Abstract
The polycyclic aromatic hydrocarbon (PAH) contamination, bacterial community, and PAH-degrading bacteria were monitored in aged PAH-contaminated soil (Neuves-Maisons [NM] soil; with a mean of 1,915 mg of 16 PAHs·kg−1 of soil dry weight) and in the same soil previously treated by thermal desorption (TD soil; with a mean of 106 mg of 16 PAHs·kg−1 of soil dry weight). This study was conducted in situ for 2 years using experimental plots of the two soils. NM soil was colonized by spontaneous vegetation (NM-SV), planted with Medicago sativa (NM-Ms), or left as bare soil (NM-BS), and the TD soil was planted with Medicago sativa (TD-Ms). The bacterial community density, structure, and diversity were estimated by real-time PCR quantification of the 16S rRNA gene copy number, temporal thermal gradient gel electrophoresis fingerprinting, and band sequencing, respectively. The density of the bacterial community increased the first year during stabilization of the system and stayed constant in the NM soil, while it continued to increase in the TD soil during the second year. The bacterial community structure diverged among all the plot types after 2 years on site. In the NM-BS plots, the bacterial community was represented mainly by Betaproteobacteria and Gammaproteobacteria. The presence of vegetation (NM-SV and NM-Ms) in the NM soil favored the development of a wider range of bacterial phyla (Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Verrucomicrobia, Actinobacteria, Firmicutes, and Chloroflexi) that, for the most part, were not closely related to known bacterial representatives. Moreover, under the influence of the same plant, the bacterial community that developed in the TD-Ms was represented by different bacterial species (Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, and Actinobacteria) than that in the NM-Ms. During the 2 years of monitoring, the PAH concentration did not evolve significantly. The abundance of gram-negative (GN) and gram-positive (GP) PAH-degrading bacteria was estimated by real-time PCR quantification of specific functional genes encoding the α subunit of PAH-ring hydroxylating dioxygenase (PAH-RHDα). The percentage of the PAH-RHDα GN bacterial genes relative to 16S rRNA gene density decreased with time in all the plots. The GP PAH-RHDα bacterial gene proportion decreased in the NM-BS plots but stayed constant or increased under vegetation influence (NM-SV, NM-Ms, and TD-Ms).
Polycyclic aromatic hydrocarbons (PAHs) are persistent organic pollutants associated with a wide range of anthropogenic activities (gas plants, wood preservation plants, waste incineration, coke production, and petrochemical industries). The intensive industrial coal mining during the 19th and 20th centuries in northern France caused contamination of large areas where the soil now needs to be remediated.
Microbiological degradation is the chief process for natural elimination of PAHs from contaminated soil (9). A wide range of bacteria are able to degrade low-molecular-weight PAHs, such as naphthalene, phenanthrene, and anthracene, while the high-molecular-weight PAHs (with four or more fused aromatic rings) are more recalcitrant, and relatively few microorganisms are able to use them as a sole carbon source (8). The first hydroxylation step of the PAH ring is crucial to initiate an efficient biodegradation. This step is performed mainly by aerobic bacteria possessing a PAH-ring hydroxylating dioxygenase (PAH-RHD) system. Homologous PAH-RHD enzymes are encoded by specific genes present in both gram-positive (GP) and gram-negative (GN) bacterial species (22). Recently, we developed real-time PCR assays to quantify the functional genes encoding the catalytic α subunit of the PAH-RHD (PAH-RHDα) enzyme. The quantifications were performed on soil DNA samples, giving important information about the PAH-degrading bacterial population present in various PAH-contaminated soils (7).
High PAH degradation rates have been observed in laboratory experiments with strains or consortia isolated from PAH-contaminated soils (6, 33). However, in situ degradation is often a slower process due to environmental constraints and low availability of PAHs in aged, polluted soils (5, 60). As a consequence, highly contaminated soils polluted by persistent organic compounds are often treated by industrial processes such as thermal desorption (short heating of the soil at a temperature close to 500°C). Such treatment generates a soil with modified characteristics, in which the resilience of biological functions has not yet been studied. Bioremediation, the use of microorganisms to clean up contaminated soil, is an environmentally safe solution for PAH removal (20, 34) that can be accelerated by the positive effect of plants via the stimulation of microbial biodegradation in the rhizosphere (27, 44, 45, 48, 54) through root exudates (12, 39). A major driving force for the rhizosphere effect is the massive input of organic substrate in soil, which can increase the bioavailability of PAHs but also induce a selection of rhizospheric communities (17, 56) and increase the total activity, diversity, and number of bacteria (45, 48, 54, 58), as well as the abundance of PAH-degrading bacteria populations (31, 56). However, the total and functional bacterial community structure and activity in a PAH-polluted rhizosphere remain poorly described. Even if numerous studies report that plants can foster the degradation of PAHs (1, 4, 21, 28, 29, 41, 44, 45, 54), others have shown no (21) or even inhibitory (31, 55) effects of plants; thus, it is important to study the potential of rhizodegradation in situ, depending on the soil and plant studied.
The aim of this study was to investigate the bacterial community density and structure, the fate of PAHs, and the PAH-degrading bacteria abundance over 2 years using a long-term in situ trial of natural and plant-assisted attenuations of PAHs. Experimental plots with contaminated soil from a former coking plant site were colonized by spontaneous vegetation (Neuves-Maisons [NM]-SV) or planted with alfalfa (Medicago sativa) (NM-Ms) and compared to the bare soil (NM-BS). Additional alfalfa-planted plots contained the same soil previously treated by thermal desorption (TD-Ms). Alfalfa was sown on the plots, since it has been shown to be effective in the removal of PAHs (46, 52, 54). Moreover, this is the first time that microbiological functions were also considered for the same soil treated by thermal desorption.
MATERIALS AND METHODS
Experimental site and soil sampling.
The in situ experiment was established in September 2005 (T0) at the site of an industrial wasteland (Homécourt, Meurthe-et-Moselle, France) within the facilities supported by Groupement d'Intérêt Scientifique sur les Friches Industrielles (www.gisfi.fr). Stainless steel tanks (each 2 by 3 by 0.4 m, length by width by height) were equipped with independent leachate water collection systems, and these plots were filled with NM soil from a former coking plant site (Neuves-Maisons, Meurthe-et-Moselle, France). The following three different treatments with four replicates were tested on NM bare soil plots: NM bare soil prevented by hand from growing vegetation (NM-BS); NM soil sown (40 g of seeds·plots−1, equivalent to about 18,000 seeds·plots−1) with Medicago sativa L. cv. Europe (alfalfa) at T0 in September 2005 (NM-Ms); and NM soil with spontaneous vegetation colonization (NM-SV). The diversity of the plants that colonized the NM-SV plots for the first year (2006; T2) has been previously described by Dazy et al. (13), who reported that 15 species colonized the plots, belonging mainly to the Asteraceae, Chenopodiaceae, and Poaceae families. In 2007 (T4), plant diversity increased (41 species identified), with Asteraceae and Poaceae remaining of the major families, but Chenopodiaceae regressed notably (our unpublished results). Four other plots were filled with the same soil that has been treated by thermal desorption (TD soil) 6 months before plot setting and kept as a pile on site. These four TD soil plots were sown with Medicago sativa (alfalfa; TD-Ms), as previously described. The TD soil treatment, different from incineration, consisted of heating the soil to 500°C, where mainly the PAHs were transferred to gas phase. Alfalfa biomass, estimated in September 2006 and 2007, was about 1,641 ± 315 and 1,820 ± 375 g·plots−1 for the NM-Ms and TD-Ms plots, respectively.
