Abstract
Tetracycline-resistant Lactococcus lactis strains originally isolated from Polish raw milk were analyzed for the ability to transfer their antibiotic resistance genes in vitro, using filter mating experiments, and in vivo, using germfree rats. Four of six analyzed L. lactis isolates were able to transfer tetracycline resistance determinants in vitro to L. lactis Bu2-60, at frequencies ranging from 10−5 to 10−7 transconjugants per recipient. Three of these four strains could also transfer resistance in vitro to Enterococcus faecalis JH2-2, whereas no transfer to Bacillus subtilis YBE01, Pseudomonas putida KT2442, Agrobacterium tumefaciens UBAPF2, or Escherichia coli JE2571 was observed. Rats were initially inoculated with the recipient E. faecalis strain JH2-2, and after a week, the L. lactis IBB477 and IBB487 donor strains were introduced. The first transconjugants were detected in fecal samples 3 days after introduction of the donors. A subtherapeutic concentration of tetracycline did not have any significant effect on the number of transconjugants, but transconjugants were observed earlier in animals dosed with this antibiotic. Molecular analysis of in vivo transconjugants containing the tet(M) gene showed that this gene was identical to tet(M) localized on the conjugative transposon Tn916. Primer-specific PCR confirmed that the Tn916 transposon was complete in all analyzed transconjugants and donors. This is the first study showing in vivo transfer of a Tn916-like antibiotic resistance transposon from L. lactis to E. faecalis. These data suggest that in certain cases food lactococci might be involved in the spread of antibiotic resistance genes to other lactic acid bacteria.
The abuse of antibiotic use is regarded as the major cause of the accumulation and dissemination of antibiotic resistance genes in the environment (33). For several decades, studies on selection and spread of antibiotic resistance genes have focused mainly on clinically relevant microbial species. Nevertheless, many investigators have recently speculated that commensal bacteria, including lactic acid bacteria (LAB), may act as reservoirs of antibiotic resistance determinants (40). Genes conferring acquired resistance to tetracycline, erythromycin, and vancomycin have been detected and characterized for Lactococcus, Enterococcus, and Lactobacillus species isolated from fermented meat and milk products (13, 18, 23, 49, 50, 56). Introduction of such bacteria into humans through ingestion of commercial food products may have negative consequences by dissemination of antibiotic resistance genes via the food chain to the resident microbiota of the human gastrointestinal tract and, in the worst case, to pathogenic bacteria (4, 17, 55). Therefore, it seems important to assess the risk of antibiotic resistance gene transmission in the environment and in the guts of animals and humans and to establish the genetic basis of the detected resistance and transmission mechanisms.
Dissemination of genetic information by horizontal gene transfer is common in the microbial world and is accomplished mainly by the following three mechanisms: natural transformation, conjugation, and transduction (14). Many antibiotic resistance genes have been detected on mobile genetic elements, such as plasmids and conjugative transposons, and it is believed that conjugation is the main mode of horizontal dissemination of antibiotic resistance determinants between bacterial species.
Conjugative transposons mediate their own transfer from a donor DNA molecule in one bacterial cell to a target molecule in another cell. Tn916, which spans about 18 kb and confers resistance to tetracycline via tet(M), belongs to the Tn916-Tn1545 family of conjugative transposons and was first identified in Enterococcus faecalis DS16 (20). It is able to be maintained in a wide range of clinically important gram-positive and gram-negative species (12, 44).
Excision of Tn916 from the donor molecule is required for conjugative transposition and results in a covalently closed circular transposon molecule that is an intermediate in conjugal transfer (10). A single strand of the covalently closed circular transposon is transferred to the recipient cell, where the complementary strand is synthesized to recreate a double-stranded circular transposon, which inserts into a target site (48).
Lactococcus lactis strains are used worldwide as starter organisms in the dairy industry and for the manufacturing of many fermented products. Conjugation has been described widely for lactococci, although mainly for exploitation of this process for development of improved starter strains (22, 38, 39, 51, 53).
The objective of the present study was to establish the ability of wild-type L. lactis isolates to transfer tetracycline resistance determinants to gram-positive bacteria, namely, L. lactis Bu2-60, E. faecalis JH2-2, and Bacillus subtilis YBE01, and to gram-negative bacteria, namely, Pseudomonas putida KT2442, Agrobacterium tumefaciens UBAPF2, and Escherichia coli JE2571, by using the filter mating approach. In order to confirm whether these donor strains were able to transfer the tetracycline resistance genes to E. faecalis JH2-2 in vivo in the gastrointestinal tract, we also used germfree rats.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains used in this study and their growth conditions are listed in Table 1. The L. lactis tetracycline-resistant strains IBB28, IBB160, IBB161, IBB224, IBB477, and IBB487, used as donor strains for mating experiments, were originally isolated from samples of Polish artisanal dairy products or raw milk (J. Zycka-Krzesinska, J. Boguslawska, and J. Bardowski, unpublished data) and were grown on GM17 plates containing tetracycline at 30°C for 24 to 48 h. The following strains were used as recipients in conjugal transfer experiments: E. faecalis JH2-2, E. faecalis JH2-SS, L. lactis subsp. lactis bv. diacetylactis Bu2-60, B. subtilis YBE01, P. putida KT2442, A. tumefaciens UBAPF2, and E. coli JE257. Transconjugants were grown on recipient-specific agar plates supplemented with tetracycline. Antibiotics (Sigma, St. Louis, MO) were used at the following concentrations: rifampin (rifampicin), 50 μg ml−1; streptomycin, 500 μg ml−1; tetracycline, 10 μg ml−1; fusidic acid, 25 μg ml−1; spectinomycin, 500 μg ml−1; chloramphenicol, 25 μg ml−1; and kanamycin, 50 μg ml−1.
TABLE 1.
Bacterial strains used in this study and their cultivation conditionsa
| Strain | Relevant properties | Cultivation conditions | Reference or origin |
|---|---|---|---|
| Gram-positive bacteria | |||
| E. faecalis strains | BHI, 42°C | ||
| JH2-2 | Rifr Fusr | 27 | |
| JH2-SS | StrrSpr | 51 | |
| L. lactis strains | |||
| IBB28 | Tetr [tet(S) gene] | GM17, 30°C | Zycka-Krzesinska et al., unpublished data |
| IBB160 | Tetr [tet(S) gene] | ||
| IBB161 | Tetr [tet(M) gene] | ||
| IBB224 | Tetr [tet(S) and tet(M) genes] | ||
| IBB477 | Tetr [tet(S) and tet(M) genes] | ||
| IBB487 | Tetr [tet(M) gene] | 35 | |
| Bu2-60 | Strr Rifr | ||
| B. subtilis YBE01 | Cmr | LB, 37°C | 20 |
| Gram-negative bacteria | |||
| P. putida KT2442 | Cmr Kmr Rifr | LB, 37°C | 10 |
| A. tumefaciens UBAPF2 | Strr Kmr Rifr | LB, 30°C | 25 |
| E. coli JE2571 | Rifr | LB, 37°C | 7 |
Abbreviations: Rifr, rifampin resistance; Fusr, fusidic acid resistance; Strr, streptomycin resistance; Spr, spectinomycin resistance; Tetr, tetracycline resistance; Cmr, chloramphenicol resistance; Kmr, kanamycin resistance; BHI, brain heart infusion medium (Oxoid); GM17, M17 medium supplemented with 0.5% glucose (Difco); LB, Luria-Bertani medium (Difco).
