Abstract
The Caribbean reef sponge Svenzea zeai was previously found to contain substantial quantities of unicellular photosynthetic and autotrophic microbes in its tissues, but the identities of these symbionts and their method of transfer from adult to progeny are largely unknown. In this study, both a 16S rRNA gene-based fingerprinting technique (denaturing gradient gel electrophoresis [DGGE]) and clone library analysis were applied to compare the bacterial communities associated with adults and embryos of S. zeai to test the hypothesis of vertical transfer across generations. In addition, the same techniques were applied to the bacterial community from the seawater adjacent to adult sponges to test the hypothesis that water column bacteria could be transferred horizontally as sponge symbionts. Results of both DGGE and clone library analysis support the vertical transfer hypothesis in that the bacterial communities associated with sponge adults and embryos were highly similar to each other but completely different from those in the surrounding seawater. Sequencing of prominent DGGE bands and of clones from the libraries revealed that the bacterial communities associated with the sponge, whether adult or embryo, consisted of a large proportion of bacteria in the phyla Chloroflexi and Acidobacteria, while most of the sequences recovered from the community in the adjacent water column belonged to the class Alphaproteobacteria. Altogether, 21 monophyletic sequence clusters, comprising sequences from both sponge adults and embryos but not from the seawater, were identified. More than half of the sponge-derived sequences fell into these clusters. Comparison of sequences recovered in this study with those deposited in GenBank revealed that more than 75% of S. zeai-derived sequences were closely related to sequences derived from other sponge species, but none of the sequences recovered from the seawater column overlapped with those from adults or embryos of S. zeai. In conclusion, there is strong evidence that a dominant proportion of sponge-specific bacteria present in the tissues of S. zeai are maintained through vertical transfer during embryogenesis rather than through acquisition from the environment (horizontal transfer).
Besides being the oldest metazoans, sponges are the simplest multicellular animals and possess a low degree of tissue differentiation and coordination (54). Sponges are sessile, filter-feeding organisms that may harbor within their tissues a remarkable array of microorganisms, including bacteria (19, 59, 64), archaea (41), zooxanthellae (22), diatoms (63), and fungi (35). In some cases, microbial consortia can make up to 40 to 60% of the sponge tissue volume (21, 61) and exceed a density of 109 microbial cells per ml of sponge tissue (62), which is several orders of magnitude higher than that found in seawater. Apart from being a source of food (43), bacterial symbionts may participate in the acquisition and transfer of nutrients inside sponges (67, 68), the recycling of insoluble protein (69), the stabilization of the sponge skeleton (44), and the processing of metabolic waste (4, 65). Many antimicrobial compounds have been isolated from sponge bacterial symbionts (24, 47, 53), suggesting the involvement of symbiotic bacteria in sponge chemical defenses. In some cases, bacterial symbionts have been found to be the source of bioactive compounds that were isolated from sponges, which has opened up new research directions in marine natural product chemistry, biotechnology, and pharmaceutical development (18, 23, 40).
Based on immunological evidence from the 1980s (66), sponge-bacterium symbioses are thought to have originated in the Precambrian, when bacteria evolved to form a single clade of sponge-specific bacteria that were distinct from isolates found in the surrounding seawater. Since then, many studies have similarly documented a high level of consistency and specificity in sponge-bacterium associations (20, 27, 59). Nevertheless, questions remain about the acquisition and maintenance of symbionts in host sponges. In general, the following two hypotheses have been proposed: (i) a recently metamorphosed sponge selectively retains specific groups of bacteria from the diverse pool of bacteria present in the water column as it begins filter feeding (horizontal transfer) or (ii) specific bacterial strains are transmitted by the maternal sponge to developing embryos and are already present in the metamorphosing sponge (vertical transfer) (58). The first hypothesis requires some recognition of specific microbes by the sponge, perhaps through an innate immune system (36) or other means to distinguish symbiont strains from food bacteria (70).
Vertical transfer of bacterial symbionts in sponges was first proposed by Lévi and Porte (29), who demonstrated the presence of bacteria inside the larvae of the sponge Oscarella lobularis. Later, in 1976, Lévi and Lévi (30) studied the transmission of bacteria in the sponge Chondrosia reniformis via sponge oocytes. Since then, vertical transmission of bacterial symbionts via eggs or larvae has been documented for several sponge species, including Tethya citrina (15), Geodia cydonium (50), Stelletta grubii (49), Hippospongia sp. (25), Spongia sp. (25), Halisarca dujardini (10), and Corticium candelabrum (8). However, all of these studies employed transmission and scanning electron microscopy and could only examine the presence of bacteria in maternal sponges, oocytes, or larvae at the morphological level, with no determination of microbial identity. With advances in molecular techniques, Enticknap et al. (9) were the first to report the successful isolation of an alphaproteobacterial symbiont, strain NW001, from both the adult sponge Mycale laxissima and its larvae. They also did a preliminary denaturing gradient gel electrophoresis (DGGE) analysis of the bacterial community in seawater and compared that with the community in the sponge larval sample. However, such a comparison was not extended to the sponge adult, and no solid conclusion can be drawn for the horizontal transfer mechanism of sponge symbionts. More recently, Sharp et al. (52) used fluorescence in situ hybridization (FISH) and clone library techniques to demonstrate the presence of proteobacteria, actinobacteria, and a clade of sponge-associated bacteria in the embryos and mesohyl of the tropical sponge Corticium sp. By clone library and DGGE analyses, Schmitt et al. (48a) identified 28 vertical-transmission clusters in five different Caribbean sponge species and demonstrated that the complex sponge adult microbial community was collectively transmitted through reproductive stages. While these recent studies support the vertical transfer hypothesis, they did not fully address the identities of microbes in the water column surrounding the sponges, which is key to determining whether horizontal transfer may also take place.
The Caribbean reef sponge Pseudaxinella zeai was reclassified into a new genus, Svenzea (Demospongiae, Halichondria, Dictyonellidae), in 2002 because it has an unusual skeleton arrangement consisting mainly of short stout styles that are arranged in an isodictyal reticulation (2). It is a viviparous sponge that produces the largest embryos (>1 mm in diameter) and larvae (6 mm long) recorded for the phylum Porifera (45). Svenzea zeai has also been classified as a bacteriosponge because it contains substantial amounts of unicellular photosynthetic and autotrophic microbial symbionts in its tissues (2, 45). Although bacteria were observed in the embryos and larvae of this sponge based on transmission electron microscopy studies (45), neither the direct linkage between the maternal sponge and the propagules nor the identity of the microbial symbionts had been established.
