Abstract
Gassericin A is a circular bacteriocin produced by Lactobacillus gasseri strain LA39. We found a 33,333-bp plasmid, designated pLgLA39, in this strain. pLgLA39 contained 44 open reading frames, including seven genes related to gassericin A production/immunity (gaa), as well as genes for replication, plasmid maintenance, and conjugative transfer. pLgLA39 was transferred from LA39 to the type strain of L. gasseri (JCM 1131) by filter mating. The transconjugant exhibited >30-fold-higher more resistance to gassericin A and produced antibacterial activity. Lactobacillus reuteri LA6, the producer of reutericin 6, was proved to harbor a plasmid indistinguishable from pLgLA39 and carrying seven genes 100% identical to gaa. This suggests that pLgLA39 might have been transferred naturally between L. gasseri LA39 and L. reuteri LA6. The seven gaa genes of pLgLA39 were cloned into a plasmid vector to construct pGAA. JCM 1131T transformed with pGAA expressed antibacterial activity and resistance to gassericin A. pGAA was segregationally more stable than a pGAA derivative plasmid from which gaaA was deleted and even was more stable than the vector. This suggests the occurrence of postsegregational host killing by the gaa genes. pLgLA39 carried a pemIK homolog, and segregational stabilization of a plasmid by the pLgLA39-type pemIK genes was also confirmed. Thus, pLgLA39 was proved to carry the genes for at least two plasmid maintenance mechanisms, i.e., gaa and pemIK. Plasmids containing a repA gene similar to pLgLA39 repA were distributed in several L. gasseri strains.
Lactobacillus species are normal inhabitants of the human gastrointestinal tract, and Lactobacillus gasseri is one of the most commonly detected of these species (37, 47). Health-promoting effects of this species, such as immunomodulation (35), suppression of Helicobacter pylori-induced interleukin-8 production (44), and improvement of intestinal conditions (34), have been reported, and some L. gasseri strains are used in commercial probiotic products.
Bacteriocins are antimicrobial peptides, proteins, or protein complexes produced by bacteria and active mainly against related bacterial species (38). Several bacteriocins also inhibit the growth of food-borne pathogens, such as Listeria, Bacillus cereus, and Clostridium perfringens. Production of bacteriocin is thought to be a desired feature for probiotic strains, since bacteriocin is believed to provide an advantage for survival in the ecological niche and to prevent the growth of pathogens. Several L. gasseri strains are known to produce bacteriocins (18). The classification of bacteriocins remains controversial. We use the definition proposed by Maqueda et al. (30), where bacteriocins are classified into class I (lantibiotics), class II (nonlantibiotics), class III (large heat-labile bacteriocins), and class IV (circular bacteriocins linked at the N- and C-terminal ends). Among these, the class IV circular bacteriocins have attracted increasing attention, since they are the simplest prokaryotic representatives of the ubiquitous circular peptides with various physiological activities (6). Enterocin AS-48 from Enterococcus faecalis strain S-48 is the first and most vigorously characterized member of the class IV bacteriocins (30). L. gasseri strain LA39 (JCM 11657) produces a 58-amino-acid (aa) circular bacteriocin, gassericin A (18). Gassericin A is a representative of the non-AS-48-like circular bacteriocin group including butyrivibriocin AR10 from Butyrivibrio fibrisolvens AR10 (15) and carnocyclin A from Carnobacterium maltaromaticum UAL307 (32), as well as reutericin 6 from Lactobacillus reuteri LA6 (17) and acidocin B from Lactobacillus acidophilus M46 (26). The last two bacteriocins have nearly identical amino acid sequences to that of gassericin A. Though the number of reported circular bacteriocins has been increasing, their primary sequences and the genes responsible for production of and immunity to them are diversified (for a review, see reference 31). Recently, we isolated and sequenced seven genes (gaaBCADITE) from LA39 deduced to be responsible for production of and immunity to gassericin A (20). The gaa genes add new information to the complex world of the class IV bacteriocin genes.
The structural gene of gassericin A, gaaA, was reported to be located on the chromosome of LA39 (19). However, the high amino acid sequence identity of gassericin A to reutericin 6 (100%) and to acidocin B (98%) suggests recent horizontal gene transfers of the relevant bacteriocin genes, possibly via mobile elements. In fact, the acidocin B genes were reported to be located on a plasmid, namely, pCV461 (26). Many Lactobacillus strains are known to harbor one or more plasmids of various sizes, and several Lactobacillus plasmids have been reported to contain genes for production of bacteriocins (48). To our knowledge, however, only three have been sequenced entirely: these are pLA103 from Lactobacillus acidophilus TK8912 (16), pRC18 from Lactobacillus curvatus (previously known as Lactobacillus casei) CRL705 (7), and pMP118 from Lactobacillus salivarius subsp. salivarius UCC118 (5). Thus, genetic information about bacteriocin-producing Lactobacillus plasmids is still limited. Furthermore, little has been known about plasmids of L. gasseri, even though the existence of plasmids in a few strains has been reported, including a 26.5-kb anonymous plasmid in strain ADH (27) and pK7 in strain K7 (28).
Here we describe a 33.3-kb plasmid, designated pLgLA39, from L. gasseri LA39. The gaa genes are located on this plasmid. pLgLA39 carries a set of genes for conjugative transfer and was shown to be transmitted to another L. gasseri strain. L. reuteri LA6 also harbors a plasmid almost identical to pLgLA39. We demonstrated that production of gassericin A increased the apparent segregational stability of a plasmid carrying the gaa genes. A pemIK homolog in pLgLA39 was also functional as a plasmid-stabilizing mechanism. This is the first report describing the entire nucleotide sequence and detailed genetic analysis of an L. gasseri plasmid, which contains functional genes for circular bacteriocin production, conjugation, and plasmid maintenance.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are shown in Table 1. Lactobacillus strains were grown statically in MRS medium (Becton Dickinson and Company, Sparks, MD) at 37°C. Lactococcus lactis subsp. lactis IL1403 (4) was grown in M17 medium (Becton Dickinson and Company) supplemented with 0.5% (wt/vol) glucose at 32°C. For plating, the media were solidified with 1.5% (wt/vol) agar. MRS-agar plates for Lactobacillus strains were incubated anaerobically by using an Anaeropack system (Mitsubishi Gas Chemical Company, Inc., Tokyo, Japan).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptionb | Reference or source |
|---|---|---|
| Strains | ||
| Lactobacillus gasseri strains | ||
| LA39 | Producer of gassericin A; isolated from feces of a human child; also known as JCM 11657 | 18 |
| JCM 1131T | Type strain; also known as ATCC 33323 | JCMa |
| JCM 1131T(pSYE2) | Electrotransformant of JCM 1131T with pSYE2 | This study |
| JCM 1131T(pLgLA39/pSYE2) | pLgLA39 conjugant of JCM 1131T(pSYE2) | This study |
| JCM 1131T(pLgLA39) | pLgLA39 conjugant of JCM 1131T obtained from JCM 1131T(pLgLA39/pSYE2) by curing pSYE2 | This study |
| JCM 1131T(pLrLA6) | pLrLA6 conjugant of JCM 1131T obtained from JCM 1131T(pLrLA6/pSYE2) by curing pSYE2 | This study |
| JCM 1131T(pGAA) | Electrotransformant of JCM 1131T with pGAA | This study |
| JCM 1131T(pGAAΔgaaA) | Electrotransformant of JCM 1131T with pGAAΔgaaA | This study |
| JCM 1131T(pSYE2+T) | Electrotransformant of JCM 1131T with pSYE2+T | This study |
| JCM 1131T(pPemIK) | Electrotransformant of JCM 1131T with pPemIK | This study |
| JCM 1131T(pPemI) | Electrotransformant of JCM 1131T with pPemI | This study |
| JCM 1130 | Also known as ATCC 19992 | JCM |
| JCM 5344 | Also known as ATCC 9857 | JCM |
| JCM 8787 | JCM | |
| OLL2728, OLL2804, OLL2842, OLL2935, OLL2970 | Laboratory collection of Meiji Dairies Corp.; isolated from human feces | This study |
| Lactobacillus reuteri LA6 | Producer of reutericin 6 | 17 |
| Lactobacillus delbrueckii subsp. bulgaricus ATCC 11842T(pX3) | Type strain transformed with pX3; used as an indicator for bacteriocin activity | This study |
| Lactococcus lactis subsp. lactis IL1403 | Plasmid-free; used as a cloning host in construction of plasmids | 4 |
| Plasmids | ||
| pLgLA39 | Isolated from L. gasseri LA39; Gaa+; conjugation positive | This study |
| pLrLA6 | Isolated from L. reuteri LA6; Reu+; conjugation positive | This study |
| pSYE2 | Cloning vector consisting of lactococcal plasmid pSY1 and ermA; Emr | 42 |
| pGAA | Contains seven gaa genes (ORF04 to ORF10) of pLgLA39 (nt 1624 to 5878) cloned into the SmaI site of pSYE2; Gaa+ Emr | This study |
| pGAAΔgaaA | Derivative of pGAA with gaaA deleted (see the text); Gaa− Emr | This study |
| pSYE2+T | Derivative of pSYE2 with the Streptococcus thermophilus ldh terminator and Escherichia coli rrnBT1T2; Emr | This study |
| pPemIK | Derivative of pSYE2+T containing pemIK; Emr | This study |
| pPemI | Derivative of pSYE2+T containing pemI; Emr | This study |
| pX3 | Derivative of Lactobacillus delbrueckii subsp. bulgaricus plasmid pBUL1 containing ermA; Emr | 36 |
Japan Collection of Microorganisms.
