Abstract
Chromium is often found as a cocontaminant at sites polluted with organic compounds. For nitrate-respiring microbes, Cr(VI) may be not only directly toxic but may also specifically interfere with N reduction. In soil microcosms amended with organic electron donors, Cr(VI), and nitrate, bacteria oxidized added carbon, but relatively low doses of Cr(VI) caused a lag and then lower rates of CO2 accumulation. Cr(VI) strongly inhibited nitrate reduction; it occurred only after soluble Cr(VI) could not be detected. However, Cr(VI) additions did not eliminate Cr-sensitive populations; after a second dose of Cr(VI), bacterial activity was strongly inhibited. Differences in microbial community composition (assayed by PCR-denaturing gradient gel electrophoresis) driven by different organic substrates (glucose and protein) were smaller than when other electron acceptors had been used. However, the selection of bacterial phylotypes was modified by Cr(VI). Nine isolated clades of facultatively anaerobic Cr(VI)-resistant bacteria were closely related to cultivated members of the phylum Actinobacteria or Firmicutes. In Bacillus cereus GNCR-4, the nature of the electron donor (fermentable or nonfermentable) affected Cr(VI) resistance level and anaerobic nitrate metabolism. Our results indicate that carbon utilization and nitrate reduction in these soils were contingent upon the reduction of added Cr(VI). The amount of Cr(VI) required to inhibit nitrate reduction was 10-fold less than for aerobic catabolism of the same organic substrate. We speculate that the resistance level of a microbial process is directly related to the diversity of microbes capable of conducting it.
Chromium(VI) is a toxic metal that can negatively affect bioremediation of organic compounds in sites where chromium and organic pollutants cooccur (36). Under oxygen-limited conditions, chromium(VI) can be reduced (biologically or chemically) to insoluble and relatively nontoxic Cr(III) (22). Despite the potential interactions between biotic and chemical components, the responses of anaerobic microbial activities to Cr(VI) have not been well studied (6, 7, 42, 43).
Under anaerobic conditions, an important factor in the catabolism of organic carbon is the availability of electron acceptors. Nitrate is of special interest because it is often found as a copollutant in contaminated soils (18). Nitrate-reducing bacteria are facultative anaerobes commonly found in environmental samples and can couple the reduction of nitrate to the oxidation of diverse organic substrates (10, 13). The effect of Cr(VI) on natural denitrifying communities or pure cultures of denitrifying bacteria is not well characterized (8, 29). The environmental effects of Cr(VI) on denitrification are of particular interest because in addition to acute toxicity to the cell, Cr(VI) may compete with nitrate as an electron acceptor (15, 30). However, in other denitrifying bacteria (for example, Staphylococcus spp.), no competitive interactions were reported (45).
The purpose of this study was to extend our work on the effects of Cr(VI) upon microbes in soil that mediate discrete chemoheterotrophic processes such as the use of O2 (30) or Fe+3 (26) as terminal electron acceptors. We examined denitrification to determine whether the putative direct impact of Cr(VI) on the biochemistry of nitrate reduction would alter community dynamics from what had been observed with other terminal electron acceptors. In addition, we can add this data set to previous work to analyze the range of sensitivities to Cr(VI) that were found across a broad array of chemoheterotrophic processes.
MATERIALS AND METHODS
Soil collection.
Soil used for the microcosm experiments was collected in September 2003 from an Indiana Department of Transportation property in Seymour, Indiana. The soils at this site have been described previously (21). They are sandy (76% sand, 13% silt, and 11% clay) and have an average organic matter content of 3.1%. For this study, we used soil that had low levels of Pb (1.2 mg g−1 soil), Cr (5.9 mg g−1 soil), and petroleum hydrocarbons (toluene and xylenes, 0.2 mg g−1 soil), contaminants that were found at levels up to 1,000-fold higher within 25 m. The soil was transported to the lab and stored at 4°C for 3 months. Before the experiments were initiated, the soil was sieved through a 2-mm sieve.
Soil microcosms.
The experimental design was analogous to that used in previous studies employing other terminal electron acceptors; an organic energy source was added to act as a driving force for community change during exposure to a toxic heavy metal. The organic substrates were glucose (representative of readily degradable organic C metabolizable by a broad diversity of microbes) and protein (representing polymeric organic C originating from biomass). Individual microcosms were constructed in sterilized 105-ml serum vials with the equivalent of 10 g (dry weight) soil to which approximately 6 ml of liquid amendments were added. The treatment variables were the additions of (i) an organic energy source (glucose or the protein gelatin) at a concentration of 3 mg g−1 (dry weight) soil to act as a driving force for community change, (ii) a terminal electron acceptor (NO3−) to a final concentration of 15 μmol NO3− g−1 soil from a sterile, anoxic stock solution of KNO3, and (iii) Cr(VI) at concentrations that produced acute reductions of microbial activity of 50, 75, or 90% [designated low (L), medium (M). or high (H) and determined in preliminary experiments to be 140, 240, and 340 μg Cr(VI) g−1 soil, respectively]. Cr(VI) was added from a sterile, anoxic stock solution of potassium chromate. There were three replicate samples for each of the seven treatments for glucose (G) and seven for protein (P). Five treatments received organic carbon: (i) G (glucose) and P (protein) received only the carbon source, (ii) GN and PN received the carbon source plus NO3−, (iii) GNL and PNL received 140 μg Cr(VI) g−1 soil in addition to the carbon source and NO3−, (iv) GNM and PNM received 240 μg Cr(VI) g−1 soil in addition to the carbon source and NO3−, (v) GNH and PNH received 340 μg Cr(VI) g−1 soil in addition to the carbon source and NO3−. The two control treatments received either NO3− only or no chemical additions at all. Some microcosms were constructed with autoclaved soil to measure abiotic Cr reduction. In treatments that did not require specific amendments, an equal volume of sterile anoxic water was added. The vials were flushed with 100% nitrogen gas and sealed with gray butyl rubber stoppers and aluminum crimps. Microcosms were incubated in the dark at 20°C.
Activity measurements.
Microcosm headspaces were sampled with a gas-lock syringe at 2- to 7-day intervals, and CO2 was determined by a gas chromatograph (HP 5890 series II) as previously described (26).
Sampling regimen.
Soils in carbon-amended microcosms were destructively sampled four times based on the pattern of CO2 evolution. For samples amended with carbon, three vials were sampled per time point. All treatments were sampled 48 h after initiation (T1). Subsequently, microcosms were sampled on a treatment-specific schedule as follows: at the inception of substrate-induced mineralization (T2), during the maximum rate of CO2 production (T3), and when CO2 production leveled off (T4). Sampling on a treatment-specific basis facilitated comparison of microbial communities at similar stages of development. Treatments that lacked added carbon were sampled in duplicate at T1 and whenever Cr-amended microcosms were sampled at T3, as well as at the end of the experiment.