In this study, our objective was to examine vertical versus horizontal transfer of bacterial symbionts in Svenzea zeai. This was achieved by comparing the bacterial community profiles of the adults and embryos of the sponge by use of a combination of molecular techniques, including DGGE and clone library analysis. More than one technique was employed to compensate for deficiencies of each technique in revealing bacterial community structure. Additionally, we used the same techniques to examine the bacterial community in the seawater that surrounded the sponge to determine whether horizontal transfer was evident.
MATERIALS AND METHODS
Sample collection and extraction of DNA.
Tissue of the adult sponge Svenzea zeai was obtained at a depth of 12 m from San Salvador Island, Bahama Islands (24°03′N, 74°32′W), in June 2007. Three sponge individuals were carefully brought to the water surface by scuba divers and flushed with autoclaved 0.22-μm-filtered seawater to remove loosely attached bacteria. The adult sponges were dissected, and embryos in the adult tissues were carefully removed. Adult sponge tissue (0.5 ml) without any embryos was cut into small pieces and frozen in 0.8 ml of extraction buffer (100 mM Tris-HCl, 100 mM sodium EDTA, 100 mM Na2HPO4, 1.5 M NaCl, 1% cetyltrimethylammonium bromide, pH 8). The embryos removed from the adult tissue were first briefly washed with 70% ethanol and then thoroughly rinsed twice with autoclaved filtered seawater. Embryo mass (0.5 ml) removed from each of the three individual sponge adults was homogenized and frozen in 0.8 ml of extraction buffer.
Bacterial communities in the surrounding seawater were collected in triplicate by filtering 1 liter of seawater onto 0.22-μm polycarbonate membranes (Osmonics). The membranes were then frozen in extraction buffer. The extraction and purification of total bacterial DNA from the samples were performed following the sodium dodecyl sulfate-based method described by Liu et al. (31). Purified DNA was dissolved in 50 μl of double-distilled water (ddH2O) and kept at −20°C until use.
DNA fingerprinting analysis of bacterial communities.
Bacterial community structure was revealed by DGGE (39). The 16S rRNA genes in the crude DNA extracts were amplified by PCR, using the universal primers 341F-GC (5′-CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC CCG CCC G CCT ACG GGA GGC AGC AG) and 907R (5′-CCG TCA ATT CMT TTG AGT TT) (39). Each PCR mixture contained 2 μl of DNA template, 1.25 U of Taq polymerase (Amersham Biosciences), a 0.25 mM concentration of each deoxynucleoside triphosphate, 0.1 μM of each primer, and 1× PCR buffer in a total volume of 50 μl. PCR was performed in a thermal cycler (MJ Research) under the following thermal conditions: initial denaturation at 95°C for 2 min; 10 touchdown cycles of denaturation at 95°C for 1 min, annealing at 65°C (reduced to 55°C in increments of 1°C cycle−1) for 1 min, and extension at 72°C for 1 min; an additional 15 cycles with a constant annealing temperature of 55°C; and a final extension at 72°C for 5 min. PCR products were mixed with loading buffer and loaded onto a 6% acrylamide gel with a denaturing gradient of 35 to 70% (100% denaturant = 7 M urea, 40% [vol/vol] formamide). Electrophoresis was performed using a D-Code system (Bio-Rad) with 1× TAE (20 mM Tris base, 10 mM sodium acetate, and 0.5 mM EDTA) at a constant temperature of 60°C and a voltage of 125 V for 18 h. The gel was stained with 1× SYBR gold (Molecular Probes) for 15 min and photographed with an Alpha Imager 2200 gel documentation system (Alpha Innotech).
Sequencing analysis of DGGE bands.
Major bands from the DGGE gel were selected and excised from the gel for sequence analysis. Excised gel cubes were first washed with ddH2O and then immersed in 50 μl of ddH2O at 4°C overnight. Two microliters of DNA from each excised band was used as the template for the same PCR-DGGE analysis to check for the band position and purity. PCR products were then purified, cloned into the pCR2.1-TOPO vector, and transformed into Escherichia coli competent cells by use of a Topo TA cloning kit (Invitrogen) according to the manufacturer's manual. Transformants were screened by blue-white selection on agar containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)-isopropyl-β-d-thiogalactopyranoside (IPTG) and 100 μg ml−1 of ampicillin. White colonies were then transferred to fresh plates and reincubated overnight. DNA was extracted from each positive clone by picking a single white colony from the plate into 100 μl of ddH2O and lysing the cells by heating at 99°C for 10 min. The lysates were used as DNA templates for subsequent PCR amplification, using the external vector primers M13F and M13R. Purified PCR products were then used as templates for cycle sequencing PCR, using either M13F or M13R primer and a DYEnamic ET dye terminator kit (Amersham Biosciences). Cycle sequencing products were separated using a MegaBACE 500 genetic analyzer (Amersham Biosciences). The nucleotide sequences obtained with the two primers were assembled using Sequencher 4.2 (Gene Codes Corporation), and the assembled sequences were compared with sequences deposited in GenBank (http://www.ncbi.nlm.nih.gov/), using BLAST, to obtain their closest phylogenetic affiliations.
Clone library construction.
Since the bacterial community structures for replicated samples were highly similar, as indicated by DGGE analysis, crude DNA extracts from the three replicates were pooled as templates for the construction of clone libraries. The 16S rRNA genes in the crude DNA extracts were PCR amplified with the universal primers 8F (5′-AGA GTT TGA TCC TGG CTC AG) (3) and 1492R (5′-GGT TAC CTT GTT ACG ACT T) (28), using the following PCR conditions: initial denaturation at 95°C for 5 min; 30 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min; and a final extension at 72°C for 10 min. Successful PCR amplicons were purified using a PCR purification mini kit (Watson Biotechnologies Inc., Shanghai, China). The quantities of purified PCR products were determined by GeneQuant (Amersham Biosciences). The same amounts of purified DNA from different samples were cloned into the pCR2.1-TOPO vector and then transformed into E. coli competent cells by use of a Topo TA cloning kit (Invitrogen) according to the manufacturer's manual. The same procedures as those mentioned above were used for screening of positive clones, extraction of DNA, and subsequent PCR amplification. Aliquots (20 μl) of successful PCR amplicons were digested individually with two restriction enzymes (MspI and HaeIII) according to the manufacturer's instructions (Invitrogen). Restriction fragment length polymorphism (RFLP) patterns of each clone upon two different restriction enzyme digestions were obtained by electrophoresis on 3% agarose gels. Clones that showed the same RFLP patterns for the two enzymes were classified into the same operational taxonomic units (OTUs). One clone from each OTU was randomly selected and subjected to sequencing using vector primers M13F and M13R and internal primers 8F and 1492R. Sequencing analysis was performed as mentioned above. Nearly the full lengths of bacterial 16S rRNA gene sequences were obtained by assembling each fragment sequence using Sequencher 4.2 (Gene Codes Corporation). Chimera Check was used to exclude chimeras (34). The assembled sequences were then compared to the GenBank entries by using BLAST to obtain the closest phylogenetic affiliation for each OTU. Phylogenetic analysis was performed with sequences retrieved for each OTU in comparison with their closest affiliations, using ARB software (33). Phylogenetic trees were then constructed based on the neighbor-joining method (46), maximum parsimony (12), and the unweighted-pair group method using average linkages (UPGMA) (56), using MEGA software (26).
