Abstract
Clostridium perfringens type A isolates carrying a chromosomal copy of the enterotoxin (cpe) gene are involved in the majority of food poisoning (FP) outbreaks, while type A isolates carrying a plasmid-borne cpe gene are involved in C. perfringens-associated non-food-borne (NFB) gastrointestinal diseases. To cause diseases, C. perfringens spores must germinate and return to active growth. Previously, we showed that only spores of FP isolates were able to germinate with K+ ions. We now found that the spores of the majority of FP isolates, but none of the NFB isolates, germinated with the cogerminants Na+ and inorganic phosphate (NaPi) at a pH of ∼6.0. Spores of gerKA-KC and gerAA mutants germinated to a lesser extent and released less dipicolinic acid (DPA) than did wild-type spores with NaPi. Although gerKB spores germinated to a similar extent as wild-type spores with NaPi, their rate of germination was lower. Similarly, gerO and gerO gerQ mutant spores germinated slower and released less DPA than did wild-type spores with NaPi. In contrast, gerQ spores germinated to a slightly lesser extent than wild-type spores but released all of their DPA during NaPi germination. In sum, this study identified NaPi as a novel nutrient germinant for spores of most FP isolates and provided evidence that proteins encoded by the gerKA-KC operon, gerAA, and gerO are required for NaPi-induced spore germination.
Clostridium perfringens is a gram-positive, anaerobic, spore-forming, pathogenic bacterium that causes a wide array of gastrointestinal (GI) diseases in both animals and humans (14, 15). However, Clostridium perfringens type A food poisoning (FP) is the most common C. perfringens-associated illness among humans and is currently ranked as the third most commonly reported food-borne disease (14). Mostly type A isolates that produce the C. perfringens enterotoxin have been associated with C. perfringens-related GI illnesses (14). C. perfringens cpe-positive isolates can carry the cpe gene on either the chromosome or a plasmid (3, 4). Interestingly, the majority of C. perfringens type A FP isolates carry a chromosomal copy of the cpe gene, while all non-food-borne (NFB) GI disease isolates carry a plasmid copy of cpe (3, 4, 11, 29). The genetic differences involved in the pathogenesis differences between C. perfringens FP and NFB isolates seem to involve more factors than the simple location of the cpe gene. For example, spores of FP isolates are strikingly more resistant than spores of NFB isolates to heat (100°C) (27), cold (4°C), and freezing (−20°C) temperatures (12) and to chemicals used in food industry settings (13), making FP spores more suited for FP environments. Under favorable environmental conditions, these dormant spores germinate to return to active growth, proliferate to high numbers, and then produce toxins to cause disease (14).
Bacterial spores germinate when they sense the presence of nutrients (termed germinants) in the environment through their cognate receptors located in the spore inner membrane (18). For C. perfringens, some nutrients that initiate germination include l-asparagine, KCl, a mixture of l-asparagine and KCl, and a 1:1 chelate of Ca2+ and dipicolinic acid (DPA) (Ca-DPA) (20). The main receptor(s) involved in sensing these compounds is the GerKA and/or GerKC receptor(s), which is required for l-asparagine and Ca-DPA and only partially required for KCl and an l-asparagine-KCl mixture (20, 21). Upon binding of the germinant to its cognate receptor, a variety of biophysical events take place, including the release of monovalent ions (i.e., Na+, K+, and Li+) followed by the release of the spore's large depot of Ca-DPA (28). In Bacillus subtilis, release of Ca-DPA acts as a signal for activation of the cortex-lytic enzyme CwlJ (17). In contrast, Ca-DPA release from the spore core has no role in triggering cortex hydrolysis during C. perfringens spore germination (19, 22, 23); instead, Ca-DPA induces germination via the GerKA and/or GerKC receptor(s) (20, 21). Degradation of the cortex in both species leads to hydration of the spore core up to levels found in growing bacteria, allowing resumption of enzymatic activity and metabolism, and consequently outgrowth (22, 28).
The ability of bacterial spores to sense different nutrients appears to be tightly regulated by their adaptation to different environmental niches. For example, spores of FP isolates, but not NFB isolates, are capable of germinating with KCl (20), an intrinsic mineral of meats that are most commonly associated with FP, suggesting an adaptation of FP isolates to FP environments. In addition, the level of inorganic phosphate (Pi) is also significant in meat products (42 to 60 mM) (USDA [http://fnic.nal.usda.gov/nal_display/index.php?info_center=4&tax_level=1&tax_subject=242]). Similarly, sodium ions are also present in meats (∼30 mM), especially in processed meat products (∼300 to 400 mM) (USDA). Consequently, in this study we found that Na+ and Pi at ∼100 mM and pH 6.0 are unique cogerminants for spores of C. perfringens type A FP isolates, act through the GerKA and/or GerKC and GerAA receptors, and also require the presence of the putative Na+/K+-H+ antiporter, GerO, for normal germination.
