Abstract
Probiotics are live microorganisms which, when administered in adequate amounts, confer a health benefit on the host. Therefore, probiotic strains should be able to survive passage through the human gastrointestinal tract. Human gastrointestinal tract survival of probiotics in a low-fat spread matrix has, however, never been tested. The objective of this randomized, double-blind, placebo-controlled human intervention study was to test the human gastrointestinal tract survival of Lactobacillus reuteri DSM 17938 and Lactobacillus rhamnosus GG after daily consumption of a low-fat probiotic spread by using traditional culturing, as well as molecular methods. Forty-two healthy human volunteers were randomly assigned to one of three treatment groups provided with 20 g of placebo spread (n = 13), 20 g of spread with a target dose of 1 × 109 CFU of L. reuteri DSM 17938 (n = 13), or 20 g of spread with a target dose of 5 × 109 CFU of L. rhamnosus GG (n = 16) daily for 3 weeks. Fecal samples were obtained before and after the intervention period. A significant increase, compared to the baseline, in the recovery of viable probiotic lactobacilli in fecal samples was demonstrated after 3 weeks of daily consumption of the spread containing either L. reuteri DSM 17938 or L. rhamnosus GG by selective enumeration. In the placebo group, no increase was detected. The results of selective enumeration were supported by quantitative PCR, detecting a significant increase in DNA resulting from the probiotics after intervention. Overall, our results indicate for the first time that low-fat spread is a suitable carrier for these probiotic strains.
The human intestinal microflora or microbiota constitutes a metabolically active microbial environment. This community is relatively stable in the guts of healthy individuals (20). Some of the microbial groups harbor species that are potentially harmful, whereas others, such as the bifidobacteria and lactobacilli, are regarded as beneficial (8). Specific members of the genera Lactobacillus and Bifidobacterium are being applied in functional foods as probiotics (25). Probiotics are live microorganisms which, when administered in adequate amounts, confer a health benefit on the host (9). The current scientific consensus is that probiotics should be alive to exert their beneficial effect in the human gastrointestinal (GI) tract (6). Consequently, probiotics should remain alive in the product, such that the daily effective dose per serving is still present at the end of the shelf life (14). Food matrices, production processes, or product usages that involve heating can affect the viability of probiotics (24).
Typically, those members selected for probiotic application are chosen for their resistance to passage through the upper GI tract and thus are able to transiently colonize the gut (25). Human GI tract survival of probiotics should lead to shedding of live cells in fecal samples. GI tract survival is, however, dependent on both the strain and the food matrix involved (27). Fecal recovery of several probiotic strains has been demonstrated in different food matrices, including fermented milk and yoghurt (10, 26, 29), fruit drinks (21), a cereal bar (22), supplements (13, 17, 27), and infant formula (29).
For this study, we have selected two well-established probiotic strains to test the suitability of a low-fat spread as a probiotic carrier, namely, Lactobacillus reuteri DSM 17938 (BioGaia, Sweden) and Lactobacillus rhamnosus GG (ATCC 53103; Valio, Finland). L. reuteri DSM 17938 was derived from L. reuteri ATCC 55730 by curing of two plasmids harboring antibiotic resistance genes (23). A series of in vitro experiments confirmed the retention of the functional properties of the daughter strain, as no differences in colony morphology, fermentation patterns, production of reuterin, generation time, mucus-binding ability, or tolerance to bovine bile were found between L. reuteri ATCC 55730 and DSM 17938. The daughter strain is somewhat more resistant to low pH and grows to a higher density in vitro (23). Several studies have been published which provide data on the survival of L. reuteri ATCC 55730 in the human GI tract at doses of 4 × 108 to 1 × 1010 CFU/day in freeze-dried matrices and chewable tablets (32-34). Furthermore, L. reuteri DSM 17938 was demonstrated to survive human GI tract passage in the same way as L. reuteri ATCC 55730 (23).
L. rhamnosus GG has been isolated from a healthy human intestinal flora by Goldin et al. (10). L. rhamnosus GG is relatively resistant to acid and bile, adheres in vitro to epithelial cells, and can produce an antimicrobial substance (10, 15). A wide range of studies have been published which provide data on the survival of L. rhamnosus GG in the human GI tract (3, 4, 10, 18, 19, 27-30), as well as transient colonization of the intestinal microbiota in healthy adults in various formats, including freeze-dried powder, capsules, and tablets or via fermented milk drinks, yoghurt, or fruit juice. Saxelin et al. (28) evaluated the dose-response effect of orally administered L. rhamnosus GG in powder form on fecal colonization in healthy adults, which indicated that consumption of approximately 1010 to 1011 CFU/day was required to reach detectable levels in fecal samples from volunteers. This was also the case when L. rhamnosus GG was administered in gelatin capsules (29). Additionally, Saxelin et al. (27) observed that milk, but possibly also other protective compounds in food, can improve the survival of L. rhamnosus GG. Fecal recovery of L. rhamnosus GG in milk-based products was shown at dose levels of around 2 × 109 CFU/day.
It is, however, not known whether probiotics can survive passage through the human GI tract after the consumption of a low-fat spread. The objective of this randomized, double-blind, placebo-controlled human intervention study was therefore to test the human GI tract survival of L. reuteri DSM 17938 and L. rhamnosus GG after daily consumption of a low-fat probiotic spread by using traditional culturing, as well as molecular methods. The primary outcome parameter of this study was a significant change from the baseline in the number of probiotic bacteria of the respective strains in fecal samples.
MATERIALS AND METHODS
Subjects.