Gaa, gassericin A production; Reu, reutericin 6 production; Emr, resistant to erythromycin.
Electroporation.
L. gasseri JCM 1131T was cultivated overnight at 37°C in modified MRS medium supplemented with 1% (wt/vol) glycine and adjusted to pH 4.5 with HCl, and preparation of the electrocompetent cells and electroporation were done by the procedure originally described for Lactobacillus delbrueckii subsp. bulgaricus (43). L. lactis IL1403 was electroporated according to the method of Holo and Nes (13). Transformants were selected by using 25 μg/ml erythromycin (Em).
DNA isolation and manipulation.
Plasmid and total DNAs were prepared from L. gasseri cells after limited cell wall hydrolysis. L. gasseri cells were incubated at 37°C for 10 min in PBE (0.3 M raffinose, 1 mM EDTA, 20 mM Tris, pH 7.0) containing 2 mg/ml lysozyme and 0.04 mg/ml mutanolysin (Sigma-Aldrich, St. Louis, MO). Afterward, plasmid DNA was prepared by the standard alkaline sodium dodecyl sulfate method (40). To prepare the total DNA, L. gasseri cells, partially lysed as described above, were suspended in TE (1 mM EDTA, 10 mM Tris, pH 8.0) and thoroughly lysed. The total DNA was purified from the lysate by phenol extraction and ethanol precipitation (40). Further purification was done with standard RNase A treatment and polyethylene glycol precipitation (40). Similar methods were used to isolate plasmid and total DNAs from L. reuteri LA6 and plasmid DNA from L. lactis, except for using 6.7% (wt/vol) sucrose, 1 mM EDTA, and 50 mM Tris (pH 8.0) in place of PBE in the limited cell wall hydrolysis step.
DNA manipulation was done according to standard methods (40) or instructions from suppliers. PCRs were performed on a PerkinElmer model 9700 thermal cycler with Ex Taq polymerase (Takara Bio Inc., Otsu, Japan) unless otherwise stated. Primers used in PCRs are listed in Table 2. All oligonucleotides were purchased from Operon Biotechnologies (Tokyo, Japan). gaaA and the 16S rRNA gene were amplified from the total DNA of LA39 by using primer sets pr01-pr02 and 27F-1492R (25), respectively, and were labeled with an AlkPhos Direct kit (GE Healthcare UK Ltd., Buckinghamshire, England) and used as probes in Southern hybridization. Detection was done using a CDPStar detection kit (GE Healthcare UK Ltd.). For Fig. 3B, restriction fragments were analyzed on a DNA 7500 LabChip, using an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA).
TABLE 2.
Primers used
| Primer name | Sequence (5′ to 3′)a | Location (nt) or descriptionb | Reference |
|---|---|---|---|
| pr01 | ATGGTTACTAAGTACGGACG | 2717-2736 | |
| pr02 | CTAGGCTGCAGTCGCTCCCA | 2992-2973 | |
| pr03 | CCGCAGATAGGCTAGGTGTC | 1624-1643 | |
| pr04 | TAATTCCTAGTATAGCACAG | 5883-5864 | |
| pr05 | AAAACTCGAGAGTATCTTCCTCCCATAGG | AAAA plus XhoI site plus nt 2721 to 2703 | |
| pr06 | AAAACTCGAGTGATTATAAAGCTTTATGAATAG | AAAA plus XhoI site plus nt 2998 to 3020 | |
| pr07 | CCTCGCAAGGGAAAAGACAG | 278-297 | |
| pr08 | GCAGAGCAACTGCTGGCGAG | 1268-1249 | |
| pr09 | CTTCGTAACAATGCTATGAC | 718-699 | |
| pr10 | ATGACAAAATCAACAGATTTC | 12632-12652 | |
| pr11 | TTTAAAAGCCCCTATTCTTC | 13841-13822 | |
| pr_q01 | CAGCTATGGGAAACGACTATGAA | 6164-6186 (ATCC 33323 genome sequence) | 29 |
| pr_q02 | CGTATTCTGTGATCTTGAGGTTG | 6382-6360 (ATCC 33323 genome sequence) | 29 |
| pr_q03 | CAACAACTGGGTTGATGAAGAAG | 12799-12821 | |
| pr_q04 | AAATAGACATCGGTTGCTTTCAC | 13026-13004 | |
| pr_q05 | TACCTTGGATATTCACCGAACAC | 501-479 (pSYE2) | 42 |
| pr_q06 | GTTGACGATATTCTCGATTGACC | 287-309 (pSYE2) | 42 |
| 27F | AGAGTTTGATC(A/C)TGGCTCAG | Used to amplify 16S rRNA gene | 25 |
| 1492R | TACGG(C/T)TACCTTGTTACGACTT | Used to amplify 16S rRNA gene | 25 |
The XhoI site is underlined.
Numbers indicate coordinates in the nucleotide sequence of pLgLA39, unless otherwise indicated.
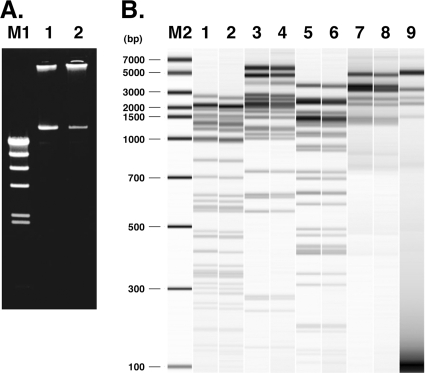
FIG. 3.
Comparison of pLgLA39 and pLrLA6. (A) Agarose gel electrophoresis. Lanes: M1, λ HindIII digest; 1, pLgLA39 prepared from L. gasseri LA39; 2, pLrLA6 prepared from L. reuteri LA6. (B) Electrophoresis of restriction enzyme digests of pLgLA39 and pLrLA6 prepared from JCM 1131T transconjugants. Lanes: M2, DNA size marker (DNA7500 ladder; Agilent Technologies); 1 and 2, pLgLA39 and pLrLA6 digested with DraI; 3 and 4, digests with EcoT14I; 5 and 6, digests with HincII; 7 and 8, digests with HindIII; 9, HindIII digest of pLgLA39 prepared from LA39.
Sequence determination and genetic analysis of pLgLA39.
The 4,100-bp sequence containing the seven gaa genes was determined previously (20). First, sequences were extended in both directions by primer walking, using the total DNA of LA39 as a template. After identification of pLgLA39 and localization of the gaa genes on pLgLA39 (Fig. 1), several primers based on the sequences of other L. gasseri plasmids we had sequenced (unpublished results) and on Lactobacillus paracasei subsp. paracasei contig 2 (8) were used to amplify partial fragments of pLgLA39. The resulting amplicons were sequenced by primer walking and assembled to give pLgLA39, a circular plasmid of 33,333 nucleotides (nt). Sequencing was performed using dye terminator sequencing reagents and an ABI3100 genetic analyzer (Applied Biosystems Japan Ltd., Tokyo, Japan). Sequence analyses were done with Genetyx software (Genetyx Corp., Tokyo, Japan). Open reading frames (ORFs) in pLgLA39 were predicted by the GeneMark.hmm program (http://opal.biology.gatech.edu/GeneMark/gmhmm2_prok.cgi), using the Lactobacillus johnsonii NCC 533 genome as a reference. Amino acid sequences of the deduced ORFs were subjected to homology searches against the NCBI nonredundant database by using the BLAST program (2) and were functionally categorized by using the COG database (45). Sequence alignments were done by using the ClustalW2 web service (http://www.ebi.ac.uk/Tools/clustalw2/index.html).
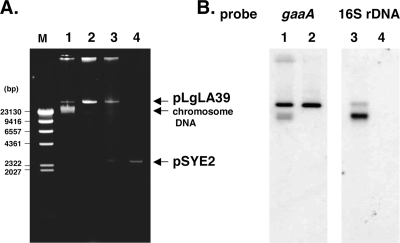
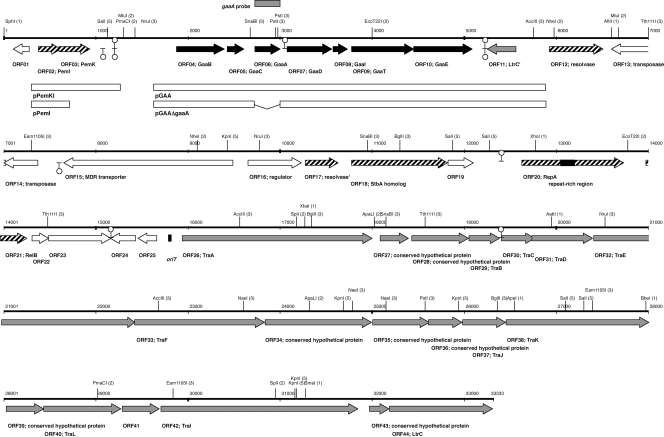
FIG. 1.
