before being frozen and subsequently stored at −20°C were used in these studies. Fecal material used in these studies was of swine origin unless indicated otherwise. When the recoveries of F-RNA coliphage from 10 g and 1 g of inoculated or naturally contaminated fecal material were compared in parallel, a 1-g portion of the fecal sample was suspended in 4 ml of peptone (0.1%)-saline (0.85%), the diluent used for the ISO 10705-1 standard method, homogenized with a vortex machine, and then enumerated. The 10-g samples were processed as described below.
Inoculated fecal material.
Batches of fecal material that had previously tested negative for F-RNA coliphages were thawed, mixed thoroughly, and divided into 10- or 11-g portions in stomacher bags. Each portion was mixed thoroughly with MS2 at levels ranging from approximately 1 × 103 to 1 × 101 PFU/g and incubated for 72 h at 4°C to facilitate the attachment of MS2.
Homogenization, elution, and precipitation of F-RNA coliphage from fecal material.
Ten-gram fecal samples were homogenized in 40 or 90 ml of eluant (10% beef extract, 1 M NaCl, pH 7.2) with a stomacher (Seward, VWR, Edmonton, Alberta, Canada) for 2 min or with a pulsifier (Microbiology International, Frederick, MD) for 15 s. Each homogenate was transferred to one or two 50-ml centrifuge tubes, depending on the initial eluant volume, and clarified by centrifugation at 4,000 × g for 30 min at 4°C. MS2 was precipitated from the supernatants with 10% PEG. The pellets obtained from fecal samples that were homogenized with 90 ml of eluant were combined into a single tube. Pellet material from each 10-g portion was resuspended in sterile PBS to a final volume of 4 ml and then enumerated.
Statistical analysis.
The % recovery for MS2 was obtained by dividing the number of PFU recovered from the reconstituted pellet, multiplied by the concentration factor (ml of pellet/initial volume), by the number of PFU in the suspension before concentration and then multiplying the value by 100. The Anderson-Darling test for normal distribution was applied to each data set for each treatment. Mean values for the data sets that followed a normal distribution were separated by the Tukey test of the general linear model procedure, and data sets that did not follow a normal distribution were tested with the nonparametric Kruskal-Wallis test. Minitab, version 14, statistical software was used for the statistical analysis (Minitab Inc., State College, PA).
RESULTS
Effects of PEG and NaCl concentrations on recovery of MS2.
The concentrations of PEG and NaCl used for the precipitation of F-RNA coliphages and other enteric viruses are highly variable (Table 1), ranging from 8 to 15% and 0.1 to 1.0 M, respectively, for F-RNA coliphages (3, 11, 15). When 8% PEG 8000 with 0.3 M NaCl was used in initial trials to precipitate the F-RNA coliphage prototype MS2 from 10% beef extract, a pellet was not observed after two cycles of centrifugation at 10,000 × g for 30 min. Therefore, various concentrations of PEG and NaCl were evaluated to optimize the recovery of MS2. The highest mean recovery of MS2 from inoculated beef extract was 92% (±12.6%) when 10% PEG and 1 M NaCl were used (Table 2), which was significantly (P < 0.05) higher than the recovery achieved when other conditions were tested. Thus, F-RNA coliphage were eluted with 10% beef extract, pH 7.2, containing 1 M NaCl and were precipitated with a final concentration of 10% PEG in subsequent trials.
TABLE 1.
