Abstract
The seasonal dynamics of the small eukaryotic fraction (cell diameter, 0.2 to 5 μm) was investigated in a mesotrophic lake by tyramide signal amplification-fluorescence in situ hybridization targeting seven different phylogenetic groups: Chlorophyceae, Chrysophyceae, Cryptophyceae, Cercozoa, LKM11, Perkinsozoa (two clades), and Fungi. The abundance of small eukaryotes ranged from 1,692 to 10,782 cells ml−1. The dominant groups were the Chrysophyceae and the Chlorophyceae, which represented 19.6% and 17.9% of small eukaryotes, respectively. The results also confirmed the quantitative importance of putative parasites, Fungi and Perkinsozoa, in the small heterotrophic eukaryotic assemblage. The relative abundances recorded for the Perkinsozoa group reached as much as 31.6% of total targeted eukaryotes during the summer. The dynamics of Perkinsozoa clade 1 coincided with abundance variations in Peridinium and Ceratium spp. (Dinoflagellates), while the dynamics of Perkinsozoa clade 2 was linked to the presence of Dinobryon spp. (Chrysophyceae). Fungi, represented by chytrids, reached maximal abundance in December (569 cells ml−1) and were mainly correlated with the dynamics of diatoms, especially Melosira varians. A further new finding of this study is the recurrent presence of Cercozoa (6.2%) and LKM11 (4.5%) cells. This quantitative approach based on newly designed probes offers a promising means of in-depth analysis of microbial food webs in lakes, especially by revealing the phylogenetic composition of the small heterotrophic flagellate assemblage, for which an important fraction of cells are generally unidentified by classical microscopy (on average, 96.8% of the small heterotrophic flagellates were identified by the specific probes we used in this study).
Recently developed molecular methods based on the amplification and sequencing of rRNA genes have made it possible to investigate picoeukaryote assemblage composition (pigmented or nonpigmented unicellular eukaryotes with cell diameters of <2 μm or <5 μm according to the studies) in various aquatic systems, independently of morphological identification and cultivation (14, 23, 27, 28, 29, 39). The essential role of picoplankton (both eukaryotic and prokaryotic) as a contributor to plankton biomass and to carbon and nutrient cycling has long been established (9), but the unexpected diversity among the smallest eukaryotes (cell diameters, <5 μm) was only recently revealed. Most of these data were obtained in oceanic systems, but a few recent studies conducted in lakes have also highlighted the broad diversity of 18S rRNA sequences affiliated with numerous phylogenetic groups: Chlorophyceae, Chrysophyceae, Cryptophyceae, Cercozoa, Fungi, Choanoflagellida, Bicosoecida, Ciliophora, Haptophyceae, Perkinsozoa, LKM11, Hyphochytridiomycota, Katablepharidaceae, Dinophyceae, and Eustigmatophyceae (22, 23, 24, 34). Thus, it has been possible to observe clear seasonal changes in small-eukaryote structure in an oligomesotrophic lake (23), and the lake-based studies generally report a dominance of heterotrophic cells within the lacustrine small-eukaryote assemblage. Moreover, the recurrent presence of sequences affiliated with parasitic groups has been highlighted in lakes of various trophic statuses (22, 23). Lepère et al. (25) reported the unexpected importance of two groups: first, fungi affiliated with two clades of chytrids known as parasites of various groups of microalgae; and second, members of the phylum Perkinsozoa belonging to two clades closely related to Perkinsus marinus and Parvilucifera infectans, which are parasites of bivalves and dinoflagellates, respectively (30), and whose systematic position has been controversial, since they are phylogenetically related to the Apicomplexa or the Dinoflagellata (6, 13).
Although these data brought new insight into the structural diversity of lacustrine small eukaryotes, the relative importance, dynamics, and functional roles of these microorganisms from various phylogenetic groups are still largely unknown. We now need to research specific in situ abundances of previously undetected taxa. In this study, specially developed oligonucleotide probes, designed on the basis of molecular data obtained from sequencing (20, 21, 22, 23, 24, 25, 34), were used for fluorescence in situ hybridization (FISH) coupled with tyramide signal amplification (TSA) to investigate the composition, abundance, and dynamics of lacustrine small eukaryotes (<5 μm) in the mesotrophic Lake Bourget over 1 year. Special attention was paid to the dynamics of putative parasitic groups (Perkinsozoa, Fungi, Cercozoa).
MATERIALS AND METHODS
Study site.
The study was conducted in the mesotrophic Lake Bourget, located in eastern France on the edge of the Alps (45°48′N, 05°49′E; altitude, 231.5 m). It is a warm, meromictic lake that, with an area of 42 km2 and a total volume of 3.5 × 109 m3, constitutes France's largest natural reservoir. It has a maximum depth of 145.4 m (mean depth, 81 m), and its water residence time is approximately 10 years.
Sampling and fixation.
Water samples were taken at a permanent station in the 0- to 20-m layer using a sampling bell to carry out integrated sampling right through the water column. Sampling was performed monthly or bimonthly from May 2006 to April 2007. From the initial water samples, a subsample (at least 100 ml) was prefiltered through 5-μm-pore-size polycarbonate filters (Millipore) at a pressure of <15,000 Pa in order to eliminate larger cells (size fraction, > 5 μm). The filtrate obtained was fixed with formaldehyde (final concentration, 4%) and incubated at 4°C for 24 h. Fixed cells were then collected on 0.2-μm-pore-size polycarbonate filters (Millipore) (pressure, <15,000 Pa). These cells (diameters, 0.2 to 5 μm) were subjected to TSA-FISH counting and were stained with 4′,6-diamidino-2-phenylindole (DAPI). The filters were preserved by dehydration in an ethanol series (50, 80, and 100% ethanol for 3 min each) and were stored at 4°C in the dark until TSA-FISH staining.
From the initial integrated water sample, a subsample (100 ml) to be used for the observation of nanophytoplankton and microphytoplankton was fixed with Lugol's iodine solution. For classical microscopic counts of heterotrophic flagellates and ciliates, flagellates were fixed with glutaraldehyde (final concentration, 1%), while ciliates were preserved with mercuric bichloride (2.5%) according to the method of Simé-Ngando et al. (36). These samples were stored at 4°C in the dark. Sampling for metazooplankton counts was carried out simultaneously at the same location, using a 200-μm-mesh zooplankton net to carry out one vertical haul from 50 m to the surface. The sample was preserved immediately in 5% formalin solution.
Determination of the main limnological parameters.
During the whole sampling period, concentrations of dissolved orthophosphates (P-PO4), nitrates (N-NO3), ammonium (N-NH4), and silicates (SiO2) in freshwater samples were measured by the chemical laboratory of the Thonon hydrobiological station according to French normalized (AFNOR) protocols (http://thononin8.win3.hebergement.com/pages/public/index.html). The temperature (°C), dissolved oxygen concentration (mg liter−1), and fluorescence-based chlorophyll a (chl a) concentration (μg liter−1) were measured using a Seabird submersible multiparametric probe with a CTD SBE 19 Seacat profiler.