Confirmation of pLgLA39. (A) Agarose gel electrophoresis. Lanes: M, molecular size marker (λ HindIII digest); 1, total DNA preparation from LA39; 2, purified pLgLA39 plasmid; 3, plasmid preparation from JCM 1131T(pLgLA39/pSYE2); 4, pSYE2. (B) Southern hybridization. Lanes: 1 and 3, total DNA preparation from LA39; 2 and 4, purified pLgLA39 plasmid. The probes used were the gaaA probe (see Fig. 2) for lanes 1 and 2 and the 16S rRNA gene probe for lanes 3 and 4.
Enrichment and measurement of gassericin A activity.
Bacteriocin activity was assayed using the stepwise dilution method. L. delbrueckii subsp. bulgaricus ATCC 11842 (type strain) transformed with pX3 (36) was used as the standard indicator strain in this study. An arbitrary unit (AU) of activity was defined as the reciprocal of the highest dilution that inhibited the growth of the indicator strain. Since pX3 conferred Em resistance on ATCC 11842T, culture supernatants containing Em were assayed using the indicator ATCC 11842T(pX3) without removing the antibiotics.
A 20× concentrate of LA39 culture supernatant was prepared as follows. Ammonium sulfate (30% [wt/vol]) was added to the overnight culture supernatant of LA39, and the precipitant was dissolved in 20 mM sodium phosphate buffer (pH 5.0) at 1/20 the volume of the initial supernatant. The activity of the concentrate was usually 3,200 to 20,480 AU per ml.
Conjugation.
Conjugation was performed using the filter mating method described for Lactobacillus plantarum (41). Cells from overnight cultures of the donor (LA39) and the recipient (JCM 1131T harboring pSYE2 [42]) were mixed at a ratio of 10:1, 1:1, or 1:10, concentrated by vacuum on a sterile membrane filter (pore size, 0.45 μm) (HA-type filter; Millipore Corp., Billerica, MA), and washed with 20 ml of 20 mM Tris (pH 7.0). The membrane filter was placed on an MRS plate and incubated at 37°C overnight. The cells on the filter were suspended in 3 ml of MRS medium, diluted stepwise in sterile saline, and plated. Transconjugants were selected on MRS plates containing gassericin A (the 20× concentrate of LA39 culture supernatant at 5% [vol/vol]) and 25 μg/ml Em. CFU on MRS plates (donor and recipient cells) and on MRS plates containing Em (recipient cells) were counted to calculate conjugation frequencies for donor cells and for recipient cells. Curing of pSYE2 from the conjugants was done by repetitively culturing the conjugant cells in Em-free MRS medium. The resulting Em-sensitive colonies were checked for the absence of pSYE2 in the plasmid preparation.
Construction of plasmids.
To construct pGAA (see Fig. 4A), the seven gaa genes were amplified from pLgLA39 by a PCR using primers pr03 and pr04 and cloned into the SmaI site of pSYE2. To delete gaaA from pGAA, an inverse PCR was done on pGAA, using primers pr05 and pr06 (see Fig. 4A), and the amplicon was digested with XhoI and recircularized. The gaaA deletant plasmid was designated pGAAΔgaaA (see Fig. 4A). pGAAΔgaaA was completely sequenced to confirm that it did not contain any mutations due to PCR. pSYE2+T (see Fig. 5A) was constructed from pSYE2 by cloning the Streptococcus thermophilus ldh terminator (14) and the Escherichia coli rrnBT1T2 terminator from pKK223-3 (GE Healthcare UK Ltd.). We had sequenced another L. gasseri plasmid that carries pemIK genes that are 99.7% identical to those of pLgLA39 (unpublished results). pemIK and pemI were amplified from the plasmid by using primer sets pr07-pr08 and pr07-pr09, respectively. These fragments were inserted into the SmaI site of pSYE2+T to obtain pPemIK and pPemI, respectively (see Fig. 5A). All plasmid constructions were done in L. lactis IL1403 as a host.
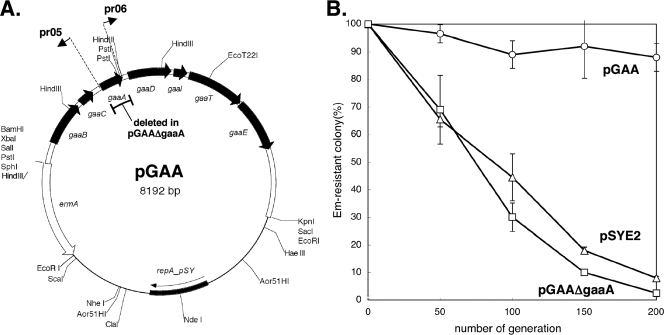
FIG. 4.
Effect of gassericin A production on apparent plasmid segregational stability. (A) gaa expression plasmid pGAA and gaaA-deletant plasmid pGAAΔgaaA. Primers used in construction of pGAAΔgaaA are shown. The double-lined region is from pLgLA39. ermA indicates the 1,111-bp HhaI fragment of pIL253 (GenBank accession number AF041239) containing the Em resistance (adenine methylase) gene. repA_pSY indicates the replication initiator protein gene of pSY1 (GenBank accession number E05086). (B) Apparent segregational stabilities of plasmids in L. gasseri JCM 1131T. Symbols: circles, pGAA; squares, pGAAΔgaaA; triangles, pSYE2.
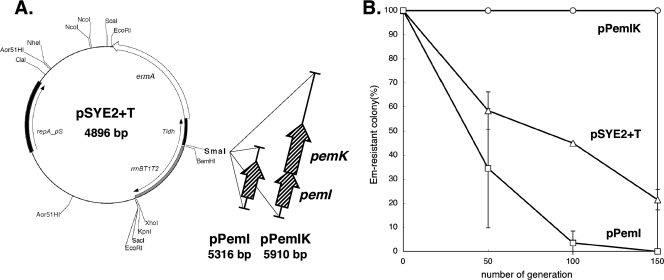
FIG. 5.
Apparent plasmid segregational stabilization by pLgLA39-type pemIK. (A) pSYE2+T, pPemIK, and pPemI. See the text for details about construction of the plasmids. Tldh indicates the S. thermophilus ldh terminator. (B) Apparent segregational stabilities of plasmids in L. gasseri JCM 1131T. Symbols: circles, pPemIK; squares, pPemI; triangles, pSYE2+T.
Segregational stability of plasmids.
L. gasseri JCM 1131T transformants harboring each plasmid were inoculated at 0.1% (vol/vol) into 1 ml MRS medium and grown. This cultivation, which corresponds to approximately 10 generations, was repeated, and the cultures were spread on MRS plates after the indicated number of times of repeated cultivation. One hundred colonies from each culture were checked for resistance to Em, indicating the presence of the plasmids. The experiments were done in duplicate.
Copy number determination.
Copy numbers of the plasmids were determined by quantitative real-time PCR (qPCR). qPCR was executed using a QuantiTect SYBR green PCR kit (Qiagen KK, Tokyo, Japan) on a Rotor-Gene RG-3000 thermal cycler (Corbett Research, Sidney, Australia) under the following conditions: initial denaturation at 95°C for 15 min; 45 cycles of denaturation (94°C for 15 s), annealing (55°C for 30 s), and extension (72°C for 20 s); and final incubation at 72°C for 60 s and 60°C for 40 s. To determine the copy number of pLgLA39 in LA39, a 219-bp fragment of gyrB on the chromosome and a 228-bp fragment of pLgLA39 repA (ORF20) were amplified from the total DNA of LA39, using primer sets pr_q01-pr_q02 and pr_q03-pr_q04, respectively. The copy numbers of pGAA, pGAAΔgaaA, pSYE2, pPemIK, pPemI, and pSYE2+T in L. gasseri JCM 1131T were determined similarly, by amplifying a 215-bp fragment of ermA, using primers pr_q05 and pr_q06, and the gyrB fragment from the total DNAs of the respective clones. The copy number relative to the chromosome equivalent was calculated from the ratio of the quantity of the repA or ermA amplicon to that of the gyrB amplicon. All of the experiments were done for two independent clones.
Nucleotide sequence accession numbers.
The nucleotide sequence of pLgLA39 was submitted to DDBJ and given the accession number AB436615. The accession numbers for the repA sequences of the plasmids from the eight L. gasseri strains are AB478699 to AB478706.
RESULTS
Genes for gassericin A production (gaa) are located on a native plasmid of L. gasseri LA39.
