Abstract
High-Arctic soils have low nutrient availability, low moisture content, and very low temperatures and, as such, they pose a particular problem in terms of hydrocarbon bioremediation. An in-depth knowledge of the microbiology involved in this process is likely to be crucial to understand and optimize the factors most influencing bioremediation. Here, we compared two distinct large-scale field bioremediation experiments, located at the Canadian high-Arctic stations of Alert (ex situ approach) and Eureka (in situ approach). Bacterial community structure and function were assessed using microarrays targeting the 16S rRNA genes of bacteria found in cold environments and hydrocarbon degradation genes as well as quantitative reverse transcriptase PCR targeting key functional genes. The results indicated a large difference between sampling sites in terms of both soil microbiology and decontamination rates. A rapid reorganization of the bacterial community structure and functional potential as well as rapid increases in the expression of alkane monooxygenases and polyaromatic hydrocarbon-ring-hydroxylating dioxygenases were observed 1 month after the bioremediation treatment commenced in the Alert soils. In contrast, no clear changes in community structure were observed in Eureka soils, while key gene expression increased after a relatively long lag period (1 year). Such discrepancies are likely caused by differences in bioremediation treatments (i.e., ex situ versus in situ), weathering of the hydrocarbons, indigenous microbial communities, and environmental factors such as soil humidity and temperature. In addition, this study demonstrates the value of molecular tools for the monitoring of polar bacteria and their associated functions during bioremediation.
With ongoing climate warming and the possible opening of the Northwest Passage for commercial shipping in the near future, human activity will increase in the Canadian high Arctic, raising the potential for environmental contamination. The settlements in the high Arctic are using fuel for transportation and to produce electricity and heating, while spills from leaking tanks or pipelines are frequent (19, 34). Bioremediation is often the only feasible cleanup option because the remoteness and unique character of these sites preclude conventional physicochemical technologies for soil treatment. Arctic soils are characterized by extremely low temperatures and the limited availability of water and nutrients, especially nitrogen (18, 26), which limit the degradation rates by indigenous soil microorganisms. One of the possible bioremediation approaches is to stimulate indigenous cold-adapted microorganisms by the application of nutrients and water. This strategy was successfully applied to diesel-contaminated soils in the Canadian high Arctic (9, 33, 34). However, these studies mostly focused on soil chemistry and general soil processes, with very few insights into the ecology and the functions of the microorganisms actually involved in the bioremediation processes. Although this is not surprising since the common criterion for decommissioning a polluted site is based on soil chemistry, the lack of knowledge of the dynamics of the microbial communities involved hampers the design of more efficient bioremediation approaches.
Common diesel fuel is composed of ∼64% saturated aliphatic hydrocarbons (alkanes), ∼1 to 2% unsaturated aliphatic hydrocarbons, and ∼35% aromatic hydrocarbons (including polycyclic aromatic hydrocarbons [PAHs]) (23). Consequently, the complete degradation of diesel requires the presence of different microorganisms with complementary enzymatic capacities. Alkanes form the part of diesel that is easier to degrade, and the first step of their aerobic degradation is catalyzed by alkane monooxygenases, for which the alkB gene is particularly well characterized (38). Although Rhodococcus was hypothesized to be the predominant alkane-degrading genus in polar soils, Pseudomonas is also thought to be enriched following hydrocarbon contamination (37). PAHs are more difficult to degrade and are therefore highly persistent in soils. The initial step of PAH degradation is carried out by multicomponent aromatic-ring-hydroxylating dioxygenases (RHD) that contain regions conserved among all the different genes encoding PAH-degrading enzymes. The typical aromatic- degrading bacteria isolated from polar soils are Pseudomonas or Sphingomonas, with Sphingomonas having a wider range of substrates than Pseudomonas (1). It appears, therefore, that the relative responses of the different bacterial species to bioremediation treatments will have a strong effect on the efficiency of pollutant removal.
Microarray analyses hold the potential to simultaneously follow functional and marker genes of hundreds of microorganisms involved in key environmental processes. Such approaches were shown to be reliable even in less well-studied polar environments (40, 41), but several issues related to the specificity and the sensitivity of microarrays warrant the use of an integrated approach. As compared to comprehensive microarray platforms that systematically target all known microorganisms (4) or most known environmentally relevant functional genes (11), smaller platforms focusing on particular processes (like hydrocarbon degradation) or specific environments (like polar soils) could yield similarly interesting information and would have the advantage of producing data that are easier to analyze and interpret. Coupled with detailed analysis of the microbial community structure and of the expression levels of key enzymes, such microarray platforms could yield a deeper functional understanding of the bioremediation of contaminated soils.
Previous studies have demonstrated the potential for indigenous Arctic soil microorganisms to degrade hydrocarbons after nutrient amendment (9, 19, 29, 33-35). The present study had two main objectives: (i) to demonstrate the utility of molecular tools (microarray and real-time PCR) to monitor bacterial communities and their associated functions in polar environments during bioremediation treatments and (ii) to observe and understand the functional dynamics of indigenous bacterial communities in diesel-contaminated soils at two Canadian high-Arctic sites and the effect of bioremediation using nutrient amendments on these bacterial communities. We used two microarray platforms to describe the bacterial community: one targeting the functional genes involved in hydrocarbon degradation and the other targeting the 16S rRNA gene of bacteria commonly found in cold environments. Quantitative reverse transcriptase PCR (qRT-PCR) was also used to evaluate the amount of transcripts related to key functional genes (alkB, PAH-RHD genes, and nirS). These results were analyzed together with complementary data such as those from soil analyses, potential mineralization, and numbers of culturable hydrocarbon-degrading bacteria.
MATERIALS AND METHODS
Site descriptions.
The two sampling sites, Alert and Eureka, are located on Ellesmere Island, Nunavut, in the Canadian high Arctic. Alert (82°31′N, 62°17′W) is the northernmost permanent human settlement in the world and is located at an elevation of 30.5 m. Annual precipitation averages 153.8 mm, while the average daily temperature is −18.0°C. The annual daily maximum is −14.7°C, and the annual daily minimum is −21.3°C. During the growing season (July to August), the daily average temperature is 2.1°C, the average daily maximum is 4.6°C, and the average daily minimum is −0.6°C. The soil under study was subjected to a large diesel spill in 2004, and the contaminated soil was moved to a contained berm area in 2005 followed by a nutrient amendment/mixing bioremediation strategy.
Eureka (79°59′N, 85°56′W) is the second-northernmost permanent human settlement and is at an elevation of 10.4 m. Eureka receives on average 75.5 mm of precipitation each year. The average daily temperature is −19.7°C, while the annual daily maximum is −16.4°C, and the annual daily minimum is −22.9°C. During the growing season (July to August), the daily average temperature is 4.2°C, the average daily maximum temperature is 6.8°C, and the average daily minimum is 1.5°C. The soil under study was subject to a 37,000-liter diesel fuel spill in 1990, resulting in the contamination of ∼3,200 m3 of soil. In situ nutrient amendment/tilling treatment was initiated in 2000.
All of the above weather data were from Canada's National Climate Archive (http://www.climate.weatheroffice.ec.gc.ca/index.html).