Microcosms were sampled by transferring their entire contents into sterile 15-ml polystyrene tubes. An aliquot of the liquid was taken and immediately used to measure the concentration of Cr(VI), NO2−, and NO3−. The remaining soil and liquid were frozen at −20°C and later used for DNA-based analysis.
Readdition of Cr(VI).
For all carbon and metal combinations (three for glucose and three for gelatin), a second set of six microcosms (designated with an R for readdition) was created. When activity in those microcosms was detectable (T2), a second dose of Cr(VI) was added. Soils in those microcosms were sampled two times in triplicate after Cr(VI) readdition—7 to 21 days after readdition (depending on activity) and when CO2 evolution in the microcosms had ceased.
Chromium and inorganic nitrogen measurements.
Chromate in solution was measured colorimetrically using the N,N-diphenylcarbazide method (12). Nitrate and nitrite were measured according to U.S. Environmental Protection Agency method 353.2 (44) on a QuickChem FIA+ 8000 series automated analyzer (Lachat Instruments, Loveland, CO).
Isolation of Cr-resistant bacteria.
Cells were extracted from GNM, GNMR, PNM, and PNMR soils in anoxic 10 mM phosphate saline buffer (pH 7.2). Small aliquots (0.1 ml) of serial dilutions were spread onto three types of solid media, 0.1× nutrient agar (Difco), soil extract agar with 0.5 g liter−1 glucose, and media for nitrate-reducing bacteria (NRB). All media contained 0.25 mM potassium chromate. Soil extract was prepared as previously described (26). NRB basal medium contained the following (all per liter): 2.5 g sodium bicarbonate, 0.25 g ammonium chloride, 0.6 g sodium phosphate monobasic, monohydrate, 0.1 g potassium chloride, 10 ml mineral solution (27), and 0.025 g yeast extract. For the isolation of NRB, the medium was supplemented with 1.36 g acetate per liter (final concentration, 20 mM) and 1.65 g potassium nitrate per liter (final concentration, 50 mM). Since denitrification is a facultative process, we did not take measures to remove oxygen from the plates prior to inoculation and incubation. The plates were incubated for 3 weeks in anaerobic jars (BBL GasPak Systems; Becton Dickinson, Franklin Lakes, NJ) at 20°C. Standard microbiological techniques were used to obtain pure cultures from single colonies on the plates. The isolates were then tested for growth on 0.5, 1, 2.5, 5, or 10 mM potassium chromate using the same solid medium that was used for isolation.
Inhibition of nitrate reduction by Cr(VI).
Isolate GNCr-4 (see Table 3) was grown in liquid NRB basal medium as described above, amended with Cr and nitrate in 30 different ratios, using a matrix of six nitrate concentrations (0, 3, 5, 10, 20, or 40 mM) and five chromate concentrations (0, 0.1, 0.25, 0.5, or 1 mM). Growth was tested with both a nonfermentable organic acid (lactate [20 mM]) that can be utilized only with concomitant nitrate reduction and with glucose (10 mM), which is a fermentable substrate that could be utilized with or without concomitant nitrate reduction. All culture experiments were carried out in triplicate for 48 h at 30°C in airtight Hungate tubes with anaerobic media and helium in the headspace. At the end of the incubation, nitrate, nitrite, and Cr(VI) in the medium were measured as described above. Biomass was estimated by measuring particulate protein. Cells were collected by centrifugation (5,000 × g for 20 min) and incubated with 1 M NaOH at 100°C for 10 min or until all biomass was dissolved. Protein was then measured using the Lowry method (28).
TABLE 3.
Summary of phylogenetic similarities determined from sequences obtained for dominant bands in DGGE profiles and isolates from microcosms amended with organic C (glucose or protein), NO3−, and 240 μg Cr(VI) g−1 soila
| Clone or isolateb | Best match | Accession no. | Similarityc (% similarity) | Phylogenetic clade |
|---|---|---|---|---|
| GNM-1(a) | Carnobacteriaceae clone BLUC-K | DQ196615 | 196/197 (99) | Firmicutes |
| GNM-2(b) | Bacterium clone FI-2F_H03 | EF220454 | 168/173 (97) | Cytophaga-Flavobacterium-Bacteroides group |
| GNM-3(b) | Uncultured Aranicola sp. strain LJEr1 | EU789569 | 197/197 (100) | Gammaproteobacteria |
| GNM-4(b) | Bacterium clone FFCH4639 | EU135403 | 169/169 (100) | Candidate division TM7 |
| GNM-5(b) | Serratia liquefaciens strain ZY-2 | EU880537 | 197/197 (100) | Gammaproteobacteria |
| GNM-6(b) | Uncultured bacterium clone 70 | DQ165123 | 165/169 (97) | Candidate division TM7 |
| GNM-7(c) | Bacterium clone FFCH10465 | EU134917 | 173/173 (89) | Candidate division OP10 |
| GNM-8(c) | Serratia sp. strain Tp5 | EU855752 | 197/197 (100) | Gammaproteobacteria |
| GNM-9(d) | Uncultured bacterium clone C08.ab1 | EU136313 | 174/174 (100) | Actinobacteria |
| GNM-10(e) | Propionibacterium freudenreichii ISU P59 | AY533300 | 171/174 (98) | Actinobacteria |
| Isolate GNCr-1 | Cellulosimicrobium sp. strain 87N50-1 | EU196469 | 455/456 (99) | Actinobacteria |
| Isolate GNCr-2 | Lactosphaera pasteurii | X87150 | 1477/1563 (94) | Firmicutes |
| Isolate GNCr-3 | Bacillus sp. strain CNJ940 PL04 | DQ448802 | 533/533 (100) | Firmicutes |
| Isolate GNCr-4 | Bacillus sp. strain cp-h51 | EU558977 | 536/536 (100) | Firmicutes |
| PNM-1(f) | Antarctic bacterium strain GA0A | EU636049 | 197/197 (100) | Gammaproteobacteria |
| PNM-2(g) | Brevibacillus brevis ZFJ-2 16S | EU931557 | 198/198 (100) | Firmicutes |
| PNM-3(h) | Aranicola clone LJEr1 | EU789569 | 197/197 (100) | Gammaproteobacteria |
| PNM-4(i) | Serratia sp. strain Tp5 | EU855752 | 197/197 (100) | Gammaproteobacteria |
| PNM-5(i) | Chlamydiales bacterium P-5 | AF364565 | 187/197 (94) | Chlamydiae |
| PNM-6(j) | Clone 37_C1_RHIZO_T7s | EF605981 | 172/172 (100) | Alphaproteobacteria |
| GNM-9(d) | Uncultured bacterium clone C08.ab1 | EU136313 | 174/174 (100) | Actinobacteria |
| PNM-8(l) | Mycobacterium sp. strain AT-3 | AF220427 | 177/177 (100) | Actinobacteria |
| Isolate PNCr-1 | Bacillus thuringiensis 2PR56-10 | EU440975 | 515/521 (98) | Firmicutes |
| Isolate PNCr-2 | Bacillus sp. strain PK-9 | EU685818 | 485/485 (100) | Firmicutes |
| Isolate PNCr-3 | Bacillus sp. strain ERI 44 | EU984074 | 422/424 (99) | Firmicutes |
| Isolate PNCr-4 | Bacterium GFCr-1 | DQ154277 | 499/504 (99) | Actinobacteria |
| Isolate PNCr-5 | Bacterium GFCr-1 | DQ154277 | 502/504 (99) | Actinobacteria |
All sequences are available in GenBank under accession numbers DQ426690 to DQ426716.