Soil samples were collected twice a year in September 2005 (T0), May 2006 (T1), September 2006 (T2), May 2007 (T3), and September 2007 (T4). Six subsamples per plot were collected with a hand auger and mixed to yield one mean sample per plot. The soil samples were directly sieved to <5 mm, part of the samples was stored at −20°C until DNA extraction, and the rest was dried, sieved at 2 mm, and then ground at 500 μm prior to chemical analyses.
Soil characteristics.
Soil sample characteristics, analyzed at the Soil Analysis Laboratory at the National Institute for Agricultural Research (Arras, France), did not vary significantly during the 2 years of study. The NM and TD soils had approximately the same texture (12.1% and 10.4% clay, 22.1% and 19.5% silt, and 65.8% and 70.1% sand for NM and TD soils, respectively). The pH was about 7.0 to 7.5 and 7.7 to 8.0 for NM and TD soils, respectively. The C/N ratio was about 23.0 to 25.1 and 55.5 to 63.2 for the NM and TD soils, respectively. The organic carbon content was 62.6 ± 3.2 and 58.4 ± 2.3 g·kg−1 soil dry weight for the NM and TD soils at T0, respectively. PAHs were extracted by dichloromethane using an automated extraction Dionex ASE 200 and further analyzed using gas chromatography-mass spectrometry (Agilent Technologies 6890N gas chromatographer coupled to an Agilent Technologies 5973 inert detector mass spectrometer). Quantifications of 16 priority PAHs from the Environmental Protection Agency list were carried out, according to the NF T 90115 Association Française de Normalisation method, as described by Biache et al. (3). The method was validated by PAH-contaminated soil/sediment CRM104-100 reference material (RT Corporation) certified by the U.S. Environmental Protection Agency SW-846 method. At T0, the 16 PAH concentrations were 1,915 ± 246 and 106 ± 4 mg·kg soil−1 for the NM and TD soils, respectively.
DNA extraction.
DNA was extracted from 0.5 g of soil using a bead beating-based method. Soil was mixed with glass beads, 800 μl of extraction buffer (100 mM Tris, 100 mM EDTA, 100 mM NaCl, 1% [wt/vol] polyvinyl-polypyrrolidone, 2% [wt/vol] sodium dodecyl sulfate [pH 8]), and 40 μl of 6% cetyltrimethylammonium bromide in 5 mM CaCl2. After being bead beaten on a horizontal grinder (Retsch; Roucaire Instruments Scientifiques, France) for 45 s and centrifuged (13,000 rpm, 5 min, 4°C), the DNA present in the supernatant was purified using GeneClean Turbo kit columns (MP Biomedicals, Illkirch, France) by following the manufacturer's instructions for genomic DNA recovery. DNA was then recovered in 100 μl of final buffer. This protocol has been chosen to prevent the coprecipitation during the isopropanol or ethanol DNA precipitation step of CaSO4, due to the high concentration of SO42− in these soils.
Temporal thermal gradient gel electrophoresis (TTGE) analysis of 16S rRNA genes.
Bacterial 16S rRNA gene fragments were amplified using the universal primer set described by Felske et al. (16), as follows: 968F-GC (5′-CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC CCG CCC GAA CGC GAA GAA CCT TAC-3′) holding a GC clamp (underlined sequence described by Muyzer et al. [47]) and 1401R (5′-CGG TGT GTA CAA GAC CC-3′). The PCR was performed using an iCycler instrument (Bio-Rad), in a final volume of 50 μl containing 1× PCR buffer (Invitrogen) supplemented with 1.5 mM MgCl2, 200 μM of each deoxynucleoside triphosphate (Fermentas), 0.2 μM of each primer (MWG-Biotech), 1.25 U of Taq DNA polymerase recombinant (Invitrogen), and 1 μl of template DNA. The temperature profile began with 5 min at 94°C. Then, a touchdown procedure followed, consisting of 30 s at 94°C, annealing for 45 s at temperatures decreasing from 63 to 54°C during the first 19 cycles (with 1°C decremental steps every 2 cycles) and at 54°C during the last 15 cycles, and ending with an extension step at 72°C for 60 s. A final extension step was carried out for 10 min at 72°C, after which the DNA was stored at 4°C. The PCR amplification product was therefore analyzed by using 1% molecular grade agarose (Euromedex) gel electrophoresis and visualized under a Gel Doc transilluminator (Bio-Rad) after staining with 0.5 μg·ml−1 ethidium bromide. The quantity of the PCR product was estimated by comparison with a concentration range of molecular markers (1-kb ladder; Fermentas), and 500 ng was used for TTGE.
16S rRNA PCR-TTGE analyses were performed for the four replicate plots from T0, T2, and T4 and for one replicate from the T1 and T3 sampling dates. TTGE was performed on a DCode system (Bio-Rad, France) by using vertical polyacrylamide gel (6% [wt/vol] acrylamide, 7 M urea, 2% [vol/vol] glycerol) in 1.25× TAE buffer (1× TAE buffer corresponds to 40 mM Tris-acetate, 20 mM acetic acid, and 1 mM EDTA [pH 8.3]). After 5 h of electrophoresis at 110 V with a temperature gradient of 57 to 67°C (rate increase of 2°C/h), gels were stained by SYBR gold (1/10,000 final dilution; Molecular Probes) and analyzed on a Gel Doc transilluminator (Bio-Rad) coupled to the Quantity One 4.0.1 software (Bio-Rad) for image treatment and band pattern analysis. Some samples were run on each TTGE gel to compare the different gel patterns.
Distinct individual bands were excised from TTGE gels, dissolved overnight in 25 μl of distilled water at 4°C, reamplified with the same primers, and reanalyzed by TTGE. The excision was repeated again when a single band was obtained, and the reamplification procedure was applied using primers without the GC clamp. The PCR products were purified using the High Pure PCR product purification kit (Roche Applied Science) and sequenced (Eurofins MWG-Biotech). About 360 bp of the 16S rRNA partial sequences were aligned using BioEdit software and compared to 16S rRNA gene sequences obtained from BLASTn in the GenBank database. A dendrogram was constructed by using the programs DNADIST, NEIGHBOR, SEQBOOT, and CONSENSE from the PHYLIP 3.65 program, and the phylogenetic tree file was analyzed by using the Tree View program.
Real-time PCR assays.