Subjects were recruited between September 2007 and January 2008. Invitation letters were sent to inhabitants of Vlaardingen (The Netherlands) and the surrounding area, and an advertisement was placed in the Erasmus University magazine. The main inclusion criteria were that the volunteers be healthy men and women 18 to 55 years old at screening with no reported medical conditions that could affect the study outcome (as judged by the study physician), have a body mass index (BMI) between 20 and 30 kg/m2, consume animal products at least twice a week, habitually consume spreads, and pass stool at least every 2 days. The main exclusion criteria were the use of antibiotics, reported dietary restrictions, alcohol abuse, very intense sporting activities, smoking, pregnancy or lactation, unstable weight, reported participation in another biomedical study 3 months before the start of this study, and reported participation in night shift work 1 month prior to or during the study. All in- and exclusion criteria were assessed by means of a screening questionnaire. Subjects who fulfilled all of the criteria were invited to participate in this study. Before the start of the run-in period, informed consent was obtained and self-reported BMIs were verified by measuring weight and height with a digital weighing and measuring station (SECA 764; SECA, The Netherlands). The study protocol was approved by the Medical Ethical Committee of Wageningen University, Wageningen, The Netherlands, in September 2007, and the study was conducted according to the WMO (Research Involving Human Subject Act).
Study design.
This study was a randomized, double-blind, placebo-controlled, parallel, three-arm study with a 2-week run-in period and a 3-week intervention period. Forty-seven study subjects were randomly assigned to three treatment groups and provided a low-fat placebo spread, a probiotic spread with at least 1 × 109 CFU of L. reuteri DSM 17938, or a probiotic spread with at least 5 × 109 CFU of L. rhamnosus GG.
Allocation to the three treatment groups was performed by means of computer-generated randomization codes stratified by gender. Two persons not involved in the conduct of the study were responsible for labeling the spread test products with the study subject identifier codes. Research staff and subjects remained blind to the type of treatment throughout the study and data analysis.
During the intervention period, subjects were instructed to consume daily 20 g of one of the three low-fat spread test products, divided into two eating occasions (breakfast and dinner). To test the human GI tract survival of probiotics, two fecal samples were collected, one at the end of the run-in period and one at the end of the intervention period. Subjects who could not deliver a fecal sample within 2 days of the sampling point were excluded from the study. During both the run-in and intervention periods, subjects were instructed to refrain from any other food products specifically claiming to contain prebiotics or probiotics. Subjects were asked to return the empty packages from the test products used, as well as the unused test products, and to register noncompliance with the test product intake and dietary background restrictions in a diary. Compliance with background diet and spread test product intake was checked at the end of the run-in period, approximately 1 week after the start of the intervention period, and at the end of the intervention period by counting the used and unused spread tubs and by checking the diaries. This study was performed at the Unilever Food and Health Research Institute in Vlaardingen, The Netherlands.
Spread test products.
The spread test products were produced and supplied by Unilever and were 20-g portion packs of low (28%)-fat spreads containing L. reuteri DSM 17938 at a target dose of 1 × 109 CFU/daily serving, L. rhamnosus GG at a target dose of 5 × 109 CFU/daily serving, or a placebo spread without any probiotic bacteria. The spread test products were identical in appearance and taste. The composition of the spread test product can be found in Table 1. Both probiotic spread test products were 10-fold overdosed during production to compensate for any potential decline in the viability of the probiotics during transport and storage and to guarantee the required minimal viable count of probiotic bacteria until the end of the intervention period. The spread test products were produced and distributed in two different batches. Subjects were instructed to store the test products at home in the refrigerator. The viable counts of probiotic bacteria in the spread test products were determined directly after production and at several occasions during the intervention period. In addition, the viable counts of probiotic bacteria in samples stored under controlled conditions (4 to 6°C) at the production center were compared to those of samples that were returned by the study subjects and had been stored at home (1 to 8°C).
TABLE 1.
Composition of 100 g of spread test producta
| Component | Amt |
|---|---|
| Total proteins | 3.0 g |
| Carbohydrates | |
| Total | 4.0 g |
| Mono- and disaccharides | 0.5 g |
| Polysaccharides | 3.5 g |
| Fats | |
| Total | 28 g |
| Saturated fatty acids | 9.0 g |
| cis-Monounsaturated fatty acids | 13 g |
| cis-Polyunsaturated fatty acids | 6 g |
| Water | 63.0 g |
| Sodium | 0.2 g |
| Vitamins | |
| A | 800 μg |
| D | 7.5 μg |
| E | 12,000 μg |
Energy, 1,200 kJ or 280 kcal.
Collection and analysis of fecal samples.
Fresh fecal samples were collected at home by defecation into a bag inserted into a bucket and stored in a 750-ml glass jar under anaerobic conditions (Anaerocult mini sachet; Merck). Samples were delivered to the investigator and processed for bacterial enumeration within 4 h of defecation. Ten grams of sample was diluted 10-fold with peptone physiological salt solution (PFZ; Tritium Microbiology, The Netherlands) and homogenized in a stomacher for 30 s by using a stomacher bag with a filter (VWR, The Netherlands). The fecal suspension was used for bacterial enumeration. The cell pellet of a 2-ml aliquot was stored at −20°C for DNA isolation and quantitative PCR (qPCR).
Bacterial enumeration of live lactobacilli in spread test products and fecal samples.
The chemicals used were purchased from Sigma-Aldrich unless indicated otherwise. Determination of the number of live probiotic bacteria in the spread test products was performed in duplicate by the viable plate count method. First, the water phase of the spread was extracted by dilution of ca. 20 g of spread in 90 ml of PFZ, which was heated at 39°C for 20 min and subsequently shaken at ambient temperature for 10 min at 200 rpm. Serial decimal dilutions in PFZ prepared from the extracted water phase of the spreads were mixed vigorously. Subsequently, appropriate dilutions were plated on MRS agar (Merck) and incubated at 37°C for 2 days under anaerobic conditions. The viable plate count of probiotic lactobacilli was expressed in CFU per 20 g of spread.