Bioremediation treatments and soil sampling.
Different bioremediation treatments were carried out at the two sites but were based on a similar rationale: stimulation of aerobic hydrocarbon-degrading microorganisms by N and P amendment and soil aeration. At Eureka, a custom-made liquid nutrient solution (minimum of 15% nitrogen, 3% phosphoric acid, 2% potash, and 1% sulfur; Oligosol Ltd., Beloeil, QC, Canada) was diluted and applied annually by sprinkler at a rate of ∼65 ml m−3 followed by tilling. At Alert, solid fertilizer in the form of NH4PO4 was thoroughly mixed with the soil at a rate of 0.3 kg m−3, which was then windrowed into 1.5-m-high biopiles within a bermed area lined with an impermeable geomembrane.
Our objective was to monitor the biodegradation of hydrocarbons over time and compare the treated soils with untreated contaminated soils and nearby uncontaminated soils. For the majority of conditions, four replicate soil samples were analyzed. For Eureka, we analyzed two samples right before the first nutrient amendment (time zero [T = 0]; August 2000), two samples 1 month after the first nutrient amendment (September 2000), four samples 1 year after the beginning of treatment (2001), four samples 4 years after the beginning of the treatment (2004), four untreated contaminated samples (two from 2000, one from 2003, and one from 2005), and four uncontaminated samples (one from 2005, two from 2006, and one from 2007). For Alert, we analyzed two samples taken right before nutrient amendment (T = 0; August 2005), four samples taken 1 month after the nutrient amendment (September 2005), four samples taken 1 year after treatment (2006), four untreated contaminated samples (one from 2007 and three from 2008), and four uncontaminated samples (two from 2005 and two from 2008).
Soil samples (1 to 2 kg) were taken and kept at 4°C and shipped to the lab. Samples were mixed thoroughly and then separated: one part was frozen (−20°C for Eureka and −80°C for Alert) for subsequent use in molecular analyses, while the remainder was kept at 4°C and processed as soon as possible for mineralization assays, viable counts, and soil analyses. At Eureka, contaminated treated soil samples were collected 50 cm below the surface at random locations. These random locations were marked and sampled throughout the time course. At Alert, contaminated treated soil samples were composite soils collected throughout the biopiles. At both locations, uncontaminated and untreated contaminated soil samples were collected randomly from a depth of 15 cm.
Soil analyses, CFU and MPN-diesel counts, and mineralization assays.
Soil petroleum hydrocarbons (C10-to-C50 fraction) were assessed by gas chromatography-flame ionization detector. Total culturable soil aerobic heterotrophic bacteria counts were carried out in triplicate at 4°C on MSM-YTS (minimum salts medium [8] plus 250 mg liter−1 each of yeast extract, tryptone, and soluble starch) using serial soil dilutions in 0.1% pyrophosphate buffer. The numbers of culturable aerobic diesel-degrading bacteria were monitored in a 96-well format by the most probable number method (MPN-diesel) at 5°C in minimum salts medium (total volume of 200 μl) supplemented with 3 μl of diesel per well (10, 39). Degradation of diesel was followed by colorimetric development following the reduction of 2-(p-iodophenyl)-3-(p-nitrophenyl)-5-phenyl tetrazolium chloride (INT), as compared to that of uninoculated controls. Mineralization was assessed following Whyte and colleagues (36) by using microcosms that contained 20 g of soil set up in triplicate and incubated at 4°C. The microcosms were not supplemented in the laboratory with additional nutrients. The microcosms were spiked with either [14C]hexadecane (alkane) or [14C]naphthalene (PAH) for final concentrations of 100 ppm and 10 ppm, respectively, and ∼100,000 dpm. The production of 14CO2 was monitored by liquid scintillation spectrometry, and mineralization was expressed as the cumulative percent relative to the initial amount injected. Sterile autoclaved control soils did not show any 14CO2 production. The mineralization extent, used in the present study, was determined after 28-day (Alert) or 40-day (Eureka) incubations.
Nucleic acid extractions.
Soil DNA was extracted from a 0.5-g soil subsample using the MoBio DNA Power soil kit (MoBio Laboratories, Carlsbad, CA), while soil RNA was extracted from a 2.0-g soil subsample using the MoBio RNA Power soil kit. Residual DNA in RNA extracts was removed using the Turbo DNA-free kit (Ambion, Austin, TX).
qRT-PCR.
The primers and PCR conditions used are summarized in Table 1. qRT-PCR was performed in 20-μl volumes using the iScript one-step RT-PCR kit with Sybr green (Bio-Rad Laboratories, Hercules, CA) on a Rotor-Gene 3000 apparatus (Corbett Life Science, Sydney, Australia). Reactions were set up as per the manufacturer's instructions, with 1 to 75 ng of total soil RNA extract. The amplification procedure was as follows: cDNA synthesis for 10 min at 50°C; RT inactivation for 5 min at 95°C; and PCR cycling and detection (40 cycles) for 30 s at 95°C, 30 s at annealing temperature (Table 1), and 30 s at 72°C (acquiring signal at the end of this step). For the assays in which primer dimers were an issue, an additional 15-s reading step was added at the end of each cycle and the fluorescence signal was acquired at the end of this step (see Table 1 for temperature). Standards were made from 10-fold dilutions of linearized plasmids containing the gene fragment of interest that was cloned from amplified soil DNA. For all reactions, several no-RT and no-template controls were carried out and yielded no detectable signals. Lambda DNA was used to correct for potential PCR inhibitors present in soil extracts (2). Equal volumes of 10× diluted soil RNA extracts and a cloned 500-bp fragment of bacteriophage lambda (105 copies per μl) were mixed. When the recovery of lambda was below 100%, quantification values for all other genes were corrected accordingly. PCR inhibition ranged from 4.6% to 75.9%.
TABLE 1.
Primers and PCR conditions used in real-time RT-PCR assays
| Target | Enzyme | Primers | Temp (°C)a
|
Reference | |
|---|---|---|---|---|---|
| Annealing | Read | ||||
| alkB | Alkane monooxygenase | alkbFd and alkbRd | 57 | 77 | 21 |
| Bacterial 16S rRNA | Eub338 and Eub518 | 53 | NA | 6 | |
| Bacteriophage lambda | lambda 7131F and lambda 7630R | 60 | NA | 2 | |
| nirS | cd1-containing nitrite reductase | cd3aF and R3cd | 57 | 83 | 30 |
| PAH-RHD GN | Gram-negative PAH-RHD α | PAH-RHD GN F and PAH-RHD GN R | 57 | 80 | 3 |
| PAH-RHD GP | Gram-positive PAH-RHD α | PAH-RHD GP F and PAH-RHD GP R | 54 | 80 | 3 |
NA, not applicable. These qRT-PCR assays did not have a reading step, and reading was performed at the end of the elongation step, at 72°C.
Microarray targeting the 16S rRNA genes of bacteria found in cold environments.