The real-time PCR quantification of 16S rRNA gene copies using the universal primers described above (without the GC clamp) and of functional PAH-RHDα genes from GN PAH-degrading bacteria (primers PAH-RHDα GN_F, 5′-GAG ATG CAT ACC ACG TKG GTT GGA-3′, and GN_R, 5′-AGC TGT TGT TCG GGA AGA YWG TGC MGT T-3′) and GP PAH-degrading bacteria (primers PAH-RHDα GP_F, 5′-CGG CGC CGA CAA YTT YGT NGG-3′, and GP_R, 5′-GGG GAA CAC GGT GCC RTG DAT RAA-3′) was performed as described previously by Cébron et al. (7). The detection limit of the method was 101 and 102 gene copies·μl−1, corresponding to about 2 × 103 and 2 × 104 gene copies·g−1 soil dry weight for PAH-RHDα and 16S rRNA genes, respectively. The quantifications were performed in triplicate with three independent real-time PCR runs using an iCycler iQ apparatus (Bio-Rad), associated with iCycler optical system interface software (version 2.3; Bio-Rad) for data collection and subsequent melting curve analysis. Amplification reactions were carried out in a volume of 25 μl containing 1× SYBR green PCR master mix (iQ SYBR green supermix; Bio-Rad, France), 0.4 μM of each primer, 15 μg of bovine serum albumin, 0.25 μl of dimethyl sulfoxide, 0.1 μl of T4 bacteriophage gene 32 product (Qbiogene, France), and 1 μl of the template DNA (soil sample DNA or 10 times the dilution series from 108 to 101 gene copies·μl−1of standard plasmids) or 1 μl of distilled water (negative control). The amplifications were carried out with the following temperature profiles: step 1 consisted of heating to 95°C (5 min), followed by four steps of 50 cycles, 30 s at 95°C, 30 s at the primer-specific annealing temperatures (56°C, 57°C, and 54°C for 16S rRNA, PAH-RHDα GN, and PAH-RHDα GP bacterial primer sets, respectively), 30 s at 72°C, and 10 s at 80°C to dissociate the primer's dimers and capture the fluorescence intensity of the SYBR green. The final step consisted of 7 min at 72°C. At the end, a melting curve analysis was performed from 51°C to 95°C, with a temperature increase of 0.5°C every 10 s.
The presence of PCR inhibitors in the DNA extracts was tested with real-time PCR by mixing 1 μl of soil DNA with 1 μl of known concentration of a plasmid standard holding a 500-bp fragment from bacteriophage T4, as described by Cébron et al. (7). These assays showed that there was no inhibition of the PCRs with any of the DNA samples.
Statistical analysis.
The 16 PAH concentrations, 16S rRNA gene copy numbers, PAH-RHDα GN and GP bacterial gene copy numbers, and gene ratios were analyzed with a two-way analysis of variance, followed by the pairwise multiple comparison test (Newman-Keuls method), with mean difference significance at the 0.05 level testing using StatView software. These statistical tests were performed to compare the NM-BS, NM-SV, and NM-Ms treatments and separately to compare the NM-Ms and TD-Ms treatments, considering the results from the four replicate plot samples. The ratios of PAH-RHDα GN and GP bacterial genes relative to 16S rRNA genes were arcsine transformed previous to statistical analysis.
Based on a binary (presence/absence) matrix of TTGE banding patterns, a dissimilarity matrix based on Jaccard's distance was calculated using XLStat 2008 software. This distance matrix was analyzed using multidimensional scaling (MDS) analysis in XLStat 2008 software, with a Kruskal stress of 0.246.
Nucleotide sequence accession numbers.
The 16S rRNA partial sequences were deposited in the GenBank database under the following accession numbers: Ms-TTGE-1 to Ms-TTGE-17 (FJ873605 to FJ873621), SV-TTGE-1 to SV-TTGE-17 (FJ873622 to FJ873638), BS-TTGE-1 to BS-TTGE-14 (FJ873639 to FJ873652), and TD-TTGE-1 to Ms-TTGE-12 (FJ873653 to FJ873664).
RESULTS
Bacterial density and diversity based on 16S rRNA analyses.
During the 2-year period at the experimental site, the density and structure of the bacterial community were studied based on 16S rRNA gene copy quantification and TTGE banding pattern analysis.
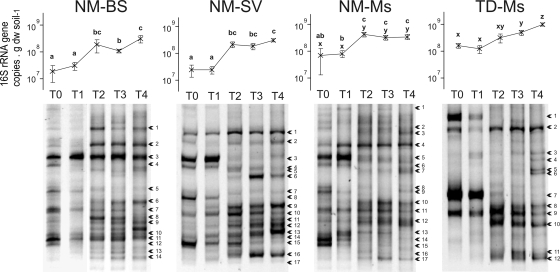
In the DNA extract from all the NM soil plots, the 16S rRNA gene copy number increased significantly (P < 0.05) 5 to 15 times during the first year and then stayed constant during the second year (Fig. 1). It ranged from 1.9 × 107 to 7.3 × 107 gene copies·g−1 soil dry weight at the setup of the experiment (T0) and from 30.9 × 107 to 35.8 × 107 gene copies·g−1 soil dry weight after 2 years (T4). The density of 16S rRNA genes in the TD-Ms was not significantly different (P > 0.05) from that of the NM-Ms at T0, but it increased from 17.1 × 107 at T0 to 103.4 × 107 gene copies·g−1 soil dry weight (mean values) at T4 and then was significantly higher than that of NM-Ms at T4 (P < 0.05). The increase in bacterial densities was accompanied by bacterial community structure modification (TTGE banding patterns) (Fig. 1).
FIG. 1.
16S rRNA gene copy numbers (mean ± standard error; n = 4) and 16S rRNA gene PCR-TTGE band patterns in the four plot types during the 2-year period (from T0 to T4). The arrows indicate the sequenced bands shown in Fig. 3.
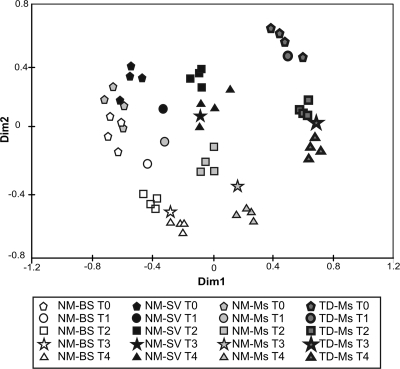
Dissimilarity matrices generated from TTGE data were analyzed by MDS, giving the spatial representation of the distance between samples (Fig. 2). The MDS distinguished the different treatments and sampling dates and highlighted the modification of the bacterial community structure in the four plot types (Fig. 2). At the setup of the experiment and before plant colonization (T0), the bacterial community structure in the NM-BS, NM-Ms, and NM-SV plots was homogeneous (closely situated dots on the MDS). After 1 and 2 years (T2 and T4, respectively), the bacterial community structure evolved differently, depending on the plot types. The TD-Ms dots were spatially separated on the MDS analysis, showing that the bacterial community structure was different from that of the NM soil and evolved over time (Fig. 2) as the TTGE patterns became more complex at T4 compared to those at T0 (Fig. 1).
FIG. 2.
MDS analysis of 16S rRNA gene PCR-TTGE band patterns, based on a dissimilarity distance matrix generated from a presence/absence of each TTGE band in the samples. TTGE analysis was performed separately on the four replicate plots for each treatment at T0, T2, and T4 (September 2005, 2006, and 2007, respectively). For the samples collected in May 2006 and 2007 (T1 and T3), only one plot was analyzed.