Soon after fixation, samples of flagellates were filtered (volume, 25 to 30 ml; pressure, <100 mm Hg) onto polycarbonate membranes (diameter, 25 mm; pore size, 0.8 μm), stained with primuline (modified from the method of Caron [8]), and preserved at −20°C (less than 1 week). Slides were examined under UV light with a Nikon Eclipse TE200 epifluorescence microscope to count the heterotrophic nanoflagellates (HNF) (on average, a total of 200 to 300 cells were counted per filter). Phytoplankton and ciliates were counted using Utermöhl's method (41) with a Zeiss Axiovert 135 inverted microscope. The metazoan zooplankton was counted by direct observation under a binocular microscope (Wild M3Z).
Design of new oligonucleotide probes.
Based on aligned sequences of eukaryotes, including environmental sequences of lacustrine small eukaryotes (22, 34, 20, 21, 23, 24, 25), oligonucleotide probes were designed using the ARB package (26). Probe specificity was checked by submitting the sequences to the nucleotide collection database of the National Center for Biotechnology Information (NCBI, BLASTn program; maximum identity, 100%). Six probes were designed to target Cercozoa, Chrysophyceae, LKM11, and two clades of Perkinsozoa, i.e., groups that had almost never before been quantified in lakes using specific probes. The probe sequences are reported in Table 1. A combination of two probes was required to efficiently stain the cells belonging to the LKM11 group.
TABLE 1.
Oligonucleotide probes used in this study
| Probe | Sequence (5′-3′) | Specificity | Reference |
|---|---|---|---|
| EUK1209R | GGG CAT CAC AGA CCT G | Eukaryota | 16 |
| CERC_02 | AAT ACG AGC ACC CCC AAC | Cercozoa | This study |
| CHLO02 | CTT CGA GCC CCC AAC TTT | Chlorophyceae | 38 |
| CRYPT 13 | CGA AAT ATA AAC GGC CCC AAC | Cryptophyceae | 25 |
| CHRYSO_01 | TTT CGG ACA AGG AAG ACT CG | Chrysophyceae | This study |
| LKM11_01 | TAC TGT CAC TAC CTC GCC | LKM11 | This study |
| LKM11_02 | TGG TCC TCA AAC CAA C | LKM11 | This study |
| MY1574 | TCC TCG TTG AAG AGC | Fungi (Eumycota) | 3 |
| PERKIN_01 | GAG GAT GCC TCG GTC AA | Perkinsozoa | This study |
| PERKIN_02 | GCC AAA CAT TG T ACT GCG | Perkinsozoa | This study |
In order to screen for any aspecific probes, these new probes were tested on eukaryotic cultures (Chlorophyceae, Chrysophyceae, Cryptophyceae, Cyanophyceae, Bacillariophyceae, Cercozoa, and Dinophyceae). When cultivated organisms were not available for positive testing (Perkinsozoa, LKM11), probes were validated using only bioinformatics tools and negative controls, as was previously done for probes targeting new stramenopiles (27).
Testing against control eukaryotic cultures showed no aspecific matches for the newly designed probes. In silico testing of these probes, performed by the ProbeCheck Web server (http://www.microbial-ecology.net/probecheck), showed that the probes targeting the monophyletic clade 1 and clade 2 of Perkinsozoa had no aspecificity for other planktonic 18S rDNA sequences. Testing of the CERC_02 probe, without mismatches, revealed that 95.6% of the eukaryotic sequences targeted were cercozoan sequences. For this probe, although no aspecificities was detected with our ARB database, the Web server ProbeCheck showed two aspecific matches with ciliate Oxytrichidae and apicomplexan Eimeriidae. The LKM11 opisthokonts, newly described by van Hannen et al. (42), in the phylogenetic neighborhood of Fungi, require a set of two specific probes. ProbeCheck analyses of these two oligonucleotide probes revealed no aspecific targets. With regard to the CHRYSO_01 probe, 99.2% of the picoplanktonic eukaryotic sequences targeted were affiliated with the Chrysophyceae. Only one aspecific match was detected, with the ciliate Glauconema trihymene. Moreover, the MY1574 probe, designed and validated by Baschien (3), has revealed its efficiency with fungal 18S rDNA sequences obtained from Lake Bourget (67% of these sequences have been matches).
Quantification of small eukaryotic organisms (cell diameter, <5 μm) using TSA-FISH.
Hybridization conditions for the FISH techniques were applied as described by Amann et al. (1). TSA-FISH was performed as described by Not et al. (31). Briefly, the hybridization filters were covered with a hybridization buffer (40% deionized formamide, 0.9 M NaCl, 20 mM Tris-HCl [pH 7.5], 0.01% sodium dodecyl sulfate, 10% blocking reagent [Roche Diagnostics/Boehringer]) and oligonucleotide probes labeled with horseradish peroxidase (50 ng μl−1). The mixture was left to hybridize at 35°C for 3 h. After two successive 20-min rinses at 37°C in a wash buffer (56 mM NaCl, 5 mM EDTA, 0.01% sodium dodecyl sulfate, 20 mM Tris-HCl [pH 7.5]), samples were equilibrated in TNT buffer (7% Tween 20, 150 mM NaCl, 100 mM Tris-HCl [pH 7.5]) at room temperature for 15 min. Tyramide amplification was performed for 30 min at room temperature in the dark in TSA mix, a mixture (1:1) of 40% dextran sulfate (Sigma-Aldrich) and 1× amplification diluent (Perkin-Elmer LAS), which provide enhanced sensitivity, to which is added fluorescein isothiocyanate coupled with tyramide (1×; Perkin-Elmer LAS) (1:50). Filters were then transferred through two successive 5-ml TNT buffer baths at 55°C for 20 min each to stop the enzymatic reaction and remove the dextran sulfate. Filters were mounted in a mixture of antifading oil AF1 (Citifluor, Biovalley, Conches, France) containing 10 μg ml−1 of propidium iodide (Sigma-Aldrich).
Hybridized cells were examined under a Nikon Eclipse TE200 epifluorescence microscope equipped with a mercury light source at ×100 magnification. The excitation/emission filter was 450 nm/490 nm for fluorescein isothiocyanate and propidium iodide. For each sample, at least 50 randomly chosen microscopic fields were analyzed and counted manually (on average, a minimum of 50 cells were counted).
Small eukaryotes were also counted by DAPI (final concentration, 5 μg ml−1) staining under UV light (350/461 nm); eukaryotic cell nuclei appeared as separate organelles, while prokaryotic organisms appeared as cells uniformly stained without visible nuclei. These counts allowed us to make comparisons between the number of eukaryotic cells targeted by DAPI and those targeted by the universal EUK1209R probe.
Statistical analysis.
