Abstract
A search for bacterium-specific biomarkers in peripheral blood following infection with Bacillus anthracis was carried out with rabbits, using a battery of specific antibodies generated by DNA vaccination against 10 preselected highly immunogenic bacterial antigens which were identified previously by a genomic/proteomic/serologic screen of the B. anthracis secretome. Detection of infection biomarkers in the circulation of infected rabbits could be achieved only after removal of highly abundant serum proteins by chromatography using a random-ligand affinity column. Besides the toxin component protective antigen, the following three secreted proteins were detected in the circulation of infected animals: the chaperone and protease HtrA (BA3660), an NlpC/P60 endopeptidase (BA1952), and a protein of unknown function harboring two SH3 (Src homology 3) domains (BA0796). The three proteins could be detected in plasma samples from infected animals exhibiting 103 to 105 CFU/ml blood and also in standard blood cultures at 3 to 6 h post-bacterial inoculation at a bacteremic level as low as 103 CFU/ml. Furthermore, the three biomarkers appear to be present only in the secretome of B. anthracis, not in those of the related pathogens B. thuringiensis and B. cereus. To the best of our knowledge, this is the first report of direct detection of B. anthracis-specific proteins, other than the toxin components, in the circulation of infected animals.
The gram-positive spore-forming bacterium Bacillus anthracis is the causative agent of anthrax, a rare fatal disease which is initiated, in its most severe form, by inhalation of spores. Due to the severity of the disease, the ease of respiratory infection, and the extreme resistance of the spores to unfavorable environmental conditions, B. anthracis is considered a potential biological warfare agent (for a review, see references 8, 10, 35, 56, and 62), and in recent years, the need for novel reliable diagnostic approaches, improved vaccination strategies, novel therapeutic targets, and a better understanding of the pathogenesis of anthrax has been widely acknowledged.
Inhaled B. anthracis spores are taken up by alveolar macrophages and germinate into vegetative bacilli which eventually invade the bloodstream, where they multiply massively and secrete toxins and virulence factors. Anthrax is toxinogenic in the sense that the bacterial binary exotoxin is necessary for the onset of the disease (54), yet other factors may be required for the colonization and expansion of bacteria in the host (15, 18, 31, 32, 37, 46, 65, 66, 70, 83). The toxin is composed of the following three proteins: protective antigen (PA), which mediates binding to the receptor on target cells and internalization of the toxin components (14, 74); lethal factor, a zinc protease targeting several mitogen-activated protein kinases (52); and edema factor (EF), a calmodulin-dependent adenylate cyclase (55, 57). The genes encoding the three exotoxin components are located on the native virulence plasmid pXO1. Genes encoding proteins with functions involved in the synthesis of the second major B. anthracis virulence determinant, an immunologically inert polyglutamyl capsule that protects bacteria from phagocytosis, are located on a second native virulence plasmid, pXO2 (56).
In humans, the initial symptoms of inhalation anthrax are nonspecific and reminiscent of influenza-like upper respiratory tract infections. The second stage is characterized by high fever, respiratory failure, dyspnea, and shock. Unless patients are treated promptly, death occurs within 24 h of the onset of the second stage, preceded by massive bacteremia (27, 34, 73, 76). The mandatory treatment for anthrax is based on administration of antibiotics (17, 76), yet study of the disease in animal models has clearly established that the time of antibiotic administration postinfection is crucial for the effectiveness of the treatment, strongly supporting the importance of rapid diagnosis (2, 47, 48). At present, due to the severity of the disease and its rapid progression, treatment is administered to each person with confirmed contact with contaminated areas (76).
Early accurate diagnosis of anthrax is complicated by the rarity of the disease and the nonspecific nature of the symptoms. Microbiologic identification of anthrax is based on the nonhemolytic nature of the typically white-gray colonies and the rod morphology of the gram-positive nonmotile bacilli detected in clinical samples or blood cultures (16, 19, 30, 73, 78). Immunofluorescence and immunohistochemistry targeted to bacterial proteins are not routinely conducted. Later in the course of the disease, seroconversion in response to the various components of the toxin may serve only as a retrospective confirmation of initial exposure. With the advent of genetic methodologies, B. anthracis in cultures inoculated with clinical and forensic samples can be detected specifically and accurately by PCR, usually designed to amplify genes located on the native virulence plasmids (30). Recently, the use of PA as a disease biomarker was suggested, owing to the presence of this protein in detectable amounts in the circulation of infected animals (53, 71). EF and lethal factor can be detected in the circulation only at later stages of infection (30).
In recent years, the search for novel biomarkers of disease, including bacterial infections, has exploited the approach of global biological inspection based on functional genomic or proteomic studies (64, 85). We previously documented such global surveys, combined with a serological study of B. anthracis (5, 6, 20, 21, 22, 38, 39), for identification of vaccine and diagnostic marker candidates among extracellular (secreted or membranal) proteins. These studies indeed revealed a list of proteins that can serve as potential biomarkers, based on their immunogenicity (which probes their in vivo expression), abundance under various culture conditions, and functional relatedness to infection. In the present study, the search was extended by directly addressing the presence of bacterial secreted proteins in the circulation of B. anthracis-infected rabbits, using specific antibodies generated by DNA vaccination against the previously selected immunogenic proteins. Visualization of bacterial proteins in the circulation of infected animals was achieved only following depletion of highly abundant serum proteins by an affinity chromatography protocol. The search enabled the successful detection, in addition to PA, of three secreted proteins uniquely expressed by B. anthracis, i.e., HtrA (BA3660), the BA1952 endopeptidase, and a protein of unknown function (BA0796). All of these proteins are potential virulence-related factors. This is the first communication of the presence of B. anthracis secreted proteins other than the bacterial toxin in the circulation of infected animals, and their identification strongly supports the validity of the reductional screening approach for selection of disease biomarkers.
MATERIALS AND METHODS
Bacterial strains.
The B. anthracis strains used in this study were the fully virulent Vollum strain (pXO1+ pXO2+) (ATCC 14578) and the attenuated strains ΔVollum (lacking pXO1 and pXO2) and Δ14185-HtrA (also lacking pXO1 and pXO2). The latter represents an engineered strain overexpressing and secreting large amounts of the serine protease HtrA (TIGR tag BA3660). Expression is driven by the Sap promoter, following transfection of the Δ14185 strain with the appropriate expression vector. The Δ14185 platform strain is a pXO1-deleted derivative of the nonproteolytic vaccine strain V770-NP1-R (84), referred to by its ATCC accession number, ATCC 14185. The platform strain, as well as the repertoire of plasmid vectors appropriate for heterologous overexpression of B. anthracis antigens, has been described previously (24, 36, 40, 60). Other bacterial cultures used were Bacillus cereus 14579, Bacillus thuringiensis subsp. israeliensis ATCC 35646, and Bacillus subtilis strain 168. All bacterial cultures were carried out in FAG medium (24) at 37°C with vigorous shaking (250 rpm).
1D-SDS-PAGE and two-dimensional electrophoresis (2DE).
For one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (1D-SDS-PAGE), samples were mixed with Laemmli sample buffer (2% SDS, 700 mM β-mercaptoethanol; Bio-Rad) and boiled for 5 min before being loaded into gels. For Western analysis of bacterial secreted proteins present in the FAG medium culture, the centrifuged (30 min, 5,000 rpm [for removal of cells]) culture supernatants were filtered through 0.45-mm polystyrene membranes (Corning).
For 2DE analysis, urea buffer, containing 8 M urea, 4% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 40 mM Tris, 2% dithiothreitol (DTT), and 0.2% (wt/vol) Bio-Lyte 3/10 (Bio-Rad), was added to the protein mixtures (1:1) and loaded onto 11-cm immobilized-pH-gradient strips (nonlinear, pH 3 to 10; Bio-Rad) overnight. First-dimension isoelectric focusing was carried out at 10,000 V to a total of 50,000 V-h, initiated by a slow step at 250 V for 30 min. Strips were then processed for the second-dimension separation by a 10-min incubation in 6 M urea, 2% SDS, 0.375 M Tris-HCl (pH 8.8), 20% glycerol, and 2% (wt/vol) DTT, followed by 10 min of incubation in a similar solution in which the DTT was replaced by 2% iodoacetamide. Strips were applied to 12.5% SDS-PAGE gels, and electrophoresis was carried out in a Bio-Rad gel running system. Gels were stained with SeeBand Forte Coomassie staining solution (GeVa, Beit Haemek, Israel) or transferred to a nitrocellulose membrane for Western blot analysis.
