Abstract
Forty-two strains of Vibrio parahaemolyticus were isolated from Bay of Bengal estuaries and, with two clinical strains, analyzed for virulence, phenotypic, and molecular traits. Serological analysis indicated O8, O3, O1, and K21 to be the major O and K serogroups, respectively, and O8:K21, O1:KUT, and O3:KUT to be predominant. The K antigen(s) was untypeable, and pandemic serogroup O3:K6 was not detected. The presence of genes toxR and tlh were confirmed by PCR in all but two strains, which also lacked toxR. A total of 18 (41%) strains possessed the virulence gene encoding thermostable direct hemolysin (TDH), and one had the TDH-related hemolysin (trh) gene, but not tdh. Ten (23%) strains exhibited Kanagawa phenomenon that surrogates virulence, of which six, including the two clinical strains, possessed tdh. Of the 18 tdh-positive strains, 17 (94%), including the two clinical strains, had the seromarker O8:K21, one was O9:KUT, and the single trh-positive strain was O1:KUT. None had the group-specific or ORF8 pandemic marker gene. DNA fingerprinting employing pulsed-field gel electrophoresis (PFGE) of SfiI-digested DNA and cluster analysis showed divergence among the strains. Dendrograms constructed using PFGE (SfiI) images from a soft database, including those of pandemic and nonpandemic strains of diverse geographic origin, however, showed that local strains formed a cluster, i.e., “clonal cluster,” as did pandemic strains of diverse origin. The demonstrated prevalence of tdh-positive and diarrheagenic serogroup O8:K21 strains in coastal villages of Bangladesh indicates a significant human health risk for inhabitants.
Vibrio parahaemolyticus, a halophilic bacterium, is a causative agent of seafood-related gastroenteritis worldwide (5, 13, 41) and one of the major causes of seafood-associated gastroenteritis in the United States, Asia, Europe, and countries where sporadic cases and outbreaks occur regularly (12, 13). The bacterium is prevalent in brackish and marine waters (43). Historically first identified as the causative agent of a gastroenteritis outbreak in Japan in 1950 (14), V. parahaemolyticus is now recognized as one of the most important food-borne pathogens in Asia, causing approximately half of food poisoning outbreaks in Taiwan, Japan, Vietnam, and Southeast Asian countries.
The gene encoding the thermostable direct hemolysin (TDH)—manifested as beta-hemolysis when V. parahaemolyticus is plated onto Wagatsuma blood agar (43), i.e., the Kanagawa phenomenon (KP)—has been shown to be present in more than 90% of clinical strains and less than 1% of environmental strains (31, 39). Some strains also possess the gene trh, encoding the TDH-related hemolysin (TRH), or both tdh and trh (18, 43). Another gene, the thermolabile hemolysin gene (tlh), was reported to be present in V. parahaemolyticus (36) and subsequently in all V. parahaemolyticus strains tested (38).
V. parahaemolyticus gastroenteritis is a multiserogroup affliction, with at least 13 O serogroups and 71 K serotypes detected (19, 42). In 1996, serogroup O3:K6 was first reported from diarrhea patients in Kolkata, India (32), and subsequently worldwide, as an increasing incidence of gastroenteritis caused by the serogroup O3:K6 was reported in many countries (41). Rapid spreading of serogroup O3:K6 infections in Asia (27, 32), and subsequently in the United States (12), Africa (3), Europe (25), and Latin America (15), indicated its potential as a pandemic pathogen (34, 43). In addition, V. parahaemolyticus serogroup O3:K6 possesses the group-specific (GS) gene sequence in the toxRS operon and ORF8, of the 10 known open reading frames (ORFs) of the O3:K6-specific filamentous phage f237. The GS gene and ORF8 provide genetic markers distinguishing O3:K6 from other serogroups (27, 29). Recent studies have shown O4:K68, O1:K25, O1:K26, O1:K untypeable (O1:KUT), and O3:K46 serogroups to share genetic markers specific for the pandemic serogroup O3:K6 (7, 10, 27, 34, 41). The non-O3:K6 serogroups with pandemic traits are increasingly found worldwide, and therefore, their pandemic potential cannot be ruled out.
In Bangladesh, strains of different serogroups having genetic markers for the serogroup O3:K6 of V. parahaemolyticus were reported to have been isolated from hospitalized gastroenteritis patients in Dhaka (7). A systematic surveillance of the coastal areas bordering the Bay of Bengal where diarrheal disease is endemic (1) has not been done. This study, the first of its kind, was undertaken to investigate virulence potential, as well as phenotypic and genotypic traits of V. parahaemolyticus strains occurring in the estuarine ecosystem of Bangladesh.
MATERIALS AND METHODS
A total of 44 V. parahaemolyticus strains, isolated between 2005 and 2006 from the estuarine ecosystem of the Bay of Bengal, were tested for serogroup, virulence genes, and phenotypic and molecular traits and were compared with pandemic and nonpandemic serogroup strains of diverse geographical origins.
Isolation of V. parahaemolyticus strains.