Redundancy analyses (RDA) were used, after forward selection of the environmental variables likely to explain a significant part (P < 0.05) of the changes in small-eukaryote abundances, by considering potential parasitic groups especially (species matrix) (5). Six different species matrices were used. The first matrix takes into account the values obtained for all putative parasites; the second matrix was composed only of abundances of Perkinsozoa groups; and the last four matrices that were tested contained, respectively, the abundances of Perkinsozoa clade 1, Perkinsozoa clade 2, Fungi, and Cercozoa, which were tested separately. The RDA conducted to detect links between these matrices and environmental parameters were always performed with the same set of explanatory variables. These environmental variables were chemical parameters (PO4-P, NH4-N, NO3-N, SiO2,), chl a concentrations, and abundances of heterotrophic flagellates, ciliates, nano- and microphytoplankton species, metazooplankton species, and bivalve larvae. These statistics were computed with R software using the Vegan package for the RDA and related methods (http://cran.r-project.org/).
RESULTS
Main abiotic parameters determined at the study site.
On average for Lake Bourget, the mean concentration of silicates was 19.7 μM (±11.4 μM), and low phosphorus concentrations were recorded in the 0- to 20-m layer, with a mean of 0.02 μM (±0.015 μM) for the year. NH4-N and NO3-N concentrations were highest at the beginning of spring, corresponding to the weakest phytoplanktonic biomass and the water homogenization period (maximum in May, with 2.15 and 9 μM, respectively). After this period, decreases in phosphorus, nitrate, ammonium, and silicate concentrations were observed during the stratification episode (between May and July 2006). Subsequently, a relative stability of these chemical parameters, characterized by lower concentrations, was recorded during the summer and autumn.
Abundance, structure, and dynamics of nano- and microplanktonic organisms. (i) Heterotrophic flagellates and ciliates.
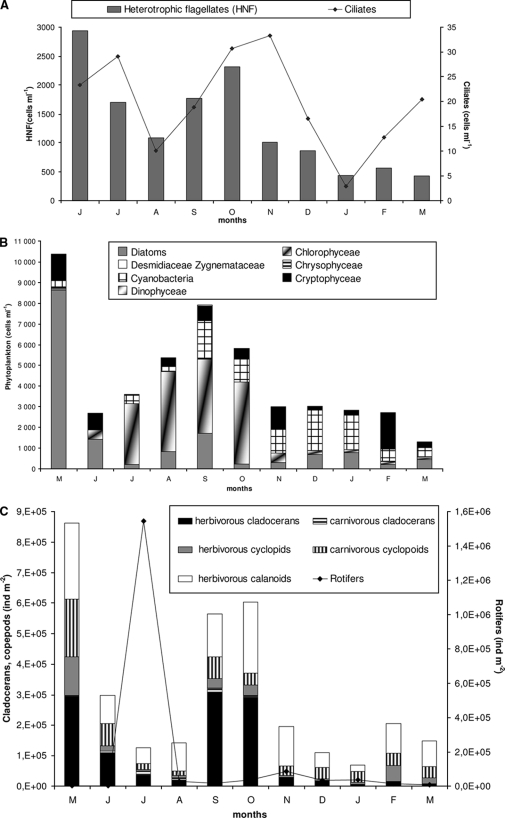
The mean abundance of HNF was 1,311 cells ml−1 in Lake Bourget from May 2006 to March 2007 (Fig. 1A), representing 26.7% of the total small eukaryotes stained by DAPI (Table 2). Microscopic observations showed that the dynamics of HNF revealed two seasonal peaks (maxima in June and October, with 2,942 cells ml−1 and 1,770 cells ml−1, respectively). Thus, HNF can represent as much as 79% of the total small eukaryotes present in Lake Bourget in June that can be counted by DAPI staining (Table 2). Among these HNF, a large fraction (92%) of the smallest cells (diameter, <5 μm) were unidentified taxa, which had similar morphotypes but could have different phylogenetic affiliations. The abundance of these unidentified <5-μm cells ranged from 425 cells ml−1 (March) to 2,942 cells ml−1 (June); they were generally less important in winter than in summer. The abundance of total ciliates had two seasonal peaks, in summer and autumn (Fig. 1A), and ranged from 3 to 33 cells ml−1 on average.
FIG. 1.
Temporal changes in abundances of the main biological parameters in Lake Bourget from May 2006 to March 2007. (A) Seasonal dynamics of heterotrophic flagellates and ciliates. (B) Temporal changes in abundances of nano- and microphytoplankton, with cell diameters of >5 μm (diatoms, Chlorophyceae, Chrysophyceae, Desmidiaceae, Zygnemataceae, Cryptophyceae, cyanobacteria, and Dinophyceae). (C) Seasonal dynamics of zooplankton (cladocerans, copepods, and rotifers).
TABLE 2.
Relative abundances of small eukaryotes in the 0- to 20-m layer of Lake Bourget
| Staining or probe | Targeted group | Mean (minimum to maximum) relative abundance (%) |
|---|---|---|
| DAPIa | Total small eukaryotes | 100 |
| EUK1209R | Total small eukaryotes | 87.2 (69.0-116.5) |
| Primuline | HNF (cell diameter, <5 μm) | 26.7 (7.9-79) |
| EUK1209Rb | Total small eukaryotes | 100 |
| CERC_02 | Cercozoa | 6.2 (1.4-11.5) |
| CHLO02 | Chlorophyceae | 17.9 (4.9-34.9) |
| CRYPT 13 | Cryptophyceae | 9.5 (1.4-15.3) |
| CHRYSO_01 | Chrysophyceae | 19.6 (10.2-34.2) |
| LKM11_01 + LKM11_02 | LKM11 | 4.5 (1.0-10.2) |
| MY1574 | Fungi | 11.4 (2.3-27.1) |
| PERKIN_01 | Perkinsozoa clade 1 | 9.0 (5.8-16.7) |
| PERKIN_02 | Perkinsozoa clade 2 | 7.3 (3.3-18.1) |
Abundances are annual means of the proportions of DAPI-stained cells (taken as 100%) either targeted by the universal probe EUK1209R or identified as heterotrophic flagellates via primuline staining.
Results are annual means of the abundances (relative to that of the total small eukaryotes targeted by EUK1209R, taken as 100%) of the eight eukaryotic groups targeted by TSA-FISH using specific oligonucleotide probes.
(ii) Chl a, nanophytoplankton, and microphytoplankton (>5 μm).
During 2006, 124 different phytoplanktonic species were detected in the 0- to 20-m layer of Lake Bourget. The seasonal dynamics of these pigmented organisms were characterized by two periods of high abundance. A spring peak was characterized by the dominance of diatoms, mainly Cyclotella spp., in May 2006 (Fig. 1B), the month during which the lowest concentration of chl a (<2 μg liter−1) was recorded. On average, a chl a concentration of 5.08 μg liter−1 (range, 2.8 μg liter−1 in July to 7.2 μg liter−1 in February) was observed in the 0- to 20-m layer of Lake Bourget. During the summer period, the phytoplanktonic populations (>5 μm) were composed largely of Chlorophyceae in July and August (mainly Chlorella spp. and Phacotus spp.), and of Chlorophyceae and cyanobacteria in September and October (mainly Planktothrix rubescens). During the winter period, pigmented eukaryotes (cell diameter, >5 μm) were represented mainly by Cryptomonas spp. and Rhodomonas spp. (Cryptophyceae), with 1,755 cells ml−1 (64%) in February. Finally, species of the Desmidiaceae and Zygnemataceae (notably Cosmarium depressum and Mougeotia gracillima) and the Dinophyceae species Ceratium hirundinella, Gymnodinium spp., and Peridinium spp. constituted the least well represented groups in Lake Bourget.