The 16S rRNA gene probe design for the microarray was as follows: (i) database searches in Greengenes (http://greengenes.lbl.gov) for 16S rRNA gene sequences with keywords “Arctic,” “Antarctic,” “Polar,” “Cold,” and “Psychro” (2,891 sequences retrieved, performed on 15 April 2008); (ii) clustering of the database sequences together with 16S rRNA gene sequences from strains isolated in our lab in previous studies of polar environments using Blastclust (NCBI, Bethesda, MD); (iii) design of five 25-mer probes for each of the 99% similarity clusters having three or more representatives using OligoPicker (32); (iv) elimination of probes having a ΔG of >−3 kJ; (v) BLAST comparison of the central 17 nucleotides of each probe against all the original sequences; and (vi) selection, for each cluster, of the probe that showed less significant hits to sequences outside the cluster and most significant hits within the cluster. The probes used and their sequences are listed in the NCBI Gene Expression Omnibus (GEO) under platform accession no. GPL8953 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL8953). Oligonucleotides (synthesized by IDT, Coralville, IA) were printed in triplicate on aminosilane-coated glass slides (Corning, Acton, MA) using a VersArray Chip Writer Pro printer (Bio-Rad, Hercules, CA). Including control spots, the microarray consisted of 528 spots. Each slide contained three identical subarrays. Total RNA extracted from all soil samples (n = 38) was submitted to an RT-PCR procedure with the F1-R13 eubacterial 16S rRNA gene primers (15) using the Qiagen OneStep RT-PCR kit (Qiagen, Valencia, CA), following the manufacturer's instruction (except that all volumes were halved), with an annealing temperature of 50°C. RT-PCR products were then chemically labeled using the Label IT nucleic acid labeling kit (Mirus Bio, Madison, WI). Slides were prehybridized for 1 h at 37°C with a DIG (digoxigenin) Easy hybridization solution (Roche Applied Science, Laval, QC, Canada) containing 5% bovine serum albumin. Samples were then assigned randomly to a subarray, and hybridization was carried out for ∼20 h at 37°C on a Slide Booster apparatus (Implen, Calabasas, CA). Slides were washed three times for 5 min at 37°C in 0.1× SSC (0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate followed by a single wash for 5 min at 37°C in 0.1× SSC. Slides were scanned in a ScanArray GX PLUS microarray scanner (PerkinElmer, Boston, MA) at a resolution of 10 μm, and images were transferred to QuantArray (PerkinElmer), where each spot was identified and quantified. Data were then exported to Excel, where net spot intensity was calculated by subtracting the background signal from the target spot intensity. A spot was scored as “present” if its net intensity was at least three times higher than the median net intensity of all spots. For a taxon to be scored as present, all of its triplicate spots had to be scored as present. The results from the 16S rRNA gene microarray were only used in the binary form (presence/absence of taxa).
Microarray targeting hydrocarbon-degrading genes.
The full description, usage, and analysis of the functional gene microarray are given in the NCBI GEO under platform accession no. GPL8960 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL8960). Hybridizations were carried out for all samples (n = 38) with 100 ng of genomic DNA labeled with Cy5 using the BioPrime Array CGH genomic labeling kit (Invitrogen, Carlsbad, CA). Following normalization (see the GEO repository for more details), intensity values were averaged over triplicate spots to give a single value per gene. The intensity of the genes for which hybridization of the probes could not be visually detected in image analysis was set to zero. Relative intensity values were then calculated by dividing the intensity of a gene by the sum of the intensity for all genes (excluding controls). This “relative abundance” represents the fraction of the total intensity that is due to a particular gene, and this value was subsequently used in statistical analyses.
Statistical analyses.
Since the bioremediation approaches were quite different, the two sites were treated separately in all analyses, except correlations. All statistical analyses were carried out in R (v 2.7.1; The R Foundation for Statistical Computing). When necessary, data was log (in most cases), square root, or cubic root transformed to meet the assumptions of parametric analysis of variance (ANOVA). Normality was tested using the “shapiro.test” function. ANOVA and subsequent Tukey's honestly significantly different (HSD) tests were carried out using the “aov” and “TukeyHSD” functions, respectively. When transformations failed to normalize data, Kruskal-Wallis and associated multiple-comparison tests were carried out using the “kruskal.test” and the “kruskalmc” functions of the “pgirmess” library, respectively. Correlation analyses were based on Spearman's r (rs) using the “cor” function. Principal coordinate analyses (PCoA) were carried out using the “cmdscale” function based on the square root of 1-Jaccard (16S rRNA gene microarray) or on the square root of Bray-Curtis (functional gene microarray) distance matrices calculated using the “vegdist” function of the “vegan” library. For PCoA of the functional gene microarrays, vectors representing the relative amount of genes related to different compounds (calculated by summing the relative signal of all genes related to this compound) were added to the ordination as supplementary variables: i.e., not involved in the calculation (28).
Microarray data accession number.
The data discussed in this publication have been deposited in NCBI GEO (5) and are accessible through GEO series accession no. GSE17533 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17533).
RESULTS
Soil analyses, CFU, MPN-diesel, and potential mineralization.
To compare and validate the molecular techniques used in this study, we measured several parameters that are traditionally used to evaluate the degradation of hydrocarbons in soils. Soils were analyzed for hydrocarbon content (C10-to-C50 fraction) to follow the reduction of the contamination under field conditions. Potential mineralization was measured under optimal conditions in the laboratory and is related to the maximum amount of particular hydrocarbons (in our case, alkane and naphthalene) that can be completely oxidized by the organisms present in a soil. CFU and MPN-diesel evaluate the numbers of culturable heterotrophic microorganisms and the numbers of culturable microorganisms that can grow with diesel as the sole carbon source, respectively. Several of the results (CFU, MPN-diesel counts, potential mineralization, and soil analyses) were not significantly different (ANOVA tests at P < 0.05), even though the averages between treatments were quite different (Table 2). This was caused by large variations in individual replicates, but also because, in some cases, not all of the replicate samples were assessed for these basic characteristics. For these reasons, we also highlighted nearly significant (0.05< P < 0.10) P values in Table 2. The C10-to-C50 hydrocarbon fraction content in surface soil decreased over the years following nutrient amendment at both Alert and Eureka. After a single year, soil C10-to-C50 content decreased by 91% at Alert and by 52% at Eureka. It took 4 years of treatment for the soil C10-to-C50 content at Eureka to decrease to the same extent (87%) as that observed at Alert after 1 year. Significant differences in total heterotrophic bacterial counts, MPN-diesel counts, and mineralization of hexadecane and naphthalene were observed between contaminated and treated and uncontaminated soils. Significant differences in hexadecane mineralization and MPN-diesel counts were also observed between treated and untreated contaminated soils at Alert.
TABLE 2.