Concentrations of PEG and NaCl and centrifugation conditions used for precipitation of viruses
| PEG molecular wt | PEG concn (%) | NaCl concn (M) | Centrifugation conditions
|
Virus(es) detected | Reference | |
|---|---|---|---|---|---|---|
| Speed (×g) | Time (min) | |||||
| 6000 | 8 | 0.3 | 10,000 | 20 | F-RNA coliphages, enterovirus | 15 |
| 15 | 0.2 | 10,000 | 20 | F-RNA coliphages, enterovirus | 15 | |
| 8 | 0.1 | 6,700 | 30 | F-RNA coliphages, somatic virus | 11 | |
| 10 | Varieda | 10,000 | 90 | Astrovirus, HAV, poliovirus | 27 | |
| NIb | 8 | 0.3 | 14,000 | 15 | Enterovirus | 24 |
| 8000 | 10 | 1.0 | 11,000 | 10 | F-RNA coliphages | 3 |
| 12 | 0.9 | 10,000 | 30 | F-RNA coliphages | 20 | |
| 12 | Varieda | 6,200 | 20 | Astrovirus, HAV, poliovirus | 27 | |
| 13 | 0.2 | 7,000 | 30 | HAV, poliovirus | 22 | |
Varied with elution buffer.
NI, not identified.
TABLE 2.
Recovery of MS2 precipitated with 10, 12, or 14% PEG from 10% beef extract, pH 7.2, containing 0.2 or 1 M NaCl
| NaCl concn (M) | PEG concn (%) | % Recoverya
|
|
|---|---|---|---|
| Mean | SD | ||
| 0.2 | 10 | 64.3A | 6.3 |
| 12 | 72.8A,B | 7.9 | |
| 14 | 73.3A,B | 6.7 | |
| 1.0 | 10 | 92.3C | 12.6 |
| 12 | 81.2B | 12.8 | |
| 14 | 73.1A,B | 7.0 | |
Values are the means for five assays analyzed in triplicate. Mean values followed by the same letter are not significantly different at a P level of 0.05.
Effect of precipitation time on recovery of MS2.
A pellet was not obtained after 2 h in two separate trials after one and two cycles of centrifugation at 10,000 × g for 30 min when MS2 was precipitated with 10% PEG and 1 M NaCl from beef extract, but a pellet was obtained after 16 h of precipitation.
Effect of ratio of eluant to fecal material on recovery of MS2.
The effect of decreasing the ratio of eluant to fecal material from 9:1 to 4:1 on the recovery of MS2 was examined to exploit the convenience of using a single disposable 50-ml centrifuge tube for 10 g of fecal material. There was no significant difference between the mean log numbers (4.48 ± 0.07 and 4.53 ± 0.08) of MS2 phage recovered from artificially inoculated fecal material with eluant ratios of 9:1 and 4:1, respectively, and thus an eluant ratio of 40 ml to 10 g of fecal material was used to elute F-RNA coliphage in subsequent studies.
Homogenization.
The log numbers of F-RNA coliphages recovered after homogenization with a stomacher or pulsifier from fecal samples that were naturally contaminated with various levels of F-RNA coliphages (Table 3) did not follow a normal distribution (data not shown). Therefore, the nonparametric Kruskal-Wallis test was applied to determine that there was no significant difference between fecal samples homogenized with a stomacher or pulsifier (P = 0.78). The average ratio of the recovery of F-RNA coliphages from samples homogenized with a pulsifier to that from samples homogenized with a stomacher was 0.96, with a range of 0.61 to 1.14 (Table 3).
TABLE 3.
Numbers of F-RNA phages recovered (log PFU/g) from an eluted PEG precipitate obtained from 10 g of naturally contaminated fecal matter homogenized in 10% beef extract, 1 M NaCl, pH 7.2, with a stomacher or pulsifier
| Sample no. | Recovery (log PFU/g)
|
P/S ratioa | |||
|---|---|---|---|---|---|
| Stomacher
|
Pulsifier
|
||||
| Meanb | SD | Meanb | SD | ||
| 1 | 4.20 | 0.10 | 4.47 | 0.03 | 1.06 |
| 2 | 2.75 | 0.09 | 2.27 | 0.09 | 0.83 |
| 3 | 3.57 | 0.02 | 4.08 | 0.14 | 1.14 |
| 4 | 1.75 | 0.09 | 1.68 | 0.04 | 0.96 |
| 5 | 1.95 | 0.21 | 1.19 | 0.36 | 0.61 |
| 6 | 2.76 | 0.04 | 2.57 | 0.37 | 0.93 |
| 7 | 3.80 | 0.05 | 3.88 | 0.10 | 1.02 |
| 8 | 1.32 | 0.07 | 1.50 | 0.02 | 1.14 |
| 9 | 1.48 | 0.04 | 1.36 | 0.05 | 0.92 |
Mean log PFU from pulsifier/mean log PFU from stomacher.