(iii) Metazooplankton.
The abundance of crustacean metazooplankton had a bimodal seasonal dynamic in Lake Bourget (Fig. 1C), with a first peak in spring (23 May 2006) and a second peak in autumn, both representing herbivorous zooplankton in the lake. In terms of abundance, rotifers (with a peak of 1.5 × 106 individuals [ind] m−2 in July), represented mainly by Asplanchna spp. and Conochilus spp., were dominant throughout the study period. Cladocerans were represented mainly by three species: Daphnia hyalina, which is responsible for the spring peak of cladocerans, and Eubosmina longispina and Diaphanosoma brachyurum, the most abundant cladocerans in September and October, respectively. Copepods comprise one calanoid species (Eudiaptomus gracilis) and seven cyclopoid species, of which, on average, 4% were at the nauplius stage.
Abundance, structure, and dynamics of the small eukaryotic assemblage (cell diameter, <5 μm).
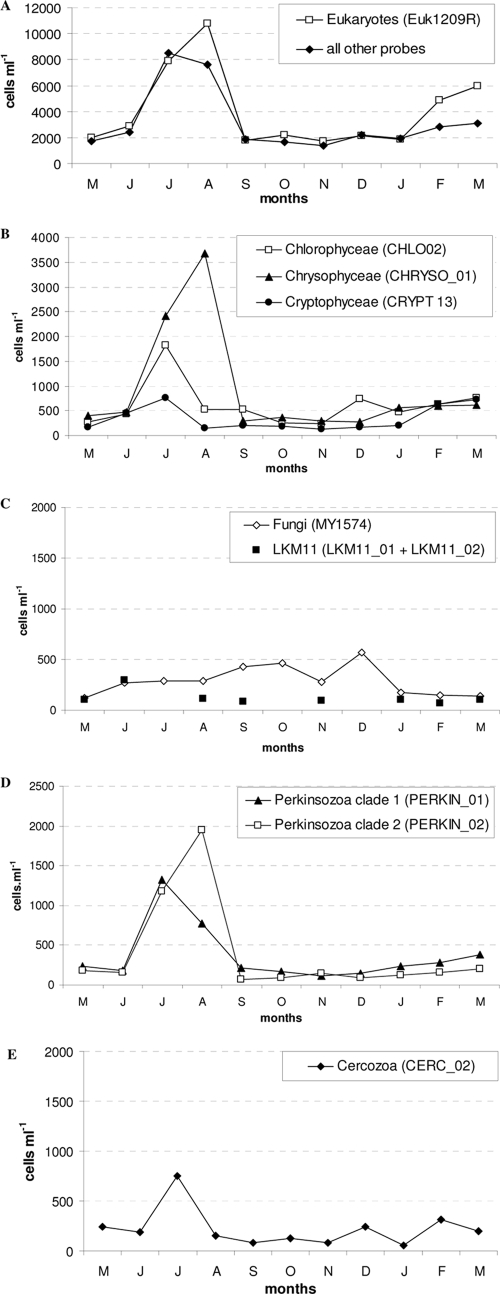
The abundance of small eukaryotes (targeted by the EUK1209R probe) ranged from 1,692 cells ml−1 (November) to 10,782 cells ml−1 (August) (Fig. 2A). For 7 of the 11 sampling dates, abundance was relatively stable and did not exceed 2,900 cells ml−1. The abundance of the small-eukaryote community increased strongly during the summer (peaking in July and August) but also in early spring (approximately 6,000 cells ml−1 in March). The mean abundance of small eukaryotes (both phototrophic and heterotrophic organisms) targeted with the EUK1209R probe was 4,007 cells ml−1 in the 0- to 20-m layer of Lake Bourget.
FIG. 2.
Dynamics of small eukaryotes targeted by TSA-FISH from May 2006 to March 2007 in Lake Bourget (0 to 20 m). (A) Abundance of cells targeted by the universal eukaryotic probe EUK1209R and total abundances of cells targeted by the other oligonucleotide probes. (B to E) Abundances of Chlorophyceae, Cryptophyceae, and Chysophyceae (B), fungi and the LKM11 group (C), Perkinsozoa clades 1 and 2 (D), and Cercozoa (E). In July, October, and December 2006, the LKM11 group could not be quantified.
In order to estimate the percentage of cells targeted by the EUK1209R probe, we compared these quantifications of small eukaryotes with those obtained by DAPI staining. The total eukaryotic abundance estimated by DAPI staining ranged from 2,478 cells ml−1 (November) to 11,036 cells ml−1 (August). On average, 87.2% of total small eukaryotes determined by DAPI staining were detected with the EUK1209R probe, and a significant correlation was recorded between the results obtained by the two methods (r = 0.98; P < 0.05).
The total abundance obtained with the specific probes (seven different eukaryotic groups targeted) ranged from 1,400 cells ml−1 (November) to 8,500 cells ml−1 (July) and was clearly correlated with the dynamics observed with the EUK1209R probe (r = 0.92; P < 0.05) (Fig. 2A). Taken together, the full set of specific probes allowed us to target, on average throughout this study, 85.5% of the small eukaryotes detected by the EUK1209R probe (Table 2). The lowest staining rates were obtained in February and March 2007, when only 55% of the total small-eukaryote community (EUK1209R) was detected with the specific probes we used.
(i) Chlorophyceae, Cryptophyceae, and Chrysophyceae.
Throughout the period studied, we were able to detect the presence of cells of the Chlorophyceae, Cryptophyceae, and Chrysophyceae in the <5-μm-cell-diameter fraction. There were large variations in the numbers of cells hybridized with the CHRYSO_01, CRYPT 13, and CHLO02 probes, with a peak in summer in particular (maxima in July and August) (Fig. 2B). Cryptophyceae were also characterized by an increase in abundance in early spring (March). These three taxonomic groups constituted, on average, 47% of total cells detected, with peak relative abundances measured at 63% and 65% of total small eukaryotes (EUK1209R probe) in July and January, respectively.
Chrysophyceae were the predominant eukaryotic group in Lake Bourget, averaging 19.6% of total targeted eukaryotic cells (Table 2). We identified a large abundance of cells targeted with the CHRYSO_01 probe in August: 3,682 cells ml−1, representing 35% of total small eukaryotes. Cryptophyceae and Chlorophyceae were also well represented, accounting for 9.5% and 17.9% of total small eukaryotes, respectively, during the study. Their highest abundances were recorded in July (752 cells ml−1 and 1,827 cells ml−1, respectively).