Statistical analysis.
Bacterial community structures, as revealed by DGGE band patterns, were compared among samples. Previous studies have suggested that major bands on DGGE gels represent the dominant bacterial species present in the respective samples and that band intensity correlates with the relative abundance of the corresponding bacterial species within the sample (13, 38). Therefore, each band in the DGGE gel was described by its position and relative intensity in the profile, using GelCompar II software (Applied Maths). Band matching was performed with 1.00% position tolerance and 1.00% optimization. Similarity matrices were calculated based on the band position and intensity of each sample. Cluster analysis was performed based on the Pearson similarity correlation, and dendrograms were constructed based on the Ward method, using GelCompar II software (Applied Maths).
For each clone library, the Chao estimator (7), the Shannon index (51), and the coverage (37) were calculated. LIBSHUFF analysis was used to compare libraries to determine if they were significantly different from each other (55). A LIBSHUFF comparison of three libraries yielded an experiment-wise critical P value of 0.0085 according to the Bonferroni correction (http://libshuff.mib.uga.edu). For each pairwise comparison, if the lower of the two P values calculated by LIBSHUFF was less than or equal to the critical P value, then there was a significant difference, with a confidence of 95%, in the composition of the communities sampled by each library.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences obtained in this study were deposited in GenBank under the following accession numbers: FJ529257 to FJ529375.
RESULTS
DGGE analysis of bacterial communities.
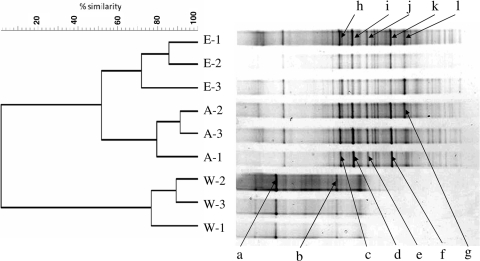
DGGE analysis of the bacterial communities associated with the adults and embryos of the sponge Svenzea zeai resulted in completely different band patterns in the DGGE gel than those associated with the bacterial communities present in the water adjacent to the adult sponges (Fig. 1, right panel). The water column bacterial communities had an average of 17 bands that concentrated at the upper area of the gel (where the denaturing concentration of the gel was low). In contrast, the average numbers of bands observed for the adult- and embryo-associated bacterial communities were as high as 29 and 30, respectively, and the bands were found mostly in the middle and lower parts of the gel (where the denaturant concentration was high). Cluster analysis using a similarity matrix based on the band position and intensity confirmed the distinctiveness of the water column bacterial community (Fig. 1, left panel). A distinct cluster was formed with replicated samples from the water column, which shared 0% similarity to the cluster formed from replicated samples from the sponge adults and embryos. In contrast, the sponge adult- and embryo-associated bacterial communities shared more than 50% similarity and formed a large cluster which consisted of two smaller clusters comprising the replicated samples from either the sponge adults or embryos.
FIG. 1.
DGGE band patterns (right) and dendrogram (left) showing the similarities of bacterial communities associated with the adults (A) and embryos (E) of the sponge Svenzea zeai and the planktonic bacterial community (W) in San Salvador, Bahama Islands. Labeled bands were excised and sequenced. Details of the excised bands are given in Table 1.
Sequence analysis of excised DGGE bands.
Altogether,12 bands from the DGGE gel (2 from the water column samples and 5 each from the sponge adults and embryos) were selected, excised, and sequenced. These bands were selected because of their prominence and uniqueness. Approximately 550 bp of sequence was retrieved from each band and compared with nucleotide sequences deposited in GenBank. The closest phylogenetic affiliation of each band sequence is listed in Table 1. All 12 sequences were affiliated with uncultured strains from clones. The 2 sequences from the water samples (bands a and b) were closely related to the phylum Alphaproteobacteria, while the other 10 sequences, from the sponge adults (bands c to g) and embryos (bands h to l), belonged to the phyla Acidobacteria and Chloroflexi. Bands excised from the same portions of the denaturing gradient (i.e., bands c versus h, d versus i, e versus j, f versus k, and g versus l) shared the same closest matches. These results indicate that strains belonging to the phylum Chloroflexi contributed large proportions of the bacterial communities associated with the adults and embryos of the sponge.
TABLE 1.
Closest phylogenetic affiliations of sequences retrieved from selected bands excised from DGGE gela
| Band | Source | Closest phylogenetic affiliation
|
||||
|---|---|---|---|---|---|---|
| Strain | Source | Phylum | GenBank accession no. | Similarity (%) | ||
| a | Water | Uncultured Roseobacter sp. clone 2_C6 | Coastal seawater from Ria de Vigo, Spain | Alphaproteobacteria | EU600651 | 94 |
| b | Water | Uncultured Rhodobacteriales bacterium clone HF70_26K06 | Seawater from Hawaii Ocean, North Pacific Subtropical Gyre | Alphaproteobacteria | EU361404 | 94 |
| c | Adult | Uncultured sponge symbiont PAUC37f | Marine sponge Theonella swinhoei from the Western Caroline Islands in the Republic of Palau | Acidobacteria | AF186413 | 97 |
| d | Adult | Uncultured Chloroflexi bacterium clone PK067 | Marine sponge Plakortis sp. from Little San Salvador Island, Bahamas | Chloroflexi | EF076103 | 96 |
| e | Adult | Uncultured Chloroflexi bacterium clone PK053 | Marine sponge Plakortis sp. from Little San Salvador Island, Bahamas | Chloroflexi | EF076110 | 95 |
| f | Adult | Uncultured Chloroflexi bacterium clone PK017 | Marine sponge Plakortis sp. from Little San Salvador Island, Bahamas | Chloroflexi | EF076100 | 99 |
| g | Adult | Uncultured Chloroflexi bacterium clone PK064 | Marine sponge Plakortis sp. from Little San Salvador Island, Bahamas | Chloroflexi | EF076111 | 96 |
| h | Embryo | Uncultured sponge symbiont PAUC37f | Marine sponge Theonella swinhoei from the Western Caroline Islands in the Republic of Palau | Acidobacteria | AF186413 | 99 |
| i | Embryo | Uncultured Chloroflexi bacterium clone PK067 | Marine sponge Plakortis sp. from Little San Salvador Island, Bahamas | Chloroflexi | EF076103 | 99 |
| j | Embryo | Uncultured Chloroflexi bacterium clone PK053 | Marine sponge Plakortis sp. from Little San Salvador Island, Bahamas | Chloroflexi | EF076110 | 99 |
| k | Embryo | Uncultured Chloroflexi bacterium clone PK017 | Marine sponge Plakortis sp. from Little San Salvador Island, Bahamas | Chloroflexi | EF076100 | 99 |
| l | Embryo | Uncultured Chloroflexi bacterium clone PK064 | Marine sponge Plakortis sp. from Little San Salvador Island, Bahamas | Chloroflexi | EF076111 | 98 |
The sequences of excised bands were compared to nucleotide sequences deposited in GenBank. The closest phylogenetic affiliation for the sequence from each band is indicated by the strain name, phylum, accession number, and similarity. Refer to Fig. 1 for band positions.