In vitro mating experiments.
The ability of the lactococcal strains to transfer the tetracycline resistance genes to E. faecalis JH2-2, E. faecalis JH2-SS, L. lactis Bu2-60, and B. subtilis YBE01 was examined by the filter mating approach. The mating procedure was performed as described earlier (32). In short, exponentially growing donor and recipient strains were mixed in a total volume of 2 ml and poured onto a sterile membrane filter (HAWP04700; Millipore, Bedford, MA), which was incubated right side up for 18 to 20 h at 30°C, 37°C, or 42°C (depending on the recipient) on nonselective recipient-specific agar plates. Control cultures of recipient and donor strains alone were treated in the same manner. After incubation, the bacteria were washed from the filters by rigorous shaking in 2 ml of PPS (8.5 g/l NaCl [Merck, Darmstadt, Germany] containing 1 g/liter neutralized bacteriological peptone [Oxoid, Hampshire, England]). Dilutions were spread onto donor-, recipient-, and transconjugant-selective agar plates. After growth at 30°C for 1 to 2 days, the numbers of transconjugant, donor, and recipient colonies were determined.
Potential transfer of tetracycline resistance to P. putida KT2442, A. tumefaciens UBAPF2, and E. coli JE2571 was examined by the filter mating approach described above, with various modifications, as follows. Exponentially growing donor and recipient strains were mixed at a 20:1 ratio (41). Aliquots of 2 ml of mating mixture were filtered by sterile membrane filters, which were then placed on brain heart infusion agar containing DNase I (100 U/ml) to hamper transfer by transformation. After 18 h of incubation at 37°C, the cells were washed out of the filters and plated on agar containing appropriate antibiotics to select for the donor, recipient, and transconjugants.
To study whether subinhibitory (sub-MIC) concentrations of tetracycline (which had no inhibitory effect on bacterial growth) would have any influence on conjugal transfer frequency, conjugation experiments between donors L. lactis IBB477 and IBB487 and recipients L. lactis Bu2-60 and E. faecalis JH2-2 were conducted as described above, with the following modifications: sterile membrane filters were incubated on nonselective recipient agar plates and on recipient agar plates containing tetracycline at the following concentrations: 0.1, 0.15, and 0.2 μg/ml.
Animal management and experimental design.
Seventeen male germfree Sprague-Dawley rats, originally supplied by Taconic (Germantown, NY), were bred at the National Food Institute, Technical University of Denmark. The rats were 3 months old at the beginning of the experiment and were housed and fed as previously described (57). The germfree status of the animals was verified by testing fecal samples for aerobic and anaerobic growth of bacteria and yeasts. The rats were caged in two isolators and grouped in the following way: group A, one rat receiving only the E. faecalis JH2-2 recipient strain, used as a control; group B, four animals dosed with the donor strain L. lactis IBB477 and with E. faecalis JH2-2; group C, four animals dosed with L. lactis IBB477, E. faecalis JH2-2, and drinking water containing tetracycline at 50 μg/ml; group D, one rat receiving only the E. faecalis JH2-2 recipient strain, used as a control; group E, four animals dosed with the donor strain L. lactis IBB487 and with E. faecalis JH2-2; and group F, three animals dosed with L. lactis IBB487, E. faecalis JH2-2, and drinking water containing tetracycline at 50 μg/ml. Isolator 1 contained groups A to C, whereas isolator 2 contained groups D to F.
At day 0, all rats received a 1-ml dose containing about 1010 CFU of the recipient strain E. faecalis JH2-2. The recipient strain was allowed to establish itself in the rats for 1 week, after which the donor strains were introduced. Each day from days 8 to 10, all rats in isolator 1 (except the control rat) received a 1-ml dose containing 108 CFU L. lactis IBB477 and all rats in isolator 2 (except the control rat) were dosed with 1 ml containing 108 CFU L. lactis IBB487. Tetracycline was added to the dosing cultures of donors and received by animals in groups C and F during the same period. Overnight cultures of all strains were washed and resuspended in phosphate-buffered saline (Oxoid), and 1 ml was given to each rat by oral gavage.
Fresh fecal samples (100 to 300 mg) were obtained directly from the rats each working day by gently pressing the abdomens of the animals. Intestinal samples from the duodenum, ileum, cecum, and colon were taken at sacrifice. The samples were initially diluted 10-fold (wt/vol) in phosphate-buffered saline, thoroughly homogenized, further diluted, and plated on the appropriate selective agar plates for enumeration of donors, recipients, and transconjugants.
DNA preparation and manipulations.
Total DNA was isolated from L. lactis cells by the following method. Cells from 2 ml of overnight culture were harvested, washed, and suspended in 0.2 ml of TES-lysozyme solution (25 mM Tris-HCl, 0.1 M EDTA, 25% [wt/vol] saccharose, pH 8, lysozyme at 8 mg/ml). After 15 min of incubation at 37°C, 15 μl of 20% sodium dodecyl sulfate solution was added, and samples were incubated for 5 min at 75°C. Cell lysates were extracted with 0.5 ml of phenol-chloroform solution (1:1 [vol/vol]). After centrifugation (15 min, 21,000 rpm), the water phase was recovered. Phenol-chloroform extraction was repeated twice. Finally, total DNA was precipitated with 3 M sodium acetate (1:10 [vol/vol]; pH 4.8) and ice-cold 96% ethanol (1:2 [vol/vol]). After centrifugation (15 min, 21,000 rpm), the pellet was washed with 70% ethanol, dried, and dissolved in 50 μl of demineralized water with RNase (100 μg/ml).
Isolation of plasmid DNA was based on the alkaline lysis method of Anderson and McKay (3).
Verification of transconjugants by PCR.
Transconjugants obtained from in vivo and in vitro conjugation assays were selected on the basis of their phenotypic resistance profile by testing their capability of growth on recipient-specific agar plates supplemented with tetracycline. In order to verify that these isolates were true transconjugants and not mutants, PCR assays with primers specific for the recipients (E. faecalis and L. lactis) and the tet(M) and tet(S) resistance genes were performed [the donor strains contain tet(S), tet(M), or both genes (Table 1)]. The PCR mixture was composed of 5 μl of total DNA template, 1 μl of Taq polymerase (Fermentas, Lithuania), and 10 pmol of each primer in a total volume of 25 μl. PCR assays were conducted in a Peltier PTC-200 thermal cycler (MJ Research, Cambridge, MA) using the following PCR conditions: initial denaturation at 94°C for 5 min; 25 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min; and a final extension step at 72°C for 10 min. Primers used for the detection of the tet(M) gene were DI (5′-GAYACNCCNGGNCAYRTNGAYTT-3′) and tetM-R (5′CACCGAGCAGGGATTTCTCCAC-3′) (24), and those for the tet(S) gene were tetS-F (5′GGAGTACAGTCACAAACTCG-3′) and tetS-R (5′-GGATATAAGGAGCAACTTTG-3′) (37). E. faecalis species-specific primers were ddl E. faecalis E1 and E. faecalis E2 (16), and L. lactis species-specific primers were 212 Fla and 1406R (46). The PCR products were run in a 0.8% agarose gel and visualized by ethidium bromide staining.