The seven gaa genes were identified by primer walking from the structural gene of gassericin A, gaaA (20). gaaA was reported to be located on the chromosome (19, 20). However, since we had revealed the presence of plasmids in several L. gasseri strains and had determined the nucleotide sequences of some of the plasmids (unpublished results), we reinvestigated the possibility that the gaa genes might be located on an as yet unidentified plasmid of L. gasseri LA39. Careful inspection of plasmid content in LA39 revealed the presence of a large plasmid (Fig. 1A). The gaaA probe (Fig. 2) hybridized to the plasmid (Fig. 1B). This indicated that gaaA and the other gaa genes were located on the plasmid, not on the chromosome. The plasmid was designated pLgLA39. The copy number of pLgLA39 in LA39 was estimated to be 21.5 ± 0.7 (average for two clones ± standard deviation) per chromosome equivalent.
FIG. 2.
Physical and genetic map of pLgLA39. Genes for gassericin A production, for conjugation, and for replication and maintenance are shown as black, shaded, and hatched arrows, respectively. Open and shaded boxes indicate the DNA regions used for construction of the plasmids and those used as the hybridization probe, respectively. A closed box indicates the supposed oriT sequence. The repeat region in repA is also indicated. Predicted terminators are shown by Ω-like symbols (upside-down ones are to terminate leftward transcriptions). Sites for rare-cutting restriction endonucleases are shown with total site numbers in parentheses.
Sequencing of pLgLA39.
We had previously found plasmids in several L. gasseri strains (see below) and had determined the entire sequences of two plasmids (unpublished results). pLgLA39 contained several regions substantially identical to these L. gasseri plasmids (unpublished results). Furthermore, pLgLA39 revealed the presence of genes highly homologous to tra genes in contig 2 of an unnamed plasmid of L. paracasei (8). Accordingly, several primers based on the nucleotide sequences of these plasmids were used in sequencing of pLgLA39. The determined entire sequence of pLgLA39 was 33,333 bp long, with a GC content of 39.5%, which was slightly higher than that (35.3%) of the genome sequence of L. gasseri ATCC 33323 (3). The first nucleotide of the unique SphI site was set to the start of the pLgLA39 sequence.
Analysis of genes in pLgLA39.
Forty-four ORFs of more than 100 bp were identified in pLgLA39. Amino acid sequences of the ORFs were subjected to homology searches against the NCBI nonredundant database and the COG database to speculate their functions. The results are shown in Table 3. Thirty-five ORFs of pLgLA39 were homologous (threshold E value, 1 × 10−5) to proteins with a known or predicted function(s), six were homologous to hypothetical proteins with no proposed functions, and three (ORF04, ORF08, and ORF19) were unique to pLgLA39. Analysis by RBSfinder predicted a ribosome binding site-like sequence 6 to 12 nt upstream of each ORF, except for ORF03, ORF05, ORF11, ORF13, ORF14, ORF17, ORF23, and ORF24. Potential ρ-independent terminators were identified between ORF03 and ORF04, ORF06 and ORF07, ORF10 and ORF11, ORF14 and ORF15, ORF17 and ORF18, ORF19 and ORF20, ORF23 and ORF24, and ORF29 and ORF30 (Fig. 2) by using the TransTermHP program (22). The sections below give a detailed description of the pLgLA39 ORFs, grouped by confirmed or predicted functions.
TABLE 3.
Characteristics of ORFs and predicted proteins in pLgLA39
| ORF | Location (nt) | Deduced function | Protein length (aa) | pI | Mol wt | COG | Cellular locationa |
|---|---|---|---|---|---|---|---|
| 01 | 269-93 | Hypothetical protein | 58 | 7.94 | 6,658.7 | Soluble | |
| 02 | 366-611 | PemI (inhibitor) | 81 | 5.43 | 9,250.5 | COG2336 | Soluble |
| 03 | 605-937 | PemK (RNase toxin) | 110 | 9.58 | 12,564.7 | COG2337 | Soluble |
| 04 | 1871-2395 | GaaB | 174 | 9.60 | 20,278.4 | Membrane (5) | |
| 05 | 2433-2615 | GaaC | 60 | 9.61 | 7,290.9 | Membrane (2) | |
| 06 | 2722-2997 | GaaA (gassericin A) | 91 | 9.40 | 9,286.0 | COG1300 | Membrane (2) |
| 07 | 3081-3569 | GaaD | 162 | 6.94 | 18,344.2 | Membrane (4) | |
| 08 | 3591-3752 | GaaI (immunity) | 53 | 11.10 | 6,134.5 | Membrane (1) | |
| 09 | 3775-4455 | GaaT (transporter) | 226 | 4.71 | 25,126.0 | COG1131 | Soluble |
| 10 | 4458-5096 | GaaE (transporter) | 212 | 9.51 | 23,665.0 | Membrane (6) | |
| 11 | 5560-5246 | LtrC (fragment) | 104 | 9.87 | 12,323.9 | Soluble | |
| 12 | 5923-6507 | Resolvase | 194 | 9.99 | 22,341.8 | COG1961 | Soluble |
| 13 | 7030-6599 | Transposase (fragment) | 143 | 8.75 | 16,163.5 | COG3293 | Membrane (1) |
| 14 | 7374-7003 | Transposase (fragment) | 123 | 9.47 | 14,365.4 | COG3293 | Soluble |
| 15 | 9491-7650 | Multidrug resistance transporter | 613 | 9.97 | 65,539.8 | COG0477 | Membrane (4) |
| 16 | 9656-10240 | Regulator | 194 | 6.07 | 22,906.3 | COG1309 | Soluble |
| 17 | 10325-10633 | Resolvase (fragment) | 102 | 10.22 | 12,215.2 | COG1961 | Soluble |
| 18 | 10765-11814 | StbA homolog | 349 | 5.16 | 39,098.0 | COG0443 | Soluble |
| 19 | 11817-12092 | Hypothetical protein | 91 | 5.82 | 10,228.7 | Soluble | |
| 20 | 12632-13741 | RepA | 369 | 6.16 | 42,999.6 | Soluble | |
| 21 | 13984-14265 | RelB antitoxin | 93 | 4.66 | 10,483.9 | COG3077 | Soluble |
| 22 | 14312-14497 | Conserved hypothetical protein | 61 | 5.29 | 7,438.5 | Soluble | |
| 23 | 14494-15174 | Conserved hypothetical protein | 226 | 9.17 | 25,968.5 | Soluble | |
| 24 | 15442-15164 | Conserved hypothetical protein | 92 | 9.94 | 11,146.7 | Soluble | |
| 25 | 15674-15465 | Conserved hypothetical protein | 69 | 9.30 | 7,971.1 | Soluble | |
| 26 | 15946-18009 | TraA (nicking enzyme) | 687 | 8.92 | 80,624.2 | Soluble | |
| 27 | 18094-18405 | Conserved hypothetical protein | 103 | 9.63 | 11,687.6 | Soluble | |
| 28 | 18442-19056 | Conserved hypothetical protein | 204 | 5.11 | 23,696.4 | Soluble | |
| 29 | 19058-19393 | TraB | 111 | 9.21 | 11,717.0 | Membrane (3) | |
| 30 | 19414-19776 | TraC | 120 | 9.95 | 13,862.6 | Membrane (2) | |
| 31 | 19745-20404 | TraD | 219 | 5.46 | 24,877.5 | Soluble | |
| 32 | 20416-22434 | TraE | 672 | 5.50 | 77,233.8 | COG1419 | Soluble |
| 33 | 22427-23845 | TraF | 472 | 9.10 | 53,277.4 | Membrane (1) | |
| 34 | 23846-25003 | Conserved hypothetical protein | 385 | 6.66 | 41,739.7 | COG0791 | Membrane (1) |
| 35 | 25017-25634 | Conserved hypothetical protein | 205 | 9.78 | 23,256.2 | Membrane (1) | |
| 36 | 25621-25989 | Conserved hypothetical protein | 122 | 9.74 | 14,199.4 | COG0526 | Membrane (1) |
| 37 | 25990-26460 | TraJ | 156 | 8.94 | 17,613.5 | Membrane (2) | |
| 38 | 26462-28018 | TraK | 518 | 5.38 | 58,268.6 | COG3505 | Soluble |
| 39 | 28033-28440 | Conserved hypothetical protein | 135 | 9.71 | 14,964.2 | Membrane (1) | |
| 40 | 28442-29281 | TraL | 279 | 8.58 | 31,155.0 | Membrane (6) | |
| 41 | 29296-29700 | Hypothetical protein | 134 | 9.27 | 16,000.8 | Membrane (2) | |
| 42 | 29715-31853 | TraI (topoisomerase) | 712 | 9.30 | 80,062.9 | COG0550 | Soluble |
| 43 | 31976-32191 | Conserved hypothetical protein | 71 | 4.14 | 7,826.8 | Soluble | |
| 44 | 32195-33319 | LtrC | 374 | 7.79 | 42,313.5 | Soluble |
Deduced by using the SOSUI web service (http://bp.nuap.nagoya-u.ac.jp/sosui/). Numbers of predicted transmembrane segments are shown in parentheses.
gaa genes.
ORF04 to ORF10 of pLgLA39 correspond to the gaa genes, gaaBCADITE (20) (Table 3). Functional cloning of the seven genes and confirmation of these genes as responsible for production of gassericin A are described below. ORF06 (gaaA) is the structural gene of gassericin A, and ORF08 (gaaI) is the immunity determinant gene. GaaT (ORF09) and GaaE (ORF10) are possible transporters of gassericin A. GaaB (ORF04), GaaC (ORF05), and GaaD (ORF07) were supposed to be membrane associated, but their roles in gassericin A production/immunity remain unknown.