Hyperimmune anti-B. anthracis antiserum and antibodies specific for the products of individual genes.
The guinea pig hyperimmune anti-B. anthracis antiserum used in this study as a probe for visualization of B. anthracis exposed antigens on Western blots was described previously (22, 38). This serum was collected from animals infected with 107 spores of the highly attenuated mntA-negative mutant derived from the Vollum strain (37).
Antibodies against HtrA (BA3660), NLP/P60 (BA1952), BA0799, BA3367, BA0796, BA4787, BA0898, BA0345, BA1295, and BA5427 proteins were generated by DNA vaccination as previously described (22, 38, 45). In brief, individual open reading frames (ORFs) were cloned into the eukaryotic expression vector pCI (Promega), which carries the eukaryotic cytomegalovirus promoter, a recombinant chimeric intron, and the simian virus 40 polyadenylation signal for efficient expression in mammalian cells, in addition to the T7 promoter for in vitro transcription and translation (38, 39, 45). Plasmid DNA for gene gun immunization was prepared by an alkaline lysis method followed by CsCl gradient centrifugation. The purified DNA preparations were solubilized in pyrogen-free water and kept frozen. Immunization protocols using gene gun vaccination (Helios gene gun system; Bio-Rad) were performed essentially as described before (38, 45). The protocol included three or four immunizations of mice (22- to 26-g females; ICR, Charles River Laboratories, Margate, United Kingdom) at 2-week intervals. Each DNA vaccination group included 10 experimental animals. Animals were bled from the tail for serum collection following each booster and by cardiac puncture for the final bleed. The specificity and potency of the immune response elicited in the animals following DNA immunization were determined by quantitative immunoprecipitation titration (data not shown) for the respective ORFs investigated, using in vitro-transcribed and -translated products as previously described (39). The antibodies were also tested for specificity by Western blot analysis using total bacterial cell extracts or fractions enriched in secreted proteins (22, 38, 39).
Generation of rabbit sera exhibiting various levels of bacteremia.
New Zealand White rabbits (2.5 to 3.5 kg; Harlan, Israel) were infected with B. anthracis strain Vollum by subcutaneous administration of 15 50% lethal doses (LD50), 150 LD50, or 1,500 LD50 (1 LD50 = 10 spores). Serum was collected at 24 h and 36 h postinfection. Blood samples were either treated with sodium citrate and then serially diluted for determination of bacteremia or centrifuged in Becton Dickinson (BD) Vacutainer tubes for removal of the cellular fraction from serum. The bacteremic level of each individual blood sample was determined by plating serial dilutions and colony counting. Sera collected from rabbits infected by the intranasal route of administration with B. anthracis spores of the Vollum strain and exhibiting a bacteremic level of 107 CFU/ml were kindly provided by Zeev Altboum (53).
Blood cultures.
Ten milliliters of rabbit whole blood was collected into a sodium citrate tube, transferred to a blood culture bottle (final volume, 25 ml) (Bactec; BD), and incubated at 37°C. B. anthracis cells (final bacteremia level, 103 CFU/ml) were inoculated into each bottle. Culture samples (1 ml) were taken every 1 h for CFU counts (100-μl samples in decimal dilutions were plated on LB plates overnight at 37°C) and for Western blot analysis (900 μl). Culture samples were centrifuged at 1,800 × g and 4°C for 2 min in Microtainer (BD) tubes for removal of red blood cells. The serum samples were subjected directly to Western blot analysis.
Enrichment of scarce proteins in serum by use of Proteominer affinity chromatography column.
The Proteominer (Bio-Rad) procedure is based on affinity chromatography of a complex mixture of proteins, using a combinatorial library of hexapeptides immobilized on beads. This procedure reduces the dynamic range of protein concentrations in the sample by means of an enhanced relative capacity for scarce proteins and rapid saturation of abundant protein ligands. One milliliter of serum was loaded on each Proteominer column, which was processed according to the manufacturer's directions. Elution of bound proteins was performed using a solution containing 300 μl 4 M urea, 1% CHAPS, and 5% acetic acid. Samples were neutralized with 4 M sodium carbonate to pH 7 to 8 and purified using a 2D cleanup kit (Bio-Rad) prior to further analysis.
Quantification of PA by ELISA.
PA levels were determined by a capture enzyme-linked immunosorbent assay (ELISA) performed in 96-well microtiter plates (Nunc, Roskilde, Denmark), with purified PA (69) as a reference standard, as previously described (36). ELISA was carried out using the following antibodies: 100 μg/ml anti-PA rabbit serum for capture (plate coating) and 100 μg/ml anti-PA mouse serum for detection. The assay was developed with alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (Sigma) and visualized using the appropriate alkaline phosphatase reagent.
Partial purification of recombinant HtrA, generation of rabbit anti-HtrA antibodies, and quantification of HtrA by capture ELISA.
Recombinant HtrA synthesized and secreted by the B. anthracis Δ14185-HtrA strain was harvested from the filtered (0.22-μm filter system; Corning, NY) supernatants of 20-h FAG medium cultures (which typically contain 40 to 60% recombinant HtrA) by 15% trichloroacetic acid precipitation carried out by incubation at 4°C for at least 8 h, followed by Sorvall centrifugation (>10,000 rpm, 25 min, 1°C; SLA-1500 rotor). The proteinaceous pellet was resuspended in a 20 times smaller volume (than the initial culture volume) of phosphate-buffered saline (PBS) by vigorous scraping and then recentrifuged for removal of undissolved material. The resulting clear supernatant, typically consisting of more than 90% recombinant HtrA at a final concentration of 0.6 to 0.8 mg/ml, was divided into aliquots and stored at −70°C until use. New Zealand White female rabbits (3 kg) were immunized by four consecutive subcutaneous injections, at 3-week intervals, of 0.5 mg HtrA, emulsified with complete Freund's adjuvant (Sigma) for the first injection and with incomplete Freund's adjuvant for the subsequent boosting. The presence of specific anti-HtrA antibodies was monitored 12 days after each booster by Western blot analysis to examine reactivity with the material used for immunization and for detection of the specific HtrA band in the supernatants of B. anthracis cultures. Rabbits were sacrificed and bled terminally by cardiac puncture for the collection of immune sera. An HtrA ELISA which may serve as a template for a future diagnostic test was established as follows. The rabbit anti-HtrA antibodies (at a 1:500 dilution) were used as the capture reagent to coat 96-well Maxisorp plates (Nunc, Denmark), in a 50 mM carbonate-bicarbonate buffer, pH 9.6 (Sigma), by overnight incubation at room temperature, followed by 1 h of blocking at 37°C with PBST buffer (Dulbecco's PBS [Biological Industries, Israel] supplemented with 0.05% Tween, 0.005% NaN3, and 2% bovine serum albumin). Serial dilutions of the samples were incubated at 37°C for 1 hour in the plate in PBST buffer. The plates were rigorously washed with PBST buffer (without bovine serum albumin), using a Nunc Immunowash apparatus. The mouse anti-HtrA antibodies obtained by DNA immunization (1:200) were then used as the detection reagent by 1 h of incubation at 37°C in PBST. The plates were washed and incubated with anti-mouse immunoglobulin antibodies (1:500) conjugated to alkaline phosphatase (Sigma) and developed with the appropriate substrate (Sigma Fast [1 mg/ml p-nitrophenyl phosphate]). The colorimetric reaction was monitored on a Versa Max ELISA reader (Molecular Devices). This quantitative ELISA requires 100 μl of sample and is appropriate for direct inspection of serum samples (does not require preremoval of abundant proteins); quantification is based on calibration curves set with accurate amounts of purified preparations of HtrA (obtained by chromatography of the secreted proteins of the HtrA overexpressor strain by use of an ion-exchange column [DEAE Macro-Prep; Bio-Rad]; the purity of HtrA in this preparation was >95%, and its concentration was determined by Bradford assay). The detection limit of the ELISA is 1 ng HtrA/ml of analyzed sample, and the linear range for accurate quantification was determined to be 5 to 200 ng/ml.
Animal experimentation.
All studies involving experimental animals were carried out according to the Guide for the Care and Use of Laboratory Animals (62a) and approved by the IIBR Animal Use Committee.
RESULTS
Detection of Bacillus anthracis antigens in blood samples collected from infected animals requires removal of abundant serum proteins.
A first attempt to identify bacterial proteins in plasmas obtained from infected animals was performed by a global comparison of proteins from infected and naïve sera, using a 2DE-PAGE approach coupled to serological analysis.