RAPD amplification.
Oligonucleotide primer M13 (45) (5′-GAGGGTGGCGGTTCT-3′) was used for amplification of randomly amplified polymorphic DNA (RAPD amplification). PCRs were performed in 20-μl reaction mixtures containing 2 μl of 10× PCR buffer (Fermentas, Lithuania), 0.5 U of Taq DNA polymerase (Fermentas), 4 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 100 μM, 4 μM primer M13, and about 10 ng of template DNA. Reactions were performed under the following conditions: 40 cycles consisting of 94°C for 1 min, 45°C for 20 s, and 72°C for 2 min and a final extension at 72°C for 5 min. The amplification products were run in a 0.8% agarose gel and visualized by ethidium bromide staining.
PFGE.
Genomic DNA was prepared in situ in agarose blocks as described previously (30) and was digested with 20 U of the restriction enzyme SmaI (Fermentas, Vilnius, Lithuania) overnight. DNA fragments were resolved by electrophoresis in 1% pulsed-field gel electrophoresis (PFGE)-certified agarose (Bio-Rad) in 0.5× TBE (45 mM Tris, 45 mM boric acid, and 1 mM EDTA) buffer by use of a CHEF DR III (Bio-Rad, Munich, Germany) apparatus. Electrophoresis was carried out at a constant voltage of 6 V/cm at 14°C for 23 h, with a 120° angle and a pulse time of 1 to 25 s. A lambda DNA ladder (New England BioLabs Inc., Medinova Scientific A/S, Denmark) was used as a molecular size marker.
Amplification of Tn916-derived sequences.
The presence of conjugative transposons of the Tn916-Tn1545 family in transconjugants was investigated with primers Int-FW and Int-FV, targeting the transposon integrase (int) gene, as previously described (15). Primers Tn916-2 and ReverseTetM-2 were used to verify the presence of tet(M) in connection with Tn916 as described by Agersø and coworkers (1). We characterized the genetic organization of the Tn916-like conjugative element harbored by transconjugants by using pairs of primers that enable the amplification of DNA segments spanning the whole of the Tn916 molecule (41). The amplicons obtained were purified and sequenced by cycle extension in an ABI 370 DNA sequencer (Applied Biosystems, Foster City, CA). Sequences obtained were then compared to those in public databases by using the BLAST program (2).
Susceptibility testing by the Etest method.
Individual colonies from Mueller-Hinton (Oxoid) plates were suspended in a sterile glass or plastic tube containing 2 to 5 ml of sterile saline (Oxoid) until a density corresponding to a McFarland standard of 1 (∼3 × 108 CFU/ml) was obtained. The suspension was used to inoculate fresh plates of the same medium with a cotton swab. The agar surface was then allowed to dry for approximately 15 min before application of tetracycline Etest strips (AB Biodisk, Solna, Sweden). Results were recorded after incubation at 28°C for 48 h, following the manufacturer's recommendations.
Statistics.
The reported values are means for 3 to 10 repetitions. Comparison of in vivo transfer frequencies between animals dosed with tetracycline and those without this antibiotic was performed using the Wilcoxon test. One-way analysis of variance was used to examine the potential influence of subinhibitory tetracycline concentrations on transfer frequencies.
RESULTS
In vitro transfer of tetracycline resistance.
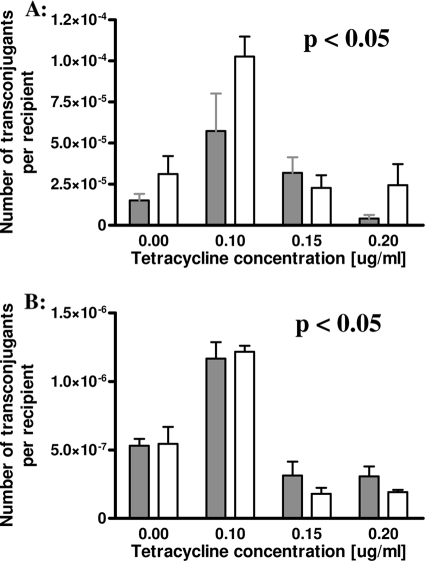
Six tetracycline-resistant L. lactis isolates (Table 1) were tested for the ability to transfer tetracycline resistance genes to L. lactis Bu2-60, E. faecalis JH2-2, B. subtilis YBE01, P. putida KT2442, A. tumefaciens UBAPF2, and E. coli JE2571 by the filter mating approach. Matings with L. lactis Bu2-60 as the recipient strain were successful for four donor strains, IBB160, IBB224, IBB477, and IBB487, at frequencies of about 10−5 transconjugants per recipient, with the highest transfer frequencies found for strains IBB477 and IBB487 (Table 2). Three L. lactis donor strains, IBB224, IBB477, and IBB487, could also transfer their tetracycline resistance determinants to E. faecalis JH2-2, at frequencies of about 10−6 transconjugants per recipient (Table 2). No transconjugants were observed after mating of the L. lactis donor strains with the other recipients (limit of detection, <10−9 transconjugants per recipient). Subinhibitory concentrations of tetracycline in the mating plates resulted in significantly higher (P < 0.05) transfer frequencies of the tetracycline resistance determinants from IBB477 and IBB487 to L. lactis Bu2-60 and E. faecalis JH2-2 when the plates contained 0.1 μg/ml tetracycline, but not when 0.15 or 0.2 μg/ml tetracycline was used (Fig. 1A and B).
TABLE 2.
In vitro conjugal transfer of tetracycline resistance from Lactococcus lactis strains
| Donor | Recipient | Transfer frequency (no. of transconjugants/recipient)a | Transferred gene |
|---|---|---|---|
| IBB160 | L. lactis Bu2-60 | 1.4 × 10−7 (1.43) | tet(S) |
| IBB224 | L. lactis Bu2-60 | 1.3 × 10−5 (0.42) | tet(M) |
| E. faecalis JH2-2 | 1.0 × 10−6 (2.0) | tet(M) | |
| IBB477 | L. lactis Bu2-60 | 5.73 × 10−5 (0.24) | tet(M) |
| E. faecalis JH2-2 | 1.3 × 10−6 (0.98) | tet(M) | |
| IBB487 | L. lactis Bu2-60 | 1.88 × 10−4 (1.16) | tet(M) |
| E. faecalis JH2-2 | 6.47 × 10−6 (0.67) | tet(M) |
Data are average values for 10 different trials, and standard deviations are given in parentheses.
FIG. 1.
(A) Influence of subinhibitory tetracycline concentrations on in vitro conjugal transfer frequency between strains L. lactis IBB477 and L. lactis Bu2-60 (gray bars) as well as between L. lactis IBB487 and L. lactis Bu2-60 (white bars). Each bar represents the average value for five independent experiments. Error bars represent standard deviations. (B) Influence of subinhibitory tetracycline concentrations on in vitro conjugal transfer frequency between strains L. lactis IBB477 and E. faecalis JH2-2 (gray bars) as well as between L. lactis IBB487 and E. faecalis JH2-2 (white bars). Each bar represents the average value for five independent experiments. Error bars represent standard deviations.
Verification and characterization of in vitro transconjugants.