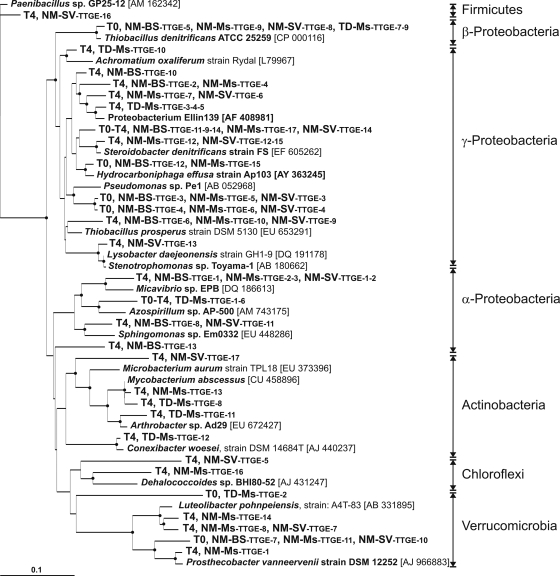
Based on TTGE band sequencing, we found that at T0, the bacterial community in the NM soil was represented by bacteria affiliated with Betaproteobacteria, Gammaproteobacteria, and Verrucomicrobia (Fig. 3) and was closely related to environmental clones found in hydrocarbons, coke, mercury, perchloroethylene, or in pesticide-contaminated environments (see Fig. S1 in the supplemental material). After 2 years (T4) at the experimental site, a wider range of bacterial phyla developed in all NM soil plot types (Fig. 3), belonging to Alphaproteobacteria (Micavibrio and Sphingomonas genera) and Gammaproteobacteria closely related to environmental clones found in a lead-zinc mine tailing site, volcanic deposits, mercury-contaminated soil, and phytoremediation of semicoke (see Fig. S1 in the supplemental material). The conditions in the NM-BS plots favored the development of bacteria belonging to Gammaproteobacteria (Fig. 3) closely related to environmental clones detected in contaminated sites under anaerobic conditions such as polychlorinated biphenyl-contaminated sediment and a benzene degradation system (see Fig. S1 in the supplemental material). Three sequences were detected only in the vegetated NM-Ms and NM-SV plots and were affiliated with Verrucomicrobia and Gammaproteobacteria (Fig. 3) closely related to environmental clones found in organic matter-rich environments such as the wheat detritusphere and soils and a bioreactor contaminated by hydrocarbons, PAHs, or heavy metals, respectively (see Fig. S1 in the supplemental material). NM-Ms plots seemed to specifically favor the development of bacteria affiliated to the Verrucomicrobia (close to the Prosthecobacter and Luteolibacter genera), the Actinobacteria (Microbacterium oxydans), and the Chloroflexi branches (Fig. 3). These sequences were closely related to environmental clones found in grassland, licorice roots, oil-contaminated seawater, and arid zone soil (see Fig. S1 in the supplemental material). Similarly, the NM-SV plots favored the development of bacteria affiliated to the Firmicutes (Paenibacillus spp.), the Gammaproteobacteria (Stenotrophomonas genera), the Actinobacteria, and the Chloroflexi branches (Fig. 3). These sequences were closely related to environmental clones found in iron-reducing enrichment, hydrocarbons and heavy metal-contaminated soil, semicoke-contaminated soil treated by phytoremediation, and mixed grass prairie, respectively (see Fig. S1 in the supplemental material).
FIG. 3.
Phylogenetic neighbor-joining tree showing 16S rRNA gene sequences of TTGE bands from the four types of plots (NM-BS, NM-Ms, NM-SV, and TD-Ms). Sequences were grouped when identity was more than 98% for clarity. The sampling dates (T0 and T4) and plot types where bands were detected are indicated. Reference strain sequences were taken from GenBank and are named by their accession numbers; to see the closest environmental clones related to our TTGE bands, refer to Fig. S1 in the supplemental material. The tree is rooted with the Paenibacillus sp. strain GP25-12 (accession number AM162342) 16S rRNA gene sequence. Bootstrap values greater than 50% derived from 100 replicates are shown (filled circles on branch junction) and were obtained using the distance matrix and neighbor-joining method within PHYLIP 3.65. The bar represents 10% sequence divergence.
The bacterial diversity identified in the TD soil plots was completely different from that in the NM soil. At T0, we identified three main bacterial species affiliated with Thiobacillus denitrificans (Betaproteobacteria), Alphaproteobacteria, and Verrucomicrobia (Fig. 3) and closely related to environmental clones detected in oil- and anaerobic toluene-contaminated environments, an anaerobic coking wastewater treatment system, and paddy soil (see Fig. S1 in the supplemental material). During the 2 years at the experimental site, a larger level of diversity of bacterial phyla developed in the TD-Ms plots. At T4, we identified sequences affiliated with Gammaproteobacteria (Achromatium), Alphaproteobacteria, and Actinobacteria (Microbacterium and Solirubrobacter spp.) (Fig. 3) and closely related to environmental clones detected in seafloor lava, mercury-contaminated soils, hydrocarbon and heavy metal-contaminated soil, acid mine drainage, and iron-sulfide and nitrate-rich anaerobe groundwater (see Fig. S1 in the supplemental material).
PAH concentration and PAH-degrading bacteria.
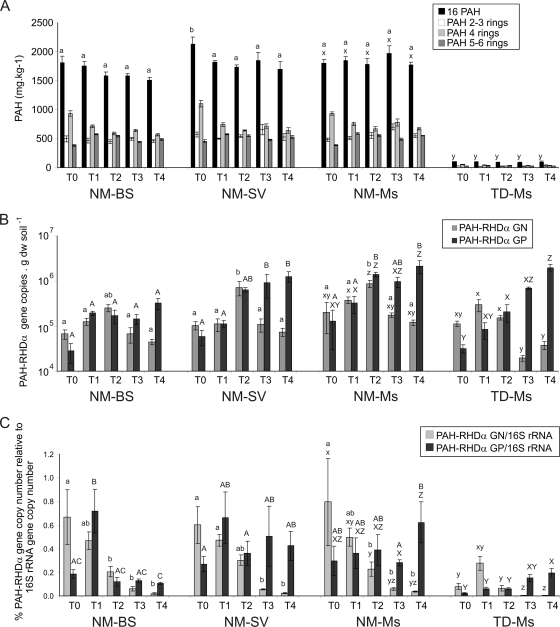
At T0, the PAH concentration ranged from 1,800 to 2,130 mg·kg−1 of soil dry weight in the NM soil and was not significantly different (P > 0.05) among plot types. At T1, after the winter period and before plant growth, the PAH concentration ranged from 1,754 to 1,850 mg·kg−1 of soil dry weight. Then, the PAH content tended to decrease (dissipation not significant; P > 0.05), reaching 1,510, 1,702, and 1,771 mg·kg−1 mean values at T4 for the NM-BS, NM-SV, and NM-Ms plots, respectively (Fig. 4A). The TD soil had 20 times less PAH content than the NM soil, and the concentration was about 106 mg·kg−1 of soil dry weight (mean value) at T0 and did not change significantly during the monitoring (Fig. 4A).