To detect the number of live probiotic bacteria in fecal samples, selective enumeration was used. Briefly, serial dilutions of fresh fecal samples were plated on modified LAMVAB agar (11) at a pH of 5.7 and supplemented with 0.05% freshly prepared cysteine and 50 mg/liter vancomycin (35). Plates were incubated for 3 days at 37°C under anaerobic conditions. The total number of lactobacilli was determined by counting the resulting colonies. L. reuteri was subsequently identified by detection of reuterin production by a method developed by BioGaia (23). For identification of L. rhamnosus GG, 20 colonies were randomly picked and tested for the inability to ferment lactose on MRS agar with 2% lactose as the sole carbon source supplemented with 100 mg/liter bromocresol purple (35). Lactose-negative colonies were identified as L. rhamnosus GG by colony PCR. Bacterial counts in feces were expressed as the log10 CFU per gram (wet weight) of feces.
Molecular identification and quantification of probiotic bacteria in fecal samples.
DNA isolation from cell pellets from fecal suspensions was done with the stool isolation kit from Qiagen according to the manufacturer's instructions, with the following modifications. One thin-walled PCR tube containing 600 to 650 mg of glass beads (Zirconia/Silica Beads, 0.1 mm; BioSpec Products) was added to the cells resuspended in the lysis buffer. The tubes were subsequently shaken for 45 s at a speed setting of 6 in a Fastprep FP120 (MP Biomedicals). The suspensions obtained were incubated for 5 min at 95°C. Isolated DNA was stored at −20°C until further use. The quantity and purity of the DNA used as a reference were determined by performing gel electrophoresis with a FlashGel system and 1.2% agarose FlashGel cassettes (Lonza) and measuring the DNA concentration with a spectrophotometer (UV mini-1240, UV-Vis spectrophotometer; Shimadzu Corporation) and the required program (Program Pack, UV mini-1240 DNA/PROTEIN; Shimadzu Corporation).
All PCRs were performed on an Applied Biosystems 7500 real-time PCR machine with the 7500 Real-Time PCR System Sequence Detection Software, version 1.3.1 (Applied Biosystems). Assays were performed in 25-μl volumes containing 12.5 μl of POWER SYBR green I PCR Master Mix, forward and reverse primers, and 2.5 μl of a 10-fold dilution of the extracted DNA. L. rhamnosus GG detection and quantification were performed with strain-specific primers Lrhamn1 (5′-CAATCTGAATGAACAGTTGTC-3′) and Lrhamn2 (5′-TATCTTGACCAAACTTGACG-3′) (7) at a final concentration of 0.4 μM. L. reuteri was quantified with species-specific primers L-reu-1 (5′-CAGACAATCTTTGATTGTTTAG-3′) and L-reu-4 (5′-GCTTGTTGGTTTGGGCTCTTC-3′) (31) at a final concentration of 0.9 μM. The amplification program consisted of 1 cycle of 95°C for 10 min and 40 cycles of amplification (95°C for 15 s, 60°C for 1 min), followed by determination of the dissociation curve. The detection limit of each assay was determined by using serial dilutions of the chromosomal DNA of each reference strain.
For quantitative analysis of the threshold cycle, each sample was compared to a standard curve made from serial DNA dilutions of the chromosomal DNA of a reference strain, either L. reuteri ATCC 55730 or L. rhamnosus GG. The number of cells equivalent to 1 μl of reference DNA was calculated by dividing the DNA concentration by the genome weight. The genome weight, in turn, was calculated by multiplying the base pair weight (607.4 g/mol) with the relevant genome size in base pairs and divided by Avogadro's number (6.02 × 1023). The genome sizes used were 2.0 Mb for L. reuteri (5) and 2.4 Mb for L. rhamnosus GG, which is based on the genome size of L. rhamnosus HN001 (16). Results were expressed as log10 cells per gram of fecal sample, taking into account the dilution steps in the DNA isolation method and assuming one genome per cell.
For colony PCR, selected colonies were resuspended in 50 μl of lysis buffer (1% Triton X-100, 20 mM Tris HCl [pH 8.0], 2 mM EDTA [pH 8.0]) and incubated for 10 min at 98°C. Debris was precipitated by centrifugation at maximum speed for 5 min. The supernatant containing DNA (20 μl) was diluted 10-fold in water and stored at −20°C. Colony PCR samples were scored as L. rhamnosus GG when the subsequent PCR displayed a threshold cycle higher than that of the negative control (L. reuteri ATCC 55730) and a dissociation peak between 80 and 84°C.
Statistics.
All statistical analyses were performed with JMP 8.0 for Windows. The Wilcoxon signed-rank test was used to test whether the changes from the baseline of both L. reuteri DSM 17938 and L. rhamnosus GG were significantly different from zero. The Mann-Whitney U test was used to compare the total number of lactobacilli in the L. reuteri DSM 17938 or L. rhamnosus GG group with that in the placebo group. Both the Wilcoxon signed-rank test and the Mann-Whitney U test were performed one sided with an alpha of 0.05 on the log-transformed values of the per-protocol population (n = 42). The statistical analysis of the intention-to-treat population was similar to the per-protocol analysis (data not shown). Differences in age and BMI between the treatment groups were evaluated with the unpaired two-sided Student t test with an alpha of 0.05.
A total of 45 subjects (15 per intervention group) were necessary in order to detect a significant increase of 1.38 × 108 in the number of probiotic bacteria in feces compared to the placebo, assuming a within-subject variance of 0.69 × 1016 CFU/g of feces, a power of 90%, and an alpha of 0.05, according to a one-sided Dunnett test correcting for multiple comparisons.