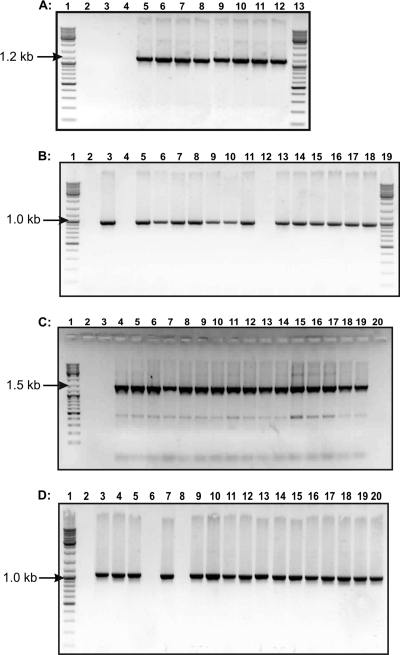
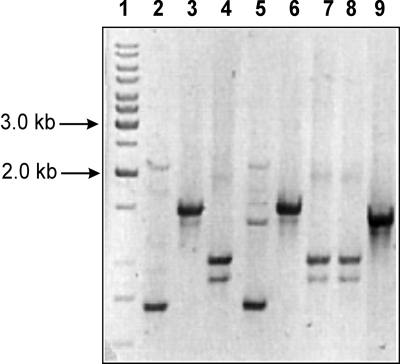
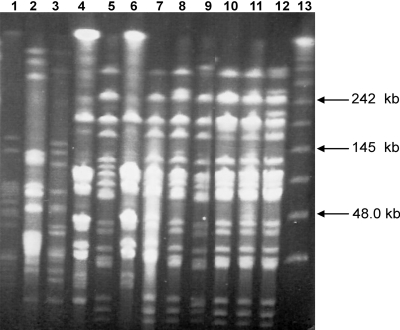
Potential transconjugant colonies (five per mating experiment) were randomly selected from selective plates of independent mating experiments, and DNAs were extracted and analyzed by PCR. They were demonstrated to be of the same species as the recipient, since they were positive by PCR with primers specific for either L. lactis or E. faecalis (see Fig. 4A and B) and they also gave proper profiles (identical with recipient profiles) in RAPD analysis with the M13 primer (Fig. 2). Furthermore, PFGE performed on donors, recipients, and transconjugants revealed that the donors had different genetic profiles, confirming that they were different strains, and the transconjugants had similar profiles to those of the corresponding recipients (Fig. 3). Thus, all of the isolated transconjugants were confirmed to originate from the recipients used.
FIG. 4.
PCR products of donors, recipients, and transconjugants. (A) PCR with primers specific for L. lactis. Lanes 1 and 13, GeneRuler ladder mix size marker (Fermentas); lane 2, E. faecalis JH2-2; lane 3, JH2-477 (in vitro transconjugant of the mating between L. lactis IBB477 and E. faecalis JH2-2); lane 4, JH2-487; lane 5, L. lactis Bu2-60; lane 6, L. lactis IBB160; lane 7, L. lactis IBB224; lane 8, L. lactis IBB477; lane 9, L. lactis IBB487; lane 10, Bu-224 (in vitro transconjugant of the mating between L. lactis IBB224 and L. lactis Bu2-60); lane 11, Bu-477; lane 12, Bu-487. (B) PCR with primers specific for E. faecalis. Lanes 1 and 19, GeneRuler ladder mix size marker (Fermentas); lane 2, L. lactis Bu2-60; lane 3, E. faecalis JH2-2; lane 4, L. lactis IBB224; lane 5, JH2-224; lane 6, JH2-477; lane 7, JH2-487; lane 8, E. faecalis JH2-SS; lane 9, JH2SS-JH2224 (in vitro transconjugant of the mating between JH2-224 and E. faecalis JH2-SS); lane 10, JH2SS-477; lane 11, JH2SS-487; lane 12, L. lactis IBB487; lanes 13 to 18, in vivo transconjugants. (C) PCR with primers specific for tet(M). Lane 1, GeneRuler ladder mix size marker (Fermentas); lane 2, L. lactis Bu2-60; lane 3, E. faecalis JH2-2; lane 4, L. lactis IBB224; lane 5, L. lactis IBB477; lane 6, L. lactis IBB487; lane 7, Bu-224; lane 8, Bu-477; lane 9, Bu-487; lane 10, JH2-224; lane 11, JH2-477; lane 12, JH2-487; lane 13, JH2SS-Bu224; lanes 14 to 19, in vivo transconjugants; lane 20, E. faecalis JH2-SS. (D) PCR with primers specific for the int gene. Lane 1, GeneRuler ladder mix size marker (Fermentas); lane 2, L. lactis Bu2-60; lane 3, L. lactis IBB224; lane 4, L. lactis IBB 477; lane 5, L. lactis IBB487; lane 6, E. faecalis JH2-2; lane 7, Bu-224; lane 8, E. faecalis JH2-SS; lane 9, Bu-477; lane 10, Bu-487; lane 11, JH2-224; lane 12, JH2-477; lane 13, JH2-487; lanes 14 to 20, in vivo transconjugants.
FIG. 2.
RAPD analysis of donors, recipients, and transconjugant strains obtained in conjugation experiments. Lane 1, GeneRuler ladder mix size marker (Fermentas); lane 2, L. lactis IBB477; lane 3, Bu-477 (in vitro transconjugant of the mating between L. lactis IBB477 and L. lactis Bu2-60); lane 4, JH2-477; lane 5, L. lactis IBB487; lane 6, Bu-487; lane 7, JH2-487; lane 8, E. faecalis JH2-2; lane 9, L. lactis Bu2-60.
FIG. 3.
PFGE profiles of donors, recipients, and selected transconjugants. Lane 1, L. lactis IBB224; lane 2, L. lactis IBB477; lane 3, L. lactis IBB487; lane 4, L. lactis Bu2-60; lane 5, E. faecalis JH2-2; lane 6, Bu-477; lanes 7 to 12, in vivo transconjugants; lane 13, lambda DNA ladder (New England BioLabs Inc., Medinova Scientific A/S, Denmark).
Furthermore, the presence of tetracycline resistance genes in the isolated transconjugants was tested. In our recent study (J. Zycka-Krzesinska, J. Boguslawska, and J. Bardowski, unpublished data), the presence of two tetracycline resistance genes, tet(S) and tet(M), was revealed in donor strains IBB224 and IBB477, but in transconjugants Bu-224, Bu-477, JH2-224, and JH2-477 (transconjugants of IBB224 and IBB477 with L. lactis Bu2-60 and E. faecalis JH2-2, respectively), only the tet(M) determinant was detected (Fig. 4C), suggesting that only this gene could be transferred (Table 2). The Bu-160 transconjugant was shown to contain the tet(S) gene [IBB160 contains only tet(S) (Zycka-Krzesinska et al. unpublished data)] by PCR analysis (Table 2). The tet(M) gene has frequently been associated with the Tn916 transposon, and therefore the presence of this transposon in the obtained transconjugants was analyzed. PCR analysis of the presence of the int gene of Tn916-Tn1545 gave positive results (Fig. 4D), and the presence of the tet(M) gene in connection with Tn916 was confirmed by using primers Tn916-2 and ReverseTetM-2 (1) (data not shown). The genetic organization of the Tn916-like conjugative element harbored by the Bu-224, Bu-477, Bu-487, JH2-224, JH2-477, and JH2-487 transconjugants as well as by donor strains was characterized by using pairs of primers that enable amplification of DNA segments spanning the whole transposon (41). The corresponding PCR products obtained were sequenced, and comparison of their nucleotide sequences against those in databases showed them to be 99% identical to Tn916 of E. faecalis DS16 (GenBank accession no. U09422). This ultimately showed that the donor strains and obtained transconjugants harbor a Tn916-like transposon.
