Abstract
In this study, 75 Shiga toxin (Stx)-producing Escherichia coli (STEC) strains originating from foods (n = 73) and drinking water (n = 2) were analyzed for their stx genotype, as well as for further chromosome-, phage-, and plasmid-encoded virulence factors. A broad spectrum of stx genes was detected. Fifty-three strains (70.7%) contained stx2 or stx2 variants, including stx2d, mucus-activatable stx2d, stx2e, and stx2g. Seven strains (9.3%) harbored stx1 or stx1c, and 15 strains (20.0%) carried both stx2 and/or stx2 variants and stx1 or stx1c. Beside stx, the most abundant accessory virulence markers in STEC food isolates were iha (57.3%), ehxA (40.0%), espP (28.0%), and subAB (25.3%). Only four strains were eae positive; three of these belonged to the serogroups O26, O103, and O157 and contained a typical enterohemorrhagic E. coli virulence spectrum. The results of this study show that a number of STEC strains that occur in foods appear to be pathogenic for humans, based on their virulence profiles. Analysis of stx subtypes and detection of additional virulence factors in eae-negative strains may help to better assess the risk of such strains for causing human infection.
Shiga toxin (Stx)-producing Escherichia coli (STEC) strains can cause a spectrum of human disease that range from watery diarrhea to bloody diarrhea (hemorrhagic colitis) and can be followed by serious sequelae, such as hemolytic-uremic syndrome (HUS) (48). STEC strains are genetically heterogeneous and, although more than 200 STEC serotypes have been described, only a limited number of serotypes has been isolated from human cases (33, 36). The most important serotypes which can cause severe human disease are O157:H7, O157:NM, and the non-O157 serotypes O26:H11, O111:NM, O103:H2, and O145:NM (12). STEC infections are mainly food-borne infections, although direct transmission from animals or from person to person has been described (58). Foods of high risk for transmission are minced meat, other meat products, produce, and dairy products (58). STEC strains are characterized by the production of one or more Stx proteins and have been isolated from a number of environmental sources (58). The Stx family consists of two major groups, Stx1 and Stx2, sharing ca. 60% sequence identity (51). The Stx1 group is more homogenous and consists of Stx1, Stx1c (72), and Stx1d (16). In contrast, the group comprised of Stx2 and related toxins is more heterogeneous. Beside Stx2, a number of variants, such as Stx2c (65), Stx2d (55), mucus-activatable Stx2d (Stx2dact) (46), Stx2e (70), Stx2f (63), and Stx2g (43), have been described. Unfortunately, the Stx nomenclature is not uniform, and despite the efforts to provide uniform nomenclature, description of new Stx variants is not sufficiently defined. Therefore, different designations for similar toxins have been published, and the methodologies for determining Stx variants also differ (6, 54, 68). The production of Stx2 and Stx2dact has been associated with high virulence, and strains producing these toxins have been isolated from patients with hemorrhagic colitis and HUS (8, 26). The activity of Stx2dact is enhanced after incubation with intestinal mucus (46). Stx2e occurs mainly in STEC isolated from pigs and foods and is only rarely detected in human feces (5, 70). Stx2f was described to occur in pigeons and has recently been described in human isolates (56). Stx2g was described originally in a bovine fecal sample but currently plays a minor role in human pathogenicity (43).
Besides the production of Stx, a number of accessory virulence factors that are probably involved in pathogenesis occur in STEC strains: one of the most prominent accessory pathomechanisms is the formation of attaching and effacing lesions (37). Enteropathogenic E. coli and enterohemorrhagic E. coli (EHEC) strains are able to cause destruction of the microvillous brush border and actin rearrangement beneath the site of bacterial attachment (25). A type III secretion system and a number of type III effector proteins are involved in this process (28, 39). They are encoded on a pathogenicity island termed locus of enterocyte effacement (LEE) (23). The genes eae and tir, encoding the bacterial outer-membrane protein intimin and the translocated intimin receptor Tir, respectively, are also located on the LEE (23). In addition to the seven LEE-encoded effectors, a number of non-LEE-encoded effectors have been described to occur in STEC strains in various alleles (69), examples of which are NleA (31) and Cif (44). Type III effector proteins exert manifold functions in eukaryotic cells (28), such as the inhibition of COPII function by NleA (41) or the inhibition of the cell cycle at the G2/M phase caused by Cif (44). Detection of eae is often used as a marker for the presence of the LEE (3, 10). Friedrich et al. (26) have shown that stx2-positive, eae-positive STEC strains are the most frequent isolates from patients with severe human disease. However, eae-negative STEC strains have been found to cause clinical symptoms (8, 26).