Average soil water and hydrocarbon concentrations, CFU counts, MPN-diesel degraders, and potential hexadecane and naphthalene mineralization at 4°C for Alert and Eureka soilsa
| Site and parameter | % Water content | Hydrocarbon (C10-C50) concn (mg kg−1) | CFU count on YTS medium (107 g−1) | MPN-diesel degraders (105 g−1) | Mineralization (% degraded)b
|
|
|---|---|---|---|---|---|---|
| Hexadecane | Naphthalene | |||||
| Alert | ||||||
| 2005 (T = 0) | 18.2 | 3,325 A | 2.47 AB | 63.7 AB | 1.21 AB | 63.42 |
| 2005 (1 mo) | 11.6 | 1,358 A | 9.32 AB | 400 A | 35.00 A | 63.91 |
| 2006 (1 yr) | 11.0 | 298 A | 14.6 A | 27.1 AB | 19.79 AB | 69.99 |
| Untreated contaminated | 14.1 | 8,607 A | 1.19 B | 23.5 BC | 0.24 B | 37.44 |
| Uncontaminated | NAc | 0 A | 0.05 B | 0.51 C | 2.97 AB | 20.80 |
| P | NA | 0.028 | 0.003 | 0.0009 | 0.009 | 0.092 |
| Eureka | ||||||
| 2000 (T = 0) | NA | 10,467 | 30.2 | 231 | 0.21 | 16.50 AB |
| 2000 (1 mo) | NA | 6,558 | 18.4 | 42.9 | 0.24 | 61.38 B |
| 2001 (1 yr) | 10.0 | 5,056 | 8.00 | 69.30 | 6.42 | 55.01 AB |
| 2004 (4 yr) | 8.1 | 1,325 | 4.89 | 4.56 | 22.64 | 57.18 B |
| Untreated contaminated | 12.7 | 6,307 | 0.76 | 0.11 | 1.35 | 25.12 AB |
| Uncontaminated | 11.1 | 0 | 0.26 | 0.07 | 0.41 | 0.53 A |
| P | NA | 0.172 | 0.151 | 0.084 | 0.054 | 0.007 |
Different letters indicate significantly (P < 0.05) different averages following Tukey's HSD or Kruskal multiple-comparison tests. For ANOVA results, P values <0.05 are in boldface and P values of <0.10 and >0.05 are in italic.
For hexadecane and naphthalene mineralization, the durations of incubation were 28 days for Alert and 40 days for Eureka.
NA, not available.
qRT-PCR quantification.
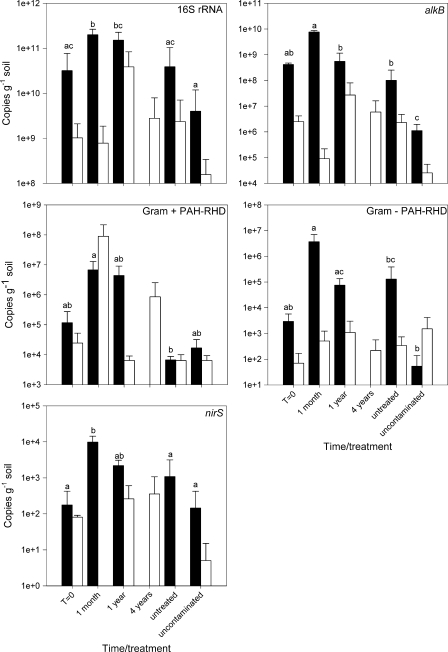
No significant differences were observed between the three Eureka sample types (contaminated and treated, contaminated and untreated, and uncontaminated) in the expression of functional genes and in the amount of 16S rRNA. The dominant trend for most of the functional genes examined in the Eureka samples was a relatively small increase within 1 month and a much larger increase within 1 year, normally followed by a decrease within 4 years (Fig. 1). Untreated contaminated and uncontaminated soils demonstrated a much lower expression of most examined genes. At Alert, several significant differences were observed (Fig. 1). Although significance varied, the general trend was the same: a large increase in the expression of the gene within 1 month of treatment followed by a decrease within 1 year. In most cases, uncontaminated soils showed significantly lower, but still detectable, expression of the examined functional genes (Fig. 1).
FIG. 1.
qRT-PCR results for the quantification of 16S rRNA and the expression of alkane monooxygenase (alkB), gram-positive and gram-negative PAH-RHDs, and cytochrome-containing nitrate reductase (nirS) in soils from Alert (▪) and Eureka (□). Different letters indicate significantly different averages at P = 0.05 following Tukey's HSD test or the Kruskal multiple-comparison test. No significant differences were observed for Eureka. The error bars represent the standard deviation.
Correlation analyses were also carried out to evaluate the consistency of the data obtained via the various methods. For this purpose, Eureka and Alert data were used together to have the largest possible data set. The expression of PAH-RHD (both gram positive and gram negative) and alkB genes was significantly positively correlated to the MPN-diesel counts (PAH-RHD gram positive, rs = 0.454 and P = 0.0133; PAH-RHD gram negative, rs = 0.570 and P = 0.0012; alkB, rs = 0.759 and P < 0.0001) and to the mineralization of hexadecane (PAH-RHD gram positive, rs = 0.563 and P = 0.0008; PAH-RHD gram negative, rs = 0.496 and P = 0.0039; alkB, rs = 0.402 and P = 0.0226) and naphthalene (PAH-RHD gram positive, rs = 0.609 and P = 0.0003; PAH-RHD gram negative, rs = 0.559 and P = 0.0009; alkB, rs = 0.498 and P = 0.0038). The amount of bacterial 16S rRNA was significantly positively correlated to CFU counts (rs = 0.549, P = 0.0036), to MPN-diesel counts (rs = 0.511, P = 0.0076), and to the mineralization of hexadecane (rs = 0.412, P = 0.0192) and naphthalene (rs = 0.555, P = 0.0012).
16S rRNA gene microarray.
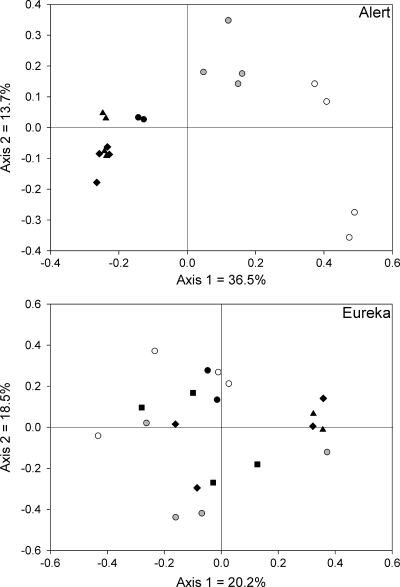
The 16S rRNA gene microarray targeting the bacteria commonly found in cold environments consisted of 525 25-mer oligonucleotides targeting 159 bacterial taxa. Out of these 159 taxa, 53 bacterial taxa were detected in our samples. On average, 12.2 taxa were detected per individual sample, ranging from 4 to 23 taxa. The taxa presence/absence data retrieved from the microarray analysis of each sample were used in a PCoA. This type of multivariate statistical analysis can be used to produce ordination graphs, in which the samples are positioned according to their similarity (samples close to each other on the graph show more similar microarray hybridization patterns). At Alert, the different treatments harbored highly contrasting bacterial communities, as can be visualized in the PCoA ordination (Fig. 2). The samples grouped tightly according to the different treatments, and these groups were clearly discriminated on the first axis of the ordination plot, which is indicative of a very strong trend in the data set. In contrast, the Eureka samples did not show such a trend in community composition (Fig. 2). There was no clear grouping of these samples according to contamination and/or treatment. Since the microarray probes were designed based on sequence clusters (see Materials and Methods), the exact phylogenetic affiliation of the different probes associated with particular treatments cannot be reported here. The relatively small numbers of probes do not make it relevant to report relative presence at the phylum level.