Clone library analysis.
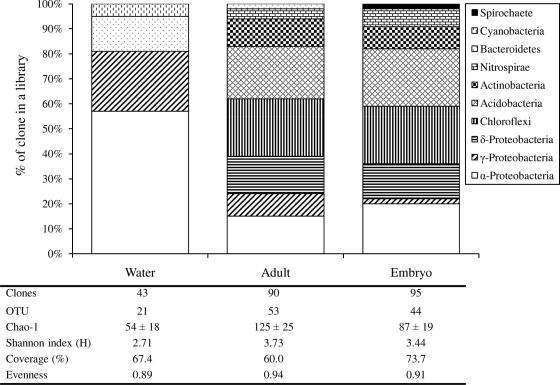
Altogether, 228 clones were retrieved from the three clone libraries, 43 of which were from the seawater library, while 90 and 95 were from the sponge adult and embryo libraries, respectively. After RFLP screening, 21, 53, and 44 OTUs were identified from the libraries of seawater and sponge adults and embryos, respectively (Fig. 2). The Chao-1 estimator (7) and the Shannon index (H) were used to estimate the species richness and to calculate the diversity of the libraries, respectively. The higher the values, the richer and more diverse was the community. The results indicated that the sponge adults had the highest species richness (Chao-1 = 125 ± 25), followed by the sponge embryos (Chao-1 = 87 ± 19), whereas the lowest species richness was found in the seawater (Chao-1 = 54 ± 18; H = 2.71).
FIG. 2.
16S rRNA gene phylotype distribution and comparison of clone libraries from water samples and adults and embryos of the sponge Svenzea zeai. Species richness was estimated using the nonparametric Chao estimator (7). The Shannon index was calculated based on the number of unique phylotypes detected by RFLP screening. Coverage of the library was expressed as a percentage and calculated from the equation C = 1 − (n1/N), where n1 was the number of clones occurring only once in the library and N was the total number of clones examined (34).
Nearly-full-length 16S rRNA gene sequences were obtained for each OTU and compared with sequences deposited in GenBank. Phylogenetic analysis of the 118 OTUs showed that only 3 of them (2.5%) were affiliated with sequences from isolates, while the remaining 115 OTUs (97.5%) were closely related to uncultured clones (Fig. 3). All clones from the seawater library, except for one (W10), were affiliated with uncultured clones from different marine environments, including seawater from Cocos Island, Monterey Bay, the Mediterranean Sea, and the Red Sea (Fig. 3). In contrast, among the 53 and 44 OTUs retrieved from the sponge adult and embryo libraries, respectively, 42 and 33 of them (a total of 77%), respectively, were closely related to sequences retrieved from other sponge species, including Plakortis sp., Corticium sp., Theonella swinhoei, Agelas dilatata, and Aplysina aerophoba (Fig. 3). Only the remaining 23% were related to sequences from other habitats, for instance, mangrove soil and seawater.
FIG. 3.
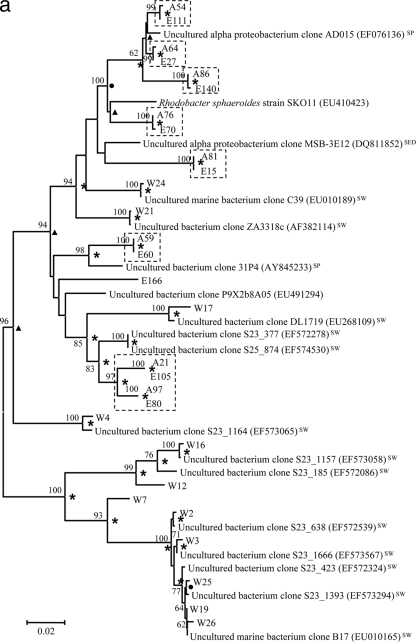
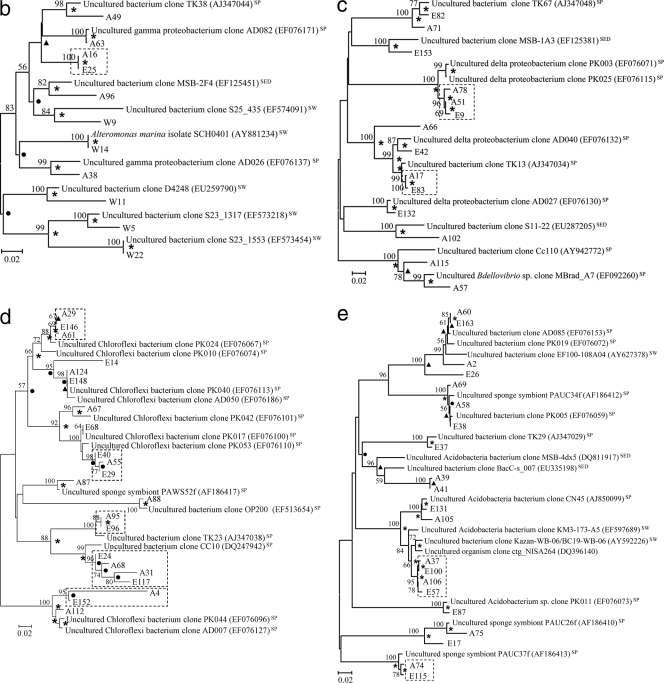
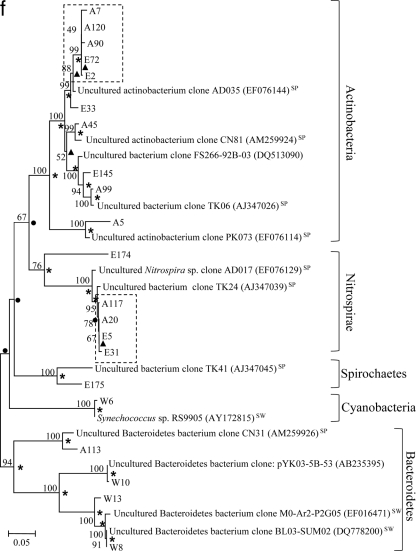
Phylogenetic trees showing genetic distances among clones retrieved from the libraries for seawater (prefixed with “W”), S. zeai adults (prefixed with “A”), and S. zeai embryos (prefixed with “E”) in reference to members of the Alphaproteobacteria (a), Gammaproteobacteria (b), Deltaproteobacteria (c), Chloroflexi (d), Acidobacteria (e), and Actinobacteria, Nitrospirae, Spirochaetes, Cyanobacteria, and Bacteroidetes (f). The trees were constructed based on the neighbor-joining method. Nodes which are observed in both maximum parsimony and UPGMA trees are marked with asterisks, while those observed in either the maximum parsimony or UPGMA tree are marked with filled circles or triangles, respectively. Reference sequences originating from seawater, sediment, and sponges are indicated with the superscripts “SW,” “SED,” and “SP,” respectively. Nucleotide accession numbers of the reference sequences are given in parentheses. Dotted boxes show monophyletic clusters consisting of sequences retrieved from both adult and embryo sponges but not from water. The scale bar represents percent substitutions per nucleotide position. Bootstrap values of >50% based on 1,000 resamplings are indicated by the numbers at the nodes.