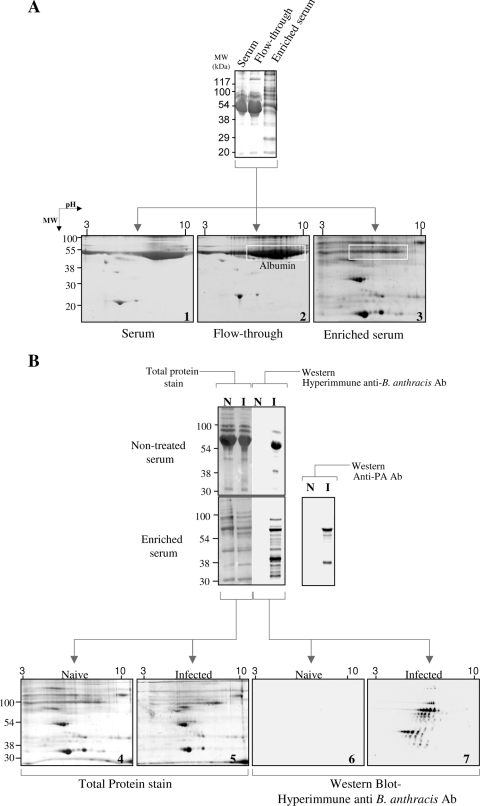
Preliminary experiments in which we attempted to visualize scarce plasma proteins established that the overwhelming presence of highly abundant proteins (such as albumin and immunoglobulins [Ig]) limits the ability to detect less abundant proteins, even when quantities as high as 200 μg protein were loaded on gels (Fig. 1A). This complication of detection, which was previously described in numerous surveys of plasma proteins in searches for disease biomarkers, is usually circumvented by depletion of abundant serum proteins (for example, see references 1, 9, 29, 44, 51, and 82). However, in our hands, different procedures, including immunodepletion and/or diminution based on affinity of Ig and albumin for various compounds (9, 29), did not result in a significant enrichment of scarce proteins (not shown). We therefore employed a newly developed procedure which enables enrichment of scarce proteins in complex mixtures by means of affinity for a random combinatorial-ligand solid-phase immobilized library (Proteominer column) (12, 13). In contrast to procedures which exploit binding of the abundant proteins to an affinity column (for their removal), the Proteominer matrix preferentially binds the scarce proteins, while the most abundant serum proteins are washed out in the chromatographic flowthrough fraction (Fig. 1A). Indeed, this affinity enrichment step enabled visualization by standard protein staining of low-quantity serum proteins whose detection was precluded by the presence of highly abundant proteins (Fig. 1A [note the almost complete removal of albumin, the most abundant serum protein]).
FIG. 1.
Visualization of scarce serum and/or bacterial proteins following removal of abundant proteins by Proteominer affinity chromatography. (A) Coomassie blue-stained 1D-SDS-PAGE gels (top) and 2DE gels (bottom) of rabbit serum before (serum) and after (enriched serum) affinity chromatography and of the corresponding flowthrough material (gels 1, 3, and 2, respectively). Note that the unbound affinity column material (flowthrough) exhibits a similar profile to that of the serum, indicating efficient removal of the abundant proteins. Protein amounts loaded were as follows: top gel, serum and flowthrough fractions, 200 μg; top gel, enriched serum, 20 μg; all 2DE gels, 200 μg. Note the almost complete depletion of albumin from the serum following the chromatographic enrichment step. (B) Plasma samples derived from naïve (N) and infected (I) rabbits, before (nontreated serum; 200 μg) or after (enriched serum; 20 μg in 1D-SDS-PAGE gels and 200 μg in the respective 2DE gels) removal of abundant proteins, were subjected to 1D-SDS-PAGE (top left) or 2DE-SDS-PAGE (gels 4 and 5) and Western analysis (top right and gels 6 and 7) with anti-B. anthracis hyperimmune serum or with specific anti-PA antibodies, as indicated.
The presence of bacterial antigens could then be investigated in highly bacteremic (107 CFU/ml) blood samples obtained from infected rabbits. The protein mixture released from the enrichment column was probed on Western blots with guinea pig total anti-B. anthracis antibodies (referred to here as “hyperimmune” serum). This hyperimmune serum was shown in previous serological studies to be efficient in identifying secreted and surface-associated B. anthracis antigens by both Western and immunoprecipitation protocols (22, 38). The data depicted in Fig. 1B clearly demonstrate that many bacterial proteins in the circulation of the infected animals reacted with the hyperimmune serum, in contrast to the case with uninfected controls. Notably, without the enrichment step, the only bacterial secreted protein which could be detected in serum samples was the toxin component PA, which before this study represented the only B. anthracis-derived secreted protein that could be detected in the circulation of infected animals at an early stage. The enriched serum samples were resolved by high-resolution 2DE, and attempts were made, using matrix-assisted laser desorption ionization-time of flight mass spectrometric tryptic fingerprinting analysis, to identify protein spots which may have represented bacterial proteins whose presence in sera obtained from infected animals is clearly established (compare Western blots 6 and 7 in Fig. 1B). Yet these attempts were not successful, given the fact that 2DE examination of plasma samples derived from infected animals and those taken from uninfected ones did not reveal the presence of bacterial proteins at levels compatible with mass spectroscopic identification (note the similarity of Coomassie blue-stained 2DE gels 4 and 5 in Fig. 1B).
Detection of B. anthracis secreted proteins in bacteremic sera by use of specific antibodies.
We recently compiled a list of B. anthracis immunogenic secreted proteins, based on extensive proteomic and genomic screens corroborated by serological analysis and inspection of patterns of expression under conditions which recapitulate the infection process in the host (21, 22, 38). This list provided a pool of potential vaccine and/or diagnostic markers; 10 proteins were selected for the generation of specific antibodies (Table 1), which were used as a tool to detect the presence of these proteins in blood samples obtained directly from infected animals or in blood cultures. To circumvent the need for laborious protein purification, the antibodies were generated by DNA vaccination of mice, using the respective ORFs cloned into an appropriate eukaryotic expression vector (39). Eight of the 10 proteins were selected on the basis of their high immunogenicity, as previously established by seroconversion observed in convalescent animals (22). These are the products of genes defining the genomic loci BA0345, BA0796, BA0799, BA0898, BA1952, BA3367, BA3660, and BA4787. InhA1, the product of the BA1295 gene, is less immunogenic, yet its high abundance (21, 23) and possible role in virulence (23, 67) have suggested that it may be expressed at early stages in infection. Finally, the product of the BA5427 gene, the putative LytE endopeptidase, was elected due to its abundance and pattern of expression, which implies induction by regulatory factors located on the pXO1 or pXO2 virulence plasmid (21). All of the proteins selected display functional domains compatible with a role in bacterial pathogenesis and/or represent putative homologs of virulence factors described for other pathogenic bacteria (6, 21).
TABLE 1.
Proteins tested as potential biomarkers in this study
| Locus taga | Protein type | Protein name | Distinctive domain(s)b | Putative function | Immunogenicitye |
|---|---|---|---|---|---|
| BA0345 | Alkylhydroxyperoxide reductase | AhpC | Oxidative stress response | ++ | |
| BA0796 | Hypothetical | S, SH3 (two), 3D | Unknown | ++ | |
| BA0799c | Hypothetical | S, HlyD | Unknown | ++ | |
| BA0898 | Alanine amidase | CwlB | S, SLH | Cell wall hydrolysis | ++ |
| BA1295 | Immune inhibitor | InhA1 | S | Zn protease | +/− |
| BA1952 | NlpC/P60 family endopeptidase | S, SH3 (two), NlpC/P60 | Cell wall hydrolysis | ++ | |
| BA3367 | γ Phage receptor | GamR | S, LPXTG | Unknown | ++ |
| BA3660 | Serine protease | HtrA | S | Chaperone, secretion, stress response | ++ |
| BA4787 | Iron-regulated determinant | IsdK | S, NEAT | Hemoglobin uptake, iron consumption | ++ |
| BA5427d | Endopeptidase | LytE | S, NlpC/P60 | Cell wall hydrolysis | +/− |
Proteins are arranged in ascending order according to genomic locus tag number (www.ncbi.nlm.nih.gov).