Furthermore, the transconjugants were examined for plasmid content, and no plasmid DNA was detected (data not shown) in transconjugants Bu-224, Bu-477, Bu-487, JH2-224, JH2-477, and JH2-487, whereas in Bu-160 plasmid DNA was detected.
Determination of MICs.
Using Etest assay, susceptibilities to tetracycline and streptomycin were compared between donors, recipients, and all verified transconjugants. The MIC for tetracycline was 256 μg/ml for all examined donors and transconjugants, with the exception of JH2-224 (transconjugant of IBB224 and E. faecalis JH2-2), where the MIC was only 125 μg/ml. The MIC for streptomycin for the L. lactis Bu2-60 recipient strain and its transconjugants was 1,024 μg/ml. Both recipient types, lactococcal and enterococcal, were susceptible to tetracycline, and the MIC was <1 μg/ml (18).
In vivo conjugation experiments in germfree rats.
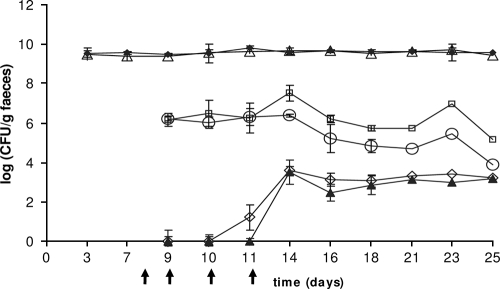
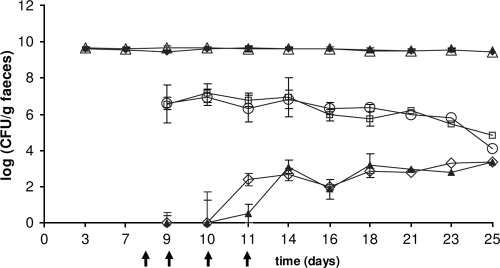
Donor strains L. lactis IBB477 and IBB487 were used to analyze the ability to transfer tetracycline resistance to the recipient strain E. faecalis JH2-2 in the gastrointestinal tract of gnotobiotic rats. The recipient strain as well as the donor strains readily colonized the guts of germfree rats, and total E. faecalis viable cell counts were approximately 5 × 109 CFU/g (wet weight) of feces and were stably maintained throughout the whole experiment (Fig. 5 and 6). The donors were introduced 1 week after the recipient and were administered to the animals each day from day 8 to day 11. The numbers of donor bacteria of strains IBB477 and IBB487 were between 105 and 107 CFU/g of feces and remained 2 to 4 orders of magnitude lower than the number of recipients; additional doses of the donors (received by animals on days 9 to 11) did not have any significant effect on their counts. Subtherapeutic treatment (dose below clinical treatment) with tetracycline did not have any detectable inhibitory effect on the number of recipients (Fig. 5 and 6).
FIG. 5.
Viable cell counts of recipient strain E. faecalis JH2-2 (closed diamonds), E. faecalis from rats dosed with tetracycline (open triangles), donor strain IBB477 (open circles), IBB477 from rats dosed with tetracycline (open squares), transconjugants isolated for tetracycline resistance (closed triangles), and transconjugants isolated from animals dosed with tetracycline (open diamonds). The arrows indicate times of donor administration. Each point represents the average for data from four animals dosed with the donor or four rats dosed with the donor and tetracycline. Error bars represent standard deviations.
FIG. 6.
Viable cell counts of recipient strain E. faecalis JH2-2 (closed diamonds), E. faecalis from rats dosed with tetracycline (open triangles), donor strain IBB487 (open circles), IBB487 from rats dosed with tetracycline (open squares), transconjugants isolated for tetracycline resistance (closed triangles), and transconjugants isolated from animals dosed with tetracycline (open diamonds). The arrows indicate times of donor administration. Each point represents the average for data from four animals dosed with the donor or three rats dosed with the donor and tetracycline. Error bars represent standard deviations.
Transconjugants were detected in fecal samples 3 days after introduction of the donors. The numbers of transconjugant cells for both analyzed mating pairs were comparable and reached stable levels of approximately 102 to 104 CFU/g feces after a few days, and no major changes in CFU per gram occurred during the rest of the time of the experiment. Tetracycline dosed concomitantly with the donor did not have any significant effect on the number of transconjugants, but transconjugants were observed earlier in animals dosed with the antibiotic (Fig. 5 and 6).
Enumeration of recipients, donors, and transconjugants in all examined segments of the gastrointestinal system after sacrifice showed that the intestinal distributions of recipients, donors, and transconjugants were relatively similar between the different rats, and also between those inoculated with different donor strains (Table 3). In all intestinal segments, the numbers of recipients were higher than the numbers of donors, which were again higher than the numbers of transconjugants, while lower bacterial concentrations were found in the upper (duodenum and ileum) than in the lower (cecum and colon) intestinal segments (Table 3).
TABLE 3.
Intestinal distributions of recipients, donors, and transconjugants
| Rat no. (treatment) | Bacteria | Bacterial concn (log CFU/g)a
|
||
|---|---|---|---|---|
| Duodenum | Ileum | Cecum | ||
| 3 (IBB477) | Donor | <LD | 3.85 | 3.70 |
| Recipient | <LD | 7.46 | 9.38 | |
| Transconjugants | <LD | 3.82 | 2.77 | |
| 7 (IBB477 + tetracycline) | Donor | <LD | 4.62 | 4.42 |
| Recipient | 6.30 | 8.15 | 8.68 | |
| Transconjugants | <LD | 3.48 | 2.30 | |
| 13 (IBB487) | Donor | <LD | 4.36 | 4.07 |
| Recipient | 5.74 | 8.32 | 9.49 | |
| Transconjugants | <LD | 3.53 | 3.64 | |
| 17 (IBB487 + tetracycline) | Donor | 3.83 | 4.41 | 4.36 |
| Recipient | 7.24 | 8.44 | 9.46 | |
| Transconjugants | 3.71 | 3.93 | 3.87 | |
<LD, below the limit of detection.
Verification of in vivo transconjugants by PCR.
DNAs from five transconjugants from each of three animals were isolated in order to verify the obtained transconjugants. All analyzed isolates were confirmed to be true transconjugants in a similar way to that for the transconjugants obtained from the in vitro experiments. Specific PCRs also confirmed that all obtained in vivo transconjugants contained complete copies of a Tn916-like transposon (Fig. 4D).
Secondary transfer of tetracycline resistance from transconjugants.