Replication and maintenance genes.
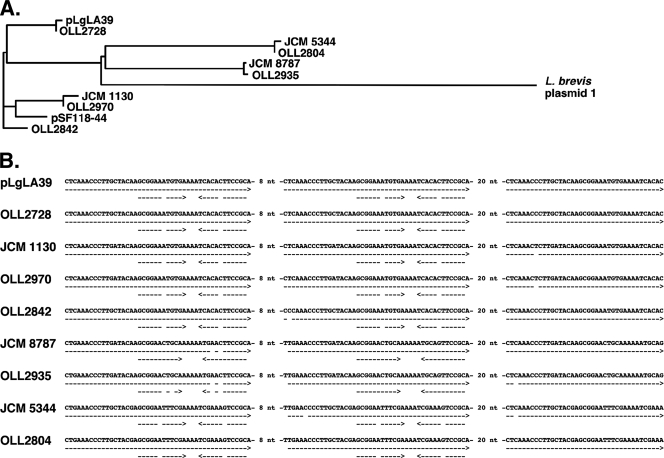
ORF20 is the replication protein gene (repA) of pLgLA39. RepA encoded by pLgLA39 contains the pfam06970 RepA_N domain in its N terminus and constitutes a distinct phylogenetic branch, together with the RepA proteins from several Lactobacillus and Lactococcus plasmids. RepA from pLgLA39 is nearly identical (96% over 364 aa) to that from L. salivarius pSF118-44 (10) and also similar to those from several Lactobacillus and Lactococcus plasmids, such as Lactobacillus brevis ATCC 367 plasmid 1 (29), L. salivarius pSF118-20 (10), L. lactis pNP40 (33), L. casei ATCC 334 plasmid 1 (29), L. lactis pCI2000 (21), and Lactobacillus sakei pSAK1 (29), with 73.1% to 52.6% identities over the entire amino acid sequence. pLgLA39 and this group of plasmids likely replicate via the θ mechanism, since pSF118-20 was reported to be θ replicating (10). pLgLA39 repA contains a long direct repeat region (black box over repA in Fig. 2; nt 422 to 580 of repA) consisting of perfect repeats of 46 nt (5′-CTCAAACCCTTGCTACAAGCGGAAATGTGAAAATCACACTTCCGCA-3′) separated by 8 nt, with an additional 39-nt stretch identical to the 5′ terminus of the 46-nt sequence located 20 nt downstream. These nucleotide repeats result in three repeats of 12 aa (QTLATSGNVKIT) in the amino acid sequence of RepA. Such long repeats in the nucleotide sequence and amino acid repeats in the translation are common in repA genes of the related plasmids shown above. The 46-nt stretch contains a 12-nt inverted repeat near the 3′ terminus (underlined in the above sequence). This feature is also conserved in most of the repA sequences of the related plasmids, except for those of pCI2000 and pSAK1.
ORF02 and ORF03 encode proteins similar to several PemI-like and PemK-like proteins, respectively, encoded on chromosomes and plasmids of various bacterial genera. pLgLA39-encoded PemI and PemK showed the highest amino acid sequence identities to SMU.172 (79.0%) and SMU.173 (84.5%), respectively, of Streptococcus mutans UA159 (1). pLgLA39-encoded PemK showed 31.5% aa identity to the original PemK protein of the E. coli R100 plasmid, although pLgLA39-encoded PemI revealed little homology to R100-encoded PemI. pemIK of R100 is known as a toxin-antitoxin (TA) system (49). The functionality of the pLgLA39-type pemIK system was confirmed experimentally (see below). ORF03 (pemK) lacks a ribosome binding site, and the 5′ end of ORF03 partially overlaps the 3′ end of ORF02 (pemI). This suggests that expression of pemK may be regulated by translational coupling to pemI to strictly control the ratio of PemK to PemI, as observed with other TA systems (11).
ORF12 encodes a putative resolvase that contains a cd03768 SR_ResInv domain (serine recombinase family catalytic domain) in the N terminus and a cd00569 HTH_Hin-like (helix-turn-helix domain of Hin and related proteins) DNA-binding motif in the C terminus. Resolvases monomerize concatemeric plasmid molecules and thereby increase the segregational stability of plasmids (24). Almost identical resolvase genes were found in pSF118-44, L. brevis ATCC 367 plasmid 1, pCD01, and L. casei ATCC 334 plasmid 1. This may suggest a contribution of this type of resolvase to stable maintenance of pLgLA39 and these plasmids. While ORF17 is 95% identical to ORF12, it is 5′ truncated and likely inactive.
tra genes.
The products of pLgLA39 ORF26 to ORF44 are similar (98.7% to 42.7% identical) to the tra gene products of L. plantarum pWCFS103 (46). These ORFs, except for ORF41, also have homologs in L. lactis subsp. lactis pMRC01 (83.2% to 37.2% identical) (9). Conjugation of pWCFS103 and pMRC01 has been reported (9, 46). The functions of the tra gene products, except for those of TraA (nicking enzyme) and TraI (topoisomerase), are not clear. A putative oriT sequence similar to those proposed for pMRC01 (9) and pWCFS103 (46) was found upstream of pLgLA39 traA (nt 15837 to 15861) (Fig. 2). Homologs of pLgLA39 ORF27, ORF28, ORF29, ORF30, ORF35, ORF37, ORF39, ORF41, and ORF43 are specific to these plasmids and an unnamed plasmid(s) of L. paracasei (8).
Other genes.
ORF15 and ORF16 of pLgLA39 are almost identical to the genes for a Qac family multidrug resistance transporter and a TetR/AcrR-type transcriptional regulator, respectively, of L. paracasei pCD01 (8). ORF13 and ORF14 of pLgLA39 putatively encode a single polypeptide transposase. The putative insertion sequence (IS) containing ORF13 and ORF14 (nt 6580 to 7440 of pLgLA39) is 99.3% identical to ISLpl3 from L. plantarum WCSF1, which belongs to the IS427 group of the IS5 family (23). ISs nearly identical to the putative ISs of pLgLA39 and ISLpl3 are also found in plasmids (pRH45II and ATCC 367 plasmid 2) and the chromosome of L. brevis (ATCC 367). These may suggest transposing activity of this IS group.
Conjugative transfer of pLgLA39 from LA39 to L. gasseri JCM 1131T.
pLgLA39 carries a set of tra genes, as described above. The conjugative ability of pLgLA39 was investigated. We succeeded in conjugative transfer of pLgLA39 from LA39 to L. gasseri JCM 1131T by filter mating. The transconjugant revealed elevated resistance to gassericin A and production of antibacterial activity.
JCM 1131T transformed with pSYE2 and thus resistant to Em was used as the recipient. The donor (LA39) and the recipient were filter mated, and transconjugants were selected on plates containing Em and gassericin A. JCM 1131T is sensitive to gassericin A (20), and the MIC for JCM 1131T of the 20×-concentrated culture supernatant of LA39 was 0.62% to 0.31% (vol/vol). To select transconjugants, 5% (vol/vol) gassericin A concentrate (approximately 10-fold the MIC) was used. The resulting clones, resistant to both Em and gassericin A, were further investigated. The clones grew in the presence of 20% (vol/vol) gassericin A concentrate. This level of resistance was >32-fold higher than that of JCM 1131T and comparable to that of the donor strain LA39. The clones harbored two plasmids: one sized similarly to pSYE2 and the other sized similarly to pLgLA39 (Fig. 1A, lane 3). Randomly amplified polymorphic DNA PCR of the clones gave a band pattern similar to that of JCM 1131T but distinct from that of LA39 (data not shown). These results indicated that the Em- and gassericin A-resistant clones were conjugants. We designated the conjugant strain JCM 1131T(pLgLA39/pSYE2). The conjugant expressed significant antimicrobial activity (40 AU/ml), while the recipient strain JCM 1131T revealed no bacteriocin activity. This indicates that the gaa genes in pLgLA39 conferred to the conjugant cells productivity of gassericin A as well as immunity to gassericin A. Frequencies of conjugation were between 2.5 × 10−8 and 1.4 × 10−5 per donor cell and between 2.8 × 10−6 and 2.3 × 10−3 per recipient cell. As the donor/recipient ratio increased, the conjugation frequency per donor decreased while that per recipient increased. Frequencies per recipient cell were much higher than those per donor cell. This was due to a drastic reduction in CFU of the recipient cells during filter mating (data not shown), probably because gassericin A produced by the donor strain LA39 killed the recipient cells sensitive to gassericin A.