MATERIALS AND METHODS
Bacterial strains and plasmids.
C. perfringens isolates used in this study included the following: six FP isolates carrying a chromosomal cpe gene, namely, SM101 (electroporatable derivative of an FP type A isolate; NCTC8798) (30), NCTC8798 (6), NCTC10239, E13, FD1041 (3), and 6263 (8); three NFB isolates carrying a plasmid copy of the cpe gene, namely, F4969, NB16, and B40 (3); one type C cpe-negative isolate, JGS1495 (from a diarrheic pig) (7); C. perfringens SM101 ger mutants DPS101 (gerKA-KC), DPS103 (gerAA) (20), and DPS108 (gerKB) (21); and C. perfringens SM101 antiporter mutants DPS113 (gerQ), DPS116 (gerO), and DPS115 (gerO gerQ) (24).
Spore preparation and purification.
Starter cultures (10 ml) of C. perfringens isolates were prepared by overnight growth at 37°C in fluid thioglycolate broth (Difco) as described previously (10). Sporulating cultures of C. perfringens were prepared by inoculating 0.2 ml of a fluid thioglycolate starter culture into 10 ml of Duncan-Strong sporulating medium (5); this culture was incubated for 24 h at 37°C to form spores, as confirmed by phase-contrast microscopy. Spore preparations were prepared by scaling up the latter procedure. Spore preparations were cleaned by repeated centrifugation and washing with sterile distilled water until spores were >99% free of sporulating cells, cell debris, and germinated spores and then suspended in distilled water at a final optical density at 600 nm (OD600) of ∼6 and stored at −20°C (20).
Germination assays.
Spores at an OD600 of ∼1.0 were routinely heat shocked (80°C; 10 min), cooled in water at ambient temperature, and germinated at 40°C and pH 6.0 with 100 mM sodium phosphate (NaPi), citric acid, sodium citrate, 3-morpholinopropanesulfonic acid (MOPS), maleic acid, acetic acid, or sodium acetate as previously described (20). Spore germination was routinely measured by monitoring the OD600 of spore cultures (Smartspec 3000 spectrophotometer; Bio-Rad Laboratories, Hercules, CA), which falls ∼60% upon complete spore germination, and levels of germination were confirmed by phase-contrast microscopy. To measure the rate at which the spore population decreased its OD, the maximum rates of spore germination were determined by measuring the OD600 of germinating cultures every 2.5 min, the maximum slopes were calculated, and maximum rates were expressed as the maximum rate of loss in OD600 of the spore suspension relative to the initial OD600 of the culture.
DPA release.
DPA release during NaPi germination was measured by incubating a heat-activated spore suspension at an OD600 of 1.0 at 40°C with 250 mM NaPi (pH 6.0). Aliquots (1 ml) of germinating cultures were centrifuged for 3 min in a microcentrifuge, and DPA in the supernatant fluid was measured by monitoring the OD270 as described previously (2, 20). To measure the total spore DPA content, a 1-ml aliquot of the germination culture was boiled for 60 min and centrifuged for 5 min in a microcentrifuge, and the OD270 of the supernatant fluid was measured (2, 20). In C. perfringens, ∼90% of the material absorbing at 270 nm comprises DPA (22).
Statistical analyses.
Student's t test was used for specific comparisons. All experiments were repeated four times with two independent spore preparations.
RESULTS
Germination of C. perfringens spores with NaPi.
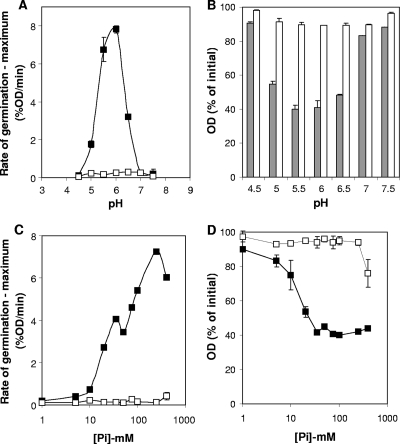
In our previous study, we observed that 25 mM NaPi (pH 7.0) was able to induce little, but significant, germination of C. perfringens spores (20). To evaluate if NaPi could indeed induce spore germination, we assayed the germination of spores of SM101 (an FP isolate) and F4969 (an NFB isolate) with various concentrations of NaPi at various pHs. Surprisingly, NaPi (100 mM) was able to induce germination of SM101 spores at a pH range of 5.0 to 6.5, but not below pH 5.0 or above pH 7.0 (Fig. 1A). Although the rates of germination at pH 5.0 and 6.5 were much lower than that at pH 6.0, SM101 spores were able to germinate to similar extents at a pH range of 5.0 to 6.5 (Fig. 1B). Indeed, phase-contrast microscopy indicated that >90% of SM101 spores had become phase dark when germinated with NaPi at a pH range of 5.0 to 6.5 (data not shown). However, NaPi was unable to induce germination of F4969 spores at all tested pHs (Fig. 1A and B). Phase-contrast microscopy showed that 100% of F4969 spores incubated with 100 mM NaPi (at various pHs) remained phase bright, indicating no germination (data not shown).