In order to determine whether the tet(M) gene present in the Tn916-like transposon in the Bu-224, Bu-477, Bu-487, JH2-224, JH2-477, and JH2-487 transconjugants (in vitro transconjugants), the TRC 1 and TRC 2 transconjugants (in vivo transconjugants of IBB477 and E. faecalis JH2-2), and the TRC 3 and TRC 4 transconjugants (in vivo transconjugants of IBB487 and E. faecalis JH2-2) was able to be retransferred to another recipient, a filter mating experiment was performed. E. faecalis JH2-SS, which is resistant to streptomycin and spectinomycin, was used as a recipient. Transfer was observed from transconjugants Bu-224, Bu-477, Bu-487, and JH2-477 as well as from transconjugants from the in vivo study. Transfer frequencies ranged from 10−5 to 10−8 transconjugants per recipient (Table 4).
TABLE 4.
Secondary transfer of antibiotic resistance genes from transconjugants to E. faecalis JH2-SS
| Donor straina | Frequency (no. of transconjugants/recipient)b |
|---|---|
| Bu-224 | 2.4 × 10−5 (0.8) |
| Bu-477 | 7.8 × 10−7 (3.2) |
| Bu-487 | 6.4 × 10−6 (2.4) |
| JH2-224 | ND |
| JH2-477 | 7.5 × 10−8 (0.1) |
| JH2-487 | 1.0 × 10−7 (0.66) |
| TRC 1 (IBB477) | 1.2 × 10−8 (1.5) |
| TRC 2 (IBB477) | 1.4 × 10−8 (0.9) |
| TRC 3 (IBB487) | 3.6 × 10−8 (4.2) |
| TRC 4 (IBB487) | 1.0 × 10−8 (0.4) |
TRC 1 and TRC 2 are in vivo transconjugants of IBB477 and E. faecalis JH2-2; TRC 3 and TRC 4 are in vivo transconjugants of IBB487 and E. faecalis JH2-2.
ND, not detected (transfer frequency of <10−9 per recipient cell). Average values for five or three (in the case of secondary transfer from in vivo transconjugants TRC 1, 2, 3, and 4) different trials are given, with standard deviations in parentheses.
DISCUSSION
Genetic studies have identified the presence of transposable elements within L. lactis strains, which are often metabolic plasmids that code for the ability to ferment carbohydrates, production of proteinases, reduced sensitivity to bacteriophages, and production of resistance to bacteriocins (36, 39, 52, 53). Conjugative transposons such as Tn5276 (42) and Tn5307 (9), coding for nisin biosynthesis and sucrose fermentation, respectively, have also been described.
In this work, the capability of intra- and interspecies conjugal transfer of tetracycline resistance determinants from L. lactis was analyzed. The filter mating experiments described in this study demonstrated that transfer of antibiotic resistance genes among LAB can occur at high frequencies under laboratory conditions of intimate cell-to-cell contact.
Transfer of the tet(S) gene from L. lactis IBB160 and of tet(M) from L. lactis IBB224, IBB477, and IBB487 donor strains was observed. The tet(S) gene harbored by donor strains IBB224 and IBB477 was neither transferred alone nor cotransferred with tet(M) by the filter mating conjugation process to any of the analyzed recipient strains. However, this finding does not exclude the possibility that this determinant is transferable to other recipients or under different mating conditions.
The tet(M) gene has often been associated with Tn916, a highly transmissible transposon. Until recently, it was suggested that L. lactis is unique among other hosts of Tn916 because the transposon can enter and insert into L. lactis following conjugative transposition from another species but is not able to mediate its own transfer from L. lactis to another host (8). It was explained that this lack of conjugative transposition in L. lactis is due to the fact that Tn916 is unable to excise from the donor DNA and is therefore unable to undergo conjugative transposition. It was also postulated that L. lactis strains are missing host factors directly involved in transcription of genes required for conjugal transfer of the excised transposon (35). In contrast, in this work the transfer of Tn916-like elements to L. lactis Bu2-60 and E. faecalis JH2-2 from several L. lactis donors, both in vitro and in vivo, was indisputably observed, for the first time. Our work sheds new light on Tn916 transposition, and further investigation of the transfer of this transposon from donor strains L. lactis IBB224, IBB477, and IBB487 may result in a better understanding of this process. Although similar results were recently presented by Florez et al. (19), they demonstrated only in vitro transfer of Tn916 from L. lactis strains isolated from a Spanish traditional starter-free cheese. The gastrointestinal tract is a very hostile environment, and in vitro transfer of antibiotic resistance determinants cannot be extrapolated to the ability to transfer these determinants in vivo. Thus, it was essential in our studies to verify experimentally if the examined Tn916-like transposon could be transferred in the gastrointestinal tract. To verify this hypothesis, germfree rats were used. In this study, in vivo transfer of the tet(M) gene from L. lactis IBB477 and IBB487 was observed. The numbers of transconjugants for both analyzed mating pairs reached stable levels of approximately 102 to 104 CFU g−1 feces and remained stable after initial formation in all groups of animals. The number of transconjugants could be dependent on the following three events: primary transfer of tet(M) determinants from donor strains to recipient strains, secondary transfer from transconjugants to recipients, and multiplication of transconjugants. It is difficult to assess the potential contribution of each of these events, but it is obvious that primary transfer of the tet(M) determinant from donor strains to recipient strains is the main source of transconjugants, especially because of high total donor counts (105 to 107 CFU/g of feces), which were quite stably maintained throughout the whole experiment. However, it is also possible that transconjugants could be donors in secondary transfers. These results were also supported by in vitro retransfer experiments in which the ability of the in vivo transconjugants to facilitate secondary transfer of the tet(M) determinants to E. faecalis JH2-SS was demonstrated. Thus, in our studies, in vivo conjugal transfer of the tet(M) gene from L. lactis donor strains was also observed. In other studies, no secondary conjugal transfer from transconjugants was detected (5, 29).
The subtherapeutic administration of tetracycline to rats was intended to promote the spread of antibiotic resistance genes, as this effect has been reported earlier (5, 34, 47). However, tetracycline stimulation of conjugal transfer was not observed in our study. Nevertheless, transconjugants were observed earlier in animals dosed with this antibiotic. Tetracycline did not have an influence on the number of transconjugants or donors. It is possible that the subtherapeutic concentration of tetracycline was too low. This assumption is supported by previous findings (5) showing that the concentration of bioavailable tetracycline within the bacterial growth habitat of the intestine represented only ca. 0.4% of the intake concentration of the antibiotic.
An interesting observation was that the two donor strains used in this study showed the ability to persist efficiently in the gastrointestinal tract of the rat, and additional doses of the donors did not have any significant effect on their counts. In contrast, it has been shown previously that L. lactis strains are quickly eliminated from the gastrointestinal tract (25, 31).
To our knowledge, this is the first study showing in vivo transfer of a Tn916-like transposon from L. lactis isolates to E. faecalis. Only a limited number of studies have investigated in vivo antibiotic resistance transfer of pAMβ1 plasmid from L. lactis donors to the human gastrointestinal microbiota (54) or to mouse intestinal bacteria (27). In vivo transfer of the Tn916 transposon was observed previously in the intestinal tract, but from E. faecalis donor strains (5).
Simultaneously, transfer of Tn916 to many gram-positive and gram-negative bacteria, including Listeria monocytogenes and E. coli, was confirmed (6), but in our study no conjugal transfer of the transposon to B. subtilis or gram-negative recipients was observed. Discrepancies between our work and previous studies could be associated with many factors, such as improper recipient strains or inadequate mating techniques.