Domains which are inherently related to the functional annotation of the proteins are not indicated. S, export signal peptide; SH3, Src homology domain; HlyD, homology domain in some proteins involved in toxin secretion in gram-negative bacteria; 3D, cation binding domain involved in protein-protein interactions; SLH, S-layer homology domain; NEAT, “near transporter” repeat domain present in ORFs adjacent to iron transporters in gram-positive bacteria; NlpC/P60, cell wall peptidase family domain; LPXTG, gram-positive sortase domain.
The product of the BA0799 gene is annotated as a hypothetical protein of unknown function in the genomic database, yet we noted that it may represent a component of an ABC transporter, based on its genomic location and sequence homology (22).
The product of the BA5427 locus is functionally annotated in the database as the B. anthracis homologue of the known LytE Bacillus cell wall hydrolase, as indicated in the table. We note that the protein indeed exhibits an LytE-like NlpC/P60 protease domain, yet it lacks the characteristic LysM domains (necessary for peptidoglycan targeting).
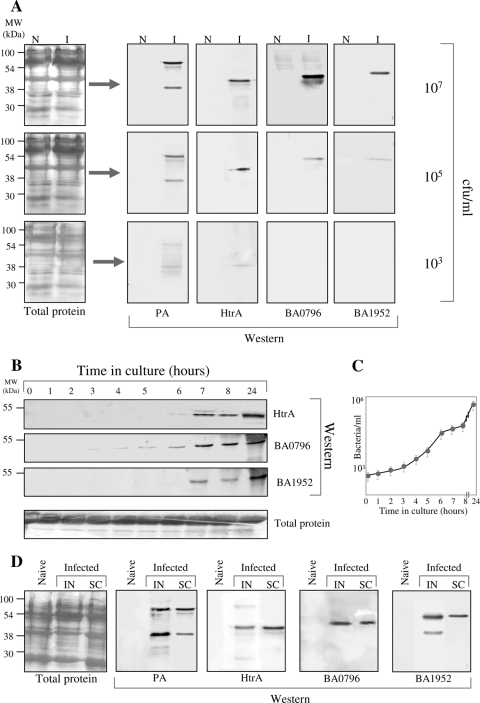
To probe for the presence of these proteins in blood samples from sick animals, enriched sera derived from infected rabbits exhibiting increasing levels of bacteremia (Fig. 2A) were subjected to Western blot analysis and compared to similarly processed sera derived from noninfected animals. As a positive control, the analysis included Western immunodetection of PA. The analysis revealed that among the 10 selected proteins, the following three antigens could be detected in significant amounts in the infected sera (Fig. 2A): the protease/chaperone HtrA (BA3660), the NlpC/P60 endopeptidase (BA1952), and a protein of unknown function specified by the BA0796 locus. These proteins could be detected in bacteremic sera, irrespective of the route of infection (Fig. 2D). It is worth noting that these three chromosomally encoded proteins are also expressed preferentially under in vitro culture conditions which are reminiscent of those encountered in the host, and furthermore, their abundance in such cultures seems to depend on the presence of the native virulence plasmids, pXO1 and pXO2 (21) (see Discussion). We note that the failure to detect other biomarkers in the circulation of infected animals does not necessarily imply their absence, and one cannot rule out the possibility that other bacterially secreted proteins in the circulation have a lower affinity for the Proteominer column and/or for the DNA immunization antibodies utilized for their detection.
FIG. 2.
Detection of HtrA, the NlpC/P60 family endopeptidase, and the product of the BA0796 gene in bacteremic sera and blood culture, using specific antibodies. (A) Thirty-microgram serum samples collected from naïve (N) or infected (I) rabbits (103 to 107 bacteria/ml, as indicated) were subjected to Proteominer affinity chromatography for removal of abundant proteins and examined by Western analysis, using 1:500 dilutions of the indicated antibodies (generated in mice by DNA vaccination [see Materials and Methods]). Western blots were developed by enhanced chemiluminescence, using horseradish peroxidase-conjugated goat anti-mouse antibodies. No specific signals could be detected in infected rabbit sera before removal of abundant proteins (not shown). A similar analysis using sera of infected animals exhibiting increasing levels of bacteremia, as indicated, was also conducted. (B) Standard (Bactec+) clinical blood culture flasks were inoculated with 10-ml rabbit blood samples (final volume in flask, 25 ml) containing 103 bacteria/ml. Serial dilutions of the 1-ml culture samples collected at the indicated times were plated for determination of bacterial counts (C). Thirty micrograms (protein) of each sample was subjected to SDS-PAGE and Western blot analysis with the indicated specific antibodies. The experiment included triplicate cultures, which served for determination of the error bars for the growth curve. (D) SDS-PAGE and Western blot analysis (similar to that shown in panel A) for immunodetection of the novel biomarkers in sera collected from rabbits infected (107 CFU/ml) by the intranasal (IN) or subcutaneous (SC) route of administration.
Detection of B. anthracis secreted biomarkers at low levels of bacteremia.
Following the initial detection of B. anthracis biomarkers in highly bacteremic sera, we addressed the question of whether these secreted proteins could be detected in sera collected from animals at early stages of infection and thus exhibiting lower levels of bacteremia. In Western blots performed directly with such sera, we could clearly detect the HtrA, BA1952, and BA796 proteins when the bacterial loads in blood were 103 to 105 CFU/ml (Fig. 2A).
Bacterial infections are usually diagnosed by tests of cultures inoculated with various clinical specimens (blood or other body fluids) obtained from sick individuals, and rarely by direct examination of the clinical samples. Accordingly, we inoculated sterile standard blood culture bottles, such as those widely used for clinical diagnosis of bacterial infection (see Materials and Methods), with blood exhibiting 103 CFU/ml. As expected, all three biomarkers identified in the blood samples were detected in the culture (Fig. 2B). The HtrA protein could be detected at 6 h postinoculation, the BA1952 endopeptidase was detected about 7 h following the onset of cultivation, and most remarkably, the BA0796 protein was detected as early as 3 h after the initiation of the blood culture (Fig. 2). None of the other screened bacterial secreted proteins addressed in this study (Table 1) could be detected. It should be noted that in blood cultures all three biomarkers could be distinguished within a time window commensurate with the accepted culture time (24 h) in clinical diagnosis routines, and the detection by Western blot analysis of the biomarkers in blood cultures did not require removal of the abundant proteins, as in the case of direct inspection of blood samples. Thus, the novel biomarkers can be detected either directly or following culture of blood samples exhibiting bacterial levels as low as 103 CFU/ml.
Detection and quantification of HtrA in blood samples by ELISA.
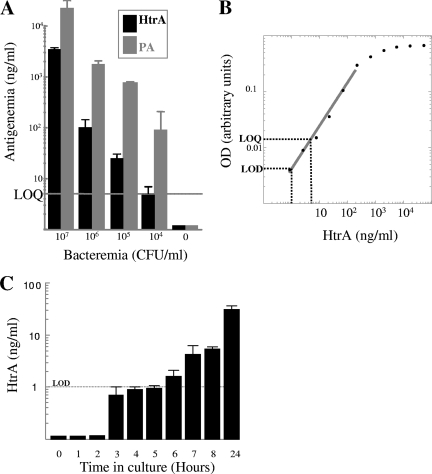
In a preliminary feasibility study aimed at probing the possible use of HtrA, one of the novel biomarkers, as a diagnostic marker for B. anthracis infection, we developed a capture ELISA test using rabbit anti-HtrA antibodies as the capture reagent and mouse anti-HtrA antibodies obtained by DNA immunization as the detection reagent (Fig. 3). Using the rapid ELISA test, HtrA could be detected directly in significant amounts in infected rabbit sera at a bacteremic level as low as 104 bacteria/ml; notably, HtrA accumulation in the circulation appeared to coincide with that of PA, the B. anthracis toxin component necessary for the onset of disease and the only biomarker described until present (Fig. 3A). Traces of HtrA could also be detected (reproducibly) in blood cultures by the ELISA test as early as 3 h after inoculation with a bacteremic blood sample exhibiting 103 CFU/ml (Fig. 3C). The assay does not require depletion of abundant serum proteins, and its high sensitivity is in line with HtrA classification as highly immunogenic in our previous seroconversion study of B. anthracis antigens (22).
FIG. 3.