L. reuteri LA6 harbors a conjugal plasmid indistinguishable from pLgLA39.
Reutericin 6 is a circular bacteriocin with an amino acid sequence identical to that of gassericin A (17). L. reuteri LA6, a producer of reutericin 6, was proved to harbor a plasmid, designated pLrLA6 (Fig. 3A). pLrLA6 proved to be quite similar to pLgLA39 in several aspects. Conjugation of pLrLA6 from L. reuteri LA6 to L. gasseri JCM 1131T was successful by the method used for conjugation of pLgLA39 from L. gasseri LA39. In a conjugation experiment, conjugal frequencies were 4.8 × 10−8 per donor cell and 4.1 × 10−7 per recipient cell. pLrLA6 and pLgLA39 were prepared from the respective transconjugants, from which pSYE2 was cured, and were digested with the restriction enzymes DraI, EcoT14I, HincII, and HindIII. All of the digests exhibited similar band patterns for the two plasmids (Fig. 3B, lanes 1 to 8). In addition, PCRs done on pLrLA6 with the pLgLA39 sequence-based primer sets, which together cover the entire pLgLA39 sequence, gave amplicons of identical lengths to the corresponding amplicons from pLgLA39 (data not shown). In those aspects, pLrLA6 was indistinguishable from pLgLA39, and the two plasmids are probably almost identical to each other. Two regions of the plasmids were compared at the nucleotide sequence level. pLrLA6 carried a locus corresponding to the gaa genes of pLgLA39. The locus of pLrLA6 thought to be responsible for reutericin 6 production was designated reu. The nucleotide sequence of the reu region was determined to be 100% identical to that of the gaa region. Thus, not only the structural gene reuA, encoding the reutericin 6 precursor, but also the other six reu genes were completely identical to the corresponding gaa genes. repA and its upstream noncoding region of pLrLA6 were also identical to those of pLgLA39, except for a 1-nt insertion (T) in pLrLA6 (at nt 12555 of the pLgLA39 sequence).
To compare the restriction enzyme digests of pLrLA6 and pLgLA39 (Fig. 3B), we used plasmids prepared from JCM 1131T, not from their original hosts. This eliminated possible unexpected effects by site-specific DNA modifications in the host cells (LA39 and LA6). Indeed, we observed site-specific modifications when HindIII was used. pLrLA6 prepared from L. reuteri LA6 was not digested at all with HindIII, while it was sensitive to AluI (data not shown). This suggests the presence of M.HindIII-like N6A-methylase activity in LA6, and actually, from LA6 we cloned restriction and modification genes homologous to those of the HindIII system (unpublished results). When pLgLA39 prepared from L. gasseri LA39 was digested with HindIII, some specific HindIII sites were resistant to digestion (Fig. 3B, lane 9). In preliminary experiments, we determined that the protection occurred at HindIII sites preceded by G (5′-GAAGCTT-3′).
Cloning and expression of the gaa genes in JCM 1131T and enhancement of plasmid segregational stability by production of gassericin A.
The seven gaa genes, gaaBCADITE, were cloned from pLgLA39 into pSYE2 to construct pGAA (Fig. 4A). L. gasseri JCM 1131T cells transformed with pGAA exhibited significant antibacterial activity (40 AU/ml). This indicates that the cloned gaa genes were active and produced gassericin A in the JCM 1131T background. Next, gaaA was removed from pGAA by inverse PCR to construct pGAAΔgaaA (Fig. 4A). JCM 1131T harboring pGAAΔgaaA produced no antibacterial activity, as expected. Both JCM 1131T(pGAA) and JCM 1131T(pGAAΔgaaA) were resistant to gassericin A, as they grew in the presence of 20% (vol/vol) gassericin A concentrate.
The apparent segregational stabilities of plasmids pGAA, pGAAΔgaaA, and pSYE2 in JCM 1131T were examined (Fig. 4B). pGAA was significantly (at a significance level of <0.05) more stable than pGAAΔgaaA and pSYE2 after 100 (P values of 0.0087 and 0.039, respectively) and 200 (P values of 0.017 and 0.025, respectively) generations. Copy numbers of the three plasmids in JCM 1131T were determined by qPCR and were 22.8 ± 7.4 for pGAA, 27.0 ± 4.1 for pGAAΔgaaA, and 47.3 ± 16.4 for pSYE2. It should be noted that pGAA was more stable than pSYE2, which exists at approximately twice the number of copies of pGAA. The observed enhancement of the apparent segregational stability of pGAA by the gaa locus is probably a result of postsegregational host killing (PSK), as discussed below.
Functionality of pLgLA39-type pemIK as a plasmid-stabilizing mechanism.
pLgLA39 ORF02 and ORF03 are pemIK homologs. The functionality of the pLgLA39-type pemIK system as a plasmid-stabilizing mechanism was confirmed experimentally. We had found pemIK in another L. gasseri plasmid, of which the translated amino acid sequences were 100% identical to those of PemIK of pLgLA39 (unpublished result). pemIK and pemI were PCR amplified from the plasmid and cloned into pSYE2+T to obtain pPemIK and pPemI, respectively (Fig. 5A). The cloned regions corresponded to nt 273 to 1263 and nt 273 to 718 of pLgLA39 (Fig. 2), respectively, and both contained a presumptive native promoter. The apparent segregational stabilities of pPemIK, pPemI, and pSYE2+T in L. gasseri JCM 1131T were compared (Fig. 5B). pPemIK was perfectly stable during the experiment and thus remarkably more stable than pPemI and pSYE2+T. Copy numbers determined for the three plasmids in JCM 1131T were nearly the same, as follows: 18.9 ± 4.3 for pPemIK, 16.5 ± 5.8 for pPemI, and 19.5 ± 1.2 for pSYE2+T. Therefore, the differences in apparent segregational stabilities were not due to differences in the quantities of the plasmids. These results indicate that the pLgLA39-type pemIK system is functional as a plasmid-stabilizing mechanism.
Distribution of pLgLA39-related plasmids in L. gasseri strains.
Of 27 L. gasseri strains examined, 17 strains exhibited possession of a plasmid(s). These plasmids varied in size and in restriction enzyme digestion patterns (data not shown). However, PCR using primers pr10 and pr11 and following DNA sequencing revealed that all of these plasmids carried a repA gene highly similar to pLgLA39 repA. This indicates a wide distribution in L. gasseri strains of plasmids belonging to the same replication type as pLgLA39. The amino acid sequences of RepAs of pLgLA39, the plasmids from eight independent L. gasseri strains, pSF118-44 from L. salivarius UCC 118, and plasmid 1 from L. brevis ATCC 367 were aligned (shown as a phylogram in Fig. 6A). The existence of the long nucleotide repeats and the short inverted repeat in the repeat unit was conserved in the repA genes from the eight L. gasseri plasmids (Fig. 6B). None of the plasmids from the 17 L. gasseri strains gave amplicons by PCR using primers pr01 and pr02 (data not shown). This indicates that these plasmids do not carry gaaA.
FIG. 6.
pLgLA39-related plasmids in L. gasseri strains. (A) Phylogram based on amino acid sequence alignment of RepAs. (B) Comparison of repeat regions in repA genes of L. gasseri plasmids. Direct repeats and inverted repeats are indicated by dashed arrows. Positions of mismatches lack a dash.
DISCUSSION
The gassericin A-producing strain L. gasseri LA39 was shown to harbor the 33,333-bp plasmid pLgLA39. This is the first report of the entire nucleotide sequence and detailed genetic analysis of an L. gasseri plasmid. Plasmids related to pLgLA39 are distributed in several L. gasseri strains as well as in other Lactobacillus and Lactococcus species. The seven gaa genes responsible for the production of gassericin A and immunity to gassericin A are located on pLgLA39, as opposed to the earlier localization of gaaA on the chromosome (19). pLgLA39 carries tra genes and was shown to conjugally transfer to L. gasseri JCM 1131T. The conjugant revealed productivity of gassericin A as well as resistance to gassericin A. Functional cloning of gaa in a small plasmid was achieved. pGAA carrying gaa and expressing gassericin A was stably maintained. Apparent segregational stabilization of a plasmid by the pLgLA39-type pemIK system was also confirmed. Thus, pLgLA39 carries at least two independent stabilization mechanisms, the gaa and pem systems.
Reutericin 6 from L. reuteri LA6 is a circular bacteriocin whose amino acid sequence is identical to that of gassericin A (17). L. reuteri LA6 was shown to harbor a plasmid, pLrLA6, almost identical to pLgLA39. Furthermore, pLrLA6 contains seven genes that are 100% identical to the gaa genes of pLgLA39. L. gasseri LA39 and L. reuteri LA6 were isolated from feces of the same human infant within a 2-month interval (17, 19). These observations strongly suggest that pLgLA39 (pLrLA6) had naturally transferred between L. gasseri LA39 and L. reuteri LA6. Since reu and gaa are 100% identical to each other, the difference in the d-alanine contents of reutericin 6 and gassericin A (17) may be dependent on an as yet unidentified host factor(s).