Of the 21 OTUs retrieved from the seawater library, 57% belonged to the phylum Alphaproteobacteria, 24% to Gammaproteobacteria, 14% to Bacteroidetes, and 5% to Cyanobacteria (Fig. 2). The sponge adult and embryo libraries comprised a wider range of phyla than the seawater library did. Apart from the clones belonging to the phyla Alphaproteobacteria, Gammaproteobacteria, and Bacteroidetes, clones closely related to six other phyla were found (Deltaproteobacteria, Spirochaetes, Nitrospirae, Actinobacteria, Acidobacteria, and Chloroflexi) (Fig. 2). For both the sponge adult and embryo libraries, the largest portion of the clones (>20% each) were affiliated with the phyla Chloroflexi and Acidobacteria, which were absent in the seawater library.
Figure 3 presents phylogenetic trees reflecting the genetic distances among clones from different libraries with reference to their closest relatives. For all phyla recovered in this study, except for Spirochaetes, Cyanobacteria, and Bacteroidetes, a large proportion of clones from the sponge adults were clustered with those from the sponge embryos, showing extremely high similarity in their 16S rRNA gene sequences. For instance, there was high similarity for clones A86 versus E140 and A76 versus E70 in the Alphaproteobacteria (Fig. 3a), clones A16 versus E25 in the Gammaproteobacteria (Fig. 3b), clones A51 versus E9 in the Deltaproteobacteria (Fig. 3c), clones A29 versus E146 and A124 versus E148 in the Chloroflexi (Fig. 3d), and clones A58 versus E38 and A74 versus E115 in the Acidobacteria (Fig. 3e). These highly similar clones were defined as vertically transmitted phylotypes and formed 21 monophyletic sequence clusters (clusters of two or more sequences retrieved from both the adult sponges and embryos but not from the seawater). More than half of the sequences derived from the sponge adults and embryos fell into these clusters. On the other hand, clones from the seawater library normally formed lineages distinct from those of the sponge libraries, as shown in the alphaproteobacterial (clones W2, W3, W12, W16, W19, W25, and W26) (Fig. 3a), gammaproteobacterial (clones W5, W11, and W22) (Fig. 3b), and Bacteroidetes (clones W8, W10, and W13) (Fig. 3f) trees.
To determine the significance of the differences between the clone libraries based on available sequence data, LIBSHUFF analysis was applied. Employing the Bonferroni correction, a LIBSHUFF comparison of three libraries yielded a critical P value of 0.0085. LIBSHUFF analysis indicated that the sponge adult and embryo libraries were not significantly different from each other, as the lower of the two P values calculated from LIBSHUFF analysis (P = 0.009) was larger than the critical P value. In contrast, the seawater library was significantly different from the libraries of the sponge adults (lower P = 0.001) and the libraries of the sponge embryos (lower P = 0.001).
DISCUSSION
The Caribbean reef sponge Svenzea zeai was categorized as a bacteriosponge on the basis of transmission electron microscopy observations because it contained substantial quantities of unicellular photosynthetic and autotrophic bacteria (45). The method by which the very large embryos of Svenzea zeai acquire their bacterial symbionts from the adult sponge remained unknown, as did the phylogenetic identities of the symbionts. This study represents the only study on the associated bacterial community in the sponge family Dictyonellidae and is one of a few detailed studies combining different molecular approaches to investigate vertical transmission as well as horizontal transfer of sponge symbionts in the sponge by incorporating both the sponge embryo-associated and the indigenous bacterioplankton communities for comparison. The results of our study corroborate the most commonly observed method of transmission, which is vertical transfer of symbionts from adult tissue to developing propagules (8, 9, 10, 48a, 52, 58, 60). Moreover, bacterial symbionts associated with both the adult sponge and sponge embryos are primarily bacteria in the phyla Chloroflexi and Acidobacteria.
The 16S rRNA gene-based DGGE fingerprinting method provided general insights into the compositions of bacterial communities associated with the adults and embryos of S. zeai and in the surrounding seawater, while the clone libraries presented a more in-depth analysis of the community composition to reveal the identities of bacterial associates likely involved in vertical transfer. Both methods yielded the same conclusion, that the bacterial communities associated with adult S. zeai sponges were highly similar to those associated with embryos but drastically different from those in the surrounding seawater. Twenty-one monophyletic sequence clusters comprising two or more sequences retrieved from the clone libraries of S. zeai adults and embryos, but none from the seawater library, were identified upon phylogenetic analysis of clones. More than half of the sponge-derived sequences fell into these clusters. Some clones from the adult and embryo libraries shared highly similar or even identical sequences (clones A59 versus E60, A81 versus E15, A16 versus E25, A95 versus E96, and A60 versus E163), indicating substantial overlap of the bacterial communities associated with S. zeai adults and embryos. Cross-contamination of the adult bacterial community with that in embryos during sample collection was ruled out because of extremely careful washing procedures. Vertical transfer of microbial symbionts has also been implicated for another Caribbean sponge, Ircinia felix (48). Interestingly, bacterial communities associated with adult I. felix sponges more closely resembled those of their own larvae than those of other adult I. felix sponges (48). This was not found to be true in the present study, as the symbiont communities found in Svenzea zeai were highly similar among adults and larvae.