Surface water samples were collected from ponds and rivers of the Bay of Bengal estuaries during the period from October 2005 to January 2006, covering four coastal districts of Bangladesh (Table 1). Water samples were collected in dark Nalgene bottles (Nalgene Nunc International) employing methods recommended by the American Public Health Association (4), transported at an ambient temperature, and processed within 8 h of collection, following aseptic techniques (2). A ca. 90-ml-volume water sample was enriched by inoculating into 10 ml of 10× alkaline peptone water and incubating at 37°C for 6 to 8 h before plating onto a suitable medium by following methods described previously (2). Ca. 2 to 3 loops full of enriched alkaline peptone water broth were streaked onto thiosulfate-citrate-bile salts-sucrose (TCBS) agar and incubated at 37°C for 18 to 24 h. Presumptive Vibrio-like colonies were selected and confirmed by biochemical tests as described elsewhere (34, 43).
TABLE 1.
Characterizations of V. parahaemolyticus strains (n = 44) isolated from the coastal aquatic ecosystem of the Bay of Bengala
| O:K serotype | Place of isolation | Yr of isolation | District | No. of strains | Presence of:
|
Result of:
|
KP | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| toxR | tdh | trh | tlh | GS-PCR | ORF8-PCR | ||||||
| O1:KUT | Karnaphooli estuary | 2005 | Chittagong | 1 | + | − | + | + | − | − | + |
| O1:KUT | Karnaphooli estuary | 2005 | Chittagong | 4 | + | − | − | + | − | − | − |
| O1:KUT | Karnaphooli estuary | 2005 | Chittagong | 1 | + | − | − | + | − | − | + |
| O1:K38 | Karnaphooli estuary | 2005 | Chittagong | 1 | − | − | − | + | − | − | − |
| O3:KUT | Karnaphooli estuary | 2005 | Chittagong | 1 | + | − | − | + | − | − | + |
| O3:KUT | Karnaphooli estuary | 2005 | Chittagong | 1 | + | − | − | + | − | − | − |
| O3:K4 | Karnaphooli estuary | 2005 | Chittagong | 1 | + | − | − | + | − | − | + |
| O3:K29 | Karnaphooli estuary | 2005 | Chittagong | 1 | + | − | − | + | − | − | − |
| O3:K30 | Karnaphooli estuary | 2005 | Chittagong | 1 | + | − | − | + | − | − | + |
| O3:K30 | Karnaphooli estuary | 2006 | Chittagong | 1 | + | − | − | + | − | − | − |
| O3:K45 | Karnaphooli estuary | 2006 | Chittagong | 1 | + | − | − | + | − | − | − |
| O4:K34 | Karnaphooli estuary | 2006 | Chittagong | 1 | + | − | − | + | − | − | − |
| O5:KUT | Karnaphooli estuary | 2006 | Chittagong | 1 | + | − | − | + | − | − | − |
| O8:K39 | Karnaphooli estuary | 2006 | Chittagong | 1 | + | − | − | + | − | − | − |
| O10:KUT | Karnaphooli estuary | 2006 | Chittagong | 2 | + | − | − | + | − | − | − |
| O11:KUT | Karnaphooli estuary | 2006 | Chittagong | 1 | + | − | − | + | − | − | − |
| OUT:KUT | Karnaphooli estuary | 2006 | Chittagong | 2 | + | − | − | + | − | − | − |
| O3:KUT | Bakergonj pond | 2006 | Barishal | 1 | + | − | − | + | − | − | − |
| O8:K21 | Bakergonj patient | 2006 | Barishal | 2 | + | + | − | + | − | − | + |
| O8:K21 | Mathbaria pond | 2006 | Pirojpur | 2 | + | + | − | + | − | − | + |
| O4:K46 | Kuakata beach* | 2006 | Potuakhali | 1 | + | − | − | + | − | − | − |
| 08:K21 | Kuakata beach | 2006 | Potuakhali | 1 | + | + | − | + | − | − | − |
| O8:K21 | Kuakata beach | 2006 | Potuakhali | 1 | + | + | − | + | − | − | + |
| O8:K21 | Kuakata beach | 2006 | Potuakhali | 11 | + | + | − | + | − | − | − |
| O9:KUT | Kuakata beach | 2006 | Potuakhali | 1 | − | − | − | + | − | − | − |
| O9:KUT | Kuakata beach | 2006 | Potuakhali | 1 | + | + | − | + | − | − | − |
| OUT:K33 | Kuakata beach | 2006 | Potuakhali | 1 | + | − | − | + | − | − | − |
+, positive; −, negative.
Storage of strains.
V. parahaemolyticus strains confirmed by biochemical methods were each subcultured onto TCBS agar, and a single representative colony from gelatinase agar was aseptically inoculated into T1N1 broth (1% Trypticase and 1% NaCl), incubated at 37°C for 3 to 4 h, and stored at −80°C with 15% glycerol.
Serogrouping.
Serogrouping of the V. parahaemolyticus isolates was done using a commercially available V. parahaemolyticus antiserum test kit (Denka Seiken, Tokyo, Japan) by following the manufacturer's instructions. Briefly, the strains were first grown on LB agar containing 3% NaCl. Following overnight incubation at 37°C, a loopful of inoculum was mixed with 1 ml normal saline. An aliquot of the cell suspension in normal saline was boiled for 2 h and used for serotyping, based on the O antigen. The remaining cell suspension (not boiled) was used for serotyping, based on the K antigen.
Hemolytic activity.
V. parahaemolyticus isolates were grown on Wagatsuma agar medium (11), containing 3 g yeast extract, 10 g peptone, 70 g NaCl, 5 g K2HPO4, 10 g mannitol, 0.001 g crystal violet, 15 g agar, 1 liter distilled water, and 50 ml sheep/human anticoagulated blood. After overnight incubation at 37°C, hemolytic activity was determined. Positive and negative controls were prepared using separate plates.