The ability to transfer tetracycline resistance from L. lactis donor strains to E. faecalis in the gastrointestinal tract of rats suggests that LAB might serve as a reservoir of resistance genes for human digestive microbiota. However, one should bear in mind that germfree animal models, like the one used in the present study, offer a less competitive environment than that found in conventional animals, and therefore the ability to transfer tetracycline determinants from L. lactis isolates in vivo cannot be conveyed to a normal intestine. To really assess the ability of L. lactis wild-type strains to transfer antibiotic resistance, further investigations should be performed, using animals with a normal microbiota.
Acknowledgments
We thank Anne Ørngren and her department for handling of the animals. We are also grateful for the excellent technical assistance given by Bodil Madsen and Kate Vibefeldt.
This work was carried out in the framework of the EC-funded project Assessment and Critical Evaluation of Antibiotic Resistance Transferability in the Food Chain (ACE-ART; grant CT-2003-506214) of the 6th Framework Program.
Footnotes
Published ahead of print on 7 August 2009.
REFERENCES
- 1.Agersø, Y., L. B. Jansen, M. Givskov, and M. C. Roberts. 2002. The identification of a tetracycline resistance gene tet(M), on a Tn916-like transposon, in the Bacillus cereus group. FEMS Microbiol. Lett. 214:251-256. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, D. G., and L. L. McKay. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andremont, A. 2003. Commensal flora may play key role in spreading antibiotic resistance. ASM News 69:601-607. [Google Scholar]
- 5.Bahl, M. I., S. J. Sorensen, L. H. Hansen, and T. R. Licht. 2004. Effect of tetracycline on transfer and establishment of the tetracycline-inducible conjugative transposon Tn916 in the guts of gnotobiotic rats. Appl. Environ. Microbiol. 70:758-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertram, J., M. Stratz, and P. Durre. 1991. Natural transfer of conjugative transposon Tn916 between gram-positive and gram-negative bacteria. J. Bacteriol. 173:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradley, D. E., D. Taylor, and D. R. Cohen. 1980. Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K-12. J. Bacteriol. 143:1466-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bringel, F., G. L. Van Alstine, and J. R. Scott. 1991. A host factor absent from Lactococcus lactis subspecies lactis MG1363 is required for conjugative transposition. Mol. Microbiol. 5:2983-2993. [DOI] [PubMed] [Google Scholar]
- 9.Broadbent, J. R., W. E. Sandine, and J. K. Kondo. 1995. Characteristics of Tn5307 exchange and intergeneric transfer of genes associated with nisin production. Appl. Microbiol. Biotechnol. 44:139-146. [DOI] [PubMed] [Google Scholar]
- 10.Caparon, M. G., and J. R. Scott. 1989. Excision and insertion of the conjugative transposon Tn916 involves a novel recombination mechanism. Cell 59:1027-1034. [DOI] [PubMed] [Google Scholar]
- 11.Christensen, B. B., C. Sternberg, J. B. Andersen, L. Eberl, S. Moller, M. Givskov, and S. Molin. 1998. Establishment of new genetic traits in a microbial biofilm community. Appl. Environ. Microbiol. 64:2247-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clewell, D. B., S. E. Flannagan, and D. D. Jaworski. 1995. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 13.Danielsen, M. 2002. Characterisation of the tetracycline resistance plasmid pMD5057 from Lactobacillus plantarum 5057 reveals a composite structure. Plasmid 48:98-103. [DOI] [PubMed] [Google Scholar]
- 14.Davison, J. 1999. Genetic exchange between bacteria in the environment. Plasmid 42:73-91. [DOI] [PubMed] [Google Scholar]
- 15.Doherty, N., K. Trzcinski, P. Pickerill, P. Zawadzki, and C. Dowson. 2000. Genetic diversity of the tet(M) gene in tetracycline-resistant clonal lineages of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutka-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.FEEDAP Panel. 2005. Opinion of the scientific panel on additives and products or substances used in animal feed on the updating of the criteria used in assessment of bacteria for resistance to antibiotics of human or veterinary importance. EFSA J. 233:1-1. [Google Scholar]
- 18.Florez, A. B., M. Danielsen, J. Korhonen, J. Zycka, A. von Wright, J. Bardowski, and B. Mayo. 2007. Antibiotic survey of Lactococcus lactis strains to six antibiotics by Etest, and establishment of new susceptibility-resistance cut-off values. J. Dairy Res. 30:1-7. [DOI] [PubMed] [Google Scholar]
- 19.Florez, A. B., M. S. Ammor, and B. Mayo. 2008. Identification of tetM in two Lactococcus lactis strains isolated from a Spanish traditional starter-free cheese made of raw milk and conjugative transfer of tetracycline resistance to lactococci and enterococci. Int. J. Food Microbiol. 121:189-194. [DOI] [PubMed] [Google Scholar]
- 20.Franke, A. E., and D. B. Clewell. 1981. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145:494-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedman, B. M., and R. E. Yasbin. 1993. The genetics and specificity of the constitutive excision repair system of Bacillus subtilis. Mol. Gen. Genet. 190:481-486. [DOI] [PubMed] [Google Scholar]
- 22.Gasson, M. 1990. In vivo genetic systems in lactic acid bacteria. FEMS Microbiol. Rev. 87:43-60. [DOI] [PubMed] [Google Scholar]
- 23.Gevers, D., G. Huys, F. Devlieghere, M. Uyttendaele, J. Debevere, and J. Swings. 2000. Isolation and identification of tetracycline resistant lactic acid bacteria from pre-packed sliced meat products. Syst. Appl. Microbiol. 23:279-284. [DOI] [PubMed] [Google Scholar]
- 24.Gevers, D., M. Danielsen, G. Huys, and J. Swings. 2003. Molecular characterization of tet(M) genes in Lactobacillus isolates from different types of fermented dry sausage. Appl. Environ. Microbiol. 69:1270-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruzza, M., M. Fons, M. F. Ouriet, Y. Duval-Iflah, and R. Ducluzeau. 1994. Study of gene transfer in vitro and in the digestive tract of gnotobiotic mice from Lactococcus lactis strains to various strains belonging to human intestinal biota. Microb. Releases 2:183-189. [PubMed] [Google Scholar]
- 26.Hynes, M. F., R. Simon, and A. Pühler. 1985. The development of plasmid-free strains of Agrobacterium tumefaciens by using incompatibility with a Rhizobium meliloti plasmid to eliminate pAtc58. Plasmid 13:99-105. [DOI] [PubMed] [Google Scholar]
- 27.Igimi, S., C. H. Ryu, S. H. Park, Y. Sasaki, T. Sasaki, and S. Kumagai. 1996. Transfer of conjugative plasmid pAMβ1 from Lactococcus lactis to mouse intestinal bacteria. Lett. Appl. Microbiol. 23:31-35. [DOI] [PubMed] [Google Scholar]
- 28.Jacobs, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobsen, L., A. Wilcks, K. Hammer, G. Huys, D. Gevers, and S. R. Andersen. 2007. Horizontal transfer of tetM and ermB resistance plasmids from food strains of Lactobacillus plantarum to Enterococcus faecalis JH2-2 in the gastrointestinal tract of gnotobiotic rats. FEMS Microbiol. Ecol. 59:158-166. [DOI] [PubMed] [Google Scholar]