Mean of triplicate analyses.
Effect of sample size on recovery of and limit of detection for F-RNA coliphages.
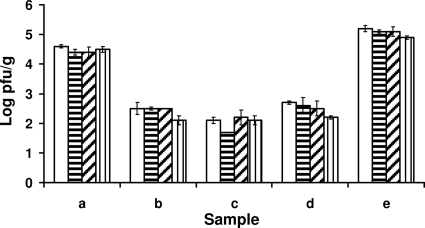
The recovery of F-RNA coliphages from 10-g samples of five naturally contaminated fecal samples that previously tested positive was examined at various steps of the elution and concentration procedure, i.e., after homogenization, after clarification, and after precipitation. There was some loss of F-RNA coliphage during the precipitation step for four of the samples (Fig. 1). Possibly due to increased handling, the numbers of F-RNA coliphages recovered from 10 g after precipitation were ≤0.5 log lower than those from 1 g for all five samples (Fig. 1). When fecal material was inoculated with approximately 1 × 102 PFU/g, MS2 was detected in 10 of 10 samples for 10-g samples but in only 4 of 10 samples for 1-g samples, and with inoculum levels of approximately 1 × 101 PFU/g, 5 of 10 samples were positive for MS2 for 10-g samples and 1 of 10 samples were positive for MS2 for 1-g samples. F-RNA coliphages were detected in 16 of 25 fecal samples from various sources when the sample size was 10 g but in 9 of 25 samples when the sample size was 1 g (Table 4). In instances where F-RNA coliphages were detected in 10-g samples but not in 1-g samples, the levels were <100 PFU/g. Furthermore, there were no occasions where F-RNA coliphages were detected in 1-g samples but not in 10-g samples. In some cases, the numbers of F-RNA coliphages recovered from 10-g samples were more than 1 log unit higher or lower than the numbers recovered from 1-g samples. One-gram portions of the 25 fecal samples used in this study were previously tested for the presence of F-RNA coliphages before they were frozen. The results were comparable before and after freezing, with a mean log PFU/g for the nine positive samples of 3.5 before freezing and 3.7 after freezing.
FIG. 1.
Numbers of F-RNA phages recovered (log PFU/g) from 1 g of naturally contaminated fecal matter after homogenization in NaCl-peptone (1:4) (white bars) and from 10 g of naturally contaminated fecal matter after homogenization in 10% beef extract, 1 M NaCl, pH 7.2 (1:4) (bars with horizontal stripes), after clarification (hatched bars), and after PEG precipitation (bars with vertical stripes).
TABLE 4.
Recovery of F-RNA coliphages (log PFU/g) from 1- or 10-g aliquots of 25 fecal samples obtained from different sources
| Source | % Recoverya
|
|
|---|---|---|
| 1-g samples | 10-g samples | |
| Cattle | — | — |
| — | — | |
| 2.1 | 2.0 | |
| — | 0.9 | |
| — | — | |
| — | 1.8 | |
| Swine | 3.5 | 3.5 |
| 2.5 | 1.2 | |
| 2.7 | 2.2 | |
| 5.1 | 5.9 | |
| — | 1.5 | |
| Horses | — | — |
| — | — | |
| — | −0.1 | |
| Sheep | — | 1.5 |
| Goats | — | — |
| Chickens | — | 1.9 |
| 5.5 | 6.6 | |
| Turkeys | 3.4 | 3.1 |
| Mink | 5.8 | 6.7 |
| Humans | 2.9 | 2.1 |
| — | — | |
| — | 0.9 | |
| — | — | |
| — | — | |
—, not detected.