RESULTS
Study subjects.
A total of 62 subjects were screened, and 57 were invited to participate in the study. Ten subjects dropped out of the study before the start of the run-in period because of illness (n = 3), voluntary withdrawal (n = 4), pregnancy (n = 1), a BMI of >30 kg/m2 (n = 1), or participation in another intervention study (n = 1). Forty-seven subjects started the study. During the study, two more subjects dropped out, one because of medicine use and one because of inability to produce a fecal sample in time. A total of 45 subjects (10 men and 35 women) completed the study. During a blind review of the results, three subjects were excluded from the per-protocol analysis because of noncompliance with the study protocol. The baseline characteristics of the per-protocol population were similar across the three intervention groups (placebo spread, n = 13; spread with L. reuteri DSM 17938, n = 13; spread with L. rhamnosus GG, n = 16) (Table 2). Compliance with spread test product intake was in the range of 95% to 100%. A total of 20 adverse events were reported: 8 during the run-in period and 12 during the intervention period. Of the adverse events in the intervention period, three occurred in the group receiving L. rhamnosus GG, five in the group receiving L. reuteri DSM 17938, and four in the placebo group. The adverse events were headache, common cold, gastroenteritis, dental caries, ankle contusion, and accident. According to the judgment of the study physician, none of the adverse events was related to spread test product intake.
TABLE 2.
Baseline characteristics of the study population
| Intervention group | Total no. of persons | No. of females/males | Mean age (yr) ± SD | Mean BMI (kg/m2) ± SD |
|---|---|---|---|---|
| L. reuteri DSM 17938 | 13 | 10/3 | 45.5 ± 8.9 | 24.8 ± 2.2 |
| L. rhamnosus GG | 16 | 12/4 | 45.3 ± 10.3 | 25.6 ± 2.5 |
| Placebo | 13 | 10/3 | 42.3 ± 10.5 | 26.1 ± 2.7 |
Probiotic bacteria in the spread test products.
All three spread test products were produced and distributed to the study subjects in two different batches. The spread test products in batch 1 were consumed 2 to 3.5 weeks after production, during the first half of the intervention period. The spread test products in batch 2 were consumed 1.5 to 3.5 weeks after production, during the last 2 weeks of the intervention period. No obvious decline in the viable count of probiotic bacteria was observed during the respective consumption periods of the different batches of both probiotic spreads (Fig. 1). In both spreads, the viable count of probiotic bacteria remained above the target level, which was at least 1 × 109 CFU for the spread with L. reuteri DSM 17938 and at least 5 × 109 CFU for the spread with L. rhamnosus GG. The actual viable counts at the time of consumption varied from 5.7 × 109 to 1.0 × 1010 CFU of L. reuteri DSM 17938/20 g of spread and from 3.3 × 1010 to 5.6 × 1010 CFU of L. rhamnosus GG/20 g of spread. The viable counts of probiotic lactobacilli at the end of the intervention in spreads stored under refrigerated conditions (1 to 8°C) at home by the volunteers were in the same range as those stored under controlled conditions (4 to 6°C) at the production center (Table 3).
FIG. 1.
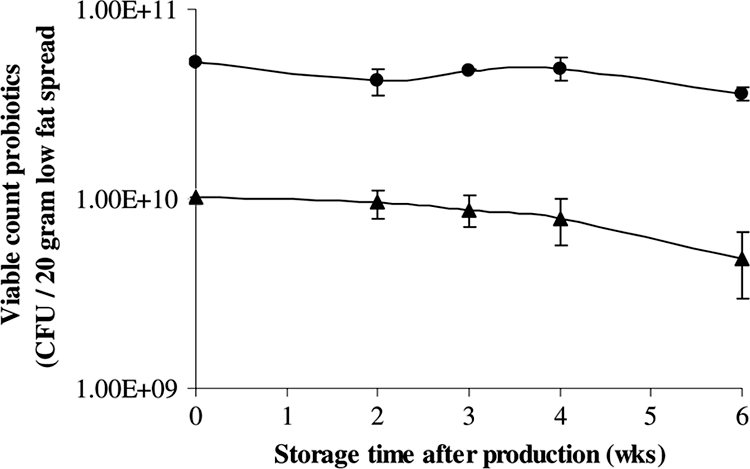
Average viable plate counts of probiotic bacteria in spread test products after production. Symbols: •, L. rhamnosus GG; ▴, L. reuteri.
TABLE 3.
End-of-intervention viable plate counts of probiotics in spread test products stored at home by the volunteers or at the production center
| Storage site | Viable plate counta of:
|
|||
|---|---|---|---|---|
|
L. reuteri DSM 17938
|
L. rhamnosus GG
|
|||
| Batch 1 | Batch 2 | Batch 1 | Batch 2 | |
| Home | 4.6 (± 2.8) × 109 | 3.6 (± 0.5) × 109 | 4.6 (± 1.3) × 1010 | 2.7 (± 0.5) × 1010 |
| Production center | 6.7 (± 2.3) × 109 | 3.1 (± 1.5) × 109 | 6.1 (± 2.5) × 1010 | 2.4 (± 0.3) × 1010 |
The data are average viable counts of lactobacilli in CFU per 20 g of spread ± the standard error of the mean.
Probiotic bacteria in fecal samples.
In Tables 4 and 5, the average fecal microbial count data analyzed by selective enumeration (Table 4) and qPCR (Table 5) are shown. All of the study subjects had detectable viable plate counts of total lactobacilli at the baseline which were on the order of about 4 to 5 log10 CFU/g of feces. Compared to that of the placebo group, the total number of lactobacilli significantly increased from the baseline after 3 weeks of intervention with the L. reuteri DSM 17938-containing spread (P = 0.0406), as well as the L. rhamnosus GG-containing spread (P = 0.0003), to a level of about 6 to 7 log10 CFU/g of feces.