FIG. 4.
PAH concentration (A), PAH-RHDα GN and GP bacterial gene copy numbers determined by real-time PCR quantification on DNA (B), and percentage of PAH-RHDα genes relative to 16S rRNA gene copy numbers (C) during the 2-year period (from T0 to T4) of the four plot type analysis. Mean values based on the four replicate plots of each treatment are presented with their corresponding standard errors. Different letters indicate significant differences (P < 0.05) between plots (letters a and b or A, B, and C for the comparison between NM-BS, NM-SV, and NM-Ms, and letters x, y, and z or X, Y, and Z for the comparison between NM-Ms and TD-Ms). In panel A, statistical analysis values were not presented for the different groups of PAHs but rather for the 16 PAHs. In panels B and C, lowercase letters refer to statistics on PAH-RHDα GN bacterial genes and the PAH-RHDα GN/16S rRNA gene ratio, and capital letters refer to statistics on PAH-RHDα GP bacterial genes and the PAH-RHDα GP/16S rRNA gene ratio.
In NM soil plots, the PAH-RHDα GN bacterial gene density significantly increased (P > 0.05) during the first year from 6.7 × 104 to 19.9 × 104 gene copies·g−1 of soil dry weight to 25.5 × 104 to 87.0 × 104 gene copies·g−1 of soil dry weight from T0 to T2, respectively (Fig. 4B). Then, it decreased during the second year down to the same level as that at T0. In TD plots, the PAH-RHDα GN bacterial gene density was similar to that in the NM soil during the first year and then reached down to the detection limit of the real-time PCR assays. The PAH-RHDα GP bacterial gene density stayed stable in the NM-BS plots, while it increased significantly (P > 0.05) over 2 years for the vegetated plots (NM-Ms, NM-SV, and TD-Ms) (Fig. 4B).
The functional gene real-time PCR quantification data were normalized to 16S rRNA gene copies (7) to compare samples and the variation during the monitoring. The percentage of PAH-RHDα GN bacterial genes varied from 0.60 to 0.79% at T0 down to 0.02 to 0.04% at T4 in NM soil plots and down to 0.004% in the TD soil plots, representing a decrease of at least 30 and 70 times for the NM and TD soil plots, respectively. The proportion of PAH-RHDα GP bacterial genes tended to decrease in the NM-BS from T1 to T4 (from 0.72 to 0.11%), while they maintained a higher level (0.28 to 0.62%) in the NM-Ms and NM-SV plots (Fig. 4C). In TD-Ms plots, the PAH-RHDα GP bacterial gene proportion increased more than 10 times in 2 years, although the percentages were lower than those in NM-Ms plots (Fig. 4C).
DISCUSSION
Bacterial community density and structure and the PAH-degrading bacterium abundance changed over the 2 years of the long-term in situ trial with aged PAH-contaminated soil and the same soil previously treated by thermal desorption. The presence of plants favored the maintenance of a higher percentage of GP PAH bacterial degraders, although no significant changes in PAH concentration were detected.
Influence of plants on soil bacterial community density and diversity.
During the first year on site (from T0 to T2), the density of 16S rRNA genes increased in all the plots, probably due to the adaptation of the bacterial community to the experimental system. After this stabilization period, the density of 16S rRNA genes did not vary significantly, and no differences were observed between bare soil (NM-BS) and the soil under plant influence (NM-Ms and NM-SV). As a plant may secrete in its root exudates 10 to 20% of the carbon assimilated during photosynthesis (14, 57), the higher availability of carbon and organic matter in the rhizosphere could increase the bacterial density (from 4- to 100-fold). Although Fan et al. (15) found 5 to 7.5 times more total heterotrophic bacteria using the CFU method in the rhizosphere of alfalfa in a pyrene-contaminated soil than in the nonrhizospheric soil, very few studies have shown such a difference using real-time PCR quantification of the 16S rRNA genes. Only Stubner (59) found a slight rice rhizospheric effect that induced a twofold increase of the 16S rRNA gene copy density compared to that of the bulk soil. If a rhizospheric gradient occurred with the root distance as described by Joner and Leyval (30), it may not be observed during in situ experiments where not only the soil adhering to the roots was sampled but rather a mixed-soil sample from the planted plot. This could explain why there was no significant rhizosphere effect on 16S rRNA gene density.
When a site gets polluted with hydrocarbons, the contamination selects for a less diverse but catabolically versatile bacterial community (40). Indeed, at the beginning of the experiment, the phylogenetic diversity was low in NM soil. Labbé et al. (38) showed that the Betaproteobacteria and the Gammaproteobacteria were dominant in hydrocarbon-contaminated soils. Similarly, at T0, a predominance of Gammaproteobacteria was observed in the NM soil. During winter and before active plant growth (T0 and T1), the microbial communities of all the NM soil plots clustered together (Fig. 2). After 2 years (T4) in the bare soil plots, the bacterial community was represented by Betaproteobacteria and Gammaproteobacteria, closely related to clone sequences detected in various anaerobic environments. As no physical mechanism could potentially reoxygenate the soil matrix, one could hypothesize that the bare soil plots could present anaerobic or microaerobic niches. Plant roots were suggested as a substitute to improve soil aeration (1) that could favor the development of larger phylogenetic diversity, as well as higher levels of the fresh carbon source input via root exudates. In the present study, the phylogenetic bacterial diversity observed in the NM-BS plots was lower than that in the NM-Ms and NM-SV plots, where the presence of vegetation favored the development of a wider range of bacterial phyla (Betaproteobacteria, Gammaproteobacteria, Verrucomicrobia, Actinobacteria, Firmicutes, and Chloroflexi), as reported by Gremion et al. (19) during phytoremediation of heavy metal-contaminated soil.
In the TD-Ms, a constant increase in 16S rRNA gene density was observed over 2 years (Fig. 1). First, during the thermal desorption treatment, residual organic matter (including killed microbial biomass) could induce a “priming effect,” as described by Fontaine et al. (18), that could intensify bacterial growth during the storage period (6 months before experiment setup). This priming effect could explain why the density of 16S rRNA genes was already high at T0. Second, the toxicity of this soil was decreased by the removal of PAH contamination, and this probably allowed the development of a higher density of bacteria compared to that of the PAH-polluted NM-Ms. However, the thermal desorption treatment resulted in a destructured soil where bacterial recolonization by diverse phyla was relatively slow. At T0, only a few main bacterial species (Alphaproteobacteria, Betaproteobacteria, and Verrucomicrobia) dominated. However, after 2 years in the plots and under alfalfa growth, the soil was recolonized by a more diversified community represented mainly by Actinobacteria, Alphaproteobacteria, and Gammaproteobacteria. The main representatives were closely related to clones detected in extremely degraded or contaminated environments, among which some were anaerobic (e.g., anaerobic toluene-degrading sediment enrichment, coking wastewater treatment, iron- and nitrate-rich water, and sulfate-reducing bioreactor treating mine drainage). Contrary to the NM soil, the presence of alfalfa did not seem to favor the oxygenation of the soil matrix, probably because the thermal desorption treatment modified the water/soil interactions and affected the oxygen transfer. The presence of the same plant on the NM-Ms and TD-Ms plots did not influence the bacterial diversity in the same way, suggesting that the bacterial community structure is driven firstly by soil characteristics (36) and by the plant favoring the development of some bacteria present initially in the soil.