Clones and isolates starting with the capital letter G were from glucose-amended microcosms; clones and isolates starting with the capital letter P were from protein-amended microcosms.
The values shown are the number of similar base pairs/number of total base pairs. Approximately 200 bp was sequenced for clones and 500 bp for isolates.
Bacterial community analysis.
Changes in community composition were determined by soil DNA extraction, PCR amplification of the V3 region of the 16S rRNA gene with primers PRBA338f (with a GC-clamp) and PRUN518r, and separation of PCR amplicons by denaturing gradient gel electrophoresis (DGGE) (D-Code apparatus; Bio-Rad, Hercules, CA) as previously described (26). As with any molecular method based on PCR, our method is subject to PCR bias (1); however, DGGE has been widely and successfully used to compare the dominant members of the microbial community, especially in time series experiments.
DGGE profiles of different treatments were compared by calculating Dice similarity coefficients based on the presence or absence of bands (17). All bands were assigned to band classes after gels were normalized in comparison to marker lanes using the BioNumerics software package (Applied Maths, Belgium). Correct assignment of band classes was confirmed by rerunning putatively similar samples in adjacent DGGE lanes. Significant differences between treatments were analyzed by performing an analysis of variance on the distance matrix. We used the function adonis in the statistical package R, which performs a permutational or nonparametric multivariate analysis of variance (MANOVA) (4, 5). In short, the adonis function partitions the sums of squares of distances in the distance matrix based on predefined groups and calculates the significance of the grouping by performing multiple permutations of the data and constructing a pseudo-F distribution. The output of the adonis function is similar to a regular MANOVA output: F values for different factors and their interactions and their corresponding P values. We analyzed glucose- and protein-amended microcosms separately.
16S rRNA gene sequence determination.
DNA extracted from microcosms was amplified with primers PRBA338f without the GC-clamp and PRUN518r (26), and the PCR product was cloned in the pGEM-T Easy vector system (Promega, Madison, WI). Inserts from 24 random clones per microcosm were amplified using the PCR conditions described above with 1 μl of a cell suspension as the template and screened by DGGE. Plasmid DNA containing inserts was extracted by alkaline lysis (37), and the inserts were sequenced at Purdue University's Genomics Facility.
Bacterial isolates that grew at 5 mM or higher Cr(VI) concentrations were differentiated using the PCR-DGGE analysis described above. Isolates that produced a unique DGGE profile were putatively identified by sequencing. The 16S rRNA genes of unique isolates were amplified using primers pA (16) and PRUN518r and sequenced as described previously (26). The nearly full-length 16S rRNA gene sequence was determined for one isolate that had 95% or less similarity to its best matches in GenBank using primers pA and pH (16) as described previously (31).
Nucleotide sequences were compared to sequences in the National Center for Biotechnology Information (NCBI) GenBank database using the BLASTn program (3).
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this study have been submitted to the GenBank database under accession numbers DQ426690 to DQ426716.
RESULTS
Microbial activity.
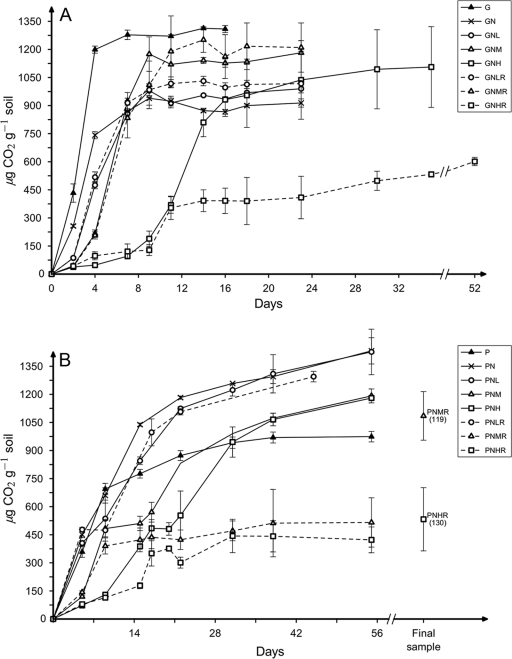
The addition of organic C and nitrate without Cr(VI) (GN and PN treatments) stimulated CO2 accumulation without an apparent lag (Fig. 1). Within 48 h, the addition of Cr inhibited CO2 production in microcosms stimulated with glucose by reducing it to 33, 17, and 15% of control [no Cr(VI)] values in GNL, GNM, and GNH microcosms, respectively. Similarly, in microcosms that received protein as a carbon and energy source, the addition of Cr(VI) at low, medium, or high levels immediately reduced CO2 production to 40, 25, and 24% of that found when no Cr(VI) was added. As Cr(VI) concentrations increased, the length of the lag phase increased in all Cr-amended microcosms, except GNL (Fig. 1). During the phases of maximum CO2 production, Cr also negatively affected the rates of CO2 accumulation in both glucose-amended (186, 152, 171, and 103 μg CO2 g−1 soil day−1 for GN, GNL, GNM, and GNH, respectively) and protein-amended (74, 58, 35 and 36 μg CO2 g−1 soil day−1 for PN, PNL, PNM, and PNH, respectively) microcosms. The maximum amount of CO2 produced in microcosms amended with organic C, nitrate, and chromium was not affected by Cr(VI) and ranged from 20 to 27% of the added C (based on the theoretical maximum mineralization of all C atoms in the added organic C source).
FIG. 1.
Cumulative CO2 accumulation in the headspace of anaerobic microcosms supplemented with glucose (G) (A) or protein (P) (B). In addition, amendments of nitrate (N) and/or one of three levels of Cr(VI) (low [L], 140 μg g−1 soil; medium [M], 240 μg g−1 soil; and high [H] 340 μg g−1 soil) were made. Some microcosms received a second dose of Cr(VI) (designated R for readdition). The microcosms subjected to PNMR and PNHR treatments were incubated for 119 and 130 days, respectively, and only the final amount of CO2 is plotted after day 56.
Chromium fate.