DISCUSSION
This systematic study of recovering F-RNA coliphages from fecal material demonstrated that the probability of recovering F-RNA coliphages from naturally contaminated fecal samples containing small coliphage numbers is increased when F-RNA coliphages are eluted from 10 g of fecal material and then precipitated with PEG, as opposed to standard methods of direct plating a suspension from a 1-g sample. A literature survey, summarized in Table 1, indicated that the conditions for the precipitation of F-RNA coliphages and other enteric viruses vary widely, from 8 to 15% PEG and from 0.1 to 1.0 M NaCl. Since the concentrated virus precipitate may subsequently be used for molecular detection assays, it is desirable to minimize the amount of interfering substances that may inhibit RT-PCR. However, initial attempts to precipitate MS2 from 10% beef extract were unsuccessful with concentrations at the low end of the range (8% PEG 8000 and 0.3 M NaCl), which led to the necessity to optimize the PEG precipitation step. The highest mean recovery rate of MS2 from inoculated beef extract was obtained with 10% PEG and 1 M NaCl. These PEG and NaCl concentrations were also used in SM buffer by Dawson et al. (3), but with a shorter time and faster speed of centrifugation for the purification of MS2 stocks.
Despite literature reports that virus yields are similar after 2 h of PEG precipitation and after overnight precipitation (1, 22), a pellet was not obtained after 2 h of precipitation with 10% PEG and 1 M NaCl. Schwab et al. (22) found no significant differences in the recovery efficiencies for poliovirus and hepatitis A virus (HAV) after 2 or 15 h of precipitation with 13% PEG 8000 and 0.2 M NaCl from a beef extract-glycine solution, while Atmar et al. (1) determined that a 2-h precipitation period with 8% PEG 6000 was satisfactory for the recovery of HAV from oysters. Although MS2 is similar in size to poliovirus and HAV, perhaps the virus type, medium composition, and % PEG influence the time required for effective precipitation. The variable results suggest that the conditions for PEG precipitation should be evaluated carefully for different viruses and elution buffers.
Guzmán et al. (7) based the optimized extraction of somatic coliphages from sludge, soil, and treated biowaste from naturally contaminated samples for enumeration by culture methods on a literature survey (Horizontal-HYG 2005 survey) (17). The minimum recommended sample size of the Horizontal-HYG 2005 survey is 10 g. The extraction method of Guzmán et al. (7) is based on a sample size of 25 g or ml and an eluant-to-sample ratio of 9:1. However, a lower eluant-to-sample ratio will reduce costs and sample processing time and may increase efficiency because more samples can be processed at one time. There was no significant difference between eluant-to-sample ratios of 1:9 and 1:4. Since some loss of F-RNA coliphage was observed during the precipitation step, there is a potential for a higher recovery rate by use of a lower eluant-to-sample ratio because increased handling of the pellet could lead to additional loss of viruses when pellets from multiple tubes are combined. In this study, there were no instances where F-RNA coliphages were detected in 1-g samples but not in 10-g samples.
Guzmán et al. (7) recommended homogenization using magnetic stirring because of lower equipment costs, but a significant difference was not observed between the homogenization methods that were evaluated, including the use of a stomacher. A pulsifier, which homogenizes samples by creating shock waves through high-speed beating action with an oscillating ring, is a recent alternative to a stomacher, which uses crushing action to homogenate samples (5). The pulsifier has been tested on food samples, and the recovery of microorganisms from foods homogenized with a pulsifier is equivalent to that from foods homogenized with a stomacher (5, 13, 23, 30). With the exception of foods that have a low level of cohesiveness, suspensions that are homogenized with a pulsifier contain less debris than that obtained with a stomacher, which is an advantage for downstream reactions such as PCR. New laboratories that work on the detection of viruses or bacteria in foods and environmental samples may thus choose to invest in a pulsifier over a stomacher, but there are neither data in the literature on the use of a pulsifier for homogenization of fecal material nor data on the recovery of F-RNA coliphages from pulsified suspensions. There was no significant difference between fecal samples homogenized with a stomacher and those homogenized with a pulsifier, and the ratios of results with the pulsifier to those with the stomacher were comparable to the findings of others (5, 13, 23, 30). Because fecal material has a loose consistency, the amount of debris generated with a pulsifier is expected to be similar to that obtained with a stomacher, and therefore there is no advantage for downstream PCRs by use of a pulsifier as opposed to a stomacher for homogenizing fecal material. This is the first study to report the use of a pulsifier for recovering F-RNA coliphages as well as for homogenizing fecal samples.