Fate of PAHs and occurrence of PAH degraders.
The functional real-time PCR assays were based on the specific and sensitive quantification of homologous genes that encode the PAH-RHDα from GN (Pseudomonas, Ralstonia, Comamonas, Burkholderia, Sphingomonas, Alcaligenes, and Polaromonas strains) and GP (Rhodococcus, Mycobacterium, Nocardioides, and Terrabacter strains) PAH bacterial degraders separately (7). If PAH-degrading bacteria possessed divergent genes or a different PAH degradation pathway, these bacteria would not be quantified; however, our primer sets allowed for targeting of a wide range of PAH-degrading genes and are an efficient tool to detect most known PAH-degrading bacteria. At T0, the densities of PAH-degrading bacteria (104 to 105 PAH-RHDα gene copies·g−1 soil dry weight) in NM and TD soils were high compared to those in soil with no contamination history, where PAH-RHDα genes could not be detected (7). This is consistent with the data from Johnsen and Karlson (23), where densities of phenanthrene and pyrene degraders reflected previous PAH exposure of the soil. After 2 years on site, the PAH concentration did not vary significantly, whatever the presence of plant, even if PAH degraders were present in the soil. However, when measuring the total PAH concentration, large variations were observed (see high standard deviation values) (Fig. 4A), which could prevent detection of any small effect such as, for example, biodegradation of the bioavailable part of the PAHs. In addition, Chaudhry et al. (10) showed no significant removal of PAHs after 29 weeks of a phytoremediation attempt in a soil with aged PAH contamination. In all types of NM soil plots, the occurrence and proportion of GN PAH bacterial degraders decreased over time (Fig. 4B). On the contrary, the density of GP PAH-RHDα bacterial genes increased by about 10 times in the vegetated plots (NM-SV and NM-Ms), with their proportions relative to those of the 16S rRNA genes, and staying constant while it decreased in the bare soil plots. Culture-dependent methods, enzymatic analyses, and molecular gene detection-based studies showed that the density of PAH-degrading bacteria increased by about 4.5- to 15-fold in rhizosphere compared to that in bulk soil (26, 27, 44-46, 48, 54) and that dioxygenase-expressing bacteria (42) and dioxygenase genes (57) increased in the PAH-contaminated rhizosphere. Our data suggest that the presence of alfalfa or spontaneous vegetation in the aged PAH-contaminated soil favored the maintenance of a larger proportion of GP bacteria capable of PAH degradation, although it is commonly accepted that rhizospheric bacteria are predominantly GN (10). A similar finding was obtained by Radwan et al. (53), who showed that GP hydrocarbon-utilizing bacteria belonging to the Cellulomonas, Rhodococcus, and Arthrobacter genera were dominant in the rhizosphere of plants grown in oil-polluted soils. Using a similar real-time PCR assay, Johnsen et al. (24) detected a high density of the GP pdo1 bacterial genes (one gene detected by the PAH-RHDα GP bacterial primer set) and did not detect GN nah and phnA bacterial genes in a soil close to a PAH-contaminated motorway pavement. The low density of the GN PAH-RHDα bacterial gene in the NM soil could be explained by the low PAH bioavailability in the NM soil. In aged PAH-contaminated soils, the strong adsorption of PAHs to soil particles, their sequestration in soil organic matter (11, 35), and their low water solubility limit their bioavailability and thus their utilization as a carbon source for microorganisms. Indeed, some authors (49, 50) found that the abundance of naphthalene dioxygenase genes (nahAc) could be increased by the addition of bioavailable hydrocarbons. GP PAH bacterial degraders may experience advantages by increasing the PAH bioavailability in aged-contaminated soils thanks to biofilm formation directly on hydrophobic pollutants (2, 25). In aged PAH-polluted soils, plants may also enhance the PAH availability via the presence of surfactant molecules in root exudates (27, 51). However, in the present study, other environmental parameters (nutrient concentration, temperature) may limit the PAH biodegradation, even if GP PAH-degrading bacteria were present in significant quantities.
Although the TD soil was treated for PAH contamination, the PAH degrader-specific genes were detected by real-time PCR at T0, with a larger proportion of GN PAH-RHDα bacterial genes. The PAH concentration in this soil was not negligible, and the “priming effect” described above could have favored the development of r-strategist, fast-growing bacteria, such as GN PAH degraders belonging to Betaproteobacteria and Gammaproteobacteria, during the 6 months of storage before plot setting. During the 2 years in situ, the GP PAH-RHDα bacterial quantity and proportion increased up to the content found in the contaminated NM soil. GP PAH bacterial degraders were shown to be involved and specialized in the degradation of a broader range of PAH molecules, mainly of high-molecular-weight PAH, than GN ones (32). Bacteria adapted to a pollutant will be able to proliferate and may become dominant in a contaminated environment (37, 43). Our study showed that the identified dominant bacteria were close to sequences identified in similar environments with high constraints of pollution (hydrocarbons or heavy metals), indicating that they were specific to extremely degraded environments.
Supplementary Material
Acknowledgments
We acknowledge GISFI (www.gisfi.fr) for providing the equipment for the field site.
We acknowledge the INSU-EC2CO program for financial support.