- 30.Jensen, L. B. 1998. Differences in the occurrence of two base pair variants of Tn1546 from vancomycin-resistant enterococci from humans, pigs, and poultry. Antimicrob. Agents Chemother. 42:2463-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klijn, N., A. H. Weekamp, and W. M. De Vos. 1995. Genetic marking of Lactococcus lactis shows its survival in the human gastrointestinal tract. Appl. Environ. Microbiol. 61:2771-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lampkowska, J., L. Feld, A. Monaghan, N. Toomey, S. Schjorring, B. Jackobsen, H. van der Voet, S. R. Andersen, D. Bolton, H. Aarts, K. A. Krogfelt, A. Wilcks, and J. Bardowski. 2008. A standardized conjugation protocol to assess antibiotic resistance transfer between lactococcal species. Int. J. Food Microbiol. 127:172-175. [DOI] [PubMed] [Google Scholar]
- 33.Levy, S. B., and B. Marshall. 2004. Antibacterial resistance worldwide: causes, challenges and responses. Nat. Med. 10:S122-S129. [DOI] [PubMed] [Google Scholar]
- 34.Licht, T. R., C. Struve, B. B. Christensen, R. L. Poulsen, S. Molin, and K. A. Krogfelt. 2003. Evidence of increased spread and establishment of plasmid RP4 in the intestine under sub-inhibitory tetracycline concentrations. FEMS Microbiol. Ecol. 44:217-223. [DOI] [PubMed] [Google Scholar]
- 35.Marra, D., G. Smith, and J. R. Scott. 1999. Excision of the conjugative transposon Tn916 in Lactococcus lactis. Appl. Environ. Microbiol. 65:2230-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neve, H., A. Geis, and M. Tauber. 1984. Conjugal transfer and characterization of bacteriocin plasmids in group N (lactic acid) streptococci. J. Bacteriol. 157:833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng, L. K., I. Martin, M. Alfa, and M. Mulvey. 2001. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 15:209-215. [DOI] [PubMed] [Google Scholar]
- 38.O'Driscoll, J., F. Glynn, G. F. Fitzgerald, and D. van Sinderen. 2006. Sequence analysis of the lactococcal plasmid pNP40: a mobile replicon for coping with environmental hazards. J. Bacteriol. 188:6629-6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Sullivan, L., M. P. Ryan, R. P. Ross, and C. Hill. 2003. Generation of food-grade lactococcal starters which produce the lantibiotics lacticin 3147 and lacticin 481. Appl. Environ. Microbiol. 69:3681-3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perreten, V., F. Schwartz, L. Cresta, M. Boeglin, G. Dasen, and M. Teuber. 1997. Antibiotic resistance spread in food. Nature 389:801-802. [DOI] [PubMed] [Google Scholar]
- 41.Poyart, C., J. Celli, and P. Trieu-Cuot. 1995. Conjugative transposition of Tn916-related elements from Enterococcus faecalis to Escherichia coli and Pseudomonas fluorescens. Antimicrob. Agents Chemother. 39:500-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poyart, C., G. Quesne, P. Acar, P. Berche, and P. Trieu-Cuot. 2000. Characterization of the Tn916-like transposon Tn3872 in a strain of Abiotrophia defectiva (Streptococcus defectivus) causing sequential episodes of endocarditis in a child. Antimicrob. Agents Chemother. 52:1285-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rauch, P. J., and W. M. De Vos. 1992. Characterization of the novel nisin-sucrose conjugative transposon Tn5276 and its insertion in Lactococcus lactis. J. Bacteriol. 174:1280-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rice, L. B. 1998. Tn916 family conjugative transposons and dissemination of antimicrobial resistance determinants. Antimicrob. Agents Chemother. 42:1871-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rossetti, L., and G. Giraffa. 2005. Rapid identification of dairy lactic acid bacteria by M13-generated, RAPD-PCR fingerprint databases. J. Microbiol. Methods 63:135-144. [DOI] [PubMed] [Google Scholar]
- 46.Salama, M., and S. Giovannoni. 1991. Development and application of oligonucleotide probes for identification of Lactococcus lactis subsp. cremoris. Appl. Environ. Microbiol. 57:1313-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salyers, A. A., and N. B. Shoemaker. 1996. Resistance gene transfer in anaerobes: new insights, new problems. Clin. Infect. Dis. 23(Suppl. 1):S36-S43. [DOI] [PubMed] [Google Scholar]
- 48.Scott, J. R., F. Bringel, D. Marra, G. Van Alstine, and C. K. Rudy. 1994. Conjugative transposition of Tn916: preferred targets and evidence for conjugative transfer of a single strand and for a double-stranded circular intermediate. Mol. Microbiol. 11:1099-1108. [DOI] [PubMed] [Google Scholar]
- 49.Teuber, M., L. Meile, and F. Schwarz. 1999. Acquired antibiotic resistance in lactic acid bacteria from food. Antonie van Leeuwenhoek 76:115-137. [PubMed] [Google Scholar]
- 50.Teuber, M., and V. Perreten. 2000. Role of milk and meat products as vehicles for antibiotic-resistant bacteria. Acta Vet. Scand. 93(Suppl.):75-87. [PubMed] [Google Scholar]
- 51.Teuber, M., F. Schwarz, and V. Perreten. 2003. Molecular structure and evolution of the conjugative multiresistance plasmid pRE25 of Enterococcus faecalis isolated from a raw-fermented sausage. Int. J. Food Microbiol. 88:325-329. [DOI] [PubMed] [Google Scholar]
- 52.Tomich, P. K., F. Y. An, and D. B. Clewell. 1980. Properties of erythromycin-inducible transposon Tn916 in Streptococcus faecalis. J. Bacteriol. 141:1366-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trotter, M., O. E. McAuliffe, G. F. Fitzgerald, C. Hill, R. P. Ross, and A. Coffey. 2004. Variable bacteriocin production in the commercial starter Lactococcus lactis DPC4275 is linked to the formation of the cointegrate plasmid pMRC02. Appl. Environ. Microbiol. 70:34-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tuohy, K., M. Davies, C. Rumney, M. R. Adams, and I. R. Rowland. 2002. Monitoring transfer of recombinant and nonrecombinant plasmids between Lactococcus lactis strains and members of the human gastrointestinal microbiota in vivo--impact of donor cell number and diet. J. Appl. Microbiol. 93:954-964. [DOI] [PubMed] [Google Scholar]
- 55.Wang, H. H., M. Manuzon, M. Lehman, K. Wan, H. Luo, T. E. Wittum, A. Yoursef, and L. O. Bakaletz. 2005. Food commensal microbes as a potentially important avenue in transmitting antibiotic resistance genes. FEMS Microbiol. Lett. 55:226-231. [DOI] [PubMed] [Google Scholar]
- 56.Wegener, H. C., M. Madsen, N. Nielsen, and F. M. Aarestrup. 1997. Isolation of vancomycin resistant Enterococcus faecium from food. Int. J. Food Microbiol. 35:57-66. [DOI] [PubMed] [Google Scholar]
- 57.Wilcks, A., A. H. A. M. van Hoek, R. G. Joosten, B. B. L. Jackobsen, and H. J. M. Aarts. 2004. Persistence of DNA studied in different ex vivo and in vivo rat model simulating the human gut situation. Food Chem. Toxicol. 42:493-502. [DOI] [PubMed] [Google Scholar]