The results obtained in this study indicate that the sensitivity in detecting F-RNA coliphages in naturally contaminated fecal samples containing <100 PFU/g is higher with the elution method using 10 g of fecal material. Overall, the prevalence of F-RNA coliphages in fecal material increased from 9 of 25 samples with 1-g samples to 16 of 25 samples with 10-g samples, and there were no occasions where F-RNA coliphages were detected in 1-g samples but not in 10-g samples. More specifically, the prevalence of F-RNA coliphages in fecal material increased from four of five samples to five of five samples for pigs, from one of six samples to three of six samples for cattle, from zero of three samples to one of three samples for horses, and from one of five samples to two of five samples for humans. The levels of F-RNA coliphage detected in fecal samples in this study are in agreement with those in other studies (2, 8, 9, 21), where swine and poultry shed higher levels of F-RNA coliphages than did cattle, horses, sheep, goats, and humans. To our knowledge, there are no previous reports on the levels of F-RNA coliphage in minks.
Since levels of F-RNA coliphages vary widely in fecal samples for the same species, it is not surprising that the log numbers in fecal material from different animals did not follow a normal distribution, and some of the variation observed between 1- and 10-g samples from the same animal could be due to the uneven distribution of F-RNA coliphages within the fecal material. Although the method improved the detection of small numbers of F-RNA coliphages in inoculated and naturally contaminated fecal samples, the method is more time-consuming and costly than direct plating of 1-g samples due to the increased sample processing time, overnight precipitation, and additional laboratory supplies, such as pulsifier bags and centrifuge tubes.
Acknowledgments
This research was supported by Agriculture and Agri-Food Canada Research Branch peer-reviewed research projects 75 and 162.
The fecal samples analyzed for Table 4 were kindly supplied by Ed Topp of Agriculture and Agri-Food Canada, London, Ontario, Canada. We thank Ken Li for technical support in processing of the samples.
Footnotes
Published ahead of print on 31 July 2009.
This is contribution 1134 from the Agriculture and Agri-Food Research Centre, Lacombe, Alberta, Canada.