TABLE 4.
Average microbial counts in fecal samples as determined by selective enumeration
| Intervention group (n) |
Lactobacillus counta
|
L. reuteri counta
|
L. rhamnosus GG counta
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Before | After | Change | Before | After | Change | Before | After | Change | |
| Placebo (13) | 4.7 ± 0.5 (13) | 4.8 ± 0.5 (13) | 0.03 ± 0.3 (13) | NDb | ND | ND | 4.4 (1) | 3.8 (1) | −0.6 (1) |
| L. reuteri DSM 17938 (13) | 5.0 ± 0.7 (13) | 6.4 ± 0.4 (13) | 1.4 ± 0.6c (13) | ND | 5.6 ± 0.3 (12) | 2.6 ± 0.7d (12) | 3.9 (1) | ND | ND |
| L. rhamnosus GG (16) | 4.6 ± 0.5 (16) | 6.8 ± 0.2 (16) | 2.2 ± 0.5c (16) | ND | ND | ND | 4.0 ± 1.8 (2) | 6.6 ± 0.3 (16) | 2.9 ± 0.5d (16) |
Data are expressed as the average log10 CFU per gram of feces ± the standard error of the mean, including the number of subjects in parentheses, for whom values were obtained and on which values the averages (log10 CFU) are based.
ND, not detected. The detection limits of the different bacterial counts determined by selective enumeration were as follows: L. reuteri, variable; L. rhamnosus GG, variable; Lactobacillus, <10.
Significantly different from the placebo based on a Mann-Whitney U test (one-sided alpha = 0.05).
Significantly different from the baseline based on a Wilcoxon signed-rank test (one-sided alpha = 0.05).
TABLE 5.
Average microbial counts in fecal samples as determined by qPCR
| Intervention group (n) |
L. reuteri counta
|
L. rhamnosus GG counta
|
||||
|---|---|---|---|---|---|---|
| Before | After | Change | Before | After | Change | |
| Placebo (13) | NDb | ND | ND | ND | ND | ND |
| L. reuteri DSM 17938 (13) | ND | 6.7 ± 0.2 (11) | 1.7 ± 0.2c (11) | ND | ND | ND |
| L. rhamnosus GG (16) | 9.2 (1) | 6.3 ± 0.7 (3) | −0.1 ± 0.8 (3) | ND | 7.3 ± 0.2 (15) | 1.6 ± 0.2c (15) |
Data are expressed as average log10 cells per gram of feces ± the standard error of the mean, including the number of subjects in parentheses, for whom values were obtained and on which values the averages (log10 cells) are based.
ND, not detected. The detection limits of the different bacterial counts determined by qPCR were as follows: L. reuteri, 5.0; L. rhamnosus GG, 5.7.
Significantly different from the baseline based on a Wilcoxon signed-rank test (one-sided alpha = 0.05).
L. reuteri was not detectable in fecal samples from any of the study subjects at the baseline by means of selective enumeration. L. reuteri counts remained undetectable by selective enumeration in the fecal samples from the study subjects in both the placebo and L. rhamnosus GG groups immediately after the 3-week intervention period. On the other hand, 12 out of the 13 study subjects who consumed the probiotic spread with L. reuteri DSM 17938 had detectable viable count numbers of L. reuteri after the 3-week intervention period and, moreover, the log10 CFU counts of L. reuteri in fecal samples increased significantly from the baseline after the 3-week intervention (P = 0.0012).
L. rhamnosus GG was detectable at the baseline by selective enumeration in fecal samples from one study subject in the placebo group, one in the L. reuteri DSM 17938 group, and two in the L. rhamnosus GG intervention group. In both the placebo and L. reuteri DSM 17938 intervention groups, the L. rhamnosus GG counts remained either low or not detectable. On the other hand, selective enumeration of isolates showed that L. rhamnosus GG was present after intervention in all of the study subjects who consumed the spread with L. rhamnosus GG and moreover the log10 CFU counts of L. rhamnosus GG in fecal samples significantly increased from the baseline after the 3-week intervention period (P = 0.0001).
Results of the qPCR analysis were almost comparable to the viable plate count data, indicating detectable levels of L. reuteri in 11 out of 13 volunteers after a 3-week intervention with the L. reuteri DSM 17938-containing spread with a smaller, but also significant, increase from the baseline (P = 0.0005). The qPCR results of the L. rhamnosus GG counts were also comparable to the viable plate count data, indicating detectable levels of L. rhamnosus GG in 15 out of the 16 volunteers after a 3-week intervention with the L. rhamnosus GG-containing spread, with also a somewhat smaller but significant increase from the baseline (P < 0.0001).
In general, values obtained by qPCR were about 1 log10 unit higher than the corresponding viable plate count data, indicating that also dead or viable but not colony-forming lactobacilli were detected by qPCR. The change from the baseline based on the qPCR results was, in general, lower than the plate count data, which was due to the higher detection limit of the qPCR analysis.
The number of subjects who showed a minimum twofold increase in fecal probiotic counts after intervention was around 80% for both interventions and by both detection methods (for L. reuteri DSM 17938, 10/13 [76.9%] by the viable plate count method and 11/13 [84.6%] by qPCR; for L. rhamnosus GG, 14/16 [87.5%] by the viable plate count method and 13/16 [81.3%] by qPCR).