FIG. 2.
PCoA based on Jaccard's similarity calculated from the presence/absence of taxa represented on the 16S rRNA gene microarray for soil samples from Eureka and Alert. •, before treatment (T = 0); ▴, 1 month after nutrient amendment; ⧫, 1 year after nutrient amendment; ▪, 4 years after nutrient amendment; gray circles, untreated contaminated; ○, uncontaminated.
Hydrocarbon degradation gene microarray.
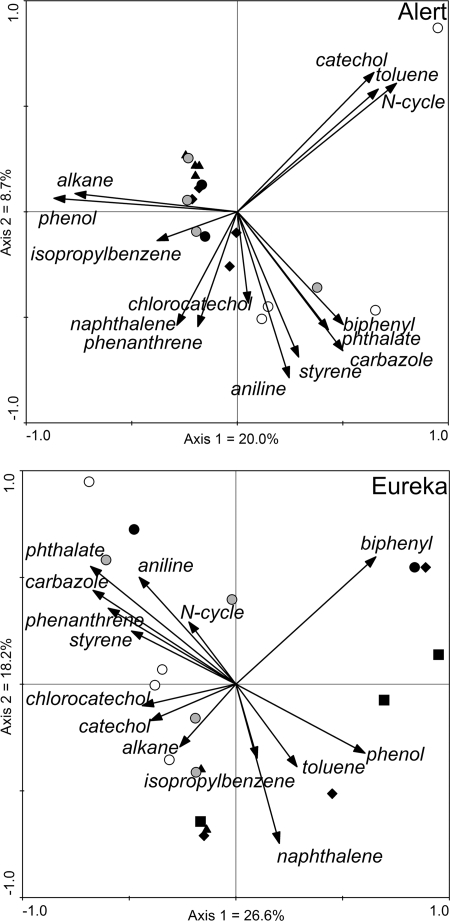
The second microarray platform used in the present study targeted functional genes involved in hydrocarbon degradation. The microarray was built from 140 PCR products amplified from cloned genes related to the degradation of alkanes (10 genes) and aromatic hydrocarbons (90 genes), to the nitrogen cycle (6 genes), to heavy metal transformation (7 genes), and to other processes (27 genes). From these 140 genes, 132 were detected in our samples. There were 57 genes detected on average in an individual soil sample, with a maximum of 99 and a minimum of 10. For Alert, the ordination patterns in hydrocarbon-degrading genes were similar to the ones observed for the 16S rRNA gene microarray. The treated soils generally grouped together tightly, especially the soils sampled 1 month after nutrient amendment (Fig. 3). The untreated samples grouped mainly with the treated samples, and the uncontaminated samples were separated from all other samples on the first ordination axis. The different probes were categorized according to their role in the degradation of different compounds. The signals from these probes were then summed for each sample, and these data were added on the PCoA ordination. The resulting compound degradation arrows (Fig. 3) point toward the samples that had the highest relative intensities for the probes related to the degradation of that particular compound. Contaminated samples from Alert were strongly associated with alkane, phenol, and isopropylbenzene degradation genes and partly to naphthalene and phenanthrene degradation genes (especially for the samples 1 year after nutrient amendment). In contrast, contaminated samples had a lower signal for biphenyl, phthalate, carbazole, styrene, and aniline degradation genes. Toluene, catechol, and N-cycle-related genes appeared to have a particularly strong association with an uncontaminated sample collected from a frost-sorted polygon. Chlorocatechol-related genes also demonstrated a stronger signal in uncontaminated samples.
FIG. 3.
PCoA based on Bray-Curtis distance calculated from the normalized relative intensities of the genes present on the hydrocarbon-degrading gene microarray for soil samples from Eureka and Alert. Arrows represent the sum of the signals due to genes that had a role in the degradation of a particular compound and were added in the analysis as supplementary variables not included in calculations. •, before treatment (T = 0); ▴, 1 month after nutrient amendment; ⧫, 1 year after nutrient amendment; ▪, 4 years after nutrient amendment; gray circles, untreated contaminated; ○, uncontaminated.
In contrast to the results from the 16S rRNA gene microarray analyses, ordination of the hydrocarbon-degrading gene microarray data revealed that a majority of the treated samples from Eureka grouped together on one side of the first axis, but this grouping was loose (Fig. 3). This indicates that these samples are showing similar levels of functional genes as compared to other samples but that there is a relatively large variation between the individual samples. Some compound degradation arrows pointed generally toward nutrient-amended samples: phenol, biphenyl, toluene, isopropylbenzene, and naphthalene degradation genes. Some other compound degradation arrows pointed in the opposite direction, indicating a lower relative abundance of the probes related to these compounds in amended samples: N-cycle, aniline, phthalate, carbazole, phenanthrene, styrene, chlorocatechol, and catechol transformation genes. The alkane degradation arrow was somewhat in between.
DISCUSSION
The results presented here indicate that the effects of pollution and nutrient amendments on Arctic soil bacteria are likely to be highly site and treatment specific. Diesel pollution was already shown to differentially affect microbial communities at two widely different Canadian Arctic sites (13). In our case, even though the basic bioremediation concept was the same and the experimental sites were similar, several site-specific factors could be responsible for the observed differences in bacterial responses. For instance, the composition and the application of the nutrient amendment, the manipulation of the soil, and the concentration and level of weathering of the contamination were different between Eureka and Alert. Similarly, the site-specific environmental and microbiological characteristics could have played a role. For instance, the indigenous bacterial communities, which subsequently carried out the biodegradation, were quite different (average of 26.9% Jaccard similarity between the taxonomic microarray community compositions between uncontaminated sites at Alert and Eureka).
Pollution and subsequent bioremediation treatments had their strongest impacts in Alert soils. In these soils, we observed a major change in bacterial community structure together with changes in the relative abundance of several functional genes and in the expression of genes involved in key degradation processes (Fig. 1 to 3). This indicates that bacteria are responding rapidly to the treatments and that they are largely responsible for the degradation of hydrocarbons in these soils. In Eureka soils, the responses to the treatments were highly variable, which precluded the observation of any significant changes. Eureka soils were left in place and mixed by tilling, which was far less efficient for homogenizing the soils than mixing the soils in a biopile, as was done at Alert. A better mixing of the Eureka soils would probably have reduced the variability between the replicates and allowed a clearer identification of the functional changes that followed bioremediation treatments. Aislabie and colleagues (1) proposed that ex situ bioremediation (e.g., biopiles) was likely to be the method of choice as compared to in situ bioremediation (e.g., soil amendment and tilling), mainly because it allows a stricter control of nutrient, moisture, and temperature conditions. Here we further demonstrated that the higher efficiency of ex situ versus in situ treatments is probably related to larger and more consistent changes in the microbial communities.