The seven gaa genes were successfully cloned into plasmid vector pSYE2 to construct pGAA. The pGAA transformant of L. gasseri JCM 1131T produced significant bacteriocin activity. In contrast, JCM 1131T transformed with pGAAΔgaaA (a gaaA-depleted derivative of pGAA) did not produce bacteriocin activity. These results indicate that the bacteriocin produced by the pGAA transformant should be gassericin A. However, since gassericin A expressed in the JCM 1131T background was not chemically analyzed, it remains unclear whether it had the same characteristics, such as circularity, specific activity, and secondary and tertiary structures, as the authentic gassericin A produced by LA39. Analyses to confirm these points are in progress. To date, detailed molecular mechanisms for production of the class IV circular bacteriocins remain unclear. The production process includes maturation (digestion of precursor molecule), secretion, and circularization (covalent binding of N- and C-terminal ends). pGAA (8.2 kb) is small enough to be amplified by one PCR. As we demonstrated in the construction of pGAAΔgaaA, the inverse PCR technique can be applied to pGAA to inactivate a specific gaa gene(s) or to introduce a specific nucleotide alteration(s) into the gaa gene(s). This could facilitate functional analyses of the gaa genes to clarify molecular mechanisms underlying production, especially intramolecular circularization, of gassericin A, as well as immunity to the bacteriocin.
TA modules and restriction-modification systems are thought to be “selfish” genes since they cause PSK upon their loss from host cells (12). Both systems are often carried by bacterial plasmids and are thought to enhance the apparent segregational stability of the plasmids. We demonstrated that pGAA expressing gassericin A was more stably maintained in JCM 1131T than pGAAΔgaaA and even than the vector pSYE2 (Fig. 4B). This apparent stabilization can be explained by PSK; JCM 1131T cells having lost pGAA, and simultaneously immunity to gassericin A, are killed by gassericin A produced by neighbor cells retaining pGAA. In this context, bacteriocin genes should also be regarded as selfish. Three general features of bacteriocins are in accordance with this proposition. First, bacteriocins are usually active against bacteria closely related to the producers of the bacteriocins. This could be plausible if the primary purpose of bacteriocins was to kill the cells that had once borne but then lost the bacteriocin genes. Second, so far as we know, no immunity mechanisms have been reported to inactivate cognate bacteriocins outside the producing cells by specifically degrading or capturing them. This may be because these mechanisms would help the sensitive cells to survive. Finally, bacteriocins are generally very stable. Above all, the class IV circular bacteriocins have no free ends and thus are even resistant to attacks by exopeptidases (6). Although the stability of immunity proteins has seldom been elucidated, they are likely less stable than cognate bacteriocins. Analogously, antitoxin proteins of TA modules are generally more fragile than toxin proteins, and the difference in the stabilities of the toxin and antitoxin is a prerequisite for PSK by TA systems.
The pLgLA39-type pemIK genes were also shown to be functional as a plasmid-stabilizing mechanism. pem in the R100 plasmid of E. coli is a well-studied TA system; PemK (toxin) is a site-specific RNase preferentially degrading mRNA at 5′-UA(C/A/U)-3′ sites, and the less stable PemI protein (antitoxin) blocks the nuclease activity of PemK (49). TA systems similar to the E. coli pem system are widely distributed among bacterial plasmids and chromosomes. We showed that the pLgLA39-type pemIK system cloned into the rolling-circle-replicating plasmid pSYE2+T drastically increased the apparent segregational stability of the resulting plasmid, pPemIK (Fig. 5A). Since the basic mechanism of TA systems is independent of the type of replicon, pemIK of pLgLA39 likely enhances the apparent segregational stability of pLgLA39. pemIK of pLgLA39 can be used as a stabilizer for unstable plasmids or for plasmids with antibiotic resistance markers in cases where the usage of antibiotics is undesirable. Recently, another pemIK homolog in pSF118-20 from L. salivarius UCC 118 was reported to function as a TA system (10). The PemK proteins of pLgLA39 and pSF118-20 are 41.6% identical to each other over the entire amino acid sequence, while the PemI proteins of the two plasmids show lower and partial similarity (33.3% identical in 63 aa over 81 aa). PemK and PemI of pLgLA39 revealed even lower amino acid sequence similarities to the proteins encoded by E. coli R100. It should be elucidated further whether PemK and PemI of pLgLA39 work via a similar mechanism to the E. coli pem system and other heterologous pem systems, such as that in pSF118-20.
We showed that several L. gasseri strains harbor a plasmid(s) with a repA gene highly homologous to repA of pLgLA39. The gaa genes, however, are not carried by these plasmids. We determined the draft genome sequence of L. gasseri strain OLL2716 (39) and found seven genes on the chromosome with significant similarities to the gaa genes (translated amino acid sequence identities of 32% to 57% [unpublished results]). The chromosomal gaa-like genes were found neither in LA39 nor in the genome sequence of L. gasseri ATCC 33323 (3). Although it remains to be elucidated whether the gaa-like genes are expressed in OLL2716, the bacteriocin might be a new member of the gassericin A group of circular bacteriocins.
pLgLA39 is the first entirely sequenced plasmid from L. gasseri. For genetic manipulation of the species, mainly plasmid vectors of lactococcal origin have been used (27). Since pLgLA39 is supposed to replicate via the θ mechanism, high segregational and structural stability is expected. The wide distribution of pLgLA39-related plasmids in Lactobacillus and Lactococcus species suggests that vectors based on pLgLA39 repA could be applicable to many lactic acid bacteria. We successfully constructed a plasmid consisting of exclusively pLgLA39 repA (nt 12604 to 13741) and ermA, and the plasmid was maintained in L. gasseri JCM 1131T and also in L. lactis IL1403 (unpublished results). Furthermore, employment of a complementary selection marker gene originating in L. gasseri would enable a self-cloning vector for the species and could be used in food-grade genetic manipulation of probiotic L. gasseri strains.
Acknowledgments
Thanks are due to M. Takeda for technical assistance in the early stage of this work. We also thank Y. Sasaki for suggestive discussions.
Footnotes
Published ahead of print on 7 August 2009.
REFERENCES
- 1.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azcarate-Peril, M. A., E. Altermann, Y. J. Goh, R. Tallon, R. B. Sanozky-Dawes, E. A. Pfeiler, S. O'Flaherty, B. L. Buck, A. Dobson, T. Duong, M. J. Miller, R. Barrangou, and T. R. Klaenhammer. 2008. Analysis of the genome sequence of Lactobacillus gasseri ATCC 33323 reveals the molecular basis of an autochthonous intestinal organism. Appl. Environ. Microbiol. 74:4610-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chopin, A., M. C. Chopin, A. Moillo-Batt, and P. Langella. 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11:260-263. [DOI] [PubMed] [Google Scholar]
- 5.Claesson, M. J., Y. Li, S. Leahy, C. Canchaya, J. P. van Pijkeren, A. M. Cerdeno-Tarraga, J. Parkhill, S. Flynn, G. C. O'Sullivan, J. K. Collins, D. Higgins, F. Shanahan, G. F. Fitzgerald, D. van Sinderen, and P. W. O'Toole. 2006. Multireplicon genome architecture of Lactobacillus salivarius. Proc. Natl. Acad. Sci. USA 103:6718-6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craik, D. J. 2006. Chemistry. Seamless proteins tie up their loose ends. Science 311:1563-1564. [DOI] [PubMed] [Google Scholar]
- 7.Cuozzo, S. A., F. Sesma, J. M. Palacios, A. P. de Ruiz Holgado, and R. R. Raya. 2000. Identification and nucleotide sequence of genes involved in the synthesis of lactocin 705, a two-peptide bacteriocin from Lactobacillus casei CRL 705. FEMS Microbiol. Lett. 185:157-161. [DOI] [PubMed] [Google Scholar]
- 8.Desmond, C., R. P. Ross, G. Fitzgerald, and C. Stanton. 2005. Sequence analysis of the plasmid genome of the probiotic strain Lactobacillus paracasei NFBC338 which includes the plasmids pCD01 and pCD02. Plasmid 54:160-175. [DOI] [PubMed] [Google Scholar]
- 9.Dougherty, B. A., C. Hill, J. F. Weidman, D. R. Richardson, J. C. Venter, and R. P. Ross. 1998. Sequence and analysis of the 60 kb conjugative, bacteriocin-producing plasmid pMRC01 from Lactococcus lactis DPC3147. Mol. Microbiol. 29:1029-1038. [DOI] [PubMed] [Google Scholar]