DISCUSSION
Probiotic bacteria are live microorganisms which, when administered in adequate amounts, confer a health benefit on the host (9). Therefore, probiotic strains should remain alive in the product and should survive passage through the human GI tract. In the present double-blind, placebo-controlled human intervention study, we have demonstrated for the first time that both L. reuteri DSM 17938 and L. rhamnosus GG can survive passage through the human GI tract after the consumption of a low-fat probiotic spread. A significant increase in the recovery of viable probiotic bacteria in fecal samples was observed after 3 weeks of daily consumption of a low-fat probiotic spread containing either L. reuteri DSM 17938 or L. rhamnosus GG. Fecal viable plate counts of L. reuteri were on the order of 5.6 log10 CFU/g of feces after 3 weeks of consumption of the L. reuteri DSM 17938 spread, which contained, on average, 5.7 × 109 to 1.0 × 1010 CFU of L. reuteri DSM 17938/20 g of spread. These levels are in line with the results of Rosander et al. (23), who found between 4.2 and 5.2 log10 CFU of L. reuteri/g of wet feces after 14 to 28 days of consumption of ∼8 × 108 CFU of freeze-dried L. reuteri DSM 17938 powder dissolved in cold water. Fecal levels were between 6.2 and 7.3 log10 CFU of L. reuteri when around 6.5 × 1010 CFU of L. reuteri DSM 17938 was consumed for 14 to 28 days (23).
Fecal viable plate counts of L. rhamnosus GG were on the order of 6.6 log10 CFU/g of feces after 3 weeks of consumption of the L. rhamnosus GG spread containing, on average, 3.3 × 1010 to 5.6 × 1010 CFU of L. rhamnosus GG/20 g of spread. These levels are also comparable to the fecal recovery results of L. rhamnosus GG in different food matrices which were previously reported in the literature. Saxelin et al. (28), for example, reported that the mean level of L. rhamnosus GG found in feces after 7 days of consumption of 1010 to 1011 CFU of L. rhamnosus GG as freeze-dried powder varied between 5 and 7 log10 CFU/g of feces. Fecal recovery values of L. rhamnosus GG when administered as fermented milk (1.2 × 1010 CFU) were also on the order of 7 log10 CFU/g of feces after a 7-day supplementation period (27).
In contrast to the studies of Saxelin et al. (27, 28) our results reveal that L. rhamnosus GG colonies were present in fecal samples from 4 out of 42 individuals at the baseline. Lactobacilli and L. rhamnosus species are present in the human diet and belong to the normal microbiota of the human GI tract. According to Ahrné et al. (2) L. rhamnosus can be isolated in 26% of individuals. It is likely that the baseline colonies result from naturally present L. rhamnosus strains that have a genotype similar to that of L. rhamnosus GG. Although it was not reported by the study subjects, we cannot exclude the possibility that the observed colonies are the result of L. rhamnosus GG consumption prior to or during the 2-week run-in period. Fecal L. reuteri levels were less abundant in the study population at the baseline, as only one individual showed a high level of 9.2 log10 cells/g of feces, which was detected by qPCR.
The total number of fecal lactobacilli at the baseline was about 4 to 5 log10 CFU/g of feces in our study. These values are within the expected range of vancomycin-resistant lactobacilli in human feces of 2.5 to 9 log10 CFU/g of feces as described by Hartemink et al. (11, 12) and <2 to 8 log10 CFU/g of wet feces as reported by Ahlroos and Tynkkynen (1). However, our values are lower than the previously reported total fecal lactobacillus counts of about 7 to 8 log10 CFU/g of feces in the studies of Saxelin et al. (28, 29). The difference between the latter studies and our study lies in the selectiveness of the medium used. Saxelin et al. used MRS, while we used MRS with a lower pH and supplemented with vancomycin, which is more selective (11). This may explain why we detected a significant increase in the change from the baseline in the total lactobacillus count after the daily consumption of a low-fat spread with either L. reuteri DSM 17938 or L. rhamnosus GG for a 3-week period, which was not the case in the L. rhamnosus GG intervention studies with healthy adult volunteers of Saxelin et al. (28, 29) and Goldin et al. (10), in which MRS agar with a higher pH and no vancomycin was used.
The fecal L. reuteri and L. rhamnosus GG levels detected by qPCR were about 1 to 2 log10 higher in our study than the fecal levels measured by viable plate counting. This is in line with results of Ahlroos and Tynkkynen (1), who also showed that L. rhamnosus GG levels in feces were 1 to 2 log10 higher when analyzed by qPCR than when analyzed by conventional plate counting on MRS-vancomycin agar. This could be related to the isolation of DNA from not only viable cells but also dead or viable but not colony-forming lactobacilli or from the fact that single colonies do not necessarily originate from single bacterial cells. Overall, our qPCR results reveal trends similar to those revealed by the selective enumeration method and we can therefore conclude that real-time qPCR is a suitable method for strain-specific quantification of probiotic bacteria in human fecal samples following probiotic interventions.
Besides human GI tract survival, probiotic bacteria need to remain alive in the product. We observed no obvious decline in the viable counts of probiotic bacteria in the spread test products throughout the intervention period of this study. Home storage conditions were sufficient to guarantee probiotic survival in the spreads. This indicates that low-fat spreads that are specifically designed to support probiotic survival can be a good alternative food matrix in which to administer live probiotic bacteria.
In conclusion, both L. reuteri DSM 17938 and L. rhamnosus GG can survive passage through the human GI tract after the consumption of a low-fat spread. Furthermore, we have demonstrated that the viable probiotic counts in both spread test products remained stable during the course of the intervention period. Overall, our results indicate for the first time that low-fat spread is a suitable carrier for L. reuteri DSM 17938 and L. rhamnosus GG, supporting the use of low-fat spread as a probiotic product.
Acknowledgments
We gratefully acknowledge the excellent assistance of the following scientists: Marjolein Toonen for fecal microbial sample analysis, Christine van der Swaluw for microbial analysis of spread test products, and Jan-Willem Sanders for microbial analysis of spread test products and critical reading of the manuscript. Furthermore, we thank Matti Zuiderwijk for preparing the spread test products and the Nutrition Intervention Study skillbase for excellent execution of the human study.