Another important difference that probably influenced the bioremediation efficiency between the two sites is the time since the contamination event at the beginning of the bioremediation experiment (10 years at Eureka versus 1 year at Alert). Diesel weathering and losses by volatilization, water leaching, and sorption can result in a significant shift toward heavier, more recalcitrant compounds over the years. Differences in the contaminants remaining in soils could explain the different response and the lower efficiency observed at Eureka. Within 1 month of treatment, Eureka soils showed relatively low expression of genes related to the easily degraded alkanes but very high expression of gram-positive PAH-RHD genes. Interestingly, gram-positive bacteria were hypothesized to be the dominant degraders of recalcitrant high-molecular weight PAH compounds (16). However, genes involved in the degradation of alkanes were expressed at both sites throughout the time course (Fig. 1), indicating that these compounds were available in every sample.
Bacterial community structure.
Two complementary microarray platforms were used in the present study: one targeting functional genes involved in hydrocarbon degradation and the other targeting bacteria commonly found in cold environments. Preliminary tests (not shown) showed that this latter microarray platform was able to generate sample clustering that was almost identical to previously published clustering based on 16S rRNA gene PCR-denaturing gradient gel electrophoresis fingerprints of another cold environment (20). This platform was therefore used as a cold-adapted bacterial community fingerprinting method.
The microarray targeting 16S rRNA genes of bacteria found in cold environments revealed a substantial shift in the bacterial community structure following hydrocarbon contamination and bioremediation at Alert (Fig. 2). The community structures were largely different between uncontaminated, untreated contaminated, and nutrient-amended contaminated soils, as these samples were clearly separated on the first axis of the ordination (Fig. 2). Similar shifts in bacterial community structure were reported following pollution and bioremediation in Antarctic and alpine soils (14, 25, 31). In contrast, bacterial community structure in Eureka soils did not show clear changes following contamination and bioremediation, with replicate samples having highly variable community structures. This lack of directional shift in bacterial community structure might be underlying the lower bioremediation efficiency and the absence of significant changes in the expression of functional genes observed at Eureka. One solution to this problem could be to introduce a more efficient bacterial community that would respond to the treatments. However, this method (bioaugmentation) was shown to have variable efficiency in polar soils (19, 29, 31, 33). Another possible solution to this problem would be to devise other bioremediation treatments that would more rapidly and strongly influence the indigenous soil bacterial community. In fact, in preliminary tests addressing Eureka bioremediation, the optimal treatment for a majority of the soils tested included moisture adjustment (34). Whyte and colleagues (34) further suggested that biodegradation would be increased by raising the temperature into the 10 to 20°C range. In addition, as mentioned above, more efficient mixing of the soil could have reduced the observed variability in the results. The 16S rRNA gene microarray described here could be useful to monitor and evaluate such alternative bioremediation approaches, as it has shown the potential to highlight shifts in bacterial communities following bioremediation. Alternatively, it could be suggested that the limited number of probes on the microarray hampered the detection of the shifts in the bacterial community in Eureka soils. This is, however, unlikely, since all of the complementary methods used in this study showed a similar, highly variable response for Eureka replicate soil samples. Increasing the number of replicates through a more elaborate sampling strategy could also have improved significance and possibly revealed trends that were not visible with the level of replication that was used in this study.
Functional gene expression.
The expression of degradation-related genes (alkB and PAH-RHD) measured by qRT-PCR showed strong correlations with other more traditional measures of hydrocarbon degradation, like MPN-diesel counts and mineralization potential. As expected, based on its successful application in other systems, this indicates that qRT-PCR is a reliable method to evaluate the degradation of hydrocarbons in polar soils. At both locations, we observed a large and rapid decrease in soil hydrocarbon concentrations within only 1 month of treatment (Alert, −59%; Eureka, −37%). This could partly be due to soil manipulation (by tilling or mixing in biopiles) that probably enhanced hydrocarbon volatilization. However, our results also indicate that at Alert this decrease in soil hydrocarbons is also caused by bacterial degradation, since the expression of the measured biodegradation genes (alkB and PAH-RHD genes) strongly increased following nutrient amendment. Hydrocarbon-degrading bacteria in this cold environment therefore seem to respond very rapidly, in <1 month, to the addition of limiting nutrients. This also indicates that Alert soil bacteria are able to actively degrade hydrocarbons at very low temperatures, since the daily average air temperature for the month following the first nutrient amendment (August) was 0.8°C (maximum, 3.3°C; minimum, −1.8°C). This rapid response might also have been possible because August is one of the wettest months, with 21.2 mm of precipitation (mainly in the form of snow). Interestingly, in Eureka soils, the expression of most of the hydrocarbon-degrading genes was maximal after 1 year, indicating that Eureka soils might be responding more slowly to nutrient amendment. Mohn and Stewart (19) reported that the lag period in laboratory mineralization experiments was mainly explained by N and hydrocarbon content. However, we found no significant correlation between the expression of any of the genes and the hydrocarbon content of the soil, probably because the concentration of hydrocarbons, although much higher in Eureka soils (Table 2), did not reach concentrations known to be inhibitory for soil microorganisms (25,000 ppm) (17). Furthermore, in our case, the composition of the hydrocarbons might be more important than the total concentration in influencing the rapidity of the response to nutrient addition. Overall, the results from gene expression analyses demonstrated that bacterial degradation activities in Arctic soils are not limited by temperature or moisture content, but mainly by nutrient availability, as was previously shown (9, 19, 33, 34).
Interestingly, within 1 month of treatment in Alert soils, we also observed significant increases in nitrite-reducing gene expression, indicating that nutrient amendment also induced denitrification. Denitrifiers are common soil microorganisms (42), and several isolates can metabolize hydrocarbons (12). Denitrifiers were previously linked to anaerobic hydrocarbon degradation in amended polar soils (22). Anaerobic degradation of hydrocarbons uses an alternative pathway, through the benzoyl coenzyme A reductase (12, 27). The genes coding for this enzyme were not assessed in the present study, since anaerobic degradation was thought to be unlikely to contribute significantly to hydrocarbon degradation in the sampled surface soils and biopiles. However, the expression of denitrification genes indicates the presence of anaerobic microsites within the surface soil and the biopiles, wherein anaerobic hydrocarbon degradation could occur. The relative contribution of anaerobic microorganisms to the hydrocarbon degradation observed here is unknown but is likely to be small. Furthermore, the expression of nitrite reductase genes could lead to N losses, which might neutralize the positive effects of nitrogen amendment on bioremediation. Denitrification in temperate, diesel-contaminated soils depends upon the form of N used (24), stressing the importance of an in-depth understanding of the functions of the microorganisms involved in bioremediation.
Hydrocarbon-degrading bacterial community.