STEC virulence factors can also be encoded by large plasmids, such as pO157, pSFO157 (13, 17), and pO113 (49). Among others, pO157 of E. coli O157:H7 strains contains genes encoding the EHEC hemolysin (ehxA) (59) and the serine protease EspP (espP) (15). The EHEC hemolysin acts as a cytolysin by building pores into cell membranes and is excreted and stabilized in membrane bubbles (1, 60). EspP is able to cleave human coagulation factor V, and its mechanism of action has been elucidated (11, 15). The plasmid-encoded subtilase cytotoxin (SubAB) was first described for STEC O113:H21 strain 98NK2, which was involved in an outbreak of HUS in Australia (52). SubAB is an AB5 toxin, the A subunit being a subtilase-like serine protease and the B subunit binding to a surface receptor. The subtilase cytotoxin is involved in the generation of disease in animal experiments (52). Beside Stx and SubAB, a phage-encoded toxin, the cytolethal distending toxin V (CDT-V), occurs in E. coli O157:H7, O157:NM, and non-O157 strains (34). It causes an arrest in the G2/M phase in human brain microvascular endothelial cells and, moreover, death of endothelial cells (9).
In addition to the production of toxins and the attaching and effacing lesions, adherence factors may contribute to STEC pathogenesis. The irgA homologue adhesion (iha) gene encodes an adherence-associated protein and has been demonstrated to be encoded on a pathogenicity island integrated in the selC locus in many strains of different serotypes (64, 66). The EHEC factor for adherence (Efa1/Lif1) mediates adherence to bovine intestinal cells and is present in a number of O157 and non-O157 STEC strains (2, 35).
Virulence profiles of STEC strains have mostly been well evaluated for clinical or veterinary isolates, whereas only limited data are available about STEC strains from foods (6, 71, 73).
The aim of our study was to characterize the virulence profiles of current STEC food isolates and to determine their stx subtypes in order to assess the risk for human infection.
MATERIALS AND METHODS
Bacterial strains and culture media.
Thirty-four STEC strains were isolated from food samples as described below and were assigned designations using the prefix TS. Forty-one current STEC strains were obtained from other sources. Seven STEC strains were received from Rohtraud Pichner, Max Rubner Institute (MRI), Kulmbach, Germany, and originated from mixed minced meat (n = 5), beef (n = 1), and sausage (n = 1). Seven strains were obtained from beef (n = 2), raw milk (n = 2), raw sausages (n = 2), and feta cheese (n = 1) from the area of Stuttgart and were a kind gift of Matthias Contzen, Chemisches und Veterinär Untersuchungsamt (CVUA), Stuttgart, Germany. Twenty-two strains were kindly provided by Ulrich Busch, Bayerisches Landesamt für Gesundheit und Lebensmittelsicherheit (LGL), Oberschleisheim, Germany, and derived from game (n = 15), raw milk (n = 2), tea (n = 2), water (n = 2), and pork (n = 1) in the area of Munich. Five strains were received from Lothar Beutin, Bundesinstitut für Risikobewertung (BfR), Berlin, Germany, and were isolated from raw milk (n = 1), raw sausages (n = 1), minced beef (n = 1), mixed minced meat (n = 1), and pork (n = 1) (6).
Detection and isolation of stx-positive E. coli strains from food samples.
Five hundred and four food samples were obtained in the years 2007 and 2008 from local retailers and supermarkets in a 50-km area surrounding Stuttgart, Germany. They originated from minced meat (n = 245), raw sausages (n = 157), and raw milk (n = 102). Samples were purchased, transported in sterile plastic bags, stored at 4°C overnight, and processed the next day. Raw-milk samples were obtained from a local dairy and a milk collection plant. The latter samples were preselected using only those with high aerobic plate counts of more than 105 CFU ml−1. Such samples indicate poor hygienic conditions and should enhance the probability of finding STEC strains.