When Cr(VI) in the liquid phase was measured 0.5 h after addition, 100% was recovered; therefore, sorption to soil particles or the added protein was negligible. Abiotic decrease in Cr(VI) was analyzed in autoclaved controls when Cr(VI) could not be detected in live treatments (Table 1). The proportion of added Cr(VI) in abiotic controls ranged from 50% (L treatments) to 82% (H treatments).
TABLE 1.
Dynamics of nitrate reduction, nitrite appearance and consumption, and Cr(VI) reduction in carbon-, nitrate-, and chromium(VI)-amended anaerobic microcosms
| Treatmenta | NO3−
|
NO2−
|
Chromium
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Reduction rateb | Final concnc | Maximum concn | Time of maximum concn (days) | Final concn | Reduction rate | Time at which Cr(VI) became undetectable (days) | Reduction rate after readdition | Final concn | |
| N | 431 ± 2 | 0 | 648 ± 161 | 2 | 0 | ||||
| GN | 307 ± 77 | 452 ± 15 | 498 ± 35 | 2 | 260 ± 10 | ||||
| GNL | 109 ± 38 | 455 ± 35 | 538 ± 45 | 7 | 192 ± 118 | 70 ± 0 | 2 | 48 ± 1 | 0 |
| GNM | 109 ± 77 | 465 ± 104 | 287 ± 128 | 7 | 149 ± 11 | 96 ± 1 | 4 | 81 ± 1 | 0 |
| GNH | 82 ± 24 | 580 ± 161 | 85 ± 48 | 35 | 85 ± 48 | 63 ± 5 | 11 | 28 ± 5 | 0 |
| PN | 309 ± 86 | 0 | 766 ± 218 | 2 | 0 | ||||
| PNL | 154 ± 15 | 0 | 1,004 ± 43 | 5 | 0 | 70 ± 0 | 2 | 52 ± 1 | 0 |
| PNM | 75 ± 55 | 0 | 755 ± 318 | 7 | 0 | 98 ± 8 | 2 | 21 ± 3 | 0 |
| PNH | 63 ± 51 | 0 | 411 ± 38 | 14 | 0 | 99 ± 2 | 2 | 1 ± 1 | 29 ± 13 |
The letters in the treatments indicate the addition of nitrate (N) (15 μmol g−1 [dry weight] soil), glucose (G) (3 mg g−1 [dry weight] soil), or protein (P) (3 mg g−1 [dry weight] soil) and one of three chromium(VI) levels, low (L) (140 μg g−1 [dry weight] soil), medium (M) (240 μg g−1 [dry weight] soil), and high (H) (340 μg g−1 [dry weight] soil).
Reduction rates calculated for the first 48 h of incubation and reported as means ± standard errors for three replicate samples. The reduction rates are reported as micrograms gram−1 soil day−1.
All final concentrations reported are means ± standard errors for three replicate samples. The final concentrations are reported as micrograms gram−1 (dry weight) soil.
Concentrations of soluble Cr(VI) in live microcosms decreased rapidly (Table 1). After 48 h of incubation, 2, 49, and 215 μg Cr(VI) g−1 soil were found in GNL, GNM, and GNH microcosms, respectively. The corresponding numbers for PNL, PNM, and PNH microcosms were 0, 46, and 142 μg Cr(VI) g−1 soil, respectively. During that time, the rate of Cr(VI) decrease was similar in all microcosms and averaged 82 ± 10 μg Cr(VI) g−1 soil day−1 (Table 1). At subsequent sampling times (T2 to T4), no more than 2% of the added Cr(VI) was detected in any microcosm that received a single dose of Cr (Table 1).
Nitrate reduction and denitrification in microcosms.
The fate of added nitrate in microcosms was affected by the nature of the added organic substrate as well as the addition of Cr(VI) (Table 1). In glucose-amended microcosms, only a fraction of added nitrate was reduced and only to the level of nitrite. In contrast, in protein-amended microcosms, all added nitrate was reduced to nitrite and then to more-reduced molecules, as by the end of the incubation (days 56 to 130) no nitrate or nitrite was detected. Even in microcosms that were not amended with organic carbon, all nitrate was reduced beyond nitrite. Chromium inhibited the rate of nitrate reduction; the inhibition was stronger as the concentration of Cr(VI) increased (Table 1).
Readdition of Cr(VI) in microcosms.
Shi et al. (40) had found that these soils contain a broad mixture of Cr-sensitive and -tolerant bacteria. A potential consequence of the initial addition of Cr(VI) could be strong selection for proliferation of Cr(VI)-tolerant bacteria. To test this, a second dose of Cr(VI) was added to a subset of microcosms. However, the rates of Cr(VI) reduction after the second dose were in fact lower than the rates after the initial Cr(VI) additions (Table 1). Nitrate/nitrite reduction was reduced in all microcosms given a second dose of Cr(VI) with the sole exception of low-Cr addition to glucose-amended microcosms (Table 1). The effect of Cr was most severe in the high-Cr microcosms (GNHR and PNHR). Not only were Cr(VI) reductions rates lower after readdition, but the rate of CO2 accumulation was reduced (9 and 4 μg CO2 g−1 soil day−1 in GNHR and PNHR treatments, respectively), as was the total amount of organic C mineralized to 14 and 10% of the theoretical maximum mineralization for GNHR and PNHR, respectively (Fig. 1).
Microbial community dynamics.
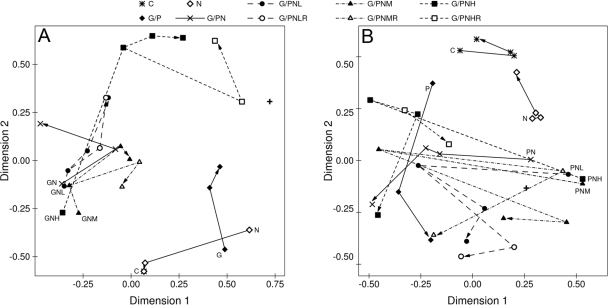
The addition of organic carbon, nitrate, and Cr(VI) resulted in a rapid change of the bacterial community structure; there were no bands in common between the initial DGGE profile and profiles of microcosms sampled after 48 h of incubation. Subsequent changes in the DGGE profiles were significantly altered by (i) the nature of the added carbon, (ii) the amount of chromium that was initially added, and (iii) the number of chromium(VI) doses that microcosms received (see Table 2 for permutational MANOVA summary statistics) (Fig. 2). In general, higher Cr(VI) concentrations led to a stronger differentiation in the microbial community, especially in glucose-amended microcosms. The immediate effect of Cr(VI) readdition was an altered bacterial community. By the end of the experiment, DGGE fingerprints of treatments that received the same single or double Cr(VI) dose converged, except in the case of GNH/GNHR and PNH/PNHR treatments. The nature of the added carbon affected the final community fingerprints; the Dice similarities between glucose- and protein-amended microcosms ranged between 0.5 and 0.6. In comparison, the average Dice similarity between replicate microcosms at sample time T4 was approximately 0.9.