REFERENCES
- 1.Atmar, R. L., T. G. Metcalf, F. H. Neill, and M. K. Estes. 1993. Detection of enteric viruses in oysters by using the polymerase chain reaction. Appl. Environ. Microbiol. 59:631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calci, K. R., W. Burkhardt III, W. D. Watkins, and S. R. Rippey. 1998. Occurrence of male-specific bacteriophage in feral and domestic animal wastes, human feces, and human associated waters. Appl. Environ. Microbiol. 64:5027-5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawson, D. J., A. Paish, L. M. Staffel, I. J. Seymour, and H. Appleton. 2005. Survival of viruses on fresh produce, using MS2 as a surrogate for norovirus. J. Appl. Microbiol. 98:203-209. [DOI] [PubMed] [Google Scholar]
- 4.Doré, W. J., K. Henshilwood, and D. N. Lees. 2000. Evaluation of F-specific RNA bacteriophage as a candidate human enteric virus indicator for bivalve molluscan shellfish. Appl. Environ. Microbiol. 66:1280-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fung, D. Y. C., A. N. Sharpe, B. C. Hart, and Y. Liu. 1998. The Pulsifier: a new instrument for preparing food suspensions for microbiological analysis. J. Rapid Methods Autom. Microbiol. 6:43-49. [Google Scholar]
- 6.Gerba, C. P., C. H. Stagg, and M. G. Abadh. 1978. Characterization of sewage solids-associated viruses and behaviour in natural waters. Water Res. 12:805-812. [Google Scholar]
- 7.Guzmán, C., J. Jofre, A. R. Blanch, and F. Lucena. 2007. Development of a feasible method to extract somatic coliphages from sludge, soil and treated biowaste. J. Virol. Methods 144:41-48. [DOI] [PubMed] [Google Scholar]
- 8.Havelaar, A. H., K. Furuse, and W. M. Hogeboom. 1986. Bacteriophages and indicator bacteria in human and animal feces. J. Appl. Bacteriol. 60:255-262. [DOI] [PubMed] [Google Scholar]
- 9.Havelaar, A. H., W. M. Pot-Hogeboom, K. Furuse, R. Pot, and M. P. Hormann. 1990. F-specific RNA bacteriophages and sensitive host strains in faeces and wastewater of human and animal origin. J. Appl. Bacteriol. 69:30-37. [DOI] [PubMed] [Google Scholar]
- 10.Hejkal, T. W., F. M. Wellings, P. A. LaRock, and A. L. Lewis. 1979. Survival of poliovirus within organic solids during chlorination. Appl. Environ. Microbiol. 38:114-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsu, F. C., Y. C. Shieh, and M. D. Sobsey. 2002. Enteric bacteriophages as potential fecal indicators in ground beef and poultry meat. J. Food Prot. 65:93-99. [DOI] [PubMed] [Google Scholar]
- 12.International Organization for Standardization. 1995. ISO 10705-1. Water quality. Detection and enumeration of bacteriophages. 1. Enumeration of F-specific RNA bacteriophages. International Organization for Standardization, Geneva, Switzerland.
- 13.Kang, D. H., R. H. Dougherty, and D. Y. C. Fung. 2001. Comparison of Pulsifier™ and Stomacher™ to detach microorganisms from lean meat tissues. J. Rapid Methods Autom. Microbiol. 9:27-32. [Google Scholar]
- 14.Leblanc, D., P. Ward, M. J. Gagné, E. Poitras, P. Müller, Y. L. Trottier, C. Simard, and A. Houde. 2007. Presence of hepatitis E virus in a naturally infected swine herd from nursery to slaughter. Int. J. Food Microbiol. 117:160-166. [DOI] [PubMed] [Google Scholar]
- 15.Lewis, G. D., A. Hough, D. H. Green, J. E. Hay, and L. R. Ferguson. 1996. Modification of the polyethylene glycol 6000 precipitation method for recovering human and indicator viruses from oysters and mussels. N. Z. J. Mar. Freshw. Res. 30:443-447. [Google Scholar]
- 16.Liu, B. L., P. R. Lambden, H. Günther, P. Otto, M. Elschner, and I. N. Clarke. 1999. Molecular characterization of a bovine-like enteric calicivirus: relationship to the Norwalk-like viruses. J. Virol. 73:819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lucena, F., A. R. Blanch, and J. Jofre. 2005. Horizontal standards on hygienic parameters for implementation of EU directives on sludge, soil and treated bio-waste. Critical review on methods for bacteriophages (and viruses) to be monitored in EU in sludges, soils and treated biowastes. SSPI-CT-2004-513660. University of Barcelona, Barcelona, Spain. http://www.ecn.nl/docs/society/horizontal/hor_desk_29_Bacteriophages_Critical_Review.pdf.