(ii) Fungi and the LKM11 group.
There was a relatively stable abundance of opisthokonts, represented by fungi (MY1574 probe) and the LKM11 group, over the course of the year. In contrast with the other eukaryotic groups targeted, these opisthokonts had abundance peaks in winter. Indeed, fungi were characterized by a small increase in December (569 cells ml−1) (Fig. 2C). Hence, during autumn and winter, the putative eukaryotic parasitic groups were essentially composed of fungi, which in September and December represented 25% and 27%, respectively, of total hybridized cells. Fungi were abundant mainly in autumn (from September to December), whereas this group represented only about 5% of total small eukaryotes during the rest of the year. Overall, fungi represented 11.4% (annual mean) of total small eukaryotes (Table 2).
The LKM11 group registered the lowest abundance among the targeted groups: a mean abundance of only 220 cells ml−1 (an average 4.5% of total small eukaryotes). However, in June, this group represented as much as 10.2% of total targeted small eukaryotes (EUK1209R probe). The LKM11 group, targeted by the LKM11_01 and LKM11_02 probes, was represented by round cells without apparent flagella.
(iii) Perkinsozoa.
Eukaryotic cells detected with the two Perkinsozoa probes (clades 1 and 2) were represented by zoosporic flagellated forms about 2 to 3 μm in diameter and had mean abundances of 366 cells ml−1 (clade 1) and 393 cells ml−1 (clade 2) (Fig. 2D), representing 9.0% and 7.3%, respectively, of total small eukaryotes (annual means) (Table 2). The dynamics of the cells hybridized with these probes was characterized by a peak in summer. Clade 1 (targeted by probe PERKIN_01) and clade 2 (targeted by probe PERKIN_02) did not show simultaneous peak abundances. The highest abundances were observed in July for clade 1, with 1,323 cells ml−1, and in August for clade 2, with 1,953 cells ml−1 (Fig. 2D). Except for these two summer dates, the Perkinsozoa group showed relatively stable abundance through the year, ranging from 229 cells ml−1 (December) to 579 cells ml−1 (March). The relative abundances recorded for these two putative parasitic groups were globally high, representing 31.6% of total targeted small eukaryotes in July (16.7% for clade 1 and 14.9% for clade 2).
(iv) Cercozoa.
Cells targeted by the CERC_02 probes presented ovoid forms about 3 to 5 μm in diameter. The abundance of Cercozoa remained relatively stable over the year, although there was a small increase during the summer, peaking in July (753 cells ml−1) (Fig. 2E). The CERC_02 probe hybridized on average 220 cells ml−1 during the study, i.e., 6.2% of the total small eukaryotes. Cercozoa represented at most 12.1% and 11.5% of the small-eukaryote assemblage in May and December, respectively, so that Cercozoa and the LKM11 group rank as the least abundant groups (Table 2).
Correlation of abiotic and biotic factors with the abundances of the different potential parasitic groups.
The results of the first RDA showed that the main biotic parameters involved in the explanation of the variance in the putative parasitic groups (Perkinsozoa and Fungi) were the abundance of Dinobryon sociale, belonging to the Chrysophyceae (53.8% of the explained variance), followed by the dynamics of Chlorophyceae and of diatoms belonging to the genera Cyclotella and Cymbella (Table 3).
TABLE 3.
Results of the six RDA used to link the dynamics of potential eukaryote parasites with biotic and abiotic environmental parametersa
| Response variable (species data) | Explanatory variable(s) (% of significantlyb explained variance) |
|---|---|
| Total putative parasites (Perkinsozoa + fungi) | Dinobryon sociale (53.8), Cyclotella bodanica (23), Cymbella sp. (12.4), Melosira varians (5.6), Chlorophyceae (4.9) |
| Total Perkinsozoa | Dinobryon sociale (80.7), Fragilaria cf. acusc (17.2) |
| Perkinsozoa clade 1 | Peridinium inconspicuum (84.4), Ceratium hirundinella (10) |
| Perkinsozoa clade 2 | Dinobryon sociale (99.5) |
| Fungi | NO3-N (64.4), Melosira varians (21.1), Chlorophyceae (7.2), Cyclotella bodanica (3.8) |
| Cercozoa | Pseudanabaena galeata (83) |
The explanatory variables tested in this analysis are chemical parameters (PO4-P, NH4-N, NO3-N, SiO2,), chl a concentration, abundances of heterotrophic flagellates and ciliates, and abundances of nano- and microphytoplankton species, metazooplankton species, and bivalve larvae. The response of small putative eukaryotic parasites to these explanatory variables was observed from six different matrixes: abundances of all putative parasites, Perkinsozoa (all clades, clade 1, and clade 2), fungi, and Cercozoa.
P < 0.05.
A Fragilaria sp. that looks like Fragilaria acus.
In order to arrive at a detailed explanation of the quantitative variations of these targeted groups, we conducted a second RDA using a species matrix containing specifically and only the Perkinsozoa group as the response variable. Repeating the analysis by focusing on this group revealed that the abundances of Perkinsozoa (clades 1 and 2) were significantly correlated with the abundance of the Chrysophyceae species Dinobryon sociale and the dynamics of some diatom species. Focusing on the two Perkinsozoa clades separately revealed that the abundance of clade 1 was related mainly to the presence in Lake Bourget of two dinoflagellate species, i.e., Ceratium hirundinella (10% of the explained variance) and Peridinium inconspicuum (84.4% of the explained variance), while the abundance of Perkinsozoa clade 2 was correlated with the presence of Dinobryon sociale (99.5% of the explained variance).
The variations in the abundance of fungi were also significantly linked to the dynamics of certain phytoplanktonic groups. Fungi were mainly and significantly correlated with the diatom Melosira varans (21.1% of explained variance), and a strong link with NO3-N concentrations (64.4% of explained variance) was also detected.
Finally, an RDA run separately on the dynamics of Cercozoa as a response variable revealed a corelation between Cercozoa and the cyanobacterium Pseudanabaena galeata (83% of the explained variance).
DISCUSSION
The emergence of the microbial-loop concept in the 1980s resulted in the use of the classical functional groups of bacteria, HNF, and ciliates to describe interactions at the microbial level. In this context, HNF have been considered a homogeneous functional group with a well-established role in the transfer of carbon from bacterioplankton up to metazooplankton (10). However, within the microbial food web, the smallest pigmented or colorless eukaryotes (cell diameters, 1 to 5 μm) generally remained unidentified. Thus, the first results obtained by sequencing of the 18S rRNA gene (22, 34), which revealed the unexpected diversity of the smallest eukaryotes in lacustrine ecosystems, have made it clear that the classical functional groups (bacteria, HNF, and ciliates), defined primarily on the basis of a predator-prey relationship, are not sufficient to describe the various interactions and pathways within the microbial food web—including parasitism (21, 25). To understand these interactions, it was necessary to target and quantify the original group of organisms whose presence had been identified by the 18S rRNA gene but which had previously been ignored by nonmolecular methods.