TABLE 2.
Results from permutational MANOVA based on Jaccard distances for bacterial communities in glucose- or protein-amended anaerobic microcosms treated with one or two doses of various concentrations of Cr(VI)
| Analysis | Factor | F value | P value |
|---|---|---|---|
| Glucose-amended microcosms | |||
| Single chromium addition | Chromium | 23.8 | 0.001 |
| Time | 43.2 | 0.001 | |
| Readdition of Cr(VI) | Chromium | 44.6 | 0.001 |
| Second addition | 3.2 | 0.05 | |
| Time | 6.0 | 0.009 | |
| Protein-amended microcosms | |||
| Single Cr(VI) addition | Chromium | 4.6 | 0.001 |
| Time | 12.8 | 0.001 | |
| Readdition of Cr(VI) | Chromium | 21.9 | 0.001 |
| Second addition | 4.3 | 0.002 | |
| Time | 18.7 | 0.001 |
FIG. 2.
Nonmetric multidimensional scaling diagram of bacterial community profiles based on denaturing gradient gel electrophoresis of the V3 region of the 16S rRNA gene amplified by PCR of DNA extracted from anaerobic microcosms. Treatments included combinations of electron acceptor (none or NO3− [N]) and one of four levels of Cr(VI) (none, low [L], medium [M], or high [H]). In addition, microcosms were amended with glucose (G) (A) or protein (P) (B) as an electron donor. Each panel also includes results from a control treatment (C) to which no amendments were made. All treatments were set up in triplicate and sampled at four times (T1 to T4). Each point on the graph represents the average score for the three replicate samples. The initial T1 sample (48 h of incubation) is identified by the position of the legend code on the graph. The trajectory of community composition through time is indicated by the line and arrow connecting treatment points. Trajectories are drawn with solid lines for treatments without chromium and dashed lines for chromium-amended treatments. For comparison, the untreated soil bacterial community used to construct the microcosms is represented by a + symbol.
16S rRNA phylogenetic analysis.
The phylogenetic similarities of dominant bands from GNM and PNM microcosms were determined by cloning and sequencing (Table 3). Cloned sequences were closely related to database sequences from members of the Proteobacteriaceae and Actinobacteriaceae families and the Firmicutes phylum. Four dominant DGGE bands that were present in most microcosms that received organic carbon and/or Cr(VI) were most closely related to sequences from members of Serratia and Propionibacterium species.
Fifty-six isolates (18 from GNM and 38 from PNM samples) were obtained on anaerobic NRB medium under a low Cr(VI) selection pressure of 0.25 mM. Genotyping (by DGGE of amplified 16S rRNA genes) revealed four unique types from GNM samples and five unique types from PNM samples (Table 3). The 16S rRNA gene sequences of all isolates except Bacillus cereus GNCr-2 exhibited more than 98% similarity to cultivated bacteria within the genus Bacillus or the phylum Actinobacteria. The strains in each of these clades were facultative anaerobes and were capable of growth in the presence of at least 10 mM Cr(VI) under aerobic conditions.
Inhibition of nitrate reduction by Cr(VI) in pure culture.
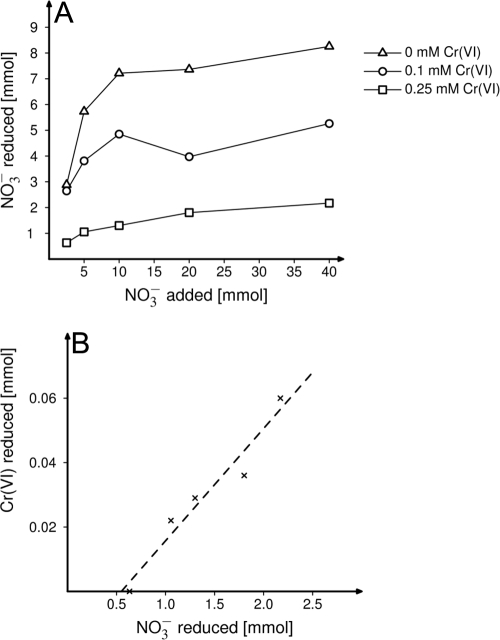
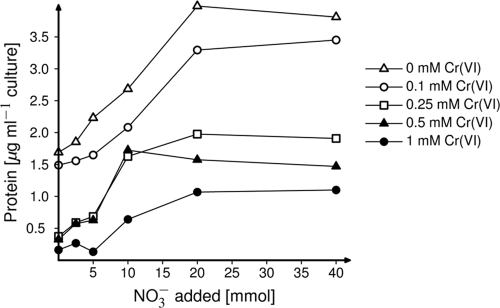
From the nine distinct clades of isolates, one strain, Bacillus cereus GNCr-4, grew in a chemically defined minimal medium and therefore was chosen for more-detailed analysis of the physiological interactions between electron donor and the reduction of Cr(VI) and nitrate. Although B. cereus GNCr-4 grew in media with up to 20 mM Cr(VI) under aerobic conditions with glucose, under anaerobic conditions with nitrate, it grew only in the presence of up to 1 mM Cr(VI) when using glucose as a carbon source and only 0.5 mM Cr(VI) when using lactate as a carbon source. When lactate was the carbon source, nitrite accumulated in either the presence or absence of Cr(VI). In contrast, when glucose was the carbon source, nitrate was reduced only in the absence of Cr(VI). Whenever nitrate reduction occurred, mass balance analyses of nitrate and nitrite concentrations demonstrated that nitrate was reduced only to the level of nitrite (data not shown). In both glucose- and lactate-amended cultures, the growth yield and amount of nitrate reduced decreased as the concentration of Cr(VI) in the medium increased (Fig. 3A and 4). In addition, the inhibitory effect of Cr(VI) was diminished at higher concentrations of nitrate (Fig. 3A and 4). The reduction of added Cr(VI) was also measured in these cultures, and strain GNCr-4 reduced Cr(VI) when lactate but not glucose was the carbon source. When 0.1 mM Cr(VI) was added to lactate-amended cultures, all Cr(VI) was reduced. At 0.25 mM Cr, the amount of Cr(VI) reduced increased at higher nitrate levels and was linearly correlated to the amount of nitrate reduction [33 μmol Cr(VI) reduced per mmol nitrate reduced; r = 0.97] (Fig. 3B).
FIG. 3.
Nitrate and chromium reduction by bacterial isolate GNCr-4 grown anaerobically in basal medium amended with various concentrations of nitrate and chromium(VI) and 20 mM lactate as a carbon source. (A) The inhibitory effect of Cr(VI) upon nitrate reduction, as illustrated by the amount of nitrate reduced as increasing amounts of nitrate were added at 0, 0.1, or 0.25 mM Cr(VI). (B) For the set of cultures that grew at the highest Cr(VI) concentration (0.25 mM), the correlation between the amounts of nitrate and chromate reduced is shown. The amounts of nitrate and Cr(VI) reduced were lowest in cultures with 2.5 mM nitrate added and highest when 40 mM nitrate was present.