Extraction and purification of chromosomal DNA.
Chromosomal DNA was extracted using the Wizard genomic DNA purification kit (Promega Corp.), according to the manufacturer's instructions. Briefly, 3 ml of 16- to 18-h culture in LB broth containing 3% NaCl was harvested by centrifugation at 13,000 × g to 16,000 × g for 2 min. Cells were lysed at 80°C in nuclei lysis solution (Promega Corp.). RNase solution was added to the cell lysate, followed by incubation at 37°C for 1 h and cooling to room temperature. Protein precipitation solution (Promega Corp.) was added to the RNase-treated cell lysate and vortexed vigorously. After incubation and centrifugation at 13,000 × g to 16,000 × g for 3 min, the DNA was precipitated by adding 0.6 volume isopropanol at room temperature. The DNA precipitate was washed with 70% ethanol, air dried, and dissolved in DNA rehydration solution (Promega Corp.). Prehydrated DNA was stored at 2 to 8°C until use.
PCR assays.
PCR assays for the species-specific gene toxR and tlh and the two virulence genes tdh and trh were performed using V. parahaemolyticus genomic DNA as a template, following methods described elsewhere (23, 35).
GS- and ORF8-PCR.
PCR assays for amplification of the GS and ORF8 pandemic marker genes were performed using specific primers previously reported to detect toxRS sequences unique to the pandemic O3:K6 clone of V. parahaemolyticus and the orf8 sequence of phage f237, respectively (23, 26, 28).
PFGE.
Pulsed-field gel electrophoresis (PFGE) of SfiI-digested DNA of Vibrio parahaemolyticus was performed using a standardized protocol, as described elsewhere (20, 37). XbaI-digested Salmonella enterica serovar Braenderup DNA was used as molecular size markers. Following electrophoresis, gels were stained with ethidium bromide (10 mg/ml) and photographed under UV transillumination.
Image analysis.
The fingerprint pattern in the gel was analyzed using the Bionumeric computer software package (Applied Maths, Belgium). After background subtraction and gel normalization, the fingerprint patterns were subjected to typing based on banding similarity and dissimilarity. Dendrograms were computed using the Bionumeric software package (Applied Maths, Belgium) the Dice similarity coefficient, and the unweighted-pair group method using average linkages (UPGMA) for PFGE profiles of V. parahaemolyticus strains. Two methods for measuring similarity were compared, one based on binary data of occurrence of the band (band-based), calculated using the Dice coefficient, and the other on the overall densitometry profile (curve-based) of the banding pattern, calculated using Pearson's product moment correlation.
RESULTS AND DISCUSSION
Vibrio parahaemolyticus, a halophilic bacterium, has in recent years emerged as a pandemic pathogen causing seafood-related gastroenteritis worldwide (27). Diarrhea caused by V. parahaemolyticus occurs with high frequency in Bangladesh and India (7, 28). Yet, systematic surveillance of V. parahaemolyticus is not done in those countries. Thus, very little is known about serogroup distribution, virulence potential, or molecular characteristics of V. parahaemolyticus present in the estuarine ecosystem of the Bay of Bengal, even though the pandemic serogroup O3:K6 that spread worldwide (27) was first isolated and reported from this region (32).
Forty-two V. parahaemolyticus strains were isolated from 119 estuarine water samples collected between October 2005 and January 2006 from four coastal districts of Bangladesh. The occurrence of V. parahaemolyticus in estuarine water is greatly influenced by a combination of temperature, salinity, and pH of water (17). A recent study carried out in a temperate region showed higher occurrence of V. parahaemolyticus in coastal water near the freshwater discharge point and at a time of the year when the salinity of the water was low (<35%) (26). Although samples were not collected on a specific sampling schedule and parameters such as temperature, salinity, and pH of the water bodies were not monitored in the present study, recovery of V. parahaemolyticus from estuarine water samples collected in the Bay of Bengal was significant.
Serogroup analysis.
As shown in Table 1, eight different O groups and 10 different K types were detected. Three strains were not recognized by specific O antisera and 15 not recognized by specific K antisera, and two of the latter did not react to O:K antisera, suggesting new variants. The predominant O group for 18 out of the 44 strains was O8, followed by O3 and O1 for eight and seven strains, respectively, followed by O4, O5, O9, O10, and O11, accounting for two strains, one strain, two strains, two strains, and one strain, respectively. In contrast, only 11 of the 44 strains were recognized by currently available K antisera representing 10 different serogroups. Of the 11 strains reacting to the 10 different K antisera, O8:K21 was predominant. Other O:K serogroups included two O3:K30 strains and one each of O1:K38, O3:K4, O3:K29, O4:K34, O3:K45, O4:K46, and O8:K39. Of 15 strains belonging to serogroup O, for which the K antigens were not typeable, i.e., none reacted to any of the available K antisera, six were serogroup O1 (O1:KUT), three were O3 (O3:KUT), and two were O10 (O10:KUT). The pandemic serogroup O:K (O3:K6) was not detected among the 44 strains tested.