Footnotes
Published ahead of print on 24 July 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aprill, W., and R. C. Sims. 1990. Evaluation of the use of prairie grasses for stimulating polycyclic aromatic hydrocarbon treatment in soil. Chemosphere 20:253-265. [Google Scholar]
- 2.Bastiaens, L., D. Springael, P. Wattiau, H. Harms, R. DeWachter, H. Verachtert, and L. Diels. 2000. Isolation of adherent polycyclic aromatic hydrocarbon (PAH)-degrading bacteria using PAH-sorbing carriers. Appl. Environ. Microbiol. 66:1834-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biache, C., L. Mansuy-Huault, P. Faure, C. Munier-Lamy, and C. Leyval. 2008. Effects of thermal desorption on the composition of two coking plant soils: impact on solvent extractable organic compounds and metal bioavailability. Environ. Pollut. 156:671-677. [DOI] [PubMed] [Google Scholar]
- 4.Binet, P., J. M. Portal, and C. Leyval. 2000. Dissipation of 3-6-ring polycyclic aromatic hydrocarbons in the rhizosphere of ryegrass. Soil Biol. Biochem. 32:2011-2017. [Google Scholar]
- 5.Boopathy, R. 2000. Factors limiting bioremediation technologies. Bioresour. Technol. 74:63-67. [Google Scholar]
- 6.Carmichael, L. M., and F. K. Pfaender. 1997. The effect of inorganic and organic supplements on the microbial degradation of phenanthrene and pyrene in soils. Biodegradation 8:1-13. [DOI] [PubMed] [Google Scholar]
- 7.Cébron, A., M.-P. Norini, T. Beguiristain, and C. Leyval. 2008. Real-time PCR quantification of PAH-ring hydroxylating dioxygenase (PAH-RHDα) genes from Gram positive and Gram negative bacteria in soil and sediment samples. J. Microbiol. Methods 73:148-159. [DOI] [PubMed] [Google Scholar]
- 8.Cerniglia, C. E. 1992. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation 3:351-368. [Google Scholar]
- 9.Cerniglia, C. E., and I. L. Allen. 1984. Microbial metabolism of polycyclic aromatic hydrocarbons, p. 31-71. In A. I. Laskin (ed.), Advances in applied microbiology, vol. 30. Academic Press, Orlando, FL. [DOI] [PubMed] [Google Scholar]
- 10.Chaudhry, Q., M. Blom-Zandstra, S. Gupta, and E. J. Joner. 2005. Utilising the synergy between plants and rhizosphere microorganisms to enhance breakdown of organic pollutants in the environment. Environ. Sci. Pollut. Res. Int. 12:34-48. [DOI] [PubMed] [Google Scholar]
- 11.Chekol, T., L. R. Vough, and R. L. Chaney. 2002. Plant-soil-contaminant specificity affects phytoremediation of organic contaminants. Int. J. Phytoremediation 4:17-26. [Google Scholar]
- 12.Chen, S. H., and M. D. Aitken. 1999. Salicylate stimulates the degradation of high-molecular weight polycyclic aromatic hydrocarbons by Pseudomonas saccharophila P15. Environ. Sci. Technol. 33:435-439. [Google Scholar]
- 13.Dazy, M., V. Jung, J.-F. Férard, and J.-F. Masfaraud. 2008. Ecological recovery of vegetation on a coke-factory soil: role of plant antioxidant enzymes and possible implications in site restoration. Chemosphere 74:57-63. [DOI] [PubMed] [Google Scholar]
- 14.Donnelly, P. K., R. S. Hegde, and J. S. Fletcher. 1994. Growth of PCB-degrading bacteria on compounds from photosynthetic plants. Chemosphere 28:981-988. [Google Scholar]
- 15.Fan, S., P. Li, Z. Gong, W. Ren, and N. He. 2008. Promotion of pyrene degradation in rhizosphere of alfalfa (Medicago sativa L.). Chemosphere 71:1593-1598. [DOI] [PubMed] [Google Scholar]
- 16.Felske, A., A. D. L. Akkermans, and W. M. De Vos. 1998. Quantification of 16S rRNAs in complex bacterial communities by multiple competitive reverse transcription-PCR in temperature gradient gel electrophoresis fingerprints. Appl. Environ. Microbiol. 64:4581-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fons, F., N. Amellal, C. Leyval, N. Saint-Martin, and M. Henry. 2003. Effects of gypsophila saponins on bacterial growth kinetics and on selection of subterranean clover rhizosphere bacteria. Can. J. Microbiol. 49:367-373. [DOI] [PubMed] [Google Scholar]
- 18.Fontaine, S., A. Mariotti, and L. Abbadie. 2003. The priming effect of organic matter: a question of microbial competition? Soil Biol. Biochem. 35:837-843. [Google Scholar]
- 19.Gremion, F., A. Chatzinotas, K. Kaufmann, W. Sigler, and H. Harms. 2004. Impacts of heavy metal contamination and phytoremediation on a microbial community during a twelve-month microcosm experiment. FEMS Microbiol. Ecol. 48:273-283. [DOI] [PubMed] [Google Scholar]
- 20.Guieysse, B., G. Viklund, A.-C. Toes, and B. Mattiasson. 2004. Combined UV-biological degradation of PAHs. Chemosphere 55:1493-1499. [DOI] [PubMed] [Google Scholar]
- 21.Günther, T., U. Dornberger, and W. Fritsche. 1996. Effects of ryegrass on biodegradation of hydrocarbons in soil. Chemosphere 33:203-215. [DOI] [PubMed] [Google Scholar]
- 22.Habe, H., and T. Omori. 2003. Genetics of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria. Biosci. Biotechnol. Biochem. 67:225-243. [DOI] [PubMed] [Google Scholar]
- 23.Johnsen, A., and U. Karlson. 2005. PAH degradation capacity of soil microbial communities—does it depend on PAH exposure? Microb. Ecol. 50:488-495. [DOI] [PubMed] [Google Scholar]
- 24.Johnsen, A. R., J. R. deLipthay, F. Reichenberg, S. J. Sorensen, O. Andersen, P. Christensen, M. L. Binderup, and C. S. Jacobsen. 2006. Biodegradation, bioaccessibility, and genotoxicity of diffuse polycyclic aromatic hydrocarbon (PAH) pollution at a motorway site. Environ. Sci. Technol. 40:3293-3298. [DOI] [PubMed] [Google Scholar]
- 25.Johnsen, A. R., and U. Karlson. 2004. Evaluation of bacterial strategies to promote the bioavailability of polycyclic aromatic hydrocarbons. Appl. Microbiol. Biotechnol. 63:452-459. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, D. L., D. R. Anderson, and S. P. McGrath. 2005. Soil microbial response during the phytoremediation of a PAH contaminated soil. Soil Biol. Biochem. 37:2334-2336. [Google Scholar]
- 27.Joner, E. J., S. C. Corgié, N. Amellal, and C. Leyval. 2002. Nutritional constraints to degradation of polycyclic aromatic hydrocarbons in a simulated rhizosphere. Soil Biol. Biochem. 34:859-864. [Google Scholar]
- 28.Joner, E. J., D. Hirmann, O. H. J. Szolar, D. Todorovic, C. Leyval, and A. P. Loibner. 2004. Priming effects on PAH degradation and ecotoxicity during a phytoremediation experiment. Environ. Pollut. 128:429-435. [DOI] [PubMed] [Google Scholar]
- 29.Joner, E. J., A. Johansen, A. P. Loibner, M. A. dela Cruz, O. H. J. Szolar, J. M. Portal, and C. Leyval. 2001. Rhizosphere effects on microbial community structure and dissipation and toxicity of polycyclic aromatic hydrocarbons (PAHs) in spiked soil. Environ. Sci. Technol. 35:2773-2777. [DOI] [PubMed] [Google Scholar]
- 30.Joner, E. J., and C. Leyval. 2003. Rhizosphere gradients of polycyclic aromatic hydrocarbon (PAH) dissipation in two industrial soils and the impact of arbuscular mycorrhiza. Environ. Sci. Technol. 37:2371-2375. [DOI] [PubMed] [Google Scholar]