FIG. 1.
Effects of pH (A and B) and NaPi concentration (C and D) on germination of C. perfringens spores. Heat-activated spores of strains SM101 (filled squares and gray bars) and F4969 (open squares and white bars) were incubated with 100 mM NaPi at various pHs (A and B), and the maximum rate (A) and extent (B) of germination after 60 min were calculated as described in Materials and Methods. (C and D) The spores were also incubated with NaPi (pH 6.0) at various concentrations, and the maximum rate (C) and extent (D) of germination after 60 min were calculated as described in Materials and Methods. Error bars denote standard deviations.
SM101 spores exhibited a maximum rate of germination with 250 mM NaPi at pH 6.0 (Fig. 1C). Interestingly, SM101 spores were able to germinate significantly after 60 min with NaPi concentrations as low as 20 mM (Fig. 1D), as phase-contrast microscopy showed that ∼85% of the spores had become phase dark. In contrast, F4969 spores showed no significant increase in their rate of germination (Fig. 1C) and no significant extent of germination, with the exception of a small but significant decrease in OD600 at 400 mM NaPi (Fig. 1D). Phase-contrast microscopy of F4969 spores with NaPi concentrations of <250 mM showed that >99% of spores remained phase bright, but with a 400 mM concentration ∼10% and ∼30% of the spores became phase dark and gray, respectively, after 60 min of incubation. Collectively, these results suggest that NaPi at pH 6.0 is a unique germinant for spores of SM101 but not for those of F4969.
Na+ and Pi are cogerminants.
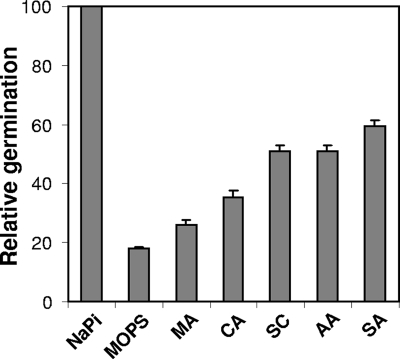
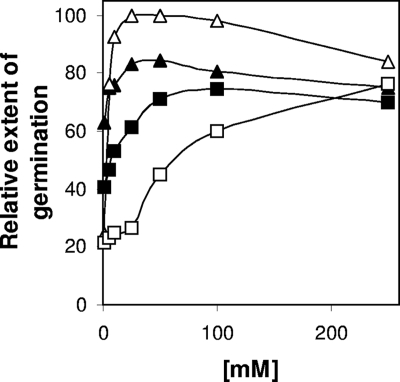
To discriminate if the ability of SM101 spores to germinate with NaPi (pH 6.0) was due to NaPi or pH 6.0 in the absence of NaPi, we evaluated various organic and inorganic buffers. As expected, SM101 spores, but not F4969 spores, germinated well with NaPi (pH 6.0) (Fig. 2 and data not shown); however, little to insignificant germination was observed when SM101 spores were incubated with a 100 mM concentration of other buffers (e.g., MOPS or maleic acid) at pH 6.0 (Fig. 2). In addition, F4969 spores also exhibited no germination with any of the buffers tested (data not shown). Phase-contrast microscopy also indicated that >90% of SM101 spores and >99% of F4969 spores remained phase bright after 60 min of incubation with the tested organic and inorganic buffers without NaPi (data not shown), suggesting that the effect is uniquely due to the presence of sodium and/or Pi. Since a previous study (20) indicated that K+ can trigger germination of C. perfringens spores, it is possible that Na+ acts as a cogerminant with Pi. Indeed, 100 mM sodium citrate (pH 6.0) and 100 mM sodium acetate (pH 6.0) induced slight increases in the extent of germination of SM101 spores compared to those with citric acid and acetic acid, respectively (Fig. 2). To evaluate further the role of Na+ in spore germination, germination assays were conducted with various concentrations of NaCl (in 25 mM NaPi to adjust the pH) at pH 6.0 and 7.0 (Fig. 3). While SM101 spores exhibited little germination at concentrations of <25 mM NaCl at pH 7.0, a significant level of germination was observed when SM101 spores were incubated at concentrations of >50 mM NaCl at pH 7.0. However, when the pH was reduced to 6.0, the addition of as little as 1 mM Na+ from NaCl, in addition to the 25 mM from the NaPi buffer, was able to induce a significant level of germination (Fig. 3). As expected, the extent of germination of SM101 spores increased when Na+ was replaced by K+ (Fig. 3). The K+ ions were able to induce higher extents of germination of SM101 spores at pH 7.0 than at pH 6.0 (Fig. 3). It was somewhat surprising that a 250 mM concentration of either Na+ or K+ induced similar extents of germination of SM101 spores at both pH 6.0 and 7.0 (Fig. 3). However, these results contradict the results from previous studies (1, 20) where the pH of 50 mM NaCl was not reported (1) and the germination assays were carried out at 30°C (20). Indeed, in this study, no significant extent of germination was observed when SM101 spores were incubated with 100 mM NaCl (pH 7.0 [adjusted with 25 mM NaPi]) at 30°C (data not shown). Collectively, these results suggest that Na+ acts as a cogerminant with Pi.