Serology, which serves as an important marker for both diarrheagenic and pandemic strains, revealed a significant diversity in sero-distribution of V. parahaemolyticus strains from the Bay of Bengal coastal region. Of the known 13 O groups and 71 K types recognized to date (19), 18 combinations of O:K serogroups were detected among the 44 V. parahaemolyticus isolates, with O8:K21 being the predominant serogroup, followed by O3:KUT and O1:KUT. A yearly variation in serogroups causing diarrhea has been reported for Bangladesh during the period from 1998 to 2000 (7). Absence of the pandemic serogroup O3:K6 among the strains tested in the present study does not rule out their presence in this region.
Detection of virulence genes by PCR.
V. parahaemolyticus strains isolated from the estuarine ecosystem of the Bay of Bengal were screened for virulence and pandemic marker genes ORF8, GS toxR, tdh, trh, and tlh by PCR (6, 27). As shown in Table 1, the species-specific gene tlh was found in all 42 isolates, but toxR was missing in two of these strains, while 18 (41%) had the major virulence gene tdh, and only one strain had trh but not tdh. Of the 18 tdh-positive strains, 17 (94%), including the two diarrheal isolates, had seromarker O8:K21, while one was O9:KUT, and a single trh-positive strain belonged to serogroup O1:KUT. Since the groundwater in coastal villages of Bangladesh is not potable, due to high salinity, and the inhabitants do not have access to salt-free freshwater, they are compelled to use surface (lagoon) water containing low salinity for drinking and other household purposes (1). Therefore, the prevalence of pathogenic V. parahaemolyticus strains in surface water indicates a significant public health problem. Also significant is that 94% of the tdh-positive strains had seromarker O8:K21, recognized as the predominant serogroup linked to diarrheal disease in the coastal villages.
Hemolytic activity.
The KP in V. parahaemolyticus strains is considered to represent the major hemolysins, TDH and/or TRH (16, 31, 39). V. parahaemolyticus strains isolated in this study were tested for KP on Wagatsuma agar. As shown in Table 1, 10 (23%) of the 44 V. parahaemolyticus isolates, including one of the two clinical strains, produced clear zones of hemolysis on Wagatsuma agar. Of the 10 KP-positive V. parahaemolyticus strains, six had tdh, including the two strains from diarrheal cases. One had trh but not tdh, while the remaining three strains lacked both tdh and trh.
KP historically has been considered to be a reliable marker for pathogenicity of V. parahaemolyticus, and KP-positive strains have been shown to cause diarrhea in volunteers (33). In contrast, a high dose of KP-negative strains can fail to cause diarrhea (33). KP reaction exhibited by the six tdh-positive V. parahaemolyticus strains was consistent with such results, since TDH is known to cause beta-hemolysis on Wagatsuma agar (8). Hemolytic activity displayed by the single trh-positive, tdh-negative isolate was unique in the present study. The trh-positive V. parahaemolyticus strains almost always display the urease function as a marker for virulence (35). In contrast, urease-positive, KP-negative V. parahaemolyticus was shown to be associated with gastroenteritis (22). V. parahaemolyticus TDH is a major virulence factor associated with pathogenicity of V. parahaemolyticus, since deletion of the tdh gene results in loss of enterotoxigenic activity, as shown in laboratory models (30). However, in the present study not all KP-positive strains possessed tdh or trh, and a majority of the tdh-positive strains were KP negative, failing to produce beta-hemolysis on Wagatsuma agar (8). This inconsistency in virulence gene content and expression and the fact that a significant number of strains lacked both tdh and trh, yet were KP positive, appear to be in agreement with results reported earlier, namely that KP is not a reliable marker for pathogenicity of V. parahaemolyticus (22). The significantly higher proportion of potentially pathogenic V. parahaemolyticus strains in estuarine water of Bangladesh is another important finding of this study. The incidence of toxigenic V. parahaemolyticus strains was low (<1%) in surface water compared to that of clinical strains (>90%) (11, 22, 39, 43).
Detection of pandemic marker genes by PCR.
Changes in the pandemic serogroup have been reported to occur over time, since GS-PCR-positive pandemic clones have been shown to be ORF8 negative (34) and an increasing number of nonpandemic serogroups, such as O4:K68, O1:K25, O1:KUT, O1:K41, O4:K12, and O3:K46, carry pandemic marker genes (7, 10, 27, 34, 41). Although non-O3:K6 serogroups are presumed to have evolved over time via alteration of the O and K antigens and by seromarker transformation of O3:K6 strains (3, 10), the isolates of V. parahaemolyticus (n = 44) tested in this study did not include only pandemic serogroup O1:KUT, which together with serogroups O3:K6 and O4:K68 has been reported previously from diarrheal cases in Bangladesh (7). Furthermore, none of the V. parahaemolyticus strains, including O1:KUT strains, possessed the GS and ORF8 pandemic marker genes, indicating no relationship with pandemic clones but, instead, evidence of local emergence of pathogenic strains. A recent study in Thailand reported O3:K46 to be a new, emergent serovar having pandemic traits, while dominance of other pandemic serogroups, such as O3:K6, O1:K25, and O1:KUT, was demonstrated in diarrheal cases (34).
PFGE and cluster analysis.