Sequencing analysis was performed with common bands obtained in DGGE gels and for all clones from the clone libraries to identify bacterial associates that might have been passed vertically from adult sponge to embryo. Our results indicated that adults and embryos of S. zeai harbored highly similar yet diverse bacterial communities covering eight different phyla, which were also encountered in gene libraries of other sponges (reviewed in references 32 and 58). The most dominant groups of sponge-derived microbial sequences were closely related to sequences from uncultured clones belonging to the phyla Acidobacteria and Chloroflexi. This is in good agreement with other studies showing abundant uncultured Acidobacteria and Chloroflexi bacteria in different sponge species (20, 58, 63). In addition, about 77% of our sponge-derived sequences were closely clustered with sequences from other sponge species, for instance, Corticium sp. (52), Theonella swinhoei, Aplysina aerophoba (20), Plakortis sp., and Agelas dilatata (58), collected from different locations around the world. Of the 77% of sponge-related sequences, nearly 70% fell into the sponge-specific clusters suggested by Hentschel et al. (20) and Taylor et al. (58). These results further strengthen the argument that there are sponge-specific clusters of bacteria present in geographically and phylogenetically diverse sponge species. Another discrepancy of the results obtained in the present study compared with previous research was that the largest and best-known sponge-specific microbial clusters, “Candidatus Synechococcus spongiarum” (60) and “Poribacteria” (11), were absent from our sponge libraries. One possible reason may be due to mismatches in the target region of the “Poribacteria” 16S rRNA genes which make our universal primers not able to amplify them (11). Another possibility is that the abundance of these microbial groups may be too low to be recovered by clone library analysis. Sequencing of all minor bands in the DGGE gels was experimentally not feasible. In the future, a metagenomic approach may be a better way to reveal the most complete picture of the sponge-associated microbial community. It would also be interesting to correlate the metabolically active bacteria in the sponge and embryo tissues by using FISH or quantitative PCR so as to understand how they are transferred between generations and their possible roles in host sponges.
An important difference between this study and others in which the vertical transfer of microbial symbionts between sponge adults and embryos has been implicated is the inclusion of a comparison with the indigenous bacterioplankton community. Analysis of the microbes in the water column adjacent to adult sponges allowed us to assess the possibility that horizontal transfer of symbionts was occurring. As indicated by the DGGE and clone library analyses, the bacterial communities in the surrounding seawater were less diverse than and completely different from the bacterial communities associated with S. zeai adults and embryos. Sequences belonging to the phyla Alpha- and Gammaproteobacteria dominated the bacterioplankton community, and most were closely related to sequences recovered from the water column in geographically distinct locations, such as the Red Sea (14), the Mediterranean Sea (1), the east coast of South Korea (5), Monterey Bay, and Cocos Island. Distinct planktonic bacterial communities were also recorded in many other studies (16, 17, 42). Hill (23) argued that considering the powerful filtration capacity of sponges, with up to 2.4 × 1013 bacterial cells filtered per day, one might expect that even an extremely rare bacterium in the water column could possibly be concentrated in a sponge to levels detectable by PCR-based methods. As such, it would be risky to completely rule out the possibility that the bacterial symbionts found in sponge samples can be present in seawater samples, even though we did not detect them in the seawater in this study. However, our observation that none of the bacteria detected in the seawater samples resembled any of the bacteria recovered from S. zeai adults or embryos strongly argues against the horizontal transfer hypothesis. Therefore, environmental acquisition of bacterial associates is probably not an important factor in maintaining the sponge-microbe association of S. zeai.
If vertical transmission of bacterial associates in sponges is the rule, then the implication for the coevolution of sponges and sponge-associated microbes is considerable (21). However, the lack of a clearly resolved phylogeny of sponges (6) still hampers our understanding of symbiont evolution in sponges. Although vertical transmission of Chloroflexi and Acidobacteria from adult to embryo was possible for S. zeai, the cellular mechanisms for the transfer remain to be explored. One technique may be to design specific probes for in situ qualitative and quantitative tracking of the presence of specific bacteria during different developmental stages of the sponge.
Acknowledgments
We are grateful to the captain and crew of the R/V Seward Johnson and to the Government of the Bahamas for allowing us to conduct scientific research in their territorial waters.
This study was supported by grants from the China Ocean Mineral Resources Research and Development Association (COMRRDA06/07.SC02) and the KAUST Global Academic Partnership Program (KAUST005-CML07/08) to P.-Y. Qian and by a U.S. National Science Foundation Biological Oceanography Program grant to J. R. Pawlik (OCE-0550468).
Footnotes
Published ahead of print on 31 July 2009.