FIG. 2.
Germination of spores of C. perfringens strain SM101 in different buffers. Heat-activated spores were incubated at 40°C and pH 6.0 in 100 mM NaPi, MOPS, maleic acid (MA), citric acid (CA), sodium citrate (SC), acetic acid (AA), or sodium acetate (SA), and the OD600 was measured after 60 min of incubation. All values are given relative to the value for NaPi, and this was set at 100. Error bars denote standard deviations.
FIG. 3.
Germination of spores of C. perfringens strain SM101 with Na+ and K+. Heat-activated spores of SM101 were incubated with various concentrations of NaCl (squares) and KCl (triangles) in 25 mM NaPi buffer at pH 6.0 (filled symbols) and pH 7.0 (open symbols) at 40°C, and the OD600 was measured after 60 min of incubation. All values are given relative to that for 50 mM KCl (pH 7.0), and this was set at 100. Standard deviations were less than 4%.
NaPi triggers germination of spores of most FP isolates.
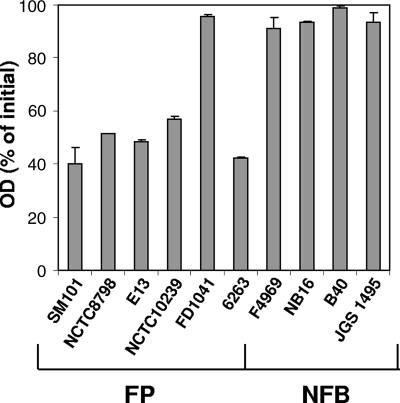
NaPi-induced germination of spores of SM101 was not due to domestication and manipulation of this laboratory strain, as spores of NCTC8798 (the parent strain of SM101) germinated similarly to SM101 spores with NaPi (Fig. 4), suggesting that NaPi (pH 6.0) might be a universal germinant for spores of C. perfringens FP isolates. To examine this hypothesis, germination experiments were extended to include spores of seven additional C. perfringens isolates, including four type A FP isolates (i.e., E13, NCTC10239, FD1041, and 6263), two NFB isolates (NB16 and B40), and one cpe-negative type C isolate (JGS1495). Strikingly, as observed with spores of SM101 (Fig. 1A to C), spores of all FP isolates, with the exception of FD1041, were able to germinate well with NaPi (pH 6.0) (Fig. 4). In contrast, spores of NFB isolates were unable to germinate with NaPi (pH 6.0) when incubated for up to 60 min (Fig. 4). Although NFB spores required a lower heat activation temperature (20), similar results were observed (i.e., no significant germination) for NaPi germination when NFB spores were heat activated at 75°C for 10 min (data not shown). Interestingly, the negative NaPi germination phenotype was not confined to cpe-positive type A NFB human isolates, as spores of strain JGS1495 (a type C cpe-negative animal isolate) were also unable to germinate with NaPi (pH 6.0) (Fig. 4). Collectively, these results clearly indicate that NaPi (pH 6.0) is a unique germinant for spores of most FP isolates.
FIG. 4.
Germination of spores from various C. perfringens isolates with NaPi. Heat-activated spores of FP type A isolates (SM101, NCTC8798, E13, NCTC10239, FD1041, and 6263), NFB type A isolates (F4969, NB16, and B40), and a cpe-negative type C isolate (JGS1495) were incubated with 100 mM NaPi (pH 6.0) for 60 min, and the OD600 was measured as described in Materials and Methods.
Role of germinant receptors in NaPi germination of C. perfringens spores.
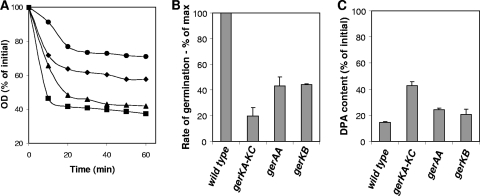
To gain insight into the mechanism of NaPi germination of C. perfringens spores, the germination of spores of strains carrying mutations in germination receptor genes gerKA-KC, gerKB, and gerAA (20, 21) was assayed using the optimum (250 mM) NaPi concentration (Fig. 1C). Although gerKB spores exhibited significantly (P < 0.05) less germination during the first 20 min than wild-type spores did, their extents of germination after 60 min were similar (Fig. 5A). This was confirmed by phase-contrast microscopy, where >95% of wild-type and gerKB spores had become phase dark after 60 min of incubation with NaPi (data not shown). In contrast, gerAA spores exhibited significantly (P < 0.001) less germination than that of wild-type and gerKB spores (Fig. 5A), as ∼50% of gerAA spores had become phase dark after 60 min of incubation (data not shown). The germination defect in gerKA-KC spores was significantly (P < 0.01) more pronounced than that in gerAA spores (Fig. 5A), with phase-contrast microscopy results showing that ∼35% of gerKA-KC spores had become phase dark after 60 min of incubation with NaPi (data not shown). These results suggest that GerKA-KC and GerAA receptors are the major receptors involved in NaPi germination and that GerKB may play an auxiliary role.