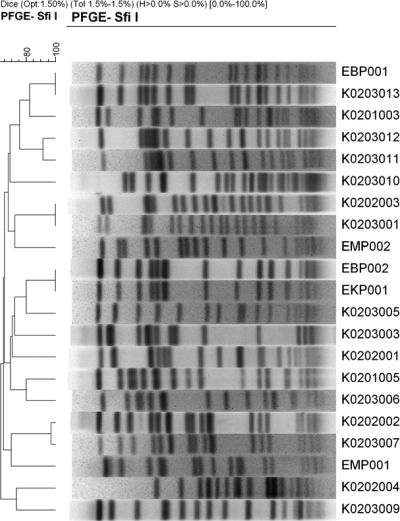
As shown in Fig. 1, 17 different PFGE patterns were obtained for 21 V. parahaemolyticus strains tested. The number of fragments generated by the SfiI restriction enzyme varied between 12 and 19 and the molecular size of the fragments ranged from 25 kb to 700 kb. The dendrogram constructed using the Dice similarity coefficient and UPGMA employing PFGE gel images showed that the various serogroup strains isolated from different coastal sites also differed in virulence gene content, but had similar PFGE patterns of the same or closely related clusters. The same serogroup strains having a similar virulence gene profile produced different PFGE patterns belonging to distinctly different clusters and, thus, are concluded to be distant clonally. The data presented here are in agreement with results reported by other investigators, namely that nonpandemic serogroups are genetically diverse. Thus, seromarkers used to categorize pandemic serogroup strains of V. parahaemolyticus (27, 40) are of limited or no use, at least for the nonpandemic strains in the Bay of Bengal, Bangladesh.
FIG. 1.
PFGE patterns of NotI-digested genomic DNA of selected (n = 26) V. parahaemolyticus isolates from the estuarine ecosystem of Bangladesh. Strain identification number, seromarker, and the place of isolation of the strains are indicated. The dendrogram was established by the Bionumeric software package (Applied Maths) using the Dice similarity coefficient and UPGMA of the PFGE profiles of the V. parahaemolyticus strains tested.
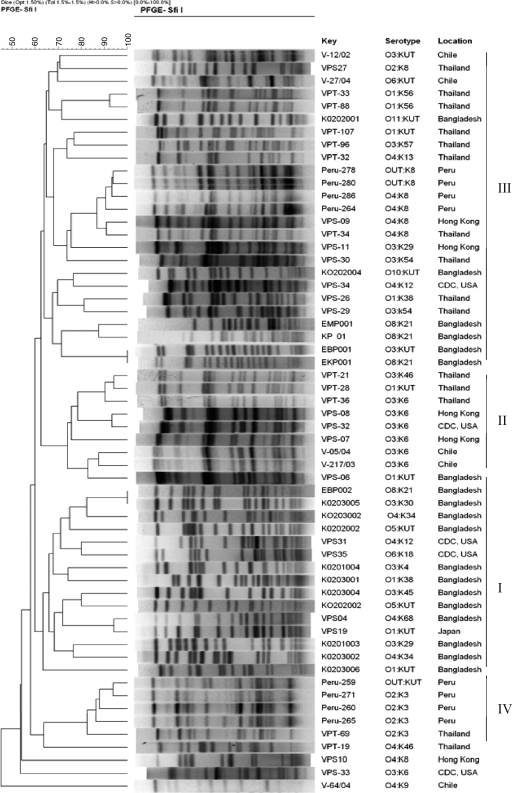
To understand how PFGE data can be applied in determining clonal relatedness of V. parahaemolyticus strains both of this region and other regions of Asia, cluster analysis was performed using PFGE images of SfiI-digested genomic DNA of local as well as diverse geographical origin available in our database. The PFGE images of clinical strains compared with those of local environmental strains by cluster analysis included different serogroup strains representing pandemic, nonpandemic but pathogenic, and nonpandemic nonpathogenic V. parahaemolyticus strains isolated worldwide, mostly after the 1996 pandemic (27). Not unexpectedly, despite a significant portion of the nonpandemic serogroup strains from the Bay of Bengal region having clustered together (Fig. 2, I), as had pandemic serogroup strains of diverse origin (Fig. 2, II), the V. parahaemolyticus strains were highly divergent, both regionally and globally. The cluster constituted mainly pandemic O3:K6 serogroup strains and also included strains of O1:KUT and a newly recognized pandemic serogroup, O3:K46 (Fig. 2, II), which was recently shown to be genetically related to pandemic serogroups O3:K6 and O1:K25 in Thailand (37). Interestingly, the lone strain of Bengal origin that clustered with the pandemic serogroup strains was of O1:KUT, which, however, lacked tdh and trh and was negative for the pandemic marker genes, confirmed by GS- and ORF8-PCR. Moreover, unlike clusters III and IV (Fig. 2), which comprised highly divergent but genetically linked nonpandemic strains of diverse geographical origin, clusters I and II included strains of either a region or a group, each having strains that are divergent but linked clonally. The nonpandemic strains of V. parahaemolyticus formed multiple secondary clusters when PFGE images were subjected to cluster analysis (20). Clusters comprising genetically dissimilar, but clonally linked, strains were designated “clonal complexes.” The overall PFGE and cluster analysis data, coupled with results of other investigators, show conclusively that DNA signatures are reliable for identifying pandemic serogroup strains (33) that expand, with respect to O:K seromarkers (9, 24, 27).
FIG. 2.