Twenty-five-gram amounts of the meat products or 25-ml amounts of the raw-milk samples were mixed with 225 ml of modified Trypticase soy broth (mTSB) and novobiocin in a stomacher bag and mixed for 2 min at 230 rpm. The mTSB medium contains 17 g liter−1 peptone from casein, 3 g liter−1 peptone from soybean flour, 2.5 g liter−1 glucose, 1.5 g liter−1 no. 3 bile salts (Becton Dickinson), 5.0 g liter−1 NaCl, and 4.0 g liter−1 K2HPO4. After the medium was autoclaved, sterile filtrated novobiocin (Sigma) was added to a final concentration of 20 mg liter−1. The mixture was removed from the bag, transferred into a 250-ml Erlenmeyer flask, and incubated at 37°C at 100 rpm for 6 h. As controls, 25-g amounts of minced meat were contaminated either with E. coli O157:H7 strain EDL933 or E. coli laboratory strain C600 to final concentrations of 1 to 5 CFU g−1. Ten-milliliter amounts of each enrichment culture were taken and centrifuged at 3,275 × g for 10 min at 4°C. The supernatant was discarded, and the pellet was resuspended in 1.5 ml of mTSB. One-hundred-fifty-microliter amounts of this suspension were transferred onto sorbitol-MacConkey agar (SMAC; Sifin) and enterohemolysin agar (Oxoid) plates with single-colony streaking. After incubation at 37°C for 18 to 24 h, half of the cell material from both plates was rubbed off of the plates with a sterile swab and resuspended in 1 ml of a 0.9% NaCl solution. An aliquot of the bacterial suspension was diluted 1:10 in sterile ultrapure water (Milli-Q plus PF; Millipore) and incubated at 100°C for 5 min. An aliquot of 2.5 μl of this suspension was mixed with 0.5 μl of primer pairs KS7 and KS8 for detection of stx1 and LP43 and LP44 for detection of stx2 and variants, respectively (Table 1), 2.5 μl Taq polymerase buffer, 0.5 μl deoxynucleoside triphosphate mix, and 0.2 μl Taq DNA polymerase (Genaxxon) and adjusted with sterile pure water to a total volume of 25 μl. The PCR protocols are described in Table 1.
TABLE 1.
Target genes, primer designations, DNA sequences, PCR conditions, and PCR product sizes used for detection of stx genes and other STEC virulence markers
| Target gene | Primer | Nucleotide sequence (5′-3′) | PCR conditions | PCR product size (bp) | Reference |
|---|---|---|---|---|---|
| stxB1 | KS7 | CCCGGATCCATGAAAAAAACATTATTAATAGC | 94°C, 30 s; 52°C, 60 s; 72°C, 60 s | 285 | 62 |
| KS8 | CCCGAATTCAGCTATTCTGAGTCAACG | ||||
| stxA2 | LP43 | ATCCTATTCCCGGGAGTTTACG | 94°C, 30 s; 57°C, 60 s; 72°C, 90 s | 584 | 19 |
| LP44 | GCGTCATCGTATACACAGGAGC | ||||
| eaea | SK1 | CCCGAATTCGGCACAAGCATAAGC | 94°C, 30 s; 52°C, 60 s; 72°C, 60 s | 863 | 61 |
| SK2 | CCCGGATCCGTCTCGCCAGTATTCG | ||||
| ehxA | HlyA1 | GGTGCAGCAGAAAAAGTTGTAG | 90°C, 30 s; 57°C, 60 s; 72°C, 90 s | 1,550 | 59 |
| HlyA4 | TCTCGCCTGATAGTGTTTGGTA | ||||
| espP | espP-A | AAACAGCAGGCACTTGAACG | 94°C, 30 s; 56°C, 60 s; 72°C, 150 s | 1,830 | 14 |
| espP-B | GGAGTCGTCAGTCAGTAGAT | ||||
| nleA | V83-rev2 | CTTCCATCGCACGTATATCAGC | 94°C, 30 s; 58°C, 60 s; 72°C, 90 s | 959-1,112 | 22 |
| V83-for2 | ACAGCAACATGCACCGGAATGC | 94°C, 30 s; 55°C, 60 s; 72°C 90s | 1,015-1,168 | ||
| V83-rev3 | GATATCGATGACCACATCTTCAGG | ||||
| subAB | RTsubABF | GCAGATAAATACCCTTCACTTG | 94°C, 30 s; 56°C, 30 s; 72°C, 42 s | 232 | 52 |
| RTsubABR | ATCACCAGTCCACTCAGCC | ||||
| cif | cif-int-s | AACAGATGGCAACAGACTGG | 94°C, 30 s; 57°C, 60 s; 72°C, 30 s | 383 | 44 |
| cif-int-as | AGTCAATGCTTTATGCGTCAT | ||||
| cdt-V | cdtV-F | TTCATTGTTCGCCTCCTG | 94°C, 60 s; 50°C, 60 s; 72°C, 60 s | 755 | 20 |
| cdtV-R | TTTATAAGCTGGTATCCTG | ||||
| efa1 | 88AT | AAGGTGTTACAGAGATTA | 94°C, 60 s; 51°C, 60 s; 72°C, 60 s | 266 | 50 |
| 88TN | TGAGGCGGCAGGATAGTT | ||||
| iha | iha-I | CAGTTCAGTTTCGCATTCACC | 94°C, 30 s; 56°C, 60 s; 72°C, 90 s | 1,305 | 64 |
| iha-II | GTATGGCTCTGATGCGATG |
Primers SK1 and SK1 are universal eae primers and detect all eae variants.