FIG. 5.
Germination of spores of C. perfringens ger receptor mutant strains with NaPi. (A) Heat-activated spores of C. perfringens strains SM101 (wild type) (filled squares), DPS101 (gerKA-KC mutant) (filled circles), DPS103 (gerAA mutant) (filled diamonds), and DPS108 (gerKB mutant) (filled triangles) were incubated at 40°C with 250 mM NaPi (pH 6.0), and the OD600 was measured as described in Materials and Methods. (B) Heat-activated spores of various C. perfringens strains were incubated at 40°C with 250 mM NaPi (pH 6.0), and maximum rates of spore germination were determined as % change in OD600/min. All values are given relative to the value for SM101 spores, and this was set at 100. (C) DPA release during C. perfringens spore germination with NaPi. Heat-activated spores of C. perfringens were germinated with 250 mM NaPi (pH 6.0) at 40°C, and after 60 min, the DPA content was measured as described in Materials and Methods.
The rate of germination was measured to determine the role of the Ger receptors in the initial triggering of germination by NaPi. Interestingly, gerKA-KC spores exhibited a rate of germination that was ∼5-fold lower than that of wild-type spores and ∼2-fold lower than those of gerKB and gerAA spores germinated with NaPi (Fig. 5B). However, gerKB and gerAA spores exhibited similar rates of germination, but these were ∼2-fold lower than that of wild-type spores (Fig. 5B). These results suggest that GerKA-KC and GerAA receptors are the major receptors involved in NaPi germination and that GerKB is required for normal NaPi-triggered germination.
For Bacillus and Clostridium spores, binding of germinants to specific receptors located in the spore's inner membrane triggers the release of the spore core's large depot of DPA as a 1:1 chelate to Ca2+ (Ca-DPA) (20, 28). Consequently, to investigate the role of each Ger receptor in DPA release during NaPi-induced germination, we measured DPA release during NaPi germination. Consistent with the similarities observed in the extent of germination between gerKB and wild-type spores, gerKB spores also released a similar level of DPA to that released from wild-type spores (Fig. 5C). However, although gerAA spores released the majority of their DPA during NaPi germination, the amount of DPA remaining in the core of gerAA spores was still slightly higher than that in wild-type spores, which is statistically significant (P < 0.01) (Fig. 5C). gerKA-KC spores released significantly (P < 0.001) less DPA than wild-type spores did. The amount of DPA released by gerKA-KC spores was also significantly (P < 0.05) less than that released by gerKB and gerAA spores. These results suggest that GerKA and/or GerKC is mainly involved in DPA release during NaPi-triggered germination, with GerAA and GerKB having an auxiliary role.
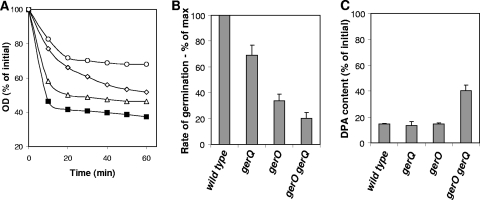
Role of antiporters in NaPi germination of C. perfringens spores.
We recently identified two putative antiporters (GerO and GerQ) involved in germination of C. perfringens spores (24). GerO, a putative Na+/K+-H+ antiporter, is required for normal germination with all known germinants for C. perfringens, while GerQ is required mainly in the absence of GerO. To evaluate their roles in NaPi-triggered germination, spores of antiporter mutants were assayed for germination in the presence of NaPi (pH 6.0). gerQ spores exhibited a slightly slower and lesser extent of germination than that of wild-type spores (Fig. 6A); gerO spores germinated to even a lesser extent. Indeed, phase-contrast microscopy indicated that only ∼75% of gerO spores had become phase dark after 60 min of germination with NaPi (pH 6.0) (data not shown). Interestingly, gerO gerQ spores had significantly (P < 0.01) slower germination than that of wild-type, gerQ, and gerO spores (Fig. 6A), and phase-contrast microscopy indicated that only ∼30% of gerO gerQ spores had become phase dark (data not shown). The gerQ, gerO, and gerO gerQ spores exhibited 30, 70, and 80% lower rates of germination, respectively, than that of wild-type spores (Fig. 6B). These results clearly suggest that GerO and GerQ, only in the absence of GerO, are essential for normal NaPi-triggered germination.