Detection of HtrA and PA by capture ELISA. (A) Histogram comparing the levels of HtrA (black bars) and PA (gray bars). HtrA was quantitated by a capture ELISA with plasma samples collected from infected rabbits exhibiting increasing levels of bacteremia. Error bars represent the variations obtained in three independent assays set up in triplicate. The experiment included three animals for the 106-CFU/ml and 107-CFU/ml groups and two animals for the 104-CFU/ml and 105-CFU/ml groups. (B) Calibration curve for HtrA-specific ELISA, obtained with pure preparations of recombinant HtrA (R = 0.996). The intercepts of the broken lines with the abscissa and ordinate axes indicate the limit of detection (LOD; twice the background level) and the limit of quantification (LOQ; the lower value of the linear range of detection). The detection limit of the assay is 1 ng HtrA/ml of analyzed sample, and the linear range for accurate quantification was determined to be 5 to 200 ng/ml. The assay requires 100 μl of serum. A PA-specific ELISA exhibiting similar detection characteristics was previously described (36). (C) Histogram showing detection of HtrA over time in Bactec cultures inoculated with rabbit blood exhibiting 103 CFU/ml B. anthracis. Values close to the limit of detection are considered to represent traces of HtrA.
Biomarkers unique to infection.
Infections caused by bacilli of the B. cereus group, which are phylogenetically related to B. anthracis, usually affect immunocompromised individuals or occur as opportunistic complications of prolonged hospitalization, traumatic or postsurgical wounds, or use of invasive clinical devices (28). In view of the fact that the B. anthracis genome shares extensive sequence similarity to those of B. cereus and B. thuringiensis (49), the use of the biomarkers as diagnostic tools may be favored by their ability to discern between B. anthracis and these closely related species.
To determine the specificities of the three biomarkers identified in this study, we conducted an analysis of the genomes of B. thuringiensis, B. cereus, Bacillus licheniformis, and B. subtilis (the last two strains are more distant species yet, invoked in sporadic rare human infections [11, 59, 87]). The genomes of 9 Bacillus cereus, 10 Bacillus thuringiensis, 4 Bacillus licheniformis, and 12 Bacillus subtilis strains available in the NCBI database at the time of this study were examined for the presence of orthologues of the biomarkers (exhibiting >80% identity across full sequence coverage [E value, ≤e−10]). No homologues of the BA0796 gene were found in the B. licheniformis and B. subtilis strains scrutinized. The functionally equivalent homologous proteins of the NlpC/P60 family endopeptidase and the HtrA protease in B. licheniformis and B. subtilis exhibit <40% identity with the respective genes of B. anthracis. The genomes of the closely related B. cereus and B. thuringiensis strains do exhibit orthologues of the three biomarkers, yet documented secretome studies involving these two related strains did not report their presence (25, 41, 42, 43). The uniqueness of the biomarkers will have to be addressed experimentally in the context of their use for the design of diagnostic tools. In a preliminary limited study aimed at verifying the uniqueness of the three biomarkers to B. anthracis, we directly tested (using Western blot analysis) cultures of B. cereus, B. thuringiensis, and B. subtilis and compared them to that of B. anthracis (Fig. 4). Indeed, the three biomarkers could be detected in significantly larger amounts in the B. anthracis culture than in those of B. cereus, B. thuringiensis, and B. subtilis.
FIG. 4.
The HtrA, BA0796, and BA1952 proteins are preferentially secreted by B. anthracis compared to the closely related strains B. cereus and B. thuringiensis. (A) Filtered supernatants collected from B. anthracis, B. cereus, B. thuringiensis, and B. subtilis rich medium cultures at 24 h postinoculation were examined by Western blotting using specific antibodies. A, C, T, and S indicate the various strains, as shown in panel B. (B) Growth profiles of the cultures of the different strains. OD600, optical density at 600 nm.
DISCUSSION
Diagnosis of bacterial disease is commonly based on distinction of characteristic clinical symptoms combined with hematological profiling and identification of microbes in cultures inoculated with various clinical specimens from the suspected patient. At later stages, detection of specific circulating antibodies often serves to confirm previous exposure to the infectious agent, and their level may correlate with the evolution of the disease. Only in a few cases can diagnosis also rely on direct immunoidentification of bacterium-borne products: for example, Yersinia pestis F1 capsular antigen was proposed as a marker for the rapid diagnosis of plague (75), and Staphylococcus aureus toxic shock syndrome toxin 1 and staphylococcal enterotoxin B are proposed markers of staphylococcal infection (26, 63, 72). PA was proposed to serve as a diagnostic marker for B. anthracis infection (53). However, some rare PA-producing B. cereus clinical isolates have been described (50), suggesting that positive identification of B. anthracis infection cannot rely only on detection of PA. In the study documented here, we demonstrate that three additional Bacillus anthracis-related secreted proteins may serve as the basis for designing diagnostic tools.
In searching for novel diagnostic biomarkers of anthrax, we relied on a comprehensive reductional survey which included (i) bioinformatic analysis of genomic ORFs for virulence-related functions (5, 6); (ii) functional genomic assessment of immunogenicity by high-throughput in vitro synthesis of selected ORF products and quantitative determination of their interaction with convalescent-phase sera (38, 39); (iii) proteomic analysis by high-resolution 2DE of bacterial proteins produced by different strains of B. anthracis under various culture conditions, including those mimicking the milieu encountered in the host during infection (21); and (iv) serological analysis of proteins resolved by 2DE (20, 22). The screen generated a pool of bacterial antigens distinguished by high immunogenicity and, in many cases, potential relatedness to pathogenesis of B. anthracis. To further probe the use of selected proteins among these immunogens as vaccine components, a large number of these candidates were selected for generation of specific antibodies by rapid DNA immunization techniques (22). Accordingly, in the present study, using this battery of antibodies, we show that three proteins—the serine protease HtrA (BA3660), the NlpC/P60 family endopeptidase (BA1952), and an SH3 domain-containing protein of unknown function (BA0796)—could be detected in blood samples drawn from B. anthracis-infected rabbits (Fig. 2). HtrA could be detected at a low level of bacteremia, compatible with postexposure prophylactic treatment. Using an ELISA test which may serve as a feasibility model for a future diagnosis tool (Fig. 3), HtrA could be detected in bacteremic blood in significant amounts (5 μg/ml) at a bacteremic level of 107 CFU/ml blood. All three biomarkers could be detected by Western analysis of clinical cultures inoculated with blood exhibiting 103 bacteria/ml after only 6 to 7 h in culture; the BA0796 protein could actually be detected as early as 3 hours following the onset of the culture, coinciding with the appearance in culture of PA, the only anthrax biomarker acknowledged before the present study (not shown). The time of detection is therefore much shorter than the routinely accepted 24-h culture for diagnosis of infection in clinical practice. Orthologues of the three biomarkers are present only in the very phylogenetically close species B. cereus and B. thuringiensis, which belong to the B. cereus group, not in more remote bacilli such as B. subtilis and B. licheniformis. Furthermore, and quite remarkably, direct examination of cultures of different bacilli clearly established that the three biomarkers are secreted preferentially by B. anthracis (Fig. 4). Actually, only traces of the biomarkers could be detected in B. cereus and B. thuringiensis by Western blotting, and they could not be detected at all in B. subtilis cultures. This observation is in line with the fact that extensive data mining of proteomic information on related bacillus species (25, 41-43, 79-81) failed to reveal the presence of orthologues of the markers in any other secretome besides that of B. anthracis. All three biomarkers appear to exhibit universality with regard to their ability to serve for detection of different B. anthracis strains, since the respective genes are present in the genomes of all B. anthracis strains sequenced to date (16 different strains are available in the NCBI database). Although the extent of universality has to be confirmed experimentally for all strains, note that proteomic data have established expression of these markers in four different strains, namely, ATCC 14185, Vollum (21), Sterne (33), and UM23C1-2 (4). Thus, it appears that the biomarkers meet both specificity and universality standards, which may facilitate their use as the basis for development of diagnostic tools.
The detection of the markers strongly supports the validity of the global inspection approach for the identification of valuable antigens with immediate applicability in diagnosis or disease progression monitoring. Furthermore, the high levels of immunogenicity of the protein targets, which served as a central parameter for their selection, may facilitate their detection based on high-affinity antibodies, as implied by the sensitive ELISA test specific for HtrA presented in this article (Fig. 3). The possibility of using individual or combinations of all three novel markers for the design of sensitive diagnostic tools is currently being explored. It is conceivable that the sensitivity of such assays may be improved significantly (compared to the ELISA presented in this study), for example, by employing highly sensitive immunoconjugates, thus lowering the detection threshold of the biomarkers and enabling diagnosis of even lower levels of bacteremia at relatively early stages of infection.