This study reports specific in situ abundances of previously unquantified taxa within the small planktonic fraction (cell diameter, <5 μm). Newly designed oligonucleotide probes have allowed us to reveal a significant fraction of the large “black box” constituted by the smallest cells (diameter, <5 μm) considered incertae sedis taxa. Indeed, by comparing the results obtained by primuline staining of <5-μm-diameter flagellates and those obtained by the TSA-FISH probes targeting heterotrophic groups with diameters of <5 μm (Cercozoa, Perkinsozoa, and Fungi), we found that on average 96.8% of the previously undetermined flagellates were identified by TSA-FISH probes and thus could be affiliated with phylogenetic groups.
The set of specific probes developed and employed for this study was able to identify, on average, 85.5% of the eukaryotes targeted by the universal probe EUK1209R. These data thus confirm the results obtained by nonquantitative molecular methods (22, 23, 24, 25) on the recurrent presence of target groups in the <5-μm-cell-diameter planktonic fraction. On average, only 14.5% of the eukaryotes present in this size fraction could not be detected and counted using the selected oligonucleotide probes. On the basis of data previously acquired through cloning and sequencing analysis of this ecosystem (25), we assume that these unidentified microorganisms likely belong to the Choanoflagellida, Ciliophora, Bicosoecida, Hyphochytridiomycota, and Haptophyceae.
The EUK1209R probe was used in this study as a general eukaryotic probe. However, we know that some species or clades, especially those belonging to Chlorophyta, may not be efficiently targeted by this probe (31). This could explain in part the differences between “DAPI counts” and “EUK1209R counts” (on average, 87.2% of total small eukaryotes determined by DAPI staining were detected with the EUK1209R probe). Therefore, a mixture of probes, consisting of this universal EUK1209R probe combined with the CHLO01-CHLO02 and NChlo01 probes (specific to the division Chlorophyta and all non-Chlorophyta algae, respectively [37, 38]) would probably provide a suitable way to target the whole eukaryotic community. The differences observed between DAPI staining and EUK1209R probe staining could also be due to the presence of cells refractory to permeabilization (31) and to a low quantity of rRNA for some cells (1).
Reports of small-eukaryote quantification by FISH in lacustrine environments have been scarce (25). The abundances obtained by the EUK1209R probe are comparable to those determined in oceanic environments (2,600 to 17,000 cells ml−1 [31, 32]).
Chrysophyceae, Cryptophyceae, and Chlorophyceae.
Our analysis revealed that the <5-μm-cell-diameter fraction included eukaryotic groups whose presence in larger fractions (nano- and microplankton) has been well established and extensively quantified, such as Chrysophyceae, Cryptophyceae, and Chlorophyceae.
Chrysophyceae and Chlorophyceae appeared as predominant groups and were characterized by strong summer abundances. The Chrysophyceae have already been pinpointed as a major component of the nanoflagellates (cell diameter, 2 to 20 μm) by conventional screening techniques (nonspecific staining with DAPI or primuline). Boenigk et al. (4) reported that Spumella/Monas-like cells (Chrysophyceae) represented 20 to 50% of pelagic HNF biomass in freshwater. Chlorophyceae, the second most abundant group in this study, are generally considered a rarer component of this size fraction (2, 10, 15). The abundances recorded were higher than what would be expected from previously described clone libraries (23, 24, 25), where the proportion of operational taxonomic units and/or clones affiliated with Chlorophyceae was always low. Although 18S rRNA gene sequences are an interesting tool for describing the composition of small eukaryotes, the technique should underestimate some taxa, especially plastid forms (22, 35), as was recently suggested by Lepère et al. (25). The low levels of these organisms in the clone libraries may be explained by a difference in the number of 18S rRNA gene copies occurring in algae or by certain PCR biases (44).
According to the results obtained previously by cloning and sequencing, showing that Chrysophyceae are represented mainly by heterotrophic taxa (25), the autotrophs, mainly composed of Chlorophyceae and Cryptophyceae, might represent around 30% of total small eukaryotes in the lacustrine systems studied. Thus, the hypotheses put forward by Lefranc et al. (22) and Lepère et al. (24) concerning the predominance of heterotrophic organisms in the <5-μm-cell-diameter eukaryotic assemblage seem to be supported by the quantitative results reported here, since other groups described below are strict heterotrophs.
LKM11 group, Cercozoa, Fungi, and Perkinsozoa.
In contrast to the groups mentioned above, which have already been characterized in this small-size fraction, the presence of Cercozoa, Fungi, the LKM11 group, and Perkinsozoa, recently revealed by 18S rRNA sequences, has never been confirmed before.
In particular, our results highlighted the recurrent presence of a group that has received little attention so far, the LKM11 group. This group, which was relatively low in abundance (on average, 4.5% of total eukaryotes) compared to the other small eukaryotes targeted during the study period, is a noncultivated set of eukaryotic organisms first defined by van Hannen et al. (42). Although the functional role of these organisms has not yet been defined, they appear to be associated with the decomposition of phytoplanktonic organisms in oligotrophic and oligomesotrophic systems (22).
Cercozoa, representing on average 6.2% of the total eukaryote community, were permanently present in the water column. The Cercozoa include numerous taxa notoriously known to feed on bacterioplankton, picoplankton, or even nanoplankton (11). The dynamics of Cercozoa in this study was strongly related to the filamentous cyanobacterium Pseudanabaena. This co-occurrence is not necessarily linked to a predator-prey relationship. Indeed, a few taxa within the Cercozoa, although proportionally rarer, are considered parasites of phytoplankton. One example is the genus Cryothecomonas, which is capable of parasitizing diatoms (18). Moreover, Tillmann et al. (40) reported the role played by Cryothecomonas spp. in the mortality of certain phytoplankton populations.
Our analysis also revealed that the composition of the small eukaryotes included the presence, in quantitatively significant numbers, of groups recognized as parasites (Perkinsozoa, Fungi).
The fungi characterized in this ecosystem were essentially chytrids affiliated with the chytridial Rhizophydium and Nowakowskiella clades (25), a parasitic group known often to be host specific and highly infectious. These organisms are able to parasitize numerous algal species (diatoms, dinoflagellates, Chrysophyceae, Chlorophyceae) at high infection rates (17). This factor may explain the fact that variance in fungal abundance is largely related to diatom dynamics (24.9% of the cumulative variances from RDA [Table 3]), particularly to Melosira varians dynamics, and to a lesser extent to the dynamics of Chlorophyceae and Chrysophyceae.