In contrast to the results of the 16S rRNA gene microarray, the hydrocarbon-degrading gene microarray indicated loose grouping of amended Eureka soil samples by ordination analyses. This might be due to subtle changes in the functional community that cannot be observed when measuring the total expression of a range of genes or the changes in 16S rRNA genes. However, here again, Alert samples showed a much clearer picture, with amended samples clustering tightly together. Both locations showed some similar sample-compound relationships (for instance, phenol, isopropylbenzene, and naphthalene with contaminated treated samples or catechol and chlorocatechol with uncontaminated and contaminated untreated samples; Fig. 3), indicating that diesel pollution and bioremediation induced a variety of consistent shifts in functional communities at both sites. Such shifts could be related to the selective degradation of hydrocarbon compounds since degradability under aerobic conditions varies between diesel compounds, in the following order: straight-chain alkanes > branched alkanes > monoaromatics > PAHs > high-molecular-weight PAHs (7, 16, 21). Supporting this hypothesis, the alkB/PAH-RHD expression ratio decreased following nutrient amendment in Alert soils from 973 (1 month) to 557 (1 year) and in Eureka soils from 1,489 (1 year) to 68 (4 years), indicating a shift from the degradation of easily degradable alkanes to more complex polyaromatic compounds. This shift in hydrocarbon-degrading communities was also observed in Antarctic soils, where a decrease in alkB gene numbers concomitant with an increase in diesel degraders was explained by a shift to bacteria able to utilize the non-alkane components of the diesel after the easily degraded alkanes had become scarce (21). Furthermore, Leys and colleagues (16) hypothesized that following contamination by PAHs, the majority of the initial degradation could be performed by gram-negative r-strategists, whereas K-strategist gram-positive bacteria could outcompete them for the degradation of more persistent high-molecular-weight PAHs. Here again, we found evidence in our data supporting this hypothesis, with a decrease following amendment in the gene expression ratio of gram-negative to gram-positive PAH-RHDs from 0.75 (1 month) to 0.13 (1 year) in Alert soils and a decrease from 0.11 (1 year) to 0.0019 (4 years) in Eureka soils.
Concluding remarks.
In this paper, we demonstrated the utility of microarrays and qRT-PCR as tools to rapidly monitor bacteria and their functions in polar environments during bioremediation. These molecular tools allowed us to confirm the potential of two Arctic soils to rapidly degrade hydrocarbons following nutrient amendment. Bioremediation efficiency was related to a rapid reorganization of the soil bacterial community and increased expression of hydrocarbon degradation genes. This efficiency differed between our two experimental sites, and site- and treatment-specific factors, like indigenous soil microbes, the extent of weathering of the contaminant, the nutrient amendment used, and the soil mixing strategy, could explain these differences. The ex situ treatment (i.e., the nutrient-amended biopiles at Alert) was more efficient than the in situ treatment (i.e., nutrient amendment followed by tilling at Eureka), the ex situ treatment being associated with less variable and larger changes in microbial indicators. Finally, the qRT-PCR strategy employed allowed us to observe successions in the expression of functional genes over time, which were most probably related to the successive degradation of different diesel compounds, starting with the most easily degraded components.
Acknowledgments
E. Yergeau was supported by a postdoctoral fellowship from NSERC. This study was supported by the Department of National Defense and Environment Canada.
Staff at HAWS-Eureka and CFS-Alert, Don Kovanen, Drew Craig, Sylvie Sanschagrin, Diane Labbé, Danielle Beaumier, Danielle Ouellette, Suzanne Labelle, and Claude Masson are gratefully acknowledged for technical support.
Footnotes
Published ahead of print on 14 August 2009.
REFERENCES
- 1.Aislabie, J., D. J. Saul, and J. M. Foght. 2006. Bioremediation of hydrocarbon-contaminated polar soils. Extremophiles 10:171-179. [DOI] [PubMed] [Google Scholar]
- 2.Beller, H. R., S. R. Kane, T. C. Legler, and P. J. J. Alvarez. 2002. A real-time polymerase chain reaction method for monitoring anaerobic, hydrocarbon-degrading bacteria based on a catabolic gene. Environ. Sci. Technol. 36:3977-3984. [DOI] [PubMed] [Google Scholar]
- 3.Cébron, A., M. P. Norini, T. Beguiristain, and C. Leyval. 2008. Real-time PCR quantification of PAH-ring hydroxylating dioxygenase (PAH-RHDα) genes from Gram positive and Gram negative bacteria in soil and sediment samples. J. Microbiol. Methods 73:148-159. [DOI] [PubMed] [Google Scholar]
- 4.DeSantis, T. Z., E. L. Brodie, J. P. Moberg, I. X. Zubieta, Y. M. Piceno, and G. L. Andersen. 2007. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb. Ecol. 53:371-383. [DOI] [PubMed] [Google Scholar]
- 5.Edgar, R., M. Domrachev, and A. E. Lash. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fierer, N., J. A. Jackson, R. Vilgalys, and R. B. Jackson. 2005. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 71:4117-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geerdink, M. J., M. C. M. van Loosdrecht, and K. C. A. M. Luyben. 1996. Biodegradability of diesel oil. Biodegradation 7:73-81. [Google Scholar]
- 8.Greer, C. W., J. Hawari, and R. Samson. 1990. Influence of environmental factors on 2,4-dichlorophenoxyacetic acid degradation by Pseudomonas cepacia isolated from peat. Arch. Microbiol. 154:317-322. [DOI] [PubMed] [Google Scholar]
- 9.Greer, C. W., C. Masson, D. Beaumier, D. Kovanen, and D. Craig. 2007. Biotreatability assessment and on site biotreatment monitoring of hydrocarbon contaminated soil at CFS-Alert, Nunavut, p. 3-11. In K. Biggar, G. Cotta, M. Nahir, A. Mullick, J. Buchko, A. Ho, and S. Guigard (ed.), Assessment and remediation of contaminated sites in Arctic and cold climates. ARCSACC, Edmonton, AB, Canada.
- 10.Haines, J. R., B. A. Wrenn, E. L. Holder, K. L. Strohmeier, R. T. Herrington, and A. D. Venosa. 1996. Measurement of hydrocarbon-degrading microbial populations by a 96-well plate most-probable-number procedure. J. Ind. Microbiol. 16:36-41. [DOI] [PubMed] [Google Scholar]
- 11.He, Z. L., T. J. Gentry, C. W. Schadt, L. Wu, J. Liebich, S. C. Chong, W. Wu, P. Jardine, C. Criddle, and J. Z. Zhou. 2007. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 1:67-77. [DOI] [PubMed] [Google Scholar]
- 12.Heider, J., A. M. Spormann, H. R. Beller, and F. Widdel. 1998. Anaerobic bacterial metabolism of hydrocarbons. FEMS Microbiol. Rev. 22:459-473. [Google Scholar]
- 13.Juck, D., T. Charles, L. G. Whyte, and C. W. Greer. 2000. Polyphasic microbial community analysis of petroleum hydrocarbon-contaminated soils from two northern Canadian communities. FEMS Microbiol. Ecol. 33:241-249. [DOI] [PubMed] [Google Scholar]
- 14.Labbé, D., R. Margesin, F. Schinner, L. G. Whyte, and C. W. Greer. 2007. Comparative phylogenetic analysis of microbial communities in pristine and hydrocarbon-contaminated Alpine soils. FEMS Microbiol. Ecol. 59:466-475. [DOI] [PubMed] [Google Scholar]
- 15.Lane, D. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acids techniques in bacterial systematics. John Wiley & Sons, West Sussex, United Kingdom.