REFERENCES
- 1.Alonso-Saez, L., V. Balague, E. L. Sa, O. Sanchez, J. M. Gonzalez, J. Pinhassi, R. Massana, J. Pernthaler, C. Pedros-Alio, and G. M. Gasol. 2007. Seasonality in bacterial diversity in north-west Mediterranean coastal waters: assessment through clone libraries, fingerprinting and FISH. FEMS Microbiol. Ecol. 60:98-112. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, B., R. W. M. van Soest, and K. Rützler. 2002. Svenzea, a new genus of Dictyonellidae (Porifera: Demospongiae) from tropical reef environments, with description of two new species. Contrib. Zool. 71:171-176. [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beer, S., and M. Ilan. 1998. In situ measurements of photosynthetic irradiance responses of two Red Sea sponges growing under dim light conditions. Mar. Biol. 131:613-617. [Google Scholar]
- 5.Bhattarai, H. D., Y. K. Lee, K. H. Cho, H. K. Lee, and H. W. Shin. 2006. The study of antagonistic interactions among pelagic bacteria: a promising way to coin environmental friendly antifouling compounds. Hydrobiologia 568:417-423. [Google Scholar]
- 6.Boury-Esnault, N. 2006. Systematics and evolution of Demospongiae. Can. J. Zool. 84:205-224. [Google Scholar]
- 7.Chao, A. 1984. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 8.de Caralt, S., M. J. Uriz, and R. H. Wijffels. 2007. Vertical transmission and successive location of symbiotic bacteria during embryo development and larva formation in Corticium candelabrum (Porifera: Demospongiae). J. Mar. Biol. Assoc. U. K. 87:1693-1699. [Google Scholar]
- 9.Enticknap, J. J., M. Kelly, O. Peraud, and R. T. Hill. 2006. Characterization of a culturable alphaproteobacterial symbiont common to many marine sponges and evidence for vertical transmission via sponge larvae. Appl. Environ. Microbiol. 72:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ereskovsky, A. V., E. Gonobobleva, and A. Vishnyakov. 2005. Morphological evidence for vertical transmission of symbiotic bacteria in the viviparous sponge Halisarca dujardini Johnston (Porifera, Demospongiae, Halisarcida). Mar. Biol. 146:869-875. [Google Scholar]
- 11.Fieseler, L., M. Horn, M. Wagner, and U. Hentschel. 2004. Discovery of the novel candidate phylum “Poribacteria” in marine sponges. Appl. Environ. Microbiol. 70:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitch, W. M. 1971. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 20:406-416. [Google Scholar]
- 13.Fromin, N., J. Hamelin, S. Tarnawski, D. Roesti, K. Jourdain-Miserez, N. Forestier, S. Teyssier-Cuvelle, F. Gillet, M. Aragno, and P. Rossi. 2002. Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns. Environ. Microbiol. 4:634-643. [DOI] [PubMed] [Google Scholar]
- 14.Fuller, N. J., D. Marie, F. Partensky, D. Vaulot, A. F. Post, and D. J. Scanlan. 2003. Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea. Appl. Environ. Microbiol. 69:2430-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaino, E., B. Burlando, P. Puffa, and M. Sara. 1987. Ultrastructural study of the mature egg of Tethya citrina Sara and Melone (Porifera, Demospongiae). Gamete Res. 16:259-265. [DOI] [PubMed] [Google Scholar]
- 16.Giovannoni, S. J., and M. S. Rappe. 2000. Evolution, diversity and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the ocean. John Wiley & Sons, Inc., New York, NY.
- 17.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 18.Haygood, M. G., E. W. Schmidt, S. K. Davidson, and D. J. Faulkner. 1999. Microbial symbionts of marine invertebrates: opportunities for microbial biotechnology. J. Mol. Microbiol. Biotechnol. 1:33-43. [PubMed] [Google Scholar]
- 19.Hentschel, U., M. Schmid, M. Wanger, L. Fieseler, C. Gernert, and J. Hacker. 2001. Isolation and phylogenetic analysis of bacteria with antimicrobial activities from the Mediterranean sponges Aplysina aerophoba and Aplysina cavernicola. FEMS Microbiol. Ecol. 35:305-312. [DOI] [PubMed] [Google Scholar]
- 20.Hentschel, U., J. Hopke, M. Horn, A. Friedrich, M. Wagner, J. Hacker, and B. Moore. 2002. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl. Environ. Microbiol. 68:4431-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hentschel, U., L. Fieseler, M. Wehrl, C. Gernert, M. Steinert, J. Hacker, and M. Horn. 2003. Microbial diversity of marine sponges. Prog. Mol. Subcell. Biol. 37:59-88. [DOI] [PubMed] [Google Scholar]
- 22.Hill, M. S. 1996. Symbiotic zooxanthellae enhance boring and growth rates of the tropical sponge Anthosigmella varians forma varians. Mar. Biol. 125:649-654. [Google Scholar]
- 23.Hill, R. T. 2004. Microbes from marine sponge: a treasure trove of biodiversity of natural products discovery, p. 177-190. In A. T. Bull (ed.), Microbial diversity and bioprospecting. ASM Press, Washington, DC.
- 24.Jayatilake, G. S., M. P. Thornton, A. C. Leonard, J. E. Grimwade, and G. J. Baker. 1996. Metabolites from an Antarctic sponge associated bacterium Pseudomonas aeruginosa. J. Nat. Prod. 59:293-296. [DOI] [PubMed] [Google Scholar]
- 25.Kaye, H. R. 1991. Sexual reproduction in four Caribbean commercial sponges. Oogenesis and transfer of bacterial symbionts. Invert. Reprod. Dev. 19:13-24. [Google Scholar]
- 26.Kumar, S., K. Tamura, and M. Nei. 1994. MEGA: molecular evolutionary genetics analysis software for microcomputers. Comput. Appl. Biosci. 10:189-191. [DOI] [PubMed] [Google Scholar]
- 27.Lafi, F. F., M. J. Garson, and J. A. Fuerst. 2005. Culturable bacterial symbionts isolated from two distinct sponge species (Pseudoceratina clavata and Rhabdastrella globostellata) from the Great Barrier Reef display similar phylogenetic diversity. Microb. Ecol. 50:213-220. [DOI] [PubMed] [Google Scholar]
- 28.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-148. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, NY.
- 29.Lévi, C., and A. Porte. 1962. Ètude au microscope électronique de l'éponge Oscarella lobularis Schmidt et de sa larve amphiblastula. Cah. Biol. Mar. 3:307-315. [Google Scholar]
- 30.Lévi, C., and P. Lévi. 1976. Embryogenese de Chondrosia reniformis (Nardo), demosponge ovipare, et transmission des bacteries symbiotiques. Ann. Sci. Nat. Zool. Biol. Anim. 18:367-380. [Google Scholar]
- 31.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez, J., A. Ledger, C. Peterson, K. Sfanos, D. Harmody, S. Pomponi, and P. McCarthy. 2006. Molecular census and comparison of cultured and uncultured microbial symbiont diversity from an ancient metazoan host, phylum Porifera, abstr. 6.15. Abstr. 5th Int. Symbiosis Soc. Congr., Vienna, Austria, 4 to 10 August 2006.