In the case of PCR-positive samples, the original bacterial suspension was diluted with 0.9% NaCl solution to obtain approximately 200 to 300 single colonies per plate after spreading on SMAC agar. In order to detect stx-positive isolates, up to 300 single colonies were analyzed for the presence of stx genes. For this purpose, pools of five colonies were subjected to PCR either with primers KS7 and KS8 or with primers LP43 and LP44. In parallel, all selected colonies were stored on agar plates. PCR-positive pools were used to identify the stx-positive strains by analyzing the single colonies with the same procedure. The species were determined with an Enterotube II ID test kit (Becton Dickinson) according to the manufacturer's instructions. The STEC strains isolated were stored in glycerin stocks (50% [vol/vol]) at −70°C.
Molecular characterization of stx variants.
The stx genes of food-borne STEC strains used in this study were subtyped according to a scheme published previously (6). In this scheme, stx2 and stx2 variants (stx2-OX392, stx2NV-206, stx2EC-1586, stx2v-ha, stx2v-hb, and stx2d) were characterized following amplification with primer pair GK5 and GK6 and subsequent restriction analysis. The stx1, stx1c, stx1d, stx2-O118, stx2e, stx2f, and stx2g alleles were identified by other PCR approaches using primers targeting the respective B subunit genes (6).
GK5/GK5-positive strains were used for further PCR with primers CKS2 and SLT-II-vc and subsequent restriction analysis to detect stx sequences with the same restriction fragment length polymorphism (RFLP) as mucus-activatable stx2d. Beutin et al. ( 6 ) renamed stx2dact, described by Zheng et al. (73), as stx2d, and the stx2d gene, which was originally described by Pierard et al. (55), as stx2-O118. With the RFLP analysis mentioned above, stx2 subfragments were restricted to define mucus-activatable genes according to the protocol of Zheng et al. (73). The whole procedure and the toxin nomenclature are described elsewhere in detail (6).
In order to characterize some stx genes in more detail, we sequenced 28 stx2 genes obtained from STEC strains isolated in our laboratory. Basically, 5 to 10 μl of the PCR product were purified using the Exo-SAP method as described previously (3). The Exo-SAP reaction mixture contained equal units of exonuclease I (Fermentas) and shrimp alkaline phosphatase (Fermentas). Twenty-five to 50 fmol of bacterial DNA were used for the setup of the sequencing reaction. The stx2 genes were amplified and initially sequenced with primer pairs stx2seq9 (5′-GCAATCGGTCACTGGTTCG-3) and stx2seq10 (5′-CAGATTACACTTGTTACCCAC-3′) or stx2seq5 (5′-CACTT ATTTTCCCTGGCTCG-3′) and stx2seq6 (5′-CACTGACCAAATTTAATCGAC-3′). The use of a second primer pair was necessary because the stx2-flanking regions differed in the strains.
Starting from the resulting sequence, the whole genes were analyzed with walking primers. The treated PCR products were sequenced with a CEQ 8000 genetic analysis system (Beckman Coulter). Precipitation of DNA with ethanol and sequence analysis were conducted according to the instructions of the manufacturer (GenomeLab dye terminator cycle sequencing with quick start kit; Beckman Coulter). The program for amplification consisted of 30 cycles of 96°C for 20 s, 50°C for 20 s, and 60°C for 360 s. In general, the lengths read were between 400 bp and 650 bp. The single fragments were assembled using BioEdit sequence alignment software (32). Using the Blast algorithm, the sequences were compared with those deposited at NCBI (http://www.ncbi.nlm.nih.gov/blast). For practical reasons, the sequences revealed were cut, and the resulting sequences included the start codon of the A subunit gene up to the stop codon of the B subunit gene. Phylogenetic analyses were performed with MEGA software version 3.1, using the unweighted pair-group method with arithmetic mean (42).