Footnotes
Published ahead of print on 14 August 2009.
REFERENCES
- 1.Ahlroos, T., and S. Tynkkynen. 2009. Quantitative strain-specific detection of Lactobacillus rhamnosus GG in human faecal samples by real-time PCR. J. Appl. Microbiol. 106:506-514. [DOI] [PubMed] [Google Scholar]
- 2.Ahrné, S., S. Nobaek, B. Jeppsson, I. Adlerberth, A. E. Wold, and G. Molin. 1998. The normal Lactobacillus flora of healthy human rectal and oral mucosa. J. Appl. Microbiol. 85:88-94. [DOI] [PubMed] [Google Scholar]
- 3.Alander, M., R. Korpela, M. Saxelin, T. Vilpponen-Salmela, T. Mattila-Sandholm, and A. von Wright. 1997. Recovery of Lactobacillus rhamnosus GG from human colonic biopsies. Lett. Appl. Microbiol. 24:361-364. [DOI] [PubMed] [Google Scholar]
- 4.Alander, M., R. Satokari, R. Korpela, M. Saxelin, T. Vilpponen-Salmela, T. Mattila-Sandholm, and A. von Wright. 1999. Persistence of colonization of human colonic mucosa by a probiotic strain, Lactobacillus rhamnosus GG, after oral consumption. Appl. Environ. Microbiol. 65:351-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Båth, K., S. Roos, T. Wall, and H. Jonsson. 2005. The cell surface of Lactobacillus reuteri ATCC 55730 highlighted by identification of 126 extracellular proteins from the genome sequence. FEMS Microbiol. Lett. 253:75-82. [DOI] [PubMed] [Google Scholar]
- 6.Boyle, R. J., R. M. Robins-Browne, and M. L. Tang. 2006. Probiotic use in clinical practice: what are the risks? Am. J. Clin. Nutr. 83:1256-1264. [DOI] [PubMed] [Google Scholar]
- 7.Brandt, K., and T. Alatossava. 2003. Specific identification of certain probiotic Lactobacillus rhamnosus strains with PCR primers based on phage-related sequences. Int. J. Food Microbiol. 84:189-196. [DOI] [PubMed] [Google Scholar]
- 8.Cummings, J. H., J. M. Antoine, F. Azpiroz, R. Bourdet-Sicard, P. Brandtzaeg, P. C. Calder, G. R. Gibson, F. Guarner, E. Isolauri, D. Pannemans, C. Shortt, S. Tuijtelaars, and B. Watzl. 2004. PASSCLAIM—gut health and immunity. Eur. J. Nutr. 43(Suppl. 2):II118-II173. [DOI] [PubMed] [Google Scholar]
- 9.Food and Agriculture Organization of the United Nations-World Health Organization Working Group. 2002. Guidelines for the evaluation of probiotics in foods. Report of a joint FAO/WHO working group on drafting guidelines for the evaluation of probiotics in food. World Health Organization, Geneva, Switzerland. ftp://ftp.fao.org/es/esn/food/wgreport2.pdf.
- 10.Goldin, B. R., S. L. Gorbach, M. Saxelin, S. Barakat, L. Gualtieri, and S. Salminen. 1992. Survival of Lactobacillus species (strain GG) in human gastrointestinal tract. Dig. Dis. Sci. 37:121-128. [DOI] [PubMed] [Google Scholar]
- 11.Hartemink, R., V. R. Domenech, and F. M. Rombouts. 1997. LAMVAB—a new selective medium for the isolation of lactobacilli from faeces. J. Microbiol. Methods 29:77-84. [Google Scholar]
- 12.Hartemink, R., and F. M. Rombouts. 1999. Comparison of media for the detection of bifidobacteria, lactobacilli and total anaerobes from faecal samples. J. Microbiol. Methods 36:181-192. [DOI] [PubMed] [Google Scholar]
- 13.Haschke, F., W. Wang, G. Ping, W. Varavithya, A. Podhipak, F. Rochat, H. Link-Amster, A. Pfeifer, E. Diallo-Ginsti, and P. Steenhout. 1998. Clinical trials prove the safety and efficacy of the probiotic strain Bifidobacterium Bb12 in follow-up formula and growing up milks. Monatsschr. Kinderheilkd. 146(Suppl. 1):S26-S30. [Google Scholar]
- 14.Holzapfel, W. H., P. Haberer, J. Snel, U. Schillinger, and J. H. Huis in't Veld. 1998. Overview of gut flora and probiotics. Int. J. Food Microbiol. 41:85-101. [DOI] [PubMed] [Google Scholar]
- 15.Jacobsen, C. N., V. Rosenfeldt Nielsen, A. E. Hayford, P. L. Møller, K. F. Michaelsen, A. Paerregaard, B. Sandström, M. Tvede, and M. Jakobsen. 1999. Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl. Environ. Microbiol. 65:4949-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klaenhammer, T., E. Altermann, F. Arigoni, A. Bolotin, F. Breidt, J. Broadbent, R. Cano, S. Chaillou, J. Deutscher, M. Gasson, M. van de Guchte, J. Guzzo, A. Hartke, T. Hawkins, P. Hols, R. Hutkins, M. Kleerebezem, J. Kok, O. Kuipers, M. Lubbers, E. Maguin, L. McKay, D. Mills, A. Nauta, R. Overbeek, H. Pel, D. Pridmore, M. Saier, D. van Sinderen, A. Sorokin, J. Steele, D. O'Sullivan, W. de Vos, B. Weimer, M. Zagorec, and R. Siezen. 2002. Discovering lactic acid bacteria by genomics. Antonie van Leeuwenhoek 82:29-58. [DOI] [PubMed] [Google Scholar]