FIG. 6.
Germination of spores of C. perfringens antiporter mutant strains with NaPi. (A) Heat-activated spores of C. perfringens strains SM101 (wild type) (filled squares), DPS113 (gerQ mutant) (open triangles), DPS116 (gerO mutant) (open diamonds), and DPS108 (gerO gerQ mutant) (open circles) were incubated at 40°C with 250 mM NaPi (pH 6.0), and the OD600 was measured as described in Materials and Methods. (B) Heat-activated spores of various C. perfringens strains were incubated at 40°C with 250 mM NaPi (pH 6.0), and maximum rates of spore germination were determined as % change in OD600/min. All values are given relative to the value for SM101 spores, and this was set at 100. (C) DPA release during C. perfringens spore germination with NaPi. Heat-activated spores of C. perfringens were germinated with 250 mM NaPi (pH 6.0) at 40°C, and after 60 min, the DPA content was measured as described in Materials and Methods.
Interestingly, gerQ spores were able to release similar amounts of DPA to that released by wild-type spores after 60 min of incubation with NaPi (Fig. 6C), consistent with the majority of the spores completing germination. To our surprise, gerO spores also released similar amounts of DPA to that released by wild-type spores (Fig. 6C), although it is unclear why the extent of germination was significantly less than that with wild-type spores (Fig. 6A; see Discussion). However, gerO gerQ spores released significantly (P < 0.001) less DPA than wild-type, gerO, and gerQ spores did (Fig. 6C). These results indicate that NaPi-induced DPA release is affected only in the absence of both GerO and GerQ antiporters, similar to the case with K+-induced germination (24).
DISCUSSION
The ability of spores of FP isolates to adapt to FP environments seems to go beyond the fact that spores of FP isolates exhibit ∼60-fold more heat resistance than spores of NFB isolates (27). The unique ability of FP isolate spores to germinate with K+ ions (20) might provide a greater advantage over spores of NFB isolates present in meat products in the ability to germinate, outgrow, and proliferate. In this respect, and perhaps offering the major conclusion of this work, Na+ and Pi are novel cogerminants of spores of C. perfringens FP isolates. Na+ and Pi, together with K+, are intrinsic minerals found in meat and processed meat products and might provide spores of FP isolates significant advantages over spores of other C. perfringens isolates in the ability to germinate, outgrow, and proliferate in meat products during inadequate processing or subsequent warming. For example, in nonprocessed meat products, where the Na+ concentration is ∼30 mM, Na+ and Pi might act as cogerminants together with the major germinant K+, triggering germination of FP spores during ripening of the meats. Contrarily, in processed meat products, with Na+ concentrations of ∼300 to 400 mM, it is likely that Na+ can become a major germinant together with K+, triggering germination of FP spores. Results from this study also suggest that spores of FP isolates might sense these ions through an evolved ionic germination mechanism not present in spores of NFB isolates. This concept has previously been proposed for some strains of Bacillus megaterium and C. perfringens strain NCTC8238 (1, 25). Several studies (9, 26) indicate that C. perfringens FP isolates belong to a different evolutionary lineage from that of all other C. perfringens isolates. Indeed, the differential germination phenotype of FP spores reinforces the theory that FP isolates have efficiently adapted to fit FP niches. However, it is unclear what genetic difference in the germination apparatuses of FP versus NFB isolates might have evolved to produce this difference in K+ and NaPi germination phenotypes, since strain F4969 (an NFB isolate) has intact copies of all four germinant receptor genes (gerAA, gerKB, and gerKA-KC) and shows >95% identity to those of SM101 (16). One possibility that deserves further research is that perhaps key residues within the receptor proteins might be essential for the unique NaPi and K+ germination of FP isolates.
A second major conclusion is that NaPi germination of FP spores requires all four Ger receptors, although to different extents. The main germinant receptor proteins involved in NaPi germination are GerKA and/or GerKC, which are also essential for l-asparagine germination and have a major role in KCl germination (20). However, gerKA-KC spores are still able to germinate significantly with KCl (20) as well as with NaPi (this study), suggesting that other Ger proteins might have a role in DPA release during NaPi germination. Indeed, in contrast to KCl germination (20), GerAA also has a role in NaPi germination, with gerAA spores germinating at a much lower rate and to a lesser extent than wild-type spores (Fig. 5A and B). In contrast, GerKB, which was previously shown to have no role in nutrient germination of FP spores (21), appears to play a role in normal germination of FP spores with NaPi. Clearly, the majority of C. perfringens Ger receptors are required for initial triggering of germination with NaPi, suggesting that all C. perfringens Ger receptors form a receptor complex and that the absence of GerKA-KC, GerAA, or GerKB has a major, medium, or small effect, respectively, on the receptor complex's function. Further biochemical studies on receptor proteins should help to clarify this issue.