Detection of STEC virulence genes and serotyping.
All STEC strains were analyzed by PCR for the presence of the virulence factor-encoding genes eae (intimin), ehxA (EHEC hemolysin), espP (serine protease), nleA and cif (type III effectors), and iha and efa1 (adhesins), as well as subAB and cdt-V (cytotoxins). Furthermore, the isolated strains were retested for the presence of stx1 and stx2 for comparison with the original results from the food samples.
The primers and conditions employed in the PCR assays for identification of these genes are listed in Table 1. The following E. coli strains were used as positive controls: E. coli O157:H7 strain EDL933 (stx1 stx2 eae ehxA espP iha efa1 nleA), O84:H4 strain 4795/97 (nleA4795) (21), O128:H2 strain CB8260 (subAB) (4), and rabbit EPEC O103:H2 strain E22 (cif) (18), as well as E. coli O157:H7 strains 5791/99 and 3010/96 (cdt-V) (27). As negative control for all genes, we used E. coli K-12 strain C600. The PCR was performed as described above with one fresh colony suspended in 50 μl of a 0.9% NaCl solution.
Serotyping was performed with antisera against E. coli O antigens 1 to 181 and E. coli H antigens 1 to 56, using a microtiter method described elsewhere (57).
Nucleotide sequence accession numbers.
The nucleotide sequences obtained by sequencing of the PCR products of each stx2 allele have been entered into the GenBank and EMBL databases under continuous accession numbers from FM998838 to FM998861 and FN182284 to FN182287.
RESULTS
In order to characterize the virulence profile of current food-borne STEC isolates and to assess their risks for human infection, we determined the stx variants, the presence of particular virulence genes, and the serotypes for 75 STEC strains. Thirty-four strains from 67 stx-positive food samples, the origins of which are listed in Table 2, were isolated by our laboratory. Interestingly, three strains could be isolated from two minced-meat samples, and in a further three samples, two strains were found. Forty-one current food-borne STEC strains were obtained from other sources (Table 2). Comparison of the virulence profiles of these isolates with those described in the literature should help to better evaluate the virulence of food-borne STEC isolates.
TABLE 2.
Results of the molecular analysis of food-borne STEC isolatesi
| Strain | Serotype | Origin |
stx type(s) as determined by:
|
eae | ehxA | nleA | subAB | cdt-V | cif | espP | iha | efa1 | Sourceh | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCRa | Sequencingb | |||||||||||||
| TS07/08 | O2:NM | Mettwurst | 2e | 2e | − | − | − | − | − | − | − | − | − | A |
| CB11637 | O2:H37 | Raw milk | 2d | ND | − | − | − | − | − | − | − | − | − | B |
| RF1a | O8:H6 | Beef | 2e | ND | − | − | − | − | − | − | − | − | − | C |
| TS04/08 | O8:H8 | Mettwurst | 2e | 2e | − | − | − | − | − | − | − | − | − | A |
| LM14822/08 | O8:H19 | Wild boar | 2e | ND | − | − | − | − | − | − | − | − | − | D |
| CB11571 | O8:H19 | Minced meat (mixed) | 2e | ND | − | − | − | − | − | − | − | − | − | B |
| TS13/08 | O8:H19 | Minced meat (pork) | 2e | 2e | − | − | − | − | − | − | − | − | − | A |
| TS16/08 | O8:NM | Minced meat (beef) | 2e | 2e | − | − | − | − | − | − | − | − | − | A |
| TS10/07 | O8:NM | Mettwurst | 2e | 2e | − | − | − | − | − | − | − | − | − | A |
| E918 | O8:HNT | Minced meat (mixed) | 2e | ND | − | − | − | − | − | − | − | − | − | C |
| E921 | O8:HNT | Minced meat (mixed) | 2e | ND | − | − | − | − | − | − | − | − | − | C |
| E922 | O54:NM | Minced meat (mixed) | 2e | ND | − | − | − | − | − | − | − | − | − | C |
| TS01/07 | O100:NM | Minced meat (mixed) | 2e | 2e | − | − | − | − | − | − | − | − | − | A |
| TS14/08 | O153:NM | Minced meat (pork) | 2 (NT) | ND | − | − | − | − | − | − | − | − | − | A |
| TS15/08 | ONT:H