Dendrogram constructed with PFGE images of NotI-digested genomic DNA of selected V. parahaemolyticus strains isolated from the estuarine ecosystems of Bangladesh to determine clonal relatedness with V. parahaemolyticus strains from different geographical regions, including different serogroups representing pandemic, nonpandemic but pathogenic, and nonpandemic nonpathogenic V. parahaemolyticus isolates available in our database (29). Strain identification number, seromarkers, and the place of isolation/origin are indicated. The dendrogram was established by the Bionumeric software package (Applied Maths) using the Dice similarity coefficient and UPGMA of the PFGE profiles of the V. parahaemolyticus strains tested.
This study is the first to show that a significantly large portion of V. parahaemolyticus strains occurring in the surface waters of coastal villages of Bangladesh is potentially virulent. Since diarrheal diseases are endemic in Bangladesh (1), where morbidity and mortality due to recurrent waterborne diseases remain a longstanding problem, it is concluded that morbidity and mortality in Bangladesh resulting each year from diarrhea, particularly in coastal villages, may be attributed, in part, to water-borne pathogens, notably V. parahaemolyticus. Despite being a halophilic bacterium (21), V. parahaemolyticus prefers low salinity for optimal growth (26) and is capable of spreading inland to freshwater, indicated by outbreaks of diarrhea caused by V. parahaemolyticus in Dhaka, Bangladesh (7), and Kolkata, India (32). The previously reported pandemic serogroups of V. parahaemolyticus were not recognized among the strains tested in this study, but their existence in this region is indicated by past records for Bangladesh (7), India (28, 31), and Thailand (34).
The high degree of divergence demonstrated by Bengal strains of V. parahaemolyticus is in agreement with many studies reporting similar results for other regions (27, 40). The rapid emergence of non-O3:K6 serogroups carrying pandemic markers provides yet another example of the predisposition of V. parahaemolyticus to genetic change (9, 24, 27, 34). Thus, like the O:K seromarkers, genetic markers widely relied upon for molecular typing appear to be highly unstable for V. parahaemolyticus. The divergence that V. parahaemolyticus exhibits when molecular tools are employed and the overall data obtained in this study of V. parahaemolyticus suggest, as has been shown by genome sequencing of Vibrio cholerae (11a), that lateral gene transfer may play a significant role in the ecology and evolution of V. parahaemolyticus. The search for pandemic strains may well be illusionary if lateral gene transfer is as extensive in V. parahaemolyticus as it is in V. cholerae.
In any case, the toxigenic V. parahaemolyticus serogroup O8:K21, recognized as a diarrheal pathogen and present in drinking water sources of coastal villages of Bangladesh, can be concluded to pose a public health risk warranting epidemiological and ecological monitoring to ensure safety.
Acknowledgments
This research was partially supported by Japan Food Hygiene Association through NIID, Tokyo, and NIH Research Grant 1RO1A13912901 under collaborative agreements between the Johns Hopkins Bloomberg School of Public Health, the University of Maryland, College Park, and the ICDDR,B. The ICDDR,B is supported by donor countries and agencies, which provide unrestricted support to the center for its operation and research.
Footnotes
Published ahead of print on 14 August 2009.
REFERENCES
- 1.Alam, M., N. A. Hasan, A. Sadique, N. A. Bhuiyan, K. U. Ahmed, S. Nusrin, G. B. Nair, A. K. Siddique, R. B. Sack, D. A. Sack, A. Huq, and R. R. Colwell. 2006. Seasonal cholera caused by Vibrio cholerae serogroups O1 and O139 in the coastal aquatic environment of Bangladesh. Appl. Environ. Microbiol. 72:4096-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alam, M. J., K. Tomochika, S. Miyoshi, and S. Shinoda. 2002. Environmental investigation of potentially pathogenic Vibrio parahaemolyticus in the Seto-Inland Sea, Japan. FEMS Microbiol. Lett. 208:83-87. [DOI] [PubMed] [Google Scholar]
- 3.Ansaruzzaman, M., M. Lucas, J. L. Deen, N. A. Bhuiyan, X. Y. Wang, A. Safa, M. Sultana, A. Chowdhury, G. B. Nair, D. A. Sack, L. von Seidlein, M. K. Puri, M. Ali, C. L. Chaignat, J. D. Clemens, and A. Barreto. 2005. Pandemic serovars (O3:K6 and O4:K68) of Vibrio parahaemolyticus associated with diarrhea in Mozambique: spread of the pandemic into the African continent. J. Clin. Microbiol. 43:2559-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.APHA. 1970. Procedures for the bacteriological examination of sea water and shellfish. American Public Health Association, Washington, DC.