- 10.Fang, F., S. Flynn, Y. Li, M. J. Claesson, J. P. van Pijkeren, J. K. Collins, D. van Sinderen, and P. W. O'Toole. 2008. Characterization of endogenous plasmids from Lactobacillus salivarius UCC118. Appl. Environ. Microbiol. 74:3216-3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerdes, K., S. K. Christensen, and A. Lobner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 12.Hayes, F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496-1499. [DOI] [PubMed] [Google Scholar]
- 13.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito, Y., and T. Sasaki. 1994. Cloning and nucleotide sequencing of l-lactate dehydrogenase gene from Streptococcus thermophilus M-192. Biosci. Biotechnol. Biochem. 58:1569-1573. [DOI] [PubMed] [Google Scholar]
- 15.Kalmokoff, M. L., T. D. Cyr, M. A. Hefford, M. F. Whitford, and R. M. Teather. 2003. Butyrivibriocin AR10, a new cyclic bacteriocin produced by the ruminal anaerobe Butyrivibrio fibrisolvens AR10: characterization of the gene and peptide. Can. J. Microbiol. 49:763-773. [DOI] [PubMed] [Google Scholar]
- 16.Kanatani, K., T. Tahara, M. Oshimura, K. Sano, and C. Umezawa. 1995. Identification of the replication region of Lactobacillus acidophilus plasmid pLA103. FEMS Microbiol. Lett. 133:127-130. [DOI] [PubMed] [Google Scholar]
- 17.Kawai, Y., Y. Ishii, K. Arakawa, K. Uemura, B. Saitoh, J. Nishimura, H. Kitazawa, Y. Yamazaki, Y. Tateno, T. Itoh, and T. Saito. 2004. Structural and functional differences in two cyclic bacteriocins with the same sequences produced by lactobacilli. Appl. Environ. Microbiol. 70:2906-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawai, Y., T. Saito, H. Kitazawa, and T. Itoh. 1998. Gassericin A; an uncommon cyclic bacteriocin produced by Lactobacillus gasseri LA39 linked at N- and C-terminal ends. Biosci. Biotechnol. Biochem. 62:2438-2440. [DOI] [PubMed] [Google Scholar]
- 19.Kawai, Y., T. Saito, M. Suzuki, and T. Itoh. 1998. Sequence analysis by cloning of the structural gene of gassericin A, a hydrophobic bacteriocin produced by Lactobacillus gasseri LA39. Biosci. Biotechnol. Biochem. 62:887-892. [DOI] [PubMed] [Google Scholar]
- 20.Kawai, Y., J. Kusnadi, R. Kemperman, J. Kok, Y. Ito, M. Endo, K. Arakawa, H. Uchida, J. Nishimura, H. Kitazawa, and T. Saito. 2009. DNA sequencing and homologous expression of a small peptide conferring immunity to gassericin A, a circular bacteriocin produced by Lactobacillus gasseri LA39. Appl. Environ. Microbiol. 75:1324-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kearney, K., G. F. Fitzgerald, and J. F. Seegers. 2000. Identification and characterization of an active plasmid partition mechanism for the novel Lactococcus lactis plasmid pCI2000. J. Bacteriol. 182:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kingsford, C. L., K. Ayanbule, and S. L. Salzberg. 2007. Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake. Genome Biol. 8:R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krause, M., and D. G. Guiney. 1991. Identification of a multimer resolution system involved in stabilization of the Salmonella dublin virulence plasmid pSDL2. J. Bacteriol. 173:5754-5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 26.Leer, R. J., J. M. van der Vossen, M. van Giezen, J. M. van Noort, and P. H. Pouwels. 1995. Genetic analysis of acidocin B, a novel bacteriocin produced by Lactobacillus acidophilus. Microbiology 141:1629-1635. [DOI] [PubMed] [Google Scholar]
- 27.Luchansky, J. B., M. C. Tennant, and T. R. Klaenhammer. 1991. Molecular cloning and deoxyribonucleic acid polymorphisms in Lactobacillus acidophilus and Lactobacillus gasseri. J. Dairy Sci. 74:3293-3302. [DOI] [PubMed] [Google Scholar]
- 28.Majhenic, A. C., B. B. Matijasic, and I. Rogelj. 2003. Chromosomal location of the genetic determinants for bacteriocins produced by Lactobacillus gasseri K7. J. Dairy Res. 70:199-203. [DOI] [PubMed] [Google Scholar]
- 29.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J. H. Lee, I. Diaz-Muniz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, J. Broadbent, R. Hutkins, D. O'Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maqueda, M., A. Galvez, M. M. Bueno, M. J. Sanchez-Barrena, C. Gonzalez, A. Albert, M. Rico, and E. Valdivia. 2004. Peptide AS-48: prototype of a new class of cyclic bacteriocins. Curr. Protein Pept. Sci. 5:399-416. [DOI] [PubMed] [Google Scholar]
- 31.Maqueda, M., M. Sáchez-Hidalgo, M. Fernádez, M. Montalbá-Lóez, E. Valdivia, and M. Martinez-Bueno. 2008. Genetic features of circular bacteriocins produced by gram-positive bacteria. FEMS Microbiol. Rev. 32:2-22. [DOI] [PubMed] [Google Scholar]
- 32.Martin-Visscher, L. A., M. J. van Belkum, S. Garneau-Tsodikova, R. M. Whittal, J. Zheng, L. M. McMullen, and J. C. Vederas. 2008. Isolation and characterization of carnocyclin A, a novel circular bacteriocin produced by Carnobacterium maltaromaticum UAL307. Appl. Environ. Microbiol. 74:4756-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Driscoll, J., F. Glynn, G. F. Fitzgerald, and D. van Sinderen. 2006. Sequence analysis of the lactococcal plasmid pNP40: a mobile replicon for coping with environmental hazards. J. Bacteriol. 188:6629-6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olivares, M., M. A. Díz-Ropero, N. Góez, F. Lara-Villoslada, S. Sierra, J. A. Maldonado, R. Martin, E. Lóez-Huertas, J. M. Rodriguez, and J. Xaus. 2006. Oral administration of two probiotic strains, Lactobacillus gasseri CECT5714 and Lactobacillus coryniformis CECT5711, enhances the intestinal function of healthy adults. Int. J. Food Microbiol. 107:104-111. [DOI] [PubMed] [Google Scholar]
- 35.Olivares, M., M. P. Díz-Ropero, N. Góez, F. Lara-Villoslada, S. Sierra, J. A. Maldonado, R. Martin, J. M. Rodriguez, and J. Xaus. 2006. The consumption of two new probiotic strains, Lactobacillus gasseri CECT 5714 and Lactobacillus coryniformis CECT 5711, boosts the immune system of healthy humans. Int. Microbiol. 9:47-52. [PubMed] [Google Scholar]
- 36.Ravin, V., T. Sasaki, L. Raisanen, K. A. Riipinen, and T. Alatossava. 2006. Effective plasmid pX3 transduction in Lactobacillus delbrueckii by bacteriophage LL-H. Plasmid 55:184-193. [DOI] [PubMed] [Google Scholar]
- 37.Reuter, G. 2001. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr. Issues Intest. Microbiol. 2:43-53. [PubMed] [Google Scholar]
- 38.Riley, M. A., and J. E. Wertz. 2002. Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie 84:357-364. [DOI] [PubMed] [Google Scholar]
- 39.Sakamoto, I., M. Igarashi, K. Kimura, A. Takagi, T. Miwa, and Y. Koga. 2001. Suppressive effect of Lactobacillus gasseri OLL 2716 (LG21) on Helicobacter pylori infection in humans. J. Antimicrob. Chemother. 47:709-710. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Sasaki, Y., N. Taketomo, and T. Sasaki. 1988. Factors affecting transfer frequency of pAMβ1 from Streptococcus faecalis to Lactobacillus plantarum. J. Bacteriol. 170:5939-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Satoh, E., Y. Ito, Y. Sasaki, and T. Sasaki. 1997. Application of the extracellular α-amylase gene from Streptococcus bovis 148 to construction of a secretion vector for yogurt starter strains. Appl. Environ. Microbiol. 63:4593-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Serror, P., T. Sasaki, S. D. Ehrlich, and E. Maguin. 2002. Electrotransformation of Lactobacillus delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis with various plasmids. Appl. Environ. Microbiol. 68:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamura, A., H. Kumai, N. Nakamichi, T. Sugiyama, R. Deguchi, A. Takagi, and Y. Koga. 2006. Suppression of Helicobacter pylori-induced interleukin-8 production in vitro and within the gastric mucosa by a live Lactobacillus strain. J. Gastroenterol. Hepatol. 21:1399-1406. [DOI] [PubMed] [Google Scholar]
- 45.Tatusov, R. L., N. D. Fedorova, J. D. Jackson, A. R. Jacobs, B. Kiryutin, E. V. Koonin, D. M. Krylov, R. Mazumder, S. L. Mekhedov, A. N. Nikolskaya, B. S. Rao, S. Smirnov, A. V. Sverdlov, S. Vasudevan, Y. I. Wolf, J. J. Yin, and D. A. Natale. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinform. 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Kranenburg, R., N. Golic, R. Bongers, R. J. Leer, W. M. de Vos, R. J. Siezen, and M. Kleerebezem. 2005. Functional analysis of three plasmids from Lactobacillus plantarum. Appl. Environ. Microbiol. 71:1223-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wall, R., G. Fitzgerald, S. Hussey, T. Ryan, B. Murphy, P. Ross, and C. Stanton. 2007. Genomic diversity of cultivable Lactobacillus populations residing in the neonatal and adult gastrointestinal tract. FEMS Microbiol. Ecol. 59:127-137. [DOI] [PubMed] [Google Scholar]
- 48.Wang, T. T., and B. H. Lee. 1997. Plasmids in Lactobacillus. Crit. Rev. Biotechnol. 17:227-272. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, J., Y. Zhang, L. Zhu, M. Suzuki, and M. Inouye. 2004. Interference of mRNA function by sequence-specific endoribonuclease PemK. J. Biol. Chem. 279:20678-20684. [DOI] [PubMed] [Google Scholar]