To date, the only bacterially derived proteins directly detected in the circulation of B. anthracis-infected animals were the bacterial toxins. The early detection of the three biomarkers in the circulation of infected animals reported in the present study and their specificity for B. anthracis infection are therefore intriguing and strongly suggest a role for these proteins in the infection process. We note that HtrA and the NlpC/P60 endopeptidase BA1952 are both proteases. Secreted proteolytic activity may facilitate colonization and bacterial spreading during infection by targeting host proteins, affecting anti-infectious activities, and/or enabling exploitation of a protein-rich environment (3, 7, 21, 23, 61, 68). HtrA is a chaperone and a component of the secretion apparatus in bacilli (77), and therefore, it may be essential in secretion of the bacterial toxins or virulence factors. Furthermore, HtrA is important for the resilience of B. anthracis under stress conditions (our unpublished data) and is therefore necessary for survival in the host. The role of the product of the BA0796 gene is obscure. This is a protein of unknown function, yet we noted and documented in the past (21, 22) that both BA0796 and BA1952 (in addition to its characteristic NlpC/P60 family domain) possess two SH3 domains; these domains are of a eukaryotic nature and therefore were invoked as potentially virulence related, mainly on the basis of their theoretical ability to interfere with phospho-relay signal transduction circuits of the host cells (58, 86).
Accurate diagnosis of anthrax has ramifications far beyond the effectiveness of the treatment for the affected individual: in cases of respiratory anthrax due to a deliberate bioterror dissemination of B. anthracis, the positive diagnosis of the disease has tremendous epidemiological and logistic implications, requiring mass postexposure prophylaxis, decontamination, and recruitment of calamity-responding authorities. Thus, the accurate and rapid identification of the causative agent in exposed individuals is of utmost importance, and efforts are continuously dedicated to the development of novel diagnostic approaches. The study presented here strongly supports the concept that novel anthrax diagnostic procedures based on identification of secreted factors are a tangible and promising possibility.
Acknowledgments
We are grateful to Z. Altboum for kindly providing the bacteremic sera collected from intranasally infected rabbits. The excellent technical support of Yossi Shlomovitch and Gila Friedman is highly appreciated.
Footnotes
Published ahead of print on 31 July 2009.
REFERENCES
- 1.Ahmed, N., G. Barker, K. Oliva, D. Garfin, K. Talmadge, H. Georgiou, M. Quinn, and G. Rice. 2003. An approach to remove albumin for the proteomic analysis of low abundance biomarkers in human serum. Proteomics 3:1980-1987. [DOI] [PubMed] [Google Scholar]
- 2.Altboum, Z., Y. Gozes, A. Barnea, A. Pass, M. White, and D. Kobiler. 2002. Postexposure prophylaxis against anthrax: evaluation of various treatment regimens in intranasally infected guinea pigs. Infect. Immun. 70:6231-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anantharaman, V., and L. Aravind. 2003. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 4:RII. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antelmann, H., R. C. Williams, M. Miethke, A. Wipat, D. Albrecht, C. R. Harwood, and M. Hecker. 2005. The extracellular and cytoplasmic proteomes of the non-virulent Bacillus anthracis strain UM23C1-2. Proteomics 5:3684-3695. [DOI] [PubMed] [Google Scholar]
- 5.Ariel, N., A. Zvi, H. Grosfeld, O. Gat, Y. Inbar, B. Velan, S. Cohen, and A. Shafferman. 2002. Search for potential vaccine candidate open reading frames in the Bacillus anthracis virulence plasmid pXO1—in silico and in vitro screening. Infect. Immun. 7:6817-6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ariel, N., A. Zvi, K. Makarova, T. Chitlaru, E. Elhanany, B. Velan, S. Cohen, A. Friedlander, and A. Shafferman. 2003. Genome-based bioinformatic selection of chromosomal Bacillus anthracis putative vaccine candidates coupled with proteomic identification of surface-associated antigens. Infect. Immun. 71:4563-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aronson, A. I., C. Bell, and B. Fulroth. 2005. Plasmid-encoded regulator of extracellular proteases in Bacillus anthracis. J. Bacteriol. 187:3133-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baillie, L., and T. D. Read. 2001. Bacillus anthracis, a bug with attitude. Curr. Opin. Microbiol. 4:78-81. [DOI] [PubMed] [Google Scholar]
- 9.Björhall, K., T. Miliotis, and P. Davidsson. 2005. Comparison of different depletion strategies for improved resolution in proteomic analysis of human serum samples. Proteomics 5:307-317. [DOI] [PubMed] [Google Scholar]
- 10.Blendon, R. J., J. M. Benson, C. M. Des Roches, W. E. Pollard, C. Parvanta, and M. J. Herrmann. 2002. The impact of anthrax attacks on the American public. Med. Gen. Med. 4:1. [PubMed] [Google Scholar]
- 11.Blue, S. R., V. R. Singh, and M. A. Saubolle. 1995. Bacillus licheniformis bacteremia: five cases associated with indwelling central venous catheters. Clin. Infect. Dis. 20:629-633. [DOI] [PubMed] [Google Scholar]
- 12.Boschetti, E., L. Lomas, A. Citterio, and P. G. Righetti. 2007. Romancing the “hidden proteome,” anno domini two zero zero seven. J. Chromatogr. A 1153:277-290. [DOI] [PubMed] [Google Scholar]
- 13.Boschetti, E., and P. G. Righetti. 2008. The ProteoMiner in the proteomic arena: a non-depleting tool for discovering low-abundance species. J. Proteomics 71:255-264. [DOI] [PubMed] [Google Scholar]
- 14.Bradley, K., J. Mogridge, M. Mourez, B. Collier, and J. Young. 2001. Identification of the cellular receptor for anthrax toxin. Nature 414:225-229. [DOI] [PubMed] [Google Scholar]
- 15.Brossier, F., M. Levy, and M. Mock. 2002. Anthrax spores make an essential contribution to vaccine efficacy. Infect. Immun. 70:661-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broussard, L. A. 2001. Biological agents: weapons of warfare and bioterrorism. Mol. Diagn. 6:323-333. [DOI] [PubMed] [Google Scholar]
- 17.Bryskier, A. 2002. Bacillus anthracis and antibacterial agents. Clin. Microbiol. Infect. 8:467-478. [DOI] [PubMed] [Google Scholar]
- 18.Cendrowski, S., W. MacArthur, and P. Hanna. 2004. Bacillus anthracis requires siderophore biosynthesis for growth in macrophages and mouse virulence. Mol. Microbiol. 551:407-417. [DOI] [PubMed] [Google Scholar]
- 19.Chensue, S. W. 2003. Exposing a killer: pathologists angle for anthrax. Am. J. Pathol. 163:1699-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chitlaru, T., N. Ariel, A. Zvi, M. Lion, B. Velan, A. Shafferman, and E. Elhanany. 2004. Identification of chromosomally encoded membranal polypeptides of Bacillus anthracis by a proteomic analysis: prevalence of proteins containing S-layer homology domains. Proteomics 4:677-691. [DOI] [PubMed] [Google Scholar]
- 21.Chitlaru, T., O. Gat, Y. Gozlan, N. Ariel, and A. Shafferman. 2006. Differential proteomic analysis of the Bacillus anthracis secretome: distinct plasmid and chromosome CO2-dependent cross-talk mechanisms modulate extracellular proteolytic activities. J. Bacteriol. 188:3551-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chitlaru, T., O. Gat, Y. Gozlan, H. Grosfeld, I. Inbar, and A. Shafferman. 2007. Identification of in vivo expressed immunogenic proteins by serological proteome analysis of Bacillus anthracis secretome. Infect. Immun. 75:2841-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung, M. C., T. G. Popova, B. A. Millis, D. V. Mukherjee, W. Zhou, L. A. Liotta, E. F. Petricoin, V. Chanhoke, C. Bailey, and S. G. Popov. 2006. Secreted neutral metalloproteases of Bacillus anthracis as candidate pathogenic factors. J. Biol. Chem. 281:31408-31418. [DOI] [PubMed] [Google Scholar]