A third notable conclusion is that the putative antiporters GerO and GerQ, only in the absence of GerO, are required for normal NaPi germination of C. perfringens spores. Our previous study showed that these antiporters are also essential for normal germination of C. perfringens spores with various germinants and that their role precedes DPA release (24). Interestingly, GerO was also essential for normal NaPi germination; however, in contrast to gerO spores releasing significantly less DPA than wild-type spores during K+ germination (24), here the gerO spores were able to release the majority of their DPA during NaPi germination, and this seems likely to occur because of the presence of GerQ. However, it is possible that the larger amount of DPA released by gerO spores in this study than in the previous study (24) was due to the nature of the novel germinant (NaPi) or to the fact that we used a twofold higher concentration of NaPi, creating a higher ionic strength in the medium that could in some fashion stimulate more DPA release. Although the rate of germination of gerQ spores was lower than that of wild-type spores, indicating reduced initial triggering of germination, they germinated to a similar extent as wild-type spores and also released the majority of their DPA. In addition, GerQ became essential only in gerO gerQ spores, which had significantly less initial triggering of germination and germinated to a lesser extent than gerO, gerQ, and wild-type spores and released about half of their DPA. These results reinforce the role of cation transporters in germination of C. perfringens spores with all known germinants (24), including NaPi (this study). However, it is not yet clear whether the effect of gerO or gerQ mutation is directly on spore germination or during spore formation. Further studies are currently being carried out to help decide between these two scenarios.
Acknowledgments
This work was supported by a grant from the N. L. Tartar Foundation of Oregon State University, by a grant from the Agricultural Research Foundation of Oregon State University, by a grant from the Army Research Office (all to M.R.S.), and by a fellowship from the Office of the Higher Education Commission (Thailand) to P.U.
Footnotes
Published ahead of print on 7 August 2009.
REFERENCES
- 1.Ando, Y. 1974. Ionic germination of spores of Clostridium perfringens type A. Jpn. J. Microbiol. 18:433-439. [DOI] [PubMed] [Google Scholar]
- 2.Cabrera-Martinez, R. M., F. Tovar-Rojo, V. R. Vepachedu, and P. Setlow. 2003. Effects of overexpression of nutrient receptors on germination of spores of Bacillus subtilis. J. Bacteriol. 185:2457-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collie, R. E., and B. A. McClane. 1998. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with non-food-borne human gastrointestinal diseases. J. Clin. Microbiol. 36:30-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cornillot, E., B. Saint-Joanis, G. Daube, S. Katayama, P. E. Granum, B. Canard, and S. T. Cole. 1995. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol. Microbiol. 15:639-647. [DOI] [PubMed] [Google Scholar]
- 5.Duncan, C. L., and D. H. Strong. 1968. Improved medium for sporulation of Clostridium perfringens. Appl. Microbiol. 16:82-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duncan, C. L., D. H. Strong, and M. Sebald. 1972. Sporulation and enterotoxin production by mutants of Clostridium perfringens. J. Bacteriol. 110:378-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garmory, H. S., N. Chanter, N. P. French, D. Bueschel, J. G. Songer, and R. W. Titball. 2000. Occurrence of Clostridium perfringens beta2-toxin amongst animals, determined using genotyping and subtyping PCR assays. Epidemiol. Infect. 124:61-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison, B., D. Raju, H. S. Garmory, M. M. Brett, R. W. Titball, and M. R. Sarker. 2005. Molecular characterization of Clostridium perfringens isolates from humans with sporadic diarrhea: evidence for transcriptional regulation of the beta2-toxin-encoding gene. Appl. Environ. Microbiol. 71:8362-8370. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Johansson, A., A. Aspan, E. Bagge, V. Baverud, B. E. Engstrom, and K. E. Johansson. 2006. Genetic diversity of Clostridium perfringens type A isolates from animals, food poisoning outbreaks and sludge. BMC Microbiol. 6:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kokai-Kun, J. F., J. G. Songer, J. R. Czeczulin, F. Chen, and B. A. McClane. 1994. Comparison of Western immunoblots and gene detection assays for identification of potentially enterotoxigenic isolates of Clostridium perfringens. J. Clin. Microbiol. 32:2533-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lahti, P., A. Heikinheimo, T. Johansson, and H. Korkeala. 2008. Clostridium perfringens type A strains carrying a plasmid-borne enterotoxin gene (genotype IS1151-cpe or IS1470-like-cpe) as a common cause of food poisoning. J. Clin. Microbiol. 46:371-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, J., and B. A. McClane. 2006. Further comparison of temperature effects on growth and survival of Clostridium perfringens type A isolates carrying a chromosomal or plasmid-borne enterotoxin gene. Appl. Environ. Microbiol. 72:4561-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, J., and B. A. McClane. 2006. Comparative effects of osmotic, sodium nitrite-induced, and pH-induced stress on growth and survival of Clostridium perfringens type A isolates carrying chromosomal or plasmid-borne enterotoxin genes. Appl. Environ. Microbiol. 72:7620-7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClane, B. A. 2007. Clostridium perfringens, p. 423-444. In M. P. Doyle and L. R. Beuchat (ed.), Food microbiology: fundamentals and frontiers, 3rd ed. ASM Press, Washington, DC.