- 5.Bag, P. K., S. Nandi, R. K. Bhadra, T. Ramamurthy, S. K. Bhattacharya, M. Nishibuchi, T. Hamabata, S. Yamasaki, Y. Takeda, and G. B. Nair. 1999. Clonal diversity among recently emerged strains of Vibrio parahaemolyticus O3:K6 associated with pandemic spread. J. Clin. Microbiol. 37:2354-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bej, A. K., D. P. Patterson, C. W. Brasher, M. C. L. Vickery, D. D. Jones, and C. Kaysner. 1999. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tlh, tdh and trh. J. Microbiol. Methods 36:215-225. [DOI] [PubMed] [Google Scholar]
- 7.Bhuiyan, N. A., M. Ansaruzzaman, M. Kamruzzaman, K. Alam, N. R. Chowdhury, M. Nishibuchi, S. M. Faruque, D. A. Sack, Y. Takeda, and G. B. Nair. 2002. Prevalence of the pandemic genotype of Vibrio parahaemolyticus in Dhaka, Bangladesh, and significance of its distribution across different serotypes. J. Clin. Microbiol. 40:284-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabrera-García, M. E., C. Vázquez-Salinas, and E. I. Quiñones-Ramírez. 2004. Serologic and molecular characterization of Vibrio parahaemolyticus strains isolated from seawater and fish products of the Gulf of Mexico. Appl. Environ. Microbiol. 70:6401-6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chowdhury, N. R., S. Chakraborty, B. Eampokalap, W. Chaicumpa, M. Chongsa-Nguan, P. Moolasart, R. Mitra, T. Ramamurthy, S. K. Bhattacharya, et al. 2000. Clonal dissemination of Vibrio parahaemolyticus displaying similar DNA fingerprint but belonging to two different serovars (O3:K6 and O4:K68) in Thailand and India. Epidemiol. Infect. 125:17-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhury, N. R., O. C. Stine, J. G. Morris, and G. B. Nair. 2004. Assessment of evolution of pandemic Vibrio parahaemolyticus by multilocus sequence typing. J. Clin. Microbiol. 42:1280-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chun, D., J. K. Chung, R. Tak, and S. Y. Seol. 1975. Nature of Kanagawa phenomenon of Vibrio parahaemolyticus. Infect. Immun. 12:81-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Chun, J., C. J. Grim, N. A. Hasan, J. H. Lee, S. Y. Choi, B. J. Haley, E. Taviani, Y.-S. Jeon, D. W. Kim, J.-H. Lee, T. S. Brettin, D. C. Bruce, J. F. Challacombe, J. C. Detter, C. S. Han, A. C. Munk, O. Chertkov, L. Meincke, E. Saunders, R. A. Walters, A. Huq, G. B. Nair, and R. R. Colwell. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 12.Daniels, N. A., B. Ray, A. Easton, N. Marano, E. Kahn, A. L. McShan, L. Del Rosario, T. Baldwin, M. A. Kingsley, N. D. Puhr, J. G. Wells, and F. J. Angulo. 2000. Emergence of a new Vibrio parahaemolyticus serotype in raw oysters. JAMA 284:1541-1545. [DOI] [PubMed] [Google Scholar]
- 13.DePaola, A., C. A. Kaysner, J. C. Bowers, and D. W. Cook. 2000. Environmental investigations of Vibrio parahaemolyticus in oysters following outbreaks in Washington, Texas, and New York (1997 and 1998). Appl. Environ. Microbiol. 66:4649-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujino, T., T. Okuno, D. Nakada, A. Aoyama, K. Fukai, T. Mukai, and T. Ueho. 1951. On the bacteriological examination of shirasu food poisoning. J. Jpn. Assoc. Infect. Dis. 25:11-12. [Google Scholar]
- 15.González-Escalona, N., V. Cachicas, C. Acevedo, M. L. Rioseco, J. A. Vergara, F. Cabello, J. Romero, and R. T. Espejo. 2005. Vibrio parahaemolyticus diarrhea, Chile, 1998 and 2004. Emerg. Infect. Dis. 11:129-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hara-Kudo, Y., T. Nishina, H. Nakagawa, H. Konuma, J. Hasegawa, and S. Kumagai. 2001. Improved method for detection of Vibrio parahaemolyticus in seafood. Appl. Environ. Microbiol. 67:5819-5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayat Mahmud, Z., A. Kassu, A. Mohammad, M. Yamato, N. A. Bhuiyan, G. Balakrish Nair, and F. Ota. 2006. Isolation and molecular characterization of toxigenic Vibrio parahaemolyticus from the Kii Channel, Japan. Microbiol. Res. 161:25-37. [DOI] [PubMed] [Google Scholar]
- 18.Honda, T., and T. Iida. 1993. The pathogenicity of Vibrio parahaemolyticus and the role of the thermostable direct haemolysin and related haemolysins. Rev. Med. Microbiol. 4:106-113. [Google Scholar]
- 19.Ishibashi, M., K. Ohta, T. Shimada, T. Honda, J. Sugiyama, T. Miwatani, and H. Yokoo. 2000. Current status of OK serotype combinations of Vibrio parahaemolyticus. Nippon Saikingaku Zasshi 55:539-541. (In Japanese.) [Google Scholar]