FIG. 4.
Biomass production (expressed as particulate protein) by cultures of Bacillus cereus GNCr-4 grown anaerobically in basal medium amended with various concentrations of nitrate and chromium(VI) and 10 mM glucose as a carbon source.
Toxicity of Cr(VI) to different microbial processes.
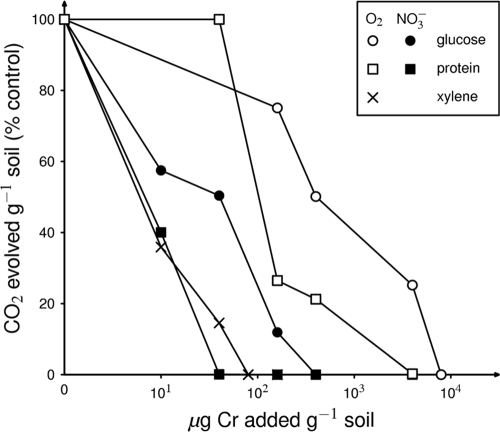
We have determined the microbial community response not only when nitrate is the terminal electron acceptor but also under aerobic and Fe-reducing conditions (26, 33) with three different organic electron donors. The dose of Cr(VI) required to produce an acute toxic effect varied about 100-fold for these different functional processes (Fig. 5). The highest concentration [about 400 μg Cr(VI) g−1 soil to achieve 50% inhibition] was needed for aerobic glucose catabolism. In contrast, aerobic xylene degradation (for which one might expect the presence of a much smaller number of functionally redundant species) was inhibited by only 10 μg Cr(VI) g−1 soil. Relatively low Cr(VI) concentrations were necessary to inhibit nitrate respiration; this may suggest that the functional diversity of this physiological group was relatively low.
FIG. 5.
Effects of increasing Cr(VI) concentrations on the initial response (2 to 7 days) of soil microbial communities to the addition of different electron acceptors (O2 [open symbols] or NO3− [closed symbols]) and carbon sources (glucose •, protein ▪, or xylene [×]).
DISCUSSION
The contaminated soils used in this experiment do not support plant growth; therefore, it is likely that bacterial communities were carbon limited, and not surprisingly, they responded very rapidly to newly added carbon (Fig. 1). Furthermore, contrasting microbial communities (Fig. 2) were selected in response to different organic amendments, as has been previously shown (25, 39). The results of these experiments are novel in that the intensity of selection imposed by different organic amendments was less when nitrate was the terminal electron acceptor (protein and glucose microcosms shared more than 50% of dominant bands in DGGE profiles) than in earlier work with Fe(III) as the terminal electron acceptor. Under those conditions, similarity coefficients between glucose and protein amendments ranged from 10 to 30% (26). Thus, nitrate modulated the capacity for an organic carbon source to drive large changes in soil community composition to a larger degree than had been observed with other terminal electron acceptors.
Dissimilatory nitrate reduction has been a subject of intense study of soil (34). However, the results from Seymour soils were striking in that the fate of nitrate was altered not only by the addition of chromate, a metal oxyanion, but also by the nature of the added organic carbon. Whereas bacteria metabolizing endogenous soil organic C or added protein reduced nitrate beyond nitrite, nitrate was not reduced beyond nitrite in glucose-amended microcosms. Nitrite accumulation has previously been observed under denitrifying conditions (24), especially when carbohydrates were the electron donors (19, 23). The fate of nitrate in microcosms with glucose was also notable in that dissimilation was incomplete; although the amount of added glucose would generate twice the number of electrons required to reduce all added nitrate to nitrogen gas, about 50% of nitrate was still present in glucose-amended microcosms at the end of the incubation. A concentration of 5 mM nitrite can inhibit nitrate reduction in pure cultures (2). We found that a nitrate-reducing bacterium isolated from the microcosms, B. cereus GNCr-4, ceased nitrate reduction when nitrite reached 10 mM.
Our results also demonstrated that chromate, an oxyanion that could potentially serve as a competing electron acceptor (15) as well as a toxic metal, modulated the composition of nitrate-reducing anaerobic bacterial communities and inhibited nitrate reduction in soil microcosms. The interaction between Cr(VI) and denitrification in intact natural communities has not been well described, but a few studies indicated that soil denitrification might be particularly sensitive to Cr(VI) inhibition (41). The acute effect of heavy metal addition to soil is often a narrowing of microbially diverse populations (due to death of sensitive organisms) and a resulting metal-tolerant community (20, 32). Over a longer time period, selective proliferation and genetic changes can occur (14). Chromium(VI) differs from many other heavy metals, because it can be reduced to less-toxic Cr(III). Furthermore, Cr(VI) bioavailability can be reduced by complexation with organic matter (35). Thus, the microbial community dynamics after chromium(VI) addition could be influenced by the survival of Cr-sensitive bacteria until Cr(VI) is detoxified. Two lines of evidence from our study indicate that organic C is utilized by a mixture of Cr-resistant and Cr-sensitive bacteria. First, at Cr(VI) levels below 240 μg g−1 soil, the similarity of DGGE profiles to unamended treatments by the end of the incubation indicates that the bacterial community in the soils may have been “protected” from the toxic effect of Cr(VI), due either to low Cr(VI) bioavailability or rapid reduction to less-toxic Cr(III) or broad biological resistance to this level. Only higher levels of Cr (340 μg g−1 soil) addition produced a subtractive effect on community composition. Second, in the case of high-Cr readdition (340 μg g−1 soil), extreme inhibition was observed, indicating that Cr-sensitive members survived the first dose of Cr(VI) and presumably were partially responsible for the oxidation of the added organic C. These responses had also been observed when Fe(III) was added as the terminal electron acceptor to these soils (26). In contrast, under aerobic conditions, carbon was mineralized in these soils even when a substantial amount of Cr(VI) remained (33).
Soil microcosms are complex systems, and our results with B. cereus GNCr-4 isolated from the microcosms, as well as previous published work with pure cultures suggest that there are multiple interactions between Cr(VI) and nitrate transformations. Cr(VI) addition has been shown to inhibit rates of nitrate and nitrite reduction in Shewanella oneidensis MR1 and several different strains of Pseudomonas (15, 29, 30). In contrast to the purely antagonistic relationship between nitrate and chromate metabolism found in Shewanella and Pseudomonas, there was a positive correlation between nitrate and Cr(VI) reduction by B. cereus GNCr-4 exposed to various ratios of nitrate to Cr(VI) (Fig. 3B). A similar relationship between Cr(VI) reduction and nitrate was reported with Staphylococcus epidermidis (45). Thus, nitrate-reducing bacteria and soil nitrate concentrations may accelerate the rate of anaerobic Cr(VI) reduction, which results in relief of Cr(VI) toxicity.