- 15.McDonnell, J. L. 1986. Toxins of Clostridium perfringens type A, B, C, D, and E, p. 477-517. In F. Dorner and J. Drews (ed.), Pharmacology of bacterial toxins. Pergamon Press, Oxford, United Kingdom.
- 16.Myers, G. S., D. A. Rasko, J. K. Cheung, J. Ravel, R. Seshadri, R. T. DeBoy, Q. Ren, J. Varga, M. M. Awad, L. M. Brinkac, S. C. Daugherty, D. H. Haft, R. J. Dodson, R. Madupu, W. C. Nelson, M. J. Rosovitz, S. A. Sullivan, H. Khouri, G. I. Dimitrov, K. L. Watkins, S. Mulligan, J. Benton, D. Radune, D. J. Fisher, H. S. Atkins, T. Hiscox, B. H. Jost, S. J. Billington, J. G. Songer, B. A. McClane, R. W. Titball, J. I. Rood, S. B. Melville, and I. T. Paulsen. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 16:1031-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paidhungat, M., K. Ragkousi, and P. Setlow. 2001. Genetic requirements for induction of germination of spores of Bacillus subtilis by Ca2+-dipicolinate. J. Bacteriol. 183:4886-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paidhungat, M., and P. Setlow. 2001. Localization of a germinant receptor protein (GerBA) to the inner membrane of Bacillus subtilis spores. J. Bacteriol. 183:3982-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paredes-Sabja, D., B. Setlow, P. Setlow, and M. R. Sarker. 2008. Characterization of Clostridium perfringens spores that lack SpoVA proteins and dipicolinic acid. J. Bacteriol. 190:4648-4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paredes-Sabja, D., J. A. Torres, P. Setlow, and M. R. Sarker. 2008. Clostridium perfringens spore germination: characterization of germinants and their receptors. J. Bacteriol. 190:1190-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paredes-Sabja, D., P. Setlow, and M. R. Sarker. 2009. Role of GerKB in germination and outgrowth of Clostridium perfringens spores. Appl. Environ. Microbiol. 75:3813-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paredes-Sabja, D., P. Setlow, and M. R. Sarker. 2009. SleC is essential for cortex peptidoglycan hydrolysis during germination of spores of the pathogenic bacterium Clostridium perfringens. J. Bacteriol. 191:2711-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paredes-Sabja, D., P. Setlow, and M. R. Sarker. 2009. The protease CspB is essential for initiation of cortex hydrolysis and DPA release during spore germination of Clostridium perfringens type A food poisoning isolates. Microbiology doi: 10.1099/mic.0.030965-0. [DOI] [PubMed]
- 24.Paredes-Sabja, D., P. Setlow, and M. R. Sarker. 2009. GerO, a putative Na+/H+-K+ antiporter, is essential for normal germination of spores of the pathogenic bacterium Clostridium perfringens. J. Bacteriol. 191:3822-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rode, L. J., and J. W. Foster. 1962. Ionic germination of spores of Bacillus megaterium QMB 1551. Arch. Mikrobiol. 43:183-200. [DOI] [PubMed] [Google Scholar]
- 26.Rooney, A. P., J. L. Swezey, R. Friedman, D. W. Hecht, and C. W. Maddox. 2006. Analysis of core housekeeping and virulence genes reveals cryptic lineages of Clostridium perfringens that are associated with distinct disease presentations. Genetics 172:2081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarker, M. R., R. P. Shivers, S. G. Sparks, V. K. Juneja, and B. A. McClane. 2000. Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfringens isolates carrying plasmid genes versus chromosomal enterotoxin genes. Appl. Environ. Microbiol. 66:3234-3240. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Setlow, P. 2003. Spore germination. Curr. Opin. Microbiol. 6:550-556. [DOI] [PubMed] [Google Scholar]
- 29.Sparks, S. G., R. J. Carman, M. R. Sarker, and B. A. McClane. 2001. Genotyping of enterotoxigenic Clostridium perfringens fecal isolates associated with antibiotic-associated diarrhea and food poisoning in North America. J. Clin. Microbiol. 39:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao, Y., and S. B. Melville. 1998. Identification and characterization of sporulation-dependent promoters upstream of the enterotoxin gene (cpe) of Clostridium perfringens. J. Bacteriol. 180:136-142. [DOI] [PMC free article] [PubMed] [Google Scholar]