- 20.Kam, K.-M., C. K. Y. Luey, M. B. Parsons, K. L. F. Cooper, G. B. Nair, M. Alam, M. A. Islam, D. T. L. Cheung, Y. W. Chu, T. Ramamurthy, G. P. Pazhzni, S. K. Bhattacharya, H. Watanabe, J. Terajima, E. Arakawa, O.-A. Ratchtrachenchai, S. Huttayananont, E. M. Ribot, P. Gerner-Smidt, and Bala Swaminathan for the Vibrio parahaemolyticus PulseNet PFGE Protocol Work Group. 2008. Evaluation and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping Vibrio parahaemolyticus: an international multicenter collaborative study. J. Clin. Microbiol. 46:2766-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaneko, T., and R. R. Colwell. 1973. Ecology of Vibrio parahaemolyticus in Chesapeake Bay. J. Bacteriol. 113:24-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly, M. T., and E. M. Stroh. 1989. Urease-positive, Kanagawa-negative Vibrio parahaemolyticus from patients and the environment in the Pacific Northwest. J. Clin. Microbiol. 27:2820-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, Y. B., J. Okuda, C. Matsumoto, N. Takahashi, S. Hashimoto, and M. Nishibuchi. 1999. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J. Clin. Microbiol. 37:1173-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laohaprertthisan, V., A. Chowdhury, U. Kongmuang, S. Kalnauwakul, M. Ishibashi, C. Matsumoto, and M. Nishibuchi. 2003. Prevalence and serodiversity of the pandemic clone among the clinical strains of Vibrio parahaemolyticus isolated in southern Thailand. Epidemiol. Infect. 130:395-406. [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez-Urtaza, J., A. Lozano-Leon, A. DePaola, M. Ishibashi, K. Shimada, M. Nishibuchi, and E. Liebana. 2004. Characterization of pathogenic Vibrio parahaemolyticus isolates from clinical sources in Spain and comparison with Asian and North American pandemic isolates. J. Clin. Microbiol. 42:4672-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Urtaza, J., A. Lozano-Leon, A. DePaola, J. Varela-Pet, J. Trinanes, Y. Pazos, and O. Gracia-Martin. 2008. Environmental determinants of the occurrence and distribution of Vibrio parahaemolyticus in the rias of Galicia, Spain. Appl. Environ. Microbiol. 74:265-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumoto C., J. Okuda, M. Ishibashi, M. Iwanaga, P. Garg, T. Rammamurthy, H. C. Wong, A. Depaola, Y. B. Kim, M. J. Albert, and M. Nishibuchi. 2000. Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nair, G. B., and J. C. Hormazabal. 2005. The Vibrio parahaemolyticus pandemic. Rev. Chilena Infectol. 22:125-130. [DOI] [PubMed] [Google Scholar]
- 29.Nasu, H., T. Iida, T. Sugahara, Y. Yamaichi, K. S. Park, K. Yokoyama, K. Makino, H. Shinagawa, and T. Honda. 2000. A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J. Clin. Microbiol. 38:2156-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishibuchi, M., A. Fasano, R. G. Russell, and J. B. Kaper. 1992. Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infect. Immun. 60:3539-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishibuchi, M., and J. B. Kaper. 1995. Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect. Immun. 63:2093-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okuda, J., M. Ishibashi, E. Hayakawa, T. Nishino, Y. Takeda, A. K. Mukhopadhyay, S. Garg, S. K. Bhattacharya, G. B. Nair, and M. Nishibuchi. 1997. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 35:3150-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanyal, S. C., and P. C. Sen. 1973. Human volunteer study on the pathogenicity of Vibrio parahaemolyticus, p. 227. In T. Fujino, G. Sakaguchi, R. Sakazaki, and Y. Takeda, (ed.), International symposium on Vibrio parahaemolyticus. Saikon Publishing Co., Ltd., Tokyo, Japan.
- 34.Serichantalergs, O., N. A. Bhuiyan, G. B. Nair, O. Chivaratanond, A. Srijan, L. Bodhidatta, S. Anuras, and C. J. Mason. 2007. The dominance of pandemic serovars of Vibrio parahaemolyticus in expatriates and sporadic cases of diarrhoea in Thailand, and a new emergent serovar (O3:K46) with pandemic traits. J. Med. Microbiol. 56:608-613. [DOI] [PubMed] [Google Scholar]
- 35.Suthienkul, O., M. Ishibashi, T. Iida, N. Nettip, S. Supavej, B. Eampokalap, M. Makino, and T. Honda. 1995. Urease production correlates with possession of the trh gene in Vibrio parahaemolyticus strains isolated in Thailand. J. Infect. Dis. 172:1405-1408. [DOI] [PubMed] [Google Scholar]
- 36.Taniguchi, H., H. Hirano, S. Kobomura, K. Higahsi, and Y. Mizuguchi. 1986. Comparison of the nucleotide sequences of the genes for the thermostable direct hemolysin and thermolabile hemolysin from Vibrio parahaemolyticus. Microb. Pathog. 1:425-432. [DOI] [PubMed] [Google Scholar]
- 37.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Versalovic, J., Koeuth, T., and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wagatsuma, S. 1974. Ecological studies on Kanagawa phenomenon positive strains of Vibrio parahaemolyticus, p. 91-96. In T. Fujino, G. Sakaguchi, R. Sakazaki, and Y. Takeda (ed.), International symposium on Vibrio parahaemolyticus. Saikon Publishing Co., Ltd., Tokyo, Japan.
- 40.Wong, H. C., S. H. Liu, and D. P. Liu. 1999. Incidence of highly genetically diversified Vibrio parahaemolyticus in seafood imported from Asian countries. Int. J. Food Microbiol. 52:181-188. [DOI] [PubMed] [Google Scholar]
- 41.Wong, H. C., S. H. Liu, T. K. Wang, C. L. Lee, C. S. Chiou, D. P. Liu, M. Nishibuchi, and B. K. Lee. 2000. Characterization of Vibrio parahaemolyticus O3:K6 from Asia. Appl. Environ. Microbiol. 66:3981-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong, H. C., and, C. H., Lin. 2001. Evaluation of typing of Vibrio parahaemolyticus by three PCR methods using specific primers. J. Clin. Microbiol. Vol. 39:4233-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong, H. C. 2003. Detecting and molecular typing of Vibrio parahaemolyticus. J. Food Drug Anal. 11:79-86. [Google Scholar]