- 16.Leys, N. M., A. Ryngaert, L. Bastiaens, P. Wattiau, E. M. Top, W. Verstraete, and D. Springael. 2005. Occurrence and community composition of fast-growing Mycobacterium in soils contaminated with polycyclic aromatic hydrocarbons. FEMS Microbiol. Ecol. 51:375-388. [DOI] [PubMed] [Google Scholar]
- 17.Long, S. C., C. M. Aelion, D. C. Dobbins, and F. K. Pfaender. 1995. A comparison of microbial community characteristics among petroleum-contaminated and uncontaminated subsurface soil samples. Microb. Ecol. 30:297-307. [DOI] [PubMed] [Google Scholar]
- 18.Marion, G. M., S. J. Hastings, S. F. Oberbauer, and W. C. Oechel. 1989. Soil-plant element relationships in a tundra ecosystem. Holarctic Ecol. 12:296-303. [Google Scholar]
- 19.Mohn, W. W., and G. R. Stewart. 2000. Limiting factors for hydrocarbon biodegradation at low temperature in Arctic soils. Soil Biol. Biochem. 32:1161-1172. [Google Scholar]
- 20.Perreault, N. N., D. T. Andersen, W. H. Pollard, C. W. Greer, and L. G. Whyte. 2007. Characterization of the prokaryotic diversity in cold saline perennial springs of the Canadian high Arctic. Appl. Environ. Microbiol. 73:1532-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powell, S. M., S. H. Ferguson, J. P. Bowman, and I. Snape. 2006. Using real-time PCR to assess changes in the hydrocarbon-degrading microbial community in Antarctic soil during bioremediation. Microb. Ecol. 52:523-532. [DOI] [PubMed] [Google Scholar]
- 22.Powell, S. M., S. H. Ferguson, I. Snape, and S. D. Siciliano. 2006. Fertilization stimulates anaerobic fuel degradation of Antarctic soils by denitrifying microorganisms. Environ. Sci. Technol. 40:2011-2017. [DOI] [PubMed] [Google Scholar]
- 23.Risher, J. F., and S. W. Rhodes. 1995. Toxicological profile for fuel oils. U.S. Department of Health and Human Services, Public Health Service, Agency for Toxic Substances and Disease Registry, Atlanta, GA. [PubMed]
- 24.Roy, R., and C. W. Greer. 2000. Hexadecane mineralization and denitrification in two diesel fuel-contaminated soils. FEMS Microbiol. Ecol. 32:17-23. [DOI] [PubMed] [Google Scholar]
- 25.Saul, D. J., J. M. Aislabie, C. E. Brown, L. Harris, and J. M. Foght. 2005. Hydrocarbon contamination changes the bacterial diversity of soil from around Scott Base, Antarctica. FEMS Microbiol. Ecol. 53:141-155. [DOI] [PubMed] [Google Scholar]
- 26.Shaver, G. R., and F. S. Chapin. 1980. Response to fertilization by various plant-growth forms in an Alaskan tundra—nutrient accumulation and growth. Ecology 61:662-675. [Google Scholar]
- 27.Song, B., and B. B. Ward. 2005. Genetic diversity of benzoyl coenzyme A reductase genes detected in denitrifying isolates and estuarine sediment communities. Appl. Environ. Microbiol. 71:2036-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ter Braak, C. J. F., and P. Šmilauer. 2002. CANOCO reference manual and CanoDraw for Windows user's guide: software for canonical community ordination (version 4.5). Microcomputer Power, Ithaca, NY.
- 29.Thomassin-Lacroix, E. J. M., M. Eriksson, K. J. Reimer, and W. W. Mohn. 2002. Biostimulation and bioaugmentation for on-site treatment of weathered diesel fuel in Arctic soil. Appl. Microbiol. Biotechnol. 59:551-556. [DOI] [PubMed] [Google Scholar]
- 30.Throbäck, I. N., K. Enwall, A. Jarvis, and S. Hallin. 2004. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 49:401-417. [DOI] [PubMed] [Google Scholar]
- 31.Vázquez, S., B. Nogales, L. Ruberto, E. Hernández, J. Christie-Oleza, A. Lo Balbo, R. Bosch, J. Lalucat, and W. Mac Cormack. 2009. Bacterial community dynamics during bioremediation of diesel oil-contaminated Antarctic soil. Microb. Ecol. 57:598-610. [DOI] [PubMed] [Google Scholar]
- 32.Wang, X. W., and B. Seed. 2003. Selection of oligonucleotide probes for protein coding sequences. Bioinformatics 19:796-802. [DOI] [PubMed] [Google Scholar]
- 33.Whyte, L. G., L. Bourbonnière, C. Bellerose, and C. W. Greer. 1999. Bioremediation assessment of hydrocarbon-contaminated soils from the High Arctic. Bioremediat. J. 3:69-79. [Google Scholar]
- 34.Whyte, L. G., B. Goalen, J. Hawari, D. Labbe, C. W. Greer, and M. Nahir. 2001. Bioremediation treatability assessment of hydrocarbon-contaminated soils from Eureka, Nunavut. Cold Regions Sci. Technol. 32:121-132. [Google Scholar]
- 35.Whyte, L. G., C. W. Greer, and W. E. Inniss. 1996. Assessment of the biodegradation potential of psychrotrophic microorganisms. Can. J. Microbiol. 42:99-106. [DOI] [PubMed] [Google Scholar]
- 36.Whyte, L. G., J. Hawari, E. Zhou, L. Bourbonnière, W. E. Inniss, and C. W. Greer. 1998. Biodegradation of variable-chain-length alkanes at low temperatures by a psychrotrophic Rhodococcus sp. Appl. Environ. Microbiol. 64:2578-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whyte, L. G., A. Schultz, J. B. van Beilen, A. P. Luz, V. Pellizari, D. Labbe, and C. W. Greer. 2002. Prevalence of alkane monooxygenase genes in Arctic and Antarctic hydrocarbon-contaminated and pristine soils. FEMS Microbiol. Ecol. 41:141-150. [DOI] [PubMed] [Google Scholar]
- 38.Whyte, L. G., T. H. M. Smits, D. Labbe, B. Witholt, C. W. Greer, and J. B. van Beilen. 2002. Gene cloning and characterization of multiple alkane hydroxylase systems in Rhodococcus strains Q15 and NRRL B-16531. Appl. Environ. Microbiol. 68:5933-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wrenn, B. A., and A. D. Venosa. 1996. Selective enumeration of aromatic and aliphatic hydrocarbon degrading bacteria by a most-probable-number procedure. Can. J. Microbiol. 42:252-258. [DOI] [PubMed] [Google Scholar]
- 40.Yergeau, E., S. Kang, Z. He, J. Zhou, and G. A. Kowalchuk. 2007. Functional microarray analysis of nitrogen and carbon cycling genes across an Antarctic latitudinal transect. ISME J. 1:163-179. [DOI] [PubMed] [Google Scholar]
- 41.Yergeau, E., S. A. Schoondermark-Stolk, E. L. Brodie, S. Déjean, T. Z. DeSantis, O. Gonçalves, Y. M. Piceno, G. L. Andersen, and G. A. Kowalchuk. 2009. Environmental microarray analyses of Antarctic soil microbial communities. ISME J. 3:340-351. [DOI] [PubMed] [Google Scholar]
- 42.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]