- 24.Cohen, S., I. Mendelson, Z. Altboum, D. Kobiler, E. Elhanany, T. Bino, M. Leitner, I. Inbar, H. Rosenberg, Y. Gozes, R. Barak, M. Fisher, C. Kronman, B. Velan, and A. Shafferman. 2000. Attenuated nontoxinogenic and nonencapsulated recombinant Bacillus anthracis spore vaccines protect against anthrax. Infect. Immun. 68:4549-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delvecchio, V. G., J. P. Connolly, T. G. Alefantis, A. Walz, M. A. Quan, G. Patra, J. M. Ashton, J. T. Whittington, R. D. Chafin, X. Liang, P. Grewal, A. S. Khan, and C. V. Mujer. 2006. Proteomic profiling and identification of immunodominant spore antigens of Bacillus anthracis, Bacillus cereus and Bacillus thuringiensis. Appl. Environ. Microbiol. 72:6355-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dinges, M., P. Orwin, and P. Schlievert. 2000. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 13:16-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dixon, T. C., M. Meselson, T. Guillemin, and P. Hanna. 1999. Anthrax. N. Engl. J. Med. 341:815-826. [DOI] [PubMed] [Google Scholar]
- 28.Drobniewski, F. A. 1993. Bacillus cereus and related species. Clin. Microbiol. Rev. 6:324-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Echan, L. A., H.-Y. Tang, N. Ali-Khan, K. Lee, and D. W. Speicher. 2005. Depletion of multiple high-abundance proteins improves protein profiling capacities of human serum and plasma. Proteomics 5:3292-3303. [DOI] [PubMed] [Google Scholar]
- 30.Edwards, K. A., H. A. Clancy, and A. J. Baeumner. 2006. Bacillus anthracis: toxicology, epidemiology and current rapid-detection methods. Anal. Bioanal. Chem. 384:73-84. [DOI] [PubMed] [Google Scholar]
- 31.Fellows, P. F., M. K. Linscott, B. E. Ivins, M. L. M. Pitt, C. A. Rossi, P. H. Gibbs, and A. M. Friedlander. 2001. Efficacy of a human anthrax vaccine in guinea pigs, rabbits, and rhesus macaques against challenge by Bacillus anthracis isolates of diverse geographical origin. Vaccine 19:3241-3247. [DOI] [PubMed] [Google Scholar]
- 32.Fisher, N., L. Shetron-Rama, A. Herring-Palmer, B. Heffernan, N. Bergman, and P. Hanna. 2006. The dltABCD operon of Bacillus anthracis Sterne is required for virulence and resistance to peptide, enzymatic, and cellular mediators of innate immunity. J. Bacteriol. 188:1301-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Francis, A. W., C. E. Ruggiero, A. T. Koppisch, J. Dong, J. Song, T. Brettin, and S. Iyer. 2005. Proteomic analysis of Bacillus anthracis Sterne vegetative cells. Biochim. Biophys. Acta 1748:191-200. [DOI] [PubMed] [Google Scholar]
- 34.Frazier, A. A., T. Franks, and J. Galvin. 2006. Inhalational anthrax. J. Thorac. Imaging 21:252-258. [DOI] [PubMed] [Google Scholar]
- 35.Friedlander, A., S. Welkos, and B. Ivins. 2002. Anthrax vaccines. Curr. Top. Microbiol. Immunol. 271:34-60. [DOI] [PubMed] [Google Scholar]
- 36.Gat, O., I. Inbar, R. Aloni-Gristein, E. Zahavy, C. Kronman, I. Mendelson, S. Cohen, B. Velan, and A. Shafferman. 2003. Use of a promoter trap system in Bacillus anthracis and Bacillus subtilis for the development of recombinant protective antigen-based vaccines. Infect. Immun. 71:801-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gat, O., I. Mendelson, T. Chitlaru, N. Ariel, Z. Altboum, H. Levy, S. Weiss, H. Grosfeld, S. Cohen, and A. Shafferman. 2005. The solute-binding component of a putative Mn(II) ABC transporter (MntA) is a novel Bacillus anthracis virulence determinant. Mol. Microbiol. 58:533-551. [DOI] [PubMed] [Google Scholar]
- 38.Gat, O., H. Grosfeld, N. Ariel, I. Inbar, G. Zaide, Y. Broder, A. Zvi, T. Chitlaru, Z. Altboum, D. Stein, S. Cohen, and A. Shafferman. 2006. Search for Bacillus anthracis potential vaccine candidates by a functional genomic-serologic screen. Infect. Immun. 74:3987-4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gat, O., H. Grosfeld, and A. Shafferman. 2007. In vitro screen of bioinformatically selected Bacillus anthracis vaccine candidates by coupled transcription, translation and immunoprecipitation analysis. Methods Mol. Biol. 375:211-233. [DOI] [PubMed] [Google Scholar]
- 40.Gat, O., G. Zaide, I. Inbar, H. Grosfeld, T. Chitlaru, H. Levy, and A. Shafferman. 2008. Characterization of Bacillus anthracis iron-regulated surface determinant (Isd) proteins containing NEAT domains. Mol. Microbiol. 70:983-999. [DOI] [PubMed] [Google Scholar]
- 41.Gohar, M., O. A. Okstad, N. Gilois, V. Sanchis, A.-B. Kolsto, and D. Lereclus. 2002. Two-dimensional electrophoresis analysis of the extracellular proteome of Bacillus cereus reveals the importance of the PlcR regulon. Proteomics 2:784-791. [DOI] [PubMed] [Google Scholar]
- 42.Gohar, M., N. Gilois, R. Graveline, C. Garreau, V. Sanchis, and D. Lereclus. 2005. A comparative study of B. cereus, B. thuringiensis and B. anthracis extracellular proteomes. Proteomics 5:3696-3711. [DOI] [PubMed] [Google Scholar]
- 43.Gohar, M., K. Faegri, S. Perchat, S. Ravnum, O. A. Økstad, M. Gominet, A.-B. Kolstø, and D. Lereclus. 2008. The PlcR virulence regulon of Bacillus cereus. PLoS ONE 3:e2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Govorukhina, N. I., A. Keizer-Gunnink, A. G. van der Zee, S. de Jong, H. W. Bruijn, and R. Bischoff. 2003. Sample preparation of human serum for the analysis of tumor markers. Comparison of different approaches for albumin and gamma-globulin depletion. J. Chromatogr. A 1009:171-178. [DOI] [PubMed] [Google Scholar]
- 45.Grosfeld, H., S. Cohen, T. Bino, Y. Flashner, R. Bar, E. Mamroud, C. Kronman, A. Shafferman, and B. Velan. 2003. Effective protective immunity to Yersinia pestis infection conferred by DNA vaccine coding for derivatives of the F1 capsular antigens. Infect. Immun. 71:374-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heffernan, B., B. Thomason, A. Herring-Palmer, L. Shaughnessy, R. McDonald, N. Fisher, G. Huffnagle, and P. Hanna. 2006. Bacillus anthracis phospholipases C facilitate macrophage-associated growth and contribute to virulence in a murine model of inhalation anthrax. Infect. Immun. 74:3756-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heine, H. S., J. Bassett, L. Miller, J. M. Hartings, B. E. Ivins, M. L. Pitt., D. Fritz, S. L. Norris, and W. R. Byrne. 2007. Determination of antibiotic efficacy against Bacillus anthracis in a mouse aerosol challenge model. Antimicrob. Agents Chemother. 51:1373-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heine, H. S., J. Bassett, L. Miller, A. Bassett, B. E. Ivins, D. Lehoux, F. F. Arhin, T. R. Parr, Jr., and G. Moeck. 2008. Efficacy of oritavancin in a murine model of Bacillus anthracis spore inhalation anthrax. Antimicrob. Agents Chemother. 52:3350-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helgason, E., O. A. Økstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A.-B. Kolstø. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis—one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoffmaster, A. R., J. Ravel, D. A. Rasko, G. D. Chapman, M. D. Chute, C. K. Marston, B. K. De, C. T. Sacchi, C. Fitzgerald, L. W. Mayer, M. C. Maiden, F. G. Priest, M. Barker, L. Jiang, R. Z. Cer, J. Rilstone, S. N. Peterson, R. S. Weyant, D. R. Galloway, T. T. Read, T. Popovic, and C. M. Fraser. 2004. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc. Natl. Acad. Sci. USA 101:8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Issaq, H., Z. Xiao, and T. D. Veenstra. 2007. Serum and plasma proteins. Chem. Rev. 107:3601-3620. [DOI] [PubMed] [Google Scholar]
- 52.Klimpel, K. R., N. Arora, and S. H. Leppla. 1994. Anthrax toxin lethal factor contains a zinc metalloprotease consensus sequence which is required for lethal toxin activity. Mol. Microbiol. 13:1093-1100. [DOI] [PubMed] [Google Scholar]
- 53.Kobiler, D., S. Weiss, H. Levy, M. Fisher, A. Mechaly, A. Pass, and Z. Altboum. 2006. Protective antigen as a correlative marker for anthrax in animal models. Infect. Immun. 74:5871-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lacy, T. M., and R. J. Collier. 2002. Structure and function of anthrax toxin. Curr. Top. Microbiol. Immunol. 271:62-85. [DOI] [PubMed] [Google Scholar]
- 55.Leppla, S. 1982. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cyclic AMP concentrations of eukaryotic cells. Proc. Natl. Acad. Sci. USA 79:3162-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leppla, S. H. 1995. Anthrax toxins, p. 543-572. In J. Moss, B. Iglewski, M. Vaughan, and A. T. Tu (ed.), Bacterial toxins and virulence factors in disease. Marcel Dekker, New York, NY.