- 18.Oka, T., K. Katayama, G. S. Hansman, T. Kageyama, S. Ogawa, F.-T. Wu, P. A. White, and N. Takeda. 2006. Detection of human sapovirus by real-time reverse transcription-polymerase chain reaction. J. Med. Virol. 78:1347-1353. [DOI] [PubMed] [Google Scholar]
- 19.Park, S. I., C. Jeong, H. H. Kim, S. H. Park, S. J. Park, B. H. Hyun, D. K. Yang, S. K. Kim, M. I. Kang, and K. O. Cho. 2007. Molecular epidemiology of bovine noroviruses in South Korea. Vet. Microbiol. 124:125-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polaczyk, A. L., J. Narayanan, T. L. Cromeans, D. Hahn, J. M. Roberts, J. E. Amburgey, and V. R. Hill. 2008. Ultrafiltration-based techniques for rapid and simultaneous concentration of multiple microbe classes from 100-L tap water samples. J. Microbiol. Methods 73:92-99. [DOI] [PubMed] [Google Scholar]
- 21.Schaper, M., J. Jofre, M. Uys, and W. O. K. Grabow. 2002. Distribution of genotypes of F-specific RNA bacteriophages in human and non-human sources of faecal pollution in South Africa and Spain. J. Appl. Microbiol. 92:657-667. [DOI] [PubMed] [Google Scholar]
- 22.Schwab, K. J., R. De Leon, and M. D. Sobsey. 1995. Concentration and purification of beef extract mock eluates from water samples for the detection of enteroviruses, hepatitis A virus, and Norwalk virus by reverse transcription-PCR. Appl. Environ. Microbiol. 61:531-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharpe, A. N., E. M. Hearn, and J. Kovacs-Nolan. 2000. Comparison of membrane filtration rates and hydrophobic grid membrane filter coliform and Escherichia coli count in food suspensions using paddle-type and pulsifier sample preparation procedures. J. Food Prot. 63:126-130. [DOI] [PubMed] [Google Scholar]
- 24.Shieh, Y. C., C. I. Wong, J. A. Krantz, and F. C. Hsu. 2008. Detection of naturally occurring enteroviruses in waters using direct RT-PCR and integrated cell culture-RT-PCR. J. Virol. Methods 149:184-189. [DOI] [PubMed] [Google Scholar]
- 25.Sobsey, M. D., D. A. Battigelli, G. A. Shin, and S. Newland. 1998. RT-PCR amplification detects inactivated viruses in water and wastewater. Water Sci. Technol. 38:91-94. [Google Scholar]
- 26.Stagg, C. H., C. Wallis, C. H. Ward, and C. P. Gerba. 1978. Chlorination of solids-associated coliphages. Prog. Water Technol. 10:381-387. [Google Scholar]
- 27.Traore, O., C. Arnal, B. Mignotte, A. Maul, H. Laveran, S. Billaudel, and L. Schwartzbrod. 1998. Reverse transcriptase PCR detection of astrovirus, hepatitis A virus, and poliovirus in experimentally contaminated mussels: comparison of several extraction and concentration methods. Appl. Environ. Microbiol. 64:3118-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woody, M. A., and D. O. Cliver. 1995. Effects of temperature and host cell growth phase on replication of F-specific RNA coliphage Qβ. Appl. Environ. Microbiol. 61:1520-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woody, M. A., and D. O. Cliver. 1997. Replication of coliphage Qβ as affected by host cell number, nutrition, competition from insusceptible cells and non-FRNA coliphages. J. Appl. Microbiol. 82:431-440. [DOI] [PubMed] [Google Scholar]
- 30.Wu, V. C. H., P. Jitareerat, and D. Y. C. Fung. 2003. Comparison of the pulsifier and the stomacher for recovering microorganisms in vegetables. J. Rapid Methods Autom. Microbiol. 11:145-152. [Google Scholar]
- 31.Yee, S. Y. F., N. Y. Fong, G. T. Fong, O. J. Tak, G. T. Hui, and Y. S. Ming. 2006. Male specific RNA coliphages detected by plaque assay and RT-PCR in tropical river waters and animal fecal matter. Int. J. Environ. Health Res. 16:59-68. [DOI] [PubMed] [Google Scholar]