- 31.Kamath, R., J. L. Schnoor, and P. J. J. Alvarez. 2004. Effect of Root-derived substrates on the expression of nah-lux genes in Pseudomonas fluorescens HK44: implications for PAH biodegradation in the rhizosphere. Environ. Sci. Technol. 38:1740-1745. [DOI] [PubMed] [Google Scholar]
- 32.Kanaly, R. A., and S. Harayama. 2000. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J. Bacteriol. 182:2059-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kästner, M., M. Breuer-Jammali, and B. Mahro. 1994. Enumeration and characterization of the soil microflora from hydrocarbon-contaminated soil sites able to mineralize polycyclic aromatic hydrocarbons (PAH). Appl. Microbiol. Biotechnol. 41:267-273. [Google Scholar]
- 34.Kästner, M., and B. Mahro. 1996. Microbial degradation of polycyclic aromatic hydrocarbons in soils affected by the organic matrix of compost. Appl. Microbiol. Biotechnol. 44:668-675. [DOI] [PubMed] [Google Scholar]
- 35.Ke, L., T. W. Y. Wong, A. H. Y. Wong, Y. S. Wong, and N. F. Y. Tam. 2003. Negative effects of humic acid addition on phytoremediation of pyrene-contaminated sediments by mangrove seedlings. Chemosphere 52:1581-1591. [DOI] [PubMed] [Google Scholar]
- 36.Kowalchuk, G., D. Buma, W. de Boer, P. Klinkhamer, and J. van Veen. 2002. Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Antonie van Leeuwenhoek 81:509-520. [DOI] [PubMed] [Google Scholar]
- 37.Kuiper, I., E. L. Lagendijk, G. V. Bloemberg, and B. J. J. Lugtenberg. 2004. Rhizoremediation: a beneficial plant-microbe interaction. Mol. Plant-Microbe Interact. 17:6-15. [DOI] [PubMed] [Google Scholar]
- 38.Labbé, D., R. Margesin, F. Schinner, L. G. Whyte, and C. W. Greer. 2007. Comparative phylogenetic analysis of microbial communities in pristine and hydrocarbon-contaminated Alpine soils. FEMS Microbiol. Ecol. 59:466-475. [DOI] [PubMed] [Google Scholar]
- 39.Leigh, M. B., J. S. Fletcher, X. Fu, and F. J. Schmitz. 2002. Root turnover: an important source of microbial substrates in rhizosphere remediation of recalcitrant contaminants. Environ. Sci. Technol. 36:1579-1583. [DOI] [PubMed] [Google Scholar]
- 40.Lindstrom, J. E., R. P. Barry, and J. F. Braddock. 1999. Long-term effects on microbial communities after a subarctic oil spill. Soil Biol. Biochem. 31:1677-1689. [Google Scholar]
- 41.Liste, H.-H., and M. Alexander. 2000. Plant-promoted pyrene degradation in soil. Chemosphere 40:7-10. [DOI] [PubMed] [Google Scholar]
- 42.Liste, H.-H., and I. Prutz. 2006. Plant performance, dioxygenase-expressing rhizosphere bacteria, and biodegradation of weathered hydrocarbons in contaminated soil. Chemosphere 62:1411-1420. [DOI] [PubMed] [Google Scholar]
- 43.Maila, M. P., P. Randima, K. Drønen, and T. E. Cloete. 2006. Soil microbial communities: influence of geographic location and hydrocarbon pollutants. Soil Biol. Biochem. 38:303-310. [Google Scholar]
- 44.Miya, R. K., and M. K. Firestone. 2001. Enhanced phenanthrene biodegradation in soil by slender oat root exudates and root debris. J. Environ. Qual. 30:1911-1918. [DOI] [PubMed] [Google Scholar]
- 45.Miya, R. K., and M. K. Firestone. 2000. Phenanthrene-degrader community dynamics in rhizosphere soil from a common annual grass. J. Environ. Qual. 29:584-592. [Google Scholar]
- 46.Muratova, A., T. Hübner, S. Tischer, O. Turkovskaya, M. Möder, and P. Kuschk. 2003. Plant-rhizosphere-microflora association during phytoremediation of PAH-contaminated soil. Int. J. Phytoremediation 5:137-151. [DOI] [PubMed] [Google Scholar]
- 47.Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schafer, and C. Wawer. 1998. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. De Bruijn (ed.), Molecular microbial ecology manual, vol. 3.4.4. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 48.Nichols, T. D., D. C. Wolf, H. B. Rogers, C. A. Beyrouty, and C. M. Reynolds. 1997. Rhizosphere microbial populations in contaminated soils. Water Air Soil Pollut. 95:165-178. [Google Scholar]
- 49.Palmroth, M., P. Koskinen, A. Kaksonen, U. Münster, J. Pichtel, and J. Puhakka. 2007. Metabolic and phylogenetic analysis of microbial communities during phytoremediation of soil contaminated with weathered hydrocarbons and heavy metals. Biodegradation 18:769-782. [DOI] [PubMed] [Google Scholar]
- 50.Park, J.-W., and D. Crowley. 2006. Dynamic changes in nahAc gene copy numbers during degradation of naphthalene in PAH-contaminated soils. Appl. Microbiol. Biotechnol. 72:1322-1329. [DOI] [PubMed] [Google Scholar]
- 51.Phillips, T. M., D. Liu, A. G. Seech, H. Lee, and J. T. Trevors. 2000. Monitoring bioremediation in creosote-contaminated soils using chemical analysis and toxicity tests. J. Ind. Microbiol. Biotechnol. 24:132-139. [Google Scholar]
- 52.Pradhan, S. P., J. R. Conrad, J. R. Paterek, and V. J. Srivastava. 1998. Potential of phytoremediation for treatment of PAHs in soil at MGP sites. Soil Sediment Contam. 7:467-480. [Google Scholar]
- 53.Radwan, S. S., H. Al-Awadhi, N. A. Sorkhoh, and I. M. El-Nemr. 1998. Rhizospheric hydrocarbon-utilizing microorganisms as potential contributors to phytoremediation for the oily Kuwaiti desert. Microbiol. Res. 153:247-251. [Google Scholar]
- 54.Reilley, K. A., M. K. Banks, and A. P. Schwab. 1996. Organic chemicals in the environment: dissipation of polycyclic aromatic hydrocarbons in the rhizosphere. J. Environ. Qual. 25:212-219. [Google Scholar]
- 55.Rentz, J. A., P. J. J. Alvarez, and J. L. Schnoor. 2004. Repression of Pseudomonas putida phenanthrene-degrading activity by plant root extracts and exudates. Environ. Microbiol. 6:574-583. [DOI] [PubMed] [Google Scholar]
- 56.Siciliano, S. D., N. Fortin, A. Mihoc, G. Wisse, S. Labelle, D. Beaumier, D. Ouellette, R. Roy, L. G. Whyte, M. K. Banks, P. Schwab, K. Lee, and C. W. Greer. 2001. Selection of specific endophytic bacterial genotypes by plants in response to soil contamination. Appl. Environ. Microbiol. 67:2469-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siciliano, S. D., J. J. Germida, K. Banks, and C. W. Greer. 2003. Changes in microbial community composition and function during a polyaromatic hydrocarbon phytoremediation field trial. Appl. Environ. Microbiol. 69:483-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smalla, K., G. Wieland, A. Buchner, A. Zock, J. Parzy, S. Kaiser, N. Roskot, H. Heuer, and G. Berg. 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67:4742-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stubner, S. 2004. Quantification of Gram-negative sulphate-reducing bacteria in rice field soil by 16S rRNA gene-targeted real-time PCR. J. Microbiol. Methods 57:219-230. [DOI] [PubMed] [Google Scholar]
- 60.Wilson, S. C., and K. C. Jones. 1993. Bioremediation of soil contaminated with polynuclear aromatic hydrocarbons (PAHs): a review. Environ. Pollut. 81:229-249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.