- 33.Ludwig, W., O. Strunk, R. Westram, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maidak, B. L., J. R. Cole, T. G. Lilbum, C. T. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (ribosomal database project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maldonado, M., N. Cortadellas, M. I. Trillas, and K. Rützler. 2005. Endosymbiotic yeast maternally transmitted in a marine sponge. Biol. Bull. 209:94-106. [DOI] [PubMed] [Google Scholar]
- 36.Müller, W. E. G., and I. M. Müller. 2003. Origin of the metazoan immune system: identification of the molecules and their functions in sponges. Integr. Comp. Biol. 144:19-29. [DOI] [PubMed] [Google Scholar]
- 37.Mullins, T. D., T. B. Britschgi, R. L. Krest, and S. J. Giovannoni. 1995. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol. Oceanogr. 40:148-158. [Google Scholar]
- 38.Murray, A. E., J. T. Hollibaugh, and C. Orrego. 1996. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl. Environ. Microbiol. 62:2676-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muyzer, G., E. C. D. Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piel, J. 2006. Bacterial symbionts: prospects for the sustainable production of invertebrate-derived pharmaceuticals. Curr. Med. Chem. 13:39-50. [PubMed] [Google Scholar]
- 41.Preston, C. M., K. Y. Wu, T. F. Molinski, and E. F. DeLong. 1996. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc. Natl. Acad. Sci. USA 93:6241-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rappe, M. S., K. Vergin, and S. J. Giovannoni. 2000. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol. Ecol. 33:219-232. [DOI] [PubMed] [Google Scholar]
- 43.Reiswig, H. M. 1975. Bacteria as food for temperate-water marine sponges. Can. J. Zool. 53:493-502. [Google Scholar]
- 44.Rützler, K. 1985. Associations between Caribbean sponges and photosynthetic organisms, p. 455-466. In K. Rützler (ed.), New perspectives in sponge biology. Smithsonian Institution Press, Washington, DC.
- 45.Rützler, K., R. W. M. van Soest, and B. Alvarez. 2003. Svenzea zeai, a Caribbean reef sponge with a giant larva, and Scopalina ruetzleri: a comparative fine structural approach to classification (Demospongiae, Halichondria, Dictyonellidae). Invert. Biol. 122:203-222. [Google Scholar]
- 46.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstruction of phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 47.Schmidt, E. W., A. Y. Obraztsova, S. K. Davidson, D. J. Faulkner, and M. G. Haygood. 2000. Identification of the antifungal peptide-containing symbionts of the marine sponge Theonella swinhoei as a novel δ-proteobacterium, “Candidatus Entotheonella palauensis.” Mar. Biol. 136:969-977. [Google Scholar]
- 48.Schmitt, S., J. B. Weisz, N. Lindquist, and U. Hentschel. 2007. Vertical transmission of a phylogenetically complex microbial consortium in the viviparous sponge Ircinia felix. Appl. Environ. Microbiol. 73:2067-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48a.Schmitt, S., H. Angermeier, R. Schiller, N. Lindquist, and U. Hentschel. 2008. Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl. Environ. Microbiol. 74:7694-7708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sciscioli, M., L. S. Liaci, E. Lepore, M. Gherardi, and T. L. Simpson. 1991. Ultrastructural study of the mature egg of the marine sponge Stelletta grubii (Porifera, Demospongiae). Mol. Reprod. Dev. 28:346-350. [DOI] [PubMed] [Google Scholar]
- 50.Sciscioli, M., E. Lepore, M. Gherardi, and L. S. Liaci. 1994. Transfer of symbiotic bacteria in the mature oocyte of Geodia cydonium (Porifera, Demospongiae): an ultrastructural study. Cah. Biol. Mar. 35:471-478. [Google Scholar]
- 51.Shannon, C. E. 1948. A mathematical theory of communication. Bell System Tech. J. 27:379-423, 623-656. [Google Scholar]
- 52.Sharp, K. H., B. Eam, D. J. Faulkner, and M. G. Haygood. 2007. Vertical transmission of diverse microbes in the tropical sponge Corticium sp. Appl. Environ. Microbiol. 73:622-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shigemori, H., M. A. Bae, K. Yazava, T. Sasaki, and J. Kobayashi. 1992. Alteramida A, a new tetracyclic alkaloid from a bacterium Alteromonas sp. associated with the marine sponge Halichondria okadai. J. Org. Chem. 57:4317-4320. [Google Scholar]
- 54.Simpson, T. L. 1984. The cell biology of sponges. Springer-Verlag, New York, NY.
- 55.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rDNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy—the principles and practice of numerical classification. W. H. Freeman, San Francisco, CA.
- 57.Reference deleted.
- 58.Taylor, M. W., R. Radax, D. Steger, and M. Wagner. 2007. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71:295-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thacker, R. W., and S. Starnes. 2003. Host specificity of the symbiotic cyanobacterium Oscillatoria spongeliae in marine sponge, Dysidea spp. Mar. Biol. 142:643-648. [Google Scholar]
- 60.Usher, K. M., S. Toze, J. Fromont, J. Kuo, and D. C. Sutton. 2004. A new species of cyanobacterial symbiont from the marine sponge, Chondrilla nucula. Symbiosis 36:183-192. [Google Scholar]
- 61.Vacelet, J., and C. Donadey. 1977. Electro microscope study of the association between sponges and bacteria. J. Exp. Mar. Biol. Ecol. 30:301-314. [Google Scholar]
- 62.Webster, N. S., and R. T. Hill. 2001. The culturable microbial community of the Great Barrier Reef sponge Rhopaloeides odorabile is dominated by an alphaproteobacterium. Mar. Biol. 138:843-851. [Google Scholar]
- 63.Webster, N. S., K. J. Wilson, L. L. Blackall, and R. T. Hill. 2001. Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeides odorabile. Appl. Environ. Microbiol. 67:434-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Webster, N. S., A. P. Negri, M. M. Munro, and C. N. Battershill. 2004. Diverse microbial communities inhabit Antarctic sponges. Environ. Microbiol. 6:288-300. [DOI] [PubMed] [Google Scholar]
- 65.Wilkinson, C. R. 1978. Microbial associations in sponges. I. Ecology, physiology and microbial populations of coral reef sponges. Mar. Biol. 49:161-167. [Google Scholar]
- 66.Wilkinson, C. R. 1984. Immunological evidence for the Precambrian origin of bacterial symbioses in marine sponges. Proc. R. Soc. Lond. B 220:509-517. [Google Scholar]
- 67.Wilkinson, C. R., and R. Garrone. 1980. Nutrition of marine sponges. Involvement of symbiotic bacteria in the uptake of dissolved carbon, p. 157-161. In D. C. Smith and Y. Tiffon (ed.), Nutrition in the lower metazoan. Pergamon Press, Oxford, United Kingdom.
- 68.Wilkinson, C. R., and J. Vacelet. 1979. Transplantation of marine sponges to different conditions of light and current. J. Exp. Mar. Biol. Ecol. 37:91-104. [Google Scholar]
- 69.Wilkinson, C. R., R. Garrone, and D. Herbage. 1979. Sponge collagen degradation in vitro by sponge-specific bacteria. Colloq. Int. CNRS 291:361-364. [Google Scholar]
- 70.Wilkinson, C. R., R. Garrone, and J. Vacelet. 1984. Marine sponges discriminate between food bacteria and bacterial symbionts: electromicroscope radioautography and in situ evidence. Proc. R. Soc. Lond. B 220:519-528. [Google Scholar]