The Perkinsozoa quantified presented profiles corresponding to small infectious cells called zoospores (6). These flagellate forms have almost certainly long figured among the heterotrophic flagellates that have remained undetermined in a large number of microbial food web studies and that are generally considered bacterivorous organisms (12, 19, 43). The seasonal dynamics highlighted for this parasitic group suggests that these Perkinsozoa may have a peak infectious impact in summer (July and August), when their proportion in relation to other eukaryotes is at its highest. Statistical analyses established a relationship between Perkinsozoa clade 1 dynamics and what were essentially phytoplanktonic taxa belonging to the dinoflagellates, more precisely to the Ceratium and Peridinium genera (94.4% of cumulative variances). The only relationship highlighted for Perkinsozoa clade 2 was with a phytoplankton taxon belonging to the Chrysophyceae, Dinobryon sociale var. americanum (99.5% of explained variance). These associations, together with the clade-specific dynamics profiles, suggest that the Perkinsozoa (if they prove to have a parasitic role) possess highly divergent host specificity depending on which of the two clades they belong to. This phylum, which is assumed to be entirely parasitic (30), has been studied mainly in marine environments. Only Brugerolle (6) has described the presence of an algal-parasitic organism belonging to Perkinsozoa in a freshwater environment (Cryptophagus) (but the18S rRNA gene has not been sequenced). In seawater environments, Perkinsus marinus is a parasite of bivalve mollusks, including Crassostrea corteziensis (7). The Perkinsozoa group also includes various protist parasites, notably Parvilucifera infectans, which is known to infect 26 different microalgal species, 17 of which belong to 10 different dinoflagellate genera (33). Our results linking the dynamics of Perkinsozoa clade 1 to the dynamics of the dinoflagellates Peridinium and Ceratium suggest that lacustrine Perkinsozoa may mirror marine Perkinsozoa in that they include a number of dinoflagellate parasites.
In conclusion, our results describe the quantitative variations in the main small-eukaryotic groups (cell diameter, <5 μm) in a lacustrine system. We were able to observe the seasonal variations in both the global abundance and the structure of the small-eukaryote assemblage. The recurrent presence of little-known phylogenetic groups such as the LKM11 group highlights the necessity to make further progress in elucidating the functional role of these groups. Moreover, this study of Lake Bourget clearly highlights the quantitative importance of putative parasitic organisms, such as Perkinsozoa, Fungi, or even some Cercozoa. This raises the question of their place and importance in lacustrine systems, particularly in terms of control over phytoplankton and zooplankton communities. Our results suggest some kind of host-parasite relationship, but further research is needed to connect the potential quantitative importance of parasites with their precise functional roles.
Acknowledgments
We thank G. Paolini and P. Perney for technical contributions to sampling and to analyses of physical and chemical parameters. The chemical parameter analyses and the microphytoplankton and metazooplankton counts were performed at the INRA's Thonon station (France). We especially acknowledge J. C. Druart and L. Lainé for the microphytoplankton and metazooplankton counts. We also thank David Bass (Oxford University) for contributing to this project through a gift of a cultivated Cercozoa strain used to validate the CERC_02 probe.
Footnotes
Published ahead of print on 7 August 2009.
REFERENCES
- 1.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arndt, H., D. Dietrich, B. Auer, E. J. Cleven, T. Grafenhan, M. Weitere, and A. P. Mylnikov. 2000. Functional diversity of heterotrophic flagellates in aquatic ecosystems, p. 240-268. In B. S. C. Leadbeater and J. C. Green (ed.), The flagellates. Taylor and Francis, London, United Kingdom.
- 3.Baschien, C. 2003. Development and evaluation of rDNA targeted in situ probes and phylogenetic relationships of freshwater fungi, p. 231. Ph.D. thesis. Technical University of Berlin, Berlin, Germany.
- 4.Boenigk, J. A., K. Pfland, P. Stadler, and A. Chatzinotas. 2005. High diversity of the “spumella-like” flagellates: an investigation based on the SSU rRNA gene sequence of isolates from habitats located in six different geographic regions. Environ. Microbiol. 7:685-697. [DOI] [PubMed] [Google Scholar]
- 5.Borcard, D., P. Legendre, and P. Drapeau. 1992. Partialling out the spatial component of ecological variation. Ecology 73:1045-1055. [Google Scholar]
- 6.Brugerolle, G. 2002. Cryptophagus subtilis: a new parasite of cryptophytes affiliated with the Perkinsozoa lineage. Eur. J. Protistol. 37:379-390. [Google Scholar]
- 7.Cáceres-Martínez, J., R. Vásquez-Yeomans, G. Padilla-Lardizábal, and M. A. del Río Portilla. 2008. Perkinsus marinus in pleasure oyster Crassostrea corteziensis from Nayarit, Pacific coast of México. J. Invertebr. Pathol. 99:66-73. [DOI] [PubMed] [Google Scholar]
- 8.Caron, D. A. 1983. Technique for enumeration of heterotrophic and phototrophic nanoplankton, using epifluorescence microscopy, and comparison with other procedures. Appl. Environ. Microbiol. 46:491-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caron, D. A., E. R. Peele, E. L. Lim, and M. R. Dennett. 1999. Picoplankton and nanoplankton and their trophic coupling in the surface waters of the Sargasso Sea south of Bermuda. Limnol. Oceanogr. 44:259-272. [Google Scholar]
- 10.Carrias, J. F., C. Amblard, and G. Bourdier. 1996. Protistan bacterivory in an oligomesotrophic lake: importance of attached ciliates and flagellates. Microb. Ecol. 31:249-268. [DOI] [PubMed] [Google Scholar]
- 11.Cavalier-Smith, T., and E. Y. Chao. 2003. Phylogeny and classification of phylum Cercozoa (Protozoa). Protist 154:341-358. [DOI] [PubMed] [Google Scholar]
- 12.Comte, J., S. Jacquet, S. Viboud, D. Fontvieille, G. Paolini, and I. Domaizon. 2006. Microbial community structure and dynamics in the largest natural French lake (Lake Bourget, Savoie, February to July 2002). Microb. Ecol. 52:72-89. [DOI] [PubMed] [Google Scholar]
- 13.Coss, C. A., J. A. Robledo, and G. R. Vasta. 2001. Fine structure of clonally propagated in vitro life stages of a Perkinsus sp. isolated from the Baltic clam Macoma balthica. J. Eukaryot. Microbiol. 487:38-51. [DOI] [PubMed] [Google Scholar]