Our primary interest in this work was upon the soil microbial community. However, limited analyses of cultures identified physiological phenomena related to Cr(VI) tolerance that have not been reported in the literature and warrant future in-depth investigations. All of the isolates were facultative anaerobes, and for most, the level of Cr(VI) tolerance was similar under aerobic and anaerobic conditions. However, in B. cereus GNCr-4, Cr(VI) resistance levels were 10-fold higher aerobically than anaerobically. Second, we found that the nature of the electron donor affected Cr(VI) resistance in this isolate; cultures fermenting glucose had higher Cr resistance than cultures that coupled nitrate reduction to lactate oxidation. The relationship between chromium and nitrate reduction in pure cultures requires subsequent investigation to determine the operative biochemical mechanisms.
Culture-independent and -dependent methods allowed us to identify some of the indigenous populations that were enriched in microcosms. The majority of Cr-resistant isolates and sequences obtained from cloning (Table 1) were closely related to known nitrate-respiring bacteria (Serratia, Propionibacterium, and Bacillus) and Cr-resistant and/or reducing bacteria (Serratia, Bacillus, and Cellulomonas) that have been previously recognized in Cr-polluted soils or sediments (9, 11, 38). In addition, several cloned sequences and one of the isolates were related to genera not previously reported to contain Cr-resistant members, including sequences that belong to candidate divisions TM7 and OP10, which have no cultivated representatives.
We have investigated the effect of Cr(VI) on microbial communities metabolizing several different electron donors and three different terminal electron acceptors (26, 33). When taken together (Fig. 5), the Cr(VI) sensitivity of these processes differed by almost 2 orders of magnitude. Although we have no quantitative data on functional redundancy in these soils, process sensitivity to acute Cr(VI) toxicity appears inversely correlated to the expected bacterial diversity for that process in soil, with the highest tolerance by aerobic glucose metabolizers and low tolerance by aromatic degraders. This is consistent with the ecological “insurance hypothesis” (46) in which functional redundancy promotes ecosystem functional stability because it increases the probability that the community contains a member resistant to a new stressor. In this regard, anaerobic nitrate respiration appeared to have a much lower level of functional redundancy than aerobic respiration of either glucose or protein.
Acknowledgments
This work was supported by a grant from the Department of Energy's Environmental Remediation Science Program (DE-FG02-98ER62681).
We thank the Indiana Department of Transport and in particular Bill Jervis for giving us access to this site.
Footnotes
Published ahead of print on 14 August 2009.
REFERENCES
- 1.Ahn, J. H., M. C. Kim, H. C. Shin, M. K. Choi, S. S. Yoon, T. Kim, H. G. Song, G. H. Lee, and J. O. Ka. 2006. Improvement of PCR amplification bias for community structure analysis of soil bacteria by denaturing gradient gel electrophoresis. J. Microbiol. Biotechnol. 16:1561-1569. [Google Scholar]
- 2.Almeida, J. S., S. M. Júlio, M. A. M. Reis, and M. J. T. Carrondo. 1995. Nitrite inhibition of denitrification in Pseudomonas fluorescens. Biotechnol. Bioeng. 46:194-201. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, M. J. 2001. A new method for non-parametric multivariate analysis of variance. Aus. Ecol. 26:32-46. [Google Scholar]
- 5.Anderson, M. J. 2001. Permutation tests for univariate or multivariate analysis of variance and regression. Can. J. Fish. Aquat. Sci. 58:626-639. [Google Scholar]
- 6.Arias, Y. M., A. Obraztsova, B. M. Tebo, and C. Green-Ruiz. 2003. Natural attenuation of Cr(VI) contamination in laboratory mesocosms. Geomicrobiol. J. 20:389-401. [Google Scholar]
- 7.Arias, Y. M., and B. M. Tebo. 2003. Cr(VI) reduction by sulfidogenic and nonsulfidogenic microbial consortia. Appl. Environ. Microbiol. 69:1847-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bardgett, R. D., T. W. Speir, D. J. Ross, G. W. Yeates, and H. A. Kettles. 1994. Impact of pasture contamination by copper, chromium, and arsenic timber preservative on soil microbial properties and nematodes. Biol. Fertil. Soils 18:71-79. [Google Scholar]
- 9.Branco, R., A.-P. Chung, A. Veríssimo, and P. V. Morais. 2005. Impact of chromium-contaminated wastewaters on the microbial community of a river. FEMS Microbiol. Ecol. 54:35-46. [DOI] [PubMed] [Google Scholar]
- 10.Burland, S. M., and E. A. Edwards. 1999. Anaerobic benzene biodegradation linked to nitrate reduction. Appl. Environ. Microbiol. 65:529-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camargo, F. A. O., F. M. Bento, B. C. Okeke, and W. T. Frankenberger. 2003. Chromate reduction by chromium-resistant bacteria isolated from soils contaminated with dichromate. J. Environ. Qual. 32:1228-1233. [DOI] [PubMed] [Google Scholar]
- 12.Clesceri, L. S., A. E. Greenberg, and A. D. Eaton (ed.). 1998. Standard methods for the examination of water and wastewater, 20th ed. American Public Health Association, Washington, DC.