- 17.Larsen, C. N., S. Nielsen, P. Kaestel, E. Brockmann, M. Bennedsen, H. R. Christensen, D. C. Eskesen, B. L. Jacobsen, and K. F. Michaelsen. 2006. Dose-response study of probiotic bacteria Bifidobacterium animalis subsp. lactis BB-12 and Lactobacillus paracasei subsp paracasei CRL-341 in healthy young adults. Eur. J. Clin. Nutr. 60:1284-1293. [DOI] [PubMed] [Google Scholar]
- 18.Ling, W. H., O. Hanninen, H. Mykkanen, M. Heikura, S. Salminen, and A. von Wright. 1992. Colonization and fecal enzyme activities after oral Lactobacillus GG administration in elderly nursing home residents. Ann. Nutr. Metab. 36:162-166. [DOI] [PubMed] [Google Scholar]
- 19.Ling, W. H., R. Korpela, H. Mykkanen, S. Salminen, and O. Hanninen. 1994. Lactobacillus strain GG supplementation decreases colonic hydrolytic and reductive enzyme activities in healthy female adults. J. Nutr. 124:18-23. [DOI] [PubMed] [Google Scholar]
- 20.Neish, A. S. 2009. Microbes in gastrointestinal health and disease. Gastroenterology 136:65-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onning, G., A. Berggren, M. Drevelius, B. Jeppsson, A. M. Lindberg, and M. L. Johansson Hagslatt. 2003. Influence of a drink containing different antioxidants and Lactobacillus plantarum 299v on plasma total antioxidant capacity, selenium status and faecal microbial flora. Int. J. Food Sci. Nutr. 54:281-289. [DOI] [PubMed] [Google Scholar]
- 22.Ouwehand, A. C., T. Kurvinen, and P. Rissanen. 2004. Use of a probiotic Bifidobacterium in a dry food matrix, an in vivo study. Int. J. Food Microbiol. 95:103-106. [DOI] [PubMed] [Google Scholar]
- 23.Rosander, A., E. Connolly, and S. Roos. 2008. Removal of antibiotic resistance gene-carrying plasmids from Lactobacillus reuteri ATCC 55730 and characterization of the resulting daughter strain, L. reuteri DSM 17938. Appl. Environ. Microbiol. 74:6032-6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross, R. P., C. Desmond, G. F. Fitzgerald, and C. Stanton. 2005. Overcoming the technological hurdles in the development of probiotic foods. J. Appl. Microbiol. 98:1410-1417. [DOI] [PubMed] [Google Scholar]
- 25.Sanders, M. E. 2003. Probiotics: considerations for human health. Nutr. Rev. 61:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satokari, R. M., E. E. Vaughan, A. D. Akkermans, M. Saarela, and W. M. de Vos. 2001. Polymerase chain reaction and denaturing gradient gel electrophoresis monitoring of fecal Bifidobacterium populations in a prebiotic and probiotic feeding trial. Syst. Appl. Microbiol. 24:227-231. [DOI] [PubMed] [Google Scholar]
- 27.Saxelin, M., M. Ahokas, and S. Salminen. 1993. Dose response on the faecal colonisation of Lactobacillus strain GG administered in two different formulations. Microb. Ecol. Health Dis. 6:119-122. [Google Scholar]
- 28.Saxelin, M., S. Elo, S. Salminen, and H. Vapaatalo. 1991. Dose response colonisation of faeces after oral administration of Lactobacillus casei strain GG. Microb. Ecol. Health Dis. 4:209-214. [Google Scholar]
- 29.Saxelin, M., T. Pessi, and S. Salminen. 1995. Fecal recovery following oral administration of Lactobacillus strain GG (ATCC 53103) in gelatine capsules to healthy volunteers. Int. J. Food Microbiol. 25:199-203. [DOI] [PubMed] [Google Scholar]
- 30.Siitonen, S., H. Vapaatalo, S. Salminen, A. Gordin, M. Saxelin, R. Wikberg, and A. L. Kirkkola. 1990. Effect of Lactobacillus GG yoghurt in prevention of antibiotic associated diarrhoea. Ann. Med. 22:57-59. [DOI] [PubMed] [Google Scholar]
- 31.Song, Y., N. Kato, C. Liu, Y. Matsumiya, H. Kato, and K. Watanabe. 2000. Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group- and species-specific primers derived from the 16S-23S rRNA intergenic spacer region and its flanking 23S rRNA. FEMS Microbiol. Lett. 187:167-173. [DOI] [PubMed] [Google Scholar]
- 32.Valeur, N., P. Engel, N. Carbajal, E. Connolly, and K. Ladefoged. 2004. Colonization and immunomodulation by Lactobacillus reuteri ATCC 55730 in the human gastrointestinal tract. Appl. Environ. Microbiol. 70:1176-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf, B. W., K. A. Galeb, D. G. Ataya, and I. A. Casas. 1995. Safety and tolerance of Lactobacillus reuteri in healthy adult subjects. Microb. Ecol. Health Dis. 8:41-50. [Google Scholar]
- 34.Wolf, B. W., K. B. Wheeler, D. G. Ataya, and K. A. Garleb. 1998. Safety and tolerance of Lactobacillus reuteri supplementation to a population infected with the human immunodeficiency virus. Food Chem. Toxicol. 36:1085-1094. [DOI] [PubMed] [Google Scholar]
- 35.Yli-Knuuttila, H., J. Snall, K. Kari, and J. H. Meurman. 2006. Colonization of Lactobacillus rhamnosus GG in the oral cavity. Oral Microbiol. Immunol. 21:129-131. [DOI] [PubMed] [Google Scholar]