- 14.Díez, B., C. Pedrós-Alió, T. L. Marsh, and R. Massana. 2001. Application of denaturing gradient gel electrophoresis (DGGE) to study the diversity of marine picoeukaryotic assemblages and comparison of DGGE with other molecular techniques. Appl. Environ. Microbiol. 67:2942-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domaizon, I., S. Viboud, and D. Fontvieille. 2003. Taxon-specific and seasonal variations in flagellates grazing on heterotrophic bacteria in the oligotrophic Lake Annecy—importance of mixotrophy. FEMS Microbiol. Ecol. 46:317-322. [DOI] [PubMed] [Google Scholar]
- 16.Giovannoni, S. J., E. F. DeLong, G. J. Olsen, and N. R. Pace. 1988. Phylogenetic group specific oligodeoxynucleotide probes for identification of single microbial cells. J. Bacteriol. 170:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ibelings, B. W., A. De Bruin, M. Kagami, M. Rijkeboer, M. Brehm, and E. Van Donk. 2004. Host parasite interactions between freshwater phytoplankton and chytrid fungi (Chytridiomycota). J. Phycol. 40:437-453. [Google Scholar]
- 18.Kühn, S., M. Lange, and L. K. Medlin. 2000. Phylogenetic position of Crythecomonas inferred from nuclear-encoded small subunit ribosomal RNA. Protist 151:337-345. [DOI] [PubMed] [Google Scholar]
- 19.Lee, W. J., and D. J. Patterson. 2002. Abundance and biomass of heterotrophic flagellates, and factors controlling their abundance and distribution in sediments of Botany Bay. Microb. Ecol. 43:467-481. [DOI] [PubMed] [Google Scholar]
- 20.Lefèvre, E., C. Bardot, C. Noël, J. F. Carrias, E. Viscogliosi, C. Amblard, and T. Simé-Ngando. 2007. Unveiling fungal zooflagellates as members of freshwater picoeukaryotes: evidence from a molecular diversity study in a deep meromictic lake. Environ. Microbiol. 9:61-71. [DOI] [PubMed] [Google Scholar]
- 21.Lefèvre, E., B. Roussel, C. Amblard, and T. Simé-Ngando. 2008. The molecular diversity of freshwater picoeukaryotes reveals high occurrence of putative parasitoids in the plankton. PLoS ONE 3:e2324. doi: 10.1371/journal.pone.0002324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lefranc, M., A. Thénot, C. Lepère, and D. Debroas. 2005. Genetic diversity of small eukaryotes in lakes differing by their trophic status. Appl. Environ. Microbiol. 71:5935-5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lepère, C., D. Boucher, L. Jardillier, I. Domaizon, and D. Debroas. 2006. Succession and regulation factors of small eukaryote community composition in a lacustrine ecosystem (Lake Pavin). Appl. Environ. Microbiol. 72:2971-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lepère, C., I. Domaizon, and D. Debroas. 2007. Community composition of lacustrine small eukaryotes in hypereutrophic conditions in relation to top-down and bottom-up factors. FEMS Microbiol. Ecol. 61:483-495. [DOI] [PubMed] [Google Scholar]
- 25.Lepère, C., I. Domaizon, and D. Debroas. 2008. Unexpected importance of potential parasites in the composition of the freshwater small-eukaryote community. Appl. Environ. Microbiol. 74:2940-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Massana, R., L. Guillou, B. Díez, and C. Pedros-Alio. 2002. Unveiling the organisms behind novel eukaryotic ribosomal DNA sequences from the ocean. Appl. Environ. Microbiol. 68:4554-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moon-van der Staay, S. Y., R. De Wachter, and D. Vaulot. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607-610. [DOI] [PubMed] [Google Scholar]
- 29.Moreira, D., and P. López-García. 2002. The molecular ecology of microbial eukaryotes unveils a hidden world. Trends Microbiol. 10:31-38. [DOI] [PubMed] [Google Scholar]
- 30.Moreira, D., and P. López-García. 2003. Are hydrothermal vents oases for parasitic protists? Trends Parasitol. 19:556-558. [DOI] [PubMed] [Google Scholar]
- 31.Not, F., N. Simon, I. C. Biegala, and D. Vaulot. 2002. Application of fluorescent in situ hybridization coupled with tyramide signal amplification (FISH-TSA) to assess eukaryotic picoplankton composition. Aquat. Microb. Ecol. 28:157-166. [Google Scholar]
- 32.Not, F., R. Massana, M. Latasa, D. Marie, C. Colson, W. Eikrem, C. Pedros-Alio, D. Vaulot, and N. Simon. 2005. Late summer community composition and abundance of photosynthetic picoeukaryotes in Norwegian and Barents Seas. Limnol. Oceanogr. 50:1677-1686. [Google Scholar]
- 33.Park, M. G., W. Yih, and D. W. Coats. 2004. Parasites and phytoplankton, with special emphasis on dinoflagellate infections. J. Eukaryot. Microbiol. 51:144-155. [DOI] [PubMed] [Google Scholar]
- 34.Richards, T. A., A. A. Vepritskiv, D. E. Gouliamova, and S. A. Nierzwicki-Bauer. 2005. The molecular diversity of freshwater picoeukaryotes from an oligotrophic lake reveals diverse, distinctive and globally dispersed lineages. Environ. Microbiol. 7:1413-1425. [DOI] [PubMed] [Google Scholar]
- 35.Savin, M. C., J. L. Martin, M. Legresley, M. Giewat, and J. Rooney-Varga. 2004. Plankton diversity in the Bay of Fundy as measured by morphological and molecular methods. Microb. Ecol. 48:51-65. [DOI] [PubMed] [Google Scholar]
- 36.Simé-Ngando, T., G. Bourdier, C. Amblard, and B. Pinel-Alloul. 1991. Short-term variations in specific biovolumes of different bacterial forms in aquatic ecosystems. Microb. Ecol. 21:211-226. [DOI] [PubMed] [Google Scholar]
- 37.Simon, N., N. Lebot, D. Marie, F. Partensky, and D. Vaulot. 1995. Fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes to identify small phytoplankton by flow cytometry. Appl. Environ. Microbiol. 61:2506-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon, N., L. Campbell, E. Ornolfsdottir, R. Groben, L. Guillou, M. Lange, and L. K. Medlin. 2000. Oligonucleotide probes for the identification of three algal groups by dot blot and fluorescent whole-cell hybridization. J. Eukaryot. Microbiol. 47:76-84. [DOI] [PubMed] [Google Scholar]
- 39.Šlapeta, J., D. Moreira, and P. López-García. 2005. The extent of protist diversity: insights from molecular ecology of freshwater eukaryotes. Proc. R. Soc. B 272:2073-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tillmann, U., K. J. Hesse, and A. Tillmann. 1999. Large-scale parasitic infection of diatoms in the Northfrisian Wadden Sea. J. Sea Res. 42:255-261. [Google Scholar]
- 41.Utermöhl, H. 1958. Zur Vervollkommnung der quantitativen Phytoplanktonmethodik, Mitt. Int. Verein. Theor. Angew. Limnol. 9:1-38. [Google Scholar]
- 42.van Hannen, E. J., W. Mooij, M. P. van Agterveld, H. J. Gons, and H. J. Laanbroek. 1999. Detritus-dependent development of the microbial community in an experimental system: qualitative analysis by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2478-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wieltschnig, C., A. K. T. Kirschner, A. Steitz, and B. Velimirov. 2001. Weak coupling between heterotrophic nanoflagellates and bacteria in a eutrophic freshwater environment. Microb. Ecol. 42:159-167. [DOI] [PubMed] [Google Scholar]
- 44.Zhu, F., R. Massana, F. Not, D. Marie, and D. Vaulot. 2005. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol. Ecol. 52:79-92. [DOI] [PubMed] [Google Scholar]