- 57.Leppla, S. 1999. The bifactorial B. anthracis lethal and oedema toxins, p. 243-263. In J. E. Alouf and J. H. Freer (ed.), Comprehensive sourcebook of bacterial protein toxins. Academic Press, London, United Kingdom.
- 58.Marino, M., M. Banerjee, R. Jonquieres, P. Cossart, and P. Ghosh. 2002. GW domains of the Listeria monocytogenes invasion protein InlB are SH3-like and mediate binding to host ligands. EMBO J. 21:5623-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matsumoto, S., H. Suenaga, K. Naito, M. Sawazaki, T. Hiramatsu, and N. Agata. 2000. Management of suspected nosocomial infection: an audit of 19 hospitalized patients with septicemia caused by Bacillus species. Jpn. J. Infect. Dis. 53:196-202. [PubMed] [Google Scholar]
- 60.Mendelson, I., O. Gat, R. Aloni-Grinstein, Z. Altboum, I. Inbar, C. Kronman, E. Bar-Haim, S. Cohen, B. Velan, and A. Shafferman. 2005. Efficacious, nontoxigenic Bacillus anthracis spore vaccines based on strains expressing mutant variants of lethal toxin components. Vaccine 23:5688-5697. [DOI] [PubMed] [Google Scholar]
- 61.Miyoshi, S., and S. Shinoda. 2000. Microbial metalloproteases in pathogenesis. Microbes Infect. 2:91-98. [DOI] [PubMed] [Google Scholar]
- 62.Mock, M., and A. Fouet. 2001. Anthrax. Annu. Rev. Microbiol. 55:647-671. [DOI] [PubMed] [Google Scholar]
- 62a.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC.
- 63.Novick, R. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 64.Omenn, G. S., and the HUPO Plasma Proteome Project Consortium. 2005. Overview of the HUPO plasma proteome project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and publicly-available database. Proteomics 5:3227-3245. [DOI] [PubMed] [Google Scholar]
- 65.Passalacqua, K. D., N. H. Bergman, A. Herring-Palmer, and P. Hanna. 2006. The superoxide dismutases of Bacillus anthracis do not cooperatively protect against endogenous superoxide stress. J. Bacteriol. 188:3837-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pezard, C., M. Weber, J.-C. Sirard, P. Berche, and M. Mock. 1995. Protective immunity induced by Bacillus anthracis toxin-deficient strains. Infect. Immunol. 63:1369-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramarao, N., and D. Lereclus. 2005. The InhA1 metalloprotease allows spores of the B. cereus group to escape macrophages. Cell. Microbiol. 7:1357-1364. [DOI] [PubMed] [Google Scholar]
- 68.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 69.Reuveny, S., M. White, Y. Adar, Y. Kafri, Z. Altboum, Y. Gozes, D. Kobiler, A. Shafferman, and B. Velan. 2002. Search for correlates of protective immunity conferred by anthrax vaccine. Infect. Immun. 69:2888-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ross, C., and T. Koehler. 2006. plcR papR-independent expression of anthrolysin O by Bacillus anthracis. J. Bacteriol. 188:7823-7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rossi, C. A., M. Ulrich, S. Norris, D. S. Reed, L. M. Pitt, and E. K. Leffel. 2008. Identification of a surrogate marker for infection in the African green monkey model of inhalation anthrax. Infect. Immun. 76:5790-5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rowe, C., S. Scruggs, M. Feldstein, J. Golden, and F. Ligler. 1999. An array immunosensor for simultaneous detection of clinical analytes. Anal. Chem. 75:433-439. [DOI] [PubMed] [Google Scholar]
- 73.Shafazand, S., R. Doyle, S. Ruoss, A. Weinacker, and T. Raffin. 1999. Inhalation anthrax: epidemiology, diagnosis and management. Chest 116:1369-1376. [DOI] [PubMed] [Google Scholar]
- 74.Singh, Y., K. Klimpel, S. Goel, P. Swain, and S. Leppla. 1999. Oligomerization of anthrax toxin protective antigen and binding of lethal factor during endocytic uptake into mammalian cells. Infect. Immun. 67:1853-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Splettstoesser, W., L. Rahalison, R. Grunow, H. Neubauer, and S. Chanteau. 2004. Evaluation of a standardized F1 capsular antigen capture ELISA test kit for the rapid diagnosis of plague. FEMS Immunol. Med. Microbiol. 41:149-155. [DOI] [PubMed] [Google Scholar]
- 76.Stern, E. J., K. B. Uhde, S. V. Shadomy, and N. Messonnier. 2008. Conference report on public health and clinical guidelines for anthrax. Emerg. Infect. Dis. 14:07-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tjalsma, H., H. Antelmann, J. D. H. Jongbloed, P. G. Braun, E. Darmon, R. Dorenbos, J.-Y. F. Dubois, H. Westers, G. Zanen, W. J. Quax, O. P. Kuipers, S. Bron, M. Hecker, and J. M. van Dijl. 2004. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68:207-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Turnbull, P. C. B. 1999. Definitive identification of Bacillus anthracis—a review. J. Appl. Microbiol. 87:237-240. [DOI] [PubMed] [Google Scholar]
- 79.Voigt, B., T. Schweder, D. Becher, A. Ehrenreich, G. Gottschalk, J. Feesche, K.-H. Maurer, and M. Hecker. 2004. A proteomic view of cell physiology of Bacillus licheniformis. Proteomics 4:1465-1490. [DOI] [PubMed] [Google Scholar]
- 80.Voigt, B., T. Schweder, M. J. J. Sibbald, D. Albrecht, A. Ehrenreich, J. J. Bernhardt, J. Feesche, K.-H. Maurer, G. Gottschalk, J. M. Van Dijl, and M. Hecker. 2005. The extracellular proteome of Bacillus licheniformis grown in different media under different nutrient starvation conditions. Proteomics 6:268-281. [DOI] [PubMed] [Google Scholar]
- 81.Voigt, B., H. Antelman, D. Albrecht, A. Ehrenreich, K.-H. Maurer, S. Evers, G. Gottschalk, J. M. van Dijl, T. Schweder, and M. Hecker. 2009. Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtillis. J. Mol. Microbiol. Biotechnol. 16:53-68. [DOI] [PubMed] [Google Scholar]
- 82.Wang, Y. Y., P. Cheng, and D. W. Chan. 2003. A simple affinity spin tube filter method for removing high-abundant common proteins or enriching low-abundant biomarkers for serum proteomic analysis. Proteomics 3:243-248. [DOI] [PubMed] [Google Scholar]
- 83.Wilson, A. C., M. Soyer, J. A. Hoch, and M. Perego. 2008. The bicarbonate transporter is essential for Bacillus anthracis lethality. PLoS Pathog. 4:e1000210. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 84.Wright, G., M. Puzzis, and B. Neely. 1962. Studies on immunity in anthrax. J. Bacteriol. 83:515-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu, H.-J., A. H.-J. Wang, and M. Jennings. 2008. Discovery of virulence factors of pathogenic bacteria. Curr. Opin. Chem. Biol. 12:93-101. [DOI] [PubMed] [Google Scholar]
- 86.Wylie, G. P., V. Ranghachari, E. A. Bienkiewicz, V. Marin, N. Bhattacharya, J. F. Love, J. R. Murphy, and T. M. Logan. 2005. Prolylpeptide binding by the prokaryotic SH3-like domain of the diphtheria toxin repressor: a regulatory switch. Biochemistry 44:40-51. [DOI] [PubMed] [Google Scholar]
- 87.Zwick, M. E., M. P. Kiley, A. C. Stewart, A. Mateczun, and T. D. Read. 2008. Genotyping of Bacillus cereus strains by microarray-based resequencing. 2008. PLoS ONE 3:e2513. [DOI] [PMC free article] [PubMed] [Google Scholar]