- 13.Cunningham, J. A., G. D. Hopkins, C. A. Lebron, and M. Reinhard. 2000. Enhanced anaerobic bioremediation of groundwater contaminated by fuel hydrocarbons at Seal Beach, California. Biodegradation 11:159-170. [DOI] [PubMed] [Google Scholar]
- 14.Díaz-Raviña, M., and E. Bååth. 1996. Development of metal tolerance in soil bacterial communities exposed to experimentally increased metal levels. Appl. Environ. Microbiol. 62:2970-2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dmitrenko, G. N., V. V. Konovalova, and T. V. Ereshko. 2006. The successive reduction of Cr(VI) and NO3− or Mn(IV) ions present in the cultivation medium of denitrifying bacteria. Microbiology 75:125-128. [PubMed] [Google Scholar]
- 16.Edwards, U., T. Rogall, H. Blocker, M. Emde, and E. C. Bottger. 1989. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feris, K. P., P. W. Ramsey, M. Rillig, J. N. Moore, J. E. Gannon, and W. E. Holben. 2004. Determining rates of change and evaluating group-level resiliency differences in hyporheic microbial communities in response to fluvial heavy-metal deposition. Appl. Environ. Microbiol. 70:4756-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fredrickson, J. K., J. M. Zachara, D. L. Balkwill, D. Kennedy, S.-M. W. Li, H. M. Kostandarithes, M. J. Daly, M. F. Romine, and F. J. Brockman. 2004. Geomicrobiology of high-level nuclear waste-contaminated vadose sediments at the Hanford site, Washington State. Appl. Environ. Microbiol. 70:4230-4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gómez, M. A., J. González-López, and E. Hontoria-García. 2000. Influence of carbon source on nitrate removal of contaminated groundwater in a denitrifying submerged filter. J. Haz. Mat. 80:69-80. [DOI] [PubMed] [Google Scholar]
- 20.Holtan-Hartwig, L., M. Bechmann, T. Risnes Hås, R. Linjordet, and L. Reier Bakken. 2002. Heavy metals tolerance of soil denitrifying communities: N2O dynamics. Soil Biol. Biochem. 34:1181-1190. [Google Scholar]
- 21.Joynt, J., M. Bischoff, R. Turco, A. Konopka, and C. H. Nakatsu. 2006. Microbial community analysis of soils contaminated with lead, chromium and petroleum hydrocarbons. Microb. Ecol. 51:209-219. [DOI] [PubMed] [Google Scholar]
- 22.Kamaludeen, S. P. B., M. Megharaj, A. L. Juhasz, N. Sethunathan, and R. Naidu. 2003. Chromium-microorganism interactions in soils: remediation implications. Rev. Environ. Contam. Toxicol. 178:93-164. [DOI] [PubMed] [Google Scholar]
- 23.Kelso, B., R. Smith, R. Laughlin, and S. Lennox. 1997. Dissimilatory nitrate reduction in anaerobic sediments leading to river nitrite accumulation. Appl. Environ. Microbiol. 63:4679-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelso, B. H. L., R. V. Smith, and R. J. Laughlin. 1999. Effects of carbon substrates on nitrite accumulation in freshwater sediments. Appl. Environ. Microbiol. 65:61-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleikemper, J., O. Pelz, M. H. Schroth, and J. Zeyer. 2002. Sulfate-reducing bacterial community response to carbon source amendments in contaminated aquifer microcosms. FEMS Microbiol. Ecol. 42:109-118. [DOI] [PubMed] [Google Scholar]
- 26.Kourtev, P. S., C. H. Nakatsu, and A. Konopka. 2006. Response of the anaerobic bacterial community responses to addition of organic C in chromium(VI)- and iron(III)-amended microcosms. Appl. Environ. Microbiol. 72:628-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lovley, D. 2006. Dissimilatory Fe(III)- and Mn(IV)-reducing prokaryotes, p. 635-658. In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, vol. 2. Ecophysiology and biochemistry. Springer-Verlag, New York, NY. [Google Scholar]
- 28.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 29.Mazierski, J. 1994. Effect of chromium (CrVI) on the growth rate of denitrifying bacteria. Water Res. 28:1981-1985. [Google Scholar]
- 30.Middleton, S. S., R. B. Latmani, M. R. Mackey, M. H. Ellisman, B. M. Tebo, and C. S. Criddle. 2003. Cometabolism of Cr(VI) by Shewanella oneidensis MR-1 produces cell-associated reduced chromium and inhibits growth. Biotechnol. Bioeng. 83:627-637. [DOI] [PubMed] [Google Scholar]
- 31.Morgan, C. A., A. Hudson, A. Konopka, and C. H. Nakatsu. 2002. Analyses of microbial activity in biomass-recycle reactors using denaturing gradient gel electrophoresis of 16S rDNA and 16S rDNA PCR products. Can. J. Microbiol. 48:331-341. [DOI] [PubMed] [Google Scholar]
- 32.Müller, A. K., K. Westergaard, S. Christensen, and S. J. Sorensen. 2001. The effect of long-term mercury pollution on the soil microbial community. FEMS Microbiol. Ecol. 36:11-19. [DOI] [PubMed] [Google Scholar]
- 33.Nakatsu, C. H., N. Carmosini, B. Baldwin, F. Beasley, P. Kourtev, and A. Konopka. 2005. Soil microbial community responses to additions of organic carbon substrates and heavy metals (Pb and Cr). Appl. Environ. Microbiol. 71:7679-7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Philippot, L., S. Hallin, and M. Schloter. 2007. Ecology of denitrifying prokaryotes in agricultural soil. Adv. Agron. 96:249-305. [Google Scholar]
- 35.Richard, F. C., and A. C. M. Bourg. 1991. Aqueous geochemistry of chromium—a review. Water Res. 25:807-816. [Google Scholar]
- 36.Said, W. A., and D. L. Lewis. 1991. Quantitative assessment of the effects of metals on microbial degradation of organic chemicals. Appl. Environ. Microbiol. 57:1498-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Sani, R. K., B. M. Peyton, W. A. Smith, W. A. Apel, and J. N. Petersen. 2002. Dissimilatory reduction of Cr(VI), Fe(III), and U(VI) by Cellulomonas isolates. Appl. Microbiol. Biotechnol. 60:192-199. [DOI] [PubMed] [Google Scholar]
- 39.Schutter, M., and R. Dick. 2001. Shifts in substrate utilization potential and structure of soil microbial communities in response to carbon substrates. Soil Biol. Biochem. 33:1481-1491. [Google Scholar]
- 40.Shi, W., M. Bischoff, R. Turco, and A. Konopka. 2002. Long-term effects of chromium and lead upon the activity of soil microbial communities. Appl. Soil Ecol. 21:169-177. [Google Scholar]
- 41.Speir, T. W., H. A. Kettles, A. Parshotam, P. L. Searle, and L. N. C. Vlaar. 1995. A simple kinetic approach to derive the ecological dose value, ED50, for the assessment of Cr(VI) toxicity to soil biological properties. Soil Biol. Biochem. 27:801-810. [Google Scholar]
- 42.Trujillo-Tapia, N., C. C. Mondragón, M. S. Vásquez-Murrieta, O. Van Cleemput, and L. Dendooven. 2008. Inorganic N dynamics and N2O production from tannery effluents irrigated soil under different water regimes and fertilizer application rates: a laboratory study. Appl. Soil Ecol. 38:279-288. [Google Scholar]
- 43.Turick, C. E., and W. A. Apel. 1997. A bioprocessing strategy that allows for the selection of Cr(VI)-reducing bacteria from soils. J. Ind. Microbiol. Biotechnol. 18:247-250. [Google Scholar]
- 44.U.S. Environmental Protection Agency. 1993. Methods for the chemical analysis of water and wastes (MCAWW) (EPA/600/4-79/020). National Exposure Research Laboratory, U.S. Environmental Protection Agency, Washington, DC. http://www.nemi.gov/apex/f?p=237:38:918307873937113::::P38_METHOD_ID:4702.
- 45.Vatsouria, A., M. Vainshtein, P. Kuschk, A. Wiessner, D. Kosolapov, and M. Kaestner. 2005. Anaerobic co-reduction of chromate and nitrate by bacterial cultures of Staphylococcus epidermidis L-02. J. Ind. Microbiol. Biotechnol. 32:409-414. [DOI] [PubMed] [Google Scholar]
- 46.Yachi, S., and M. Loreau. 1999. Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proc. Natl. Acad. Sci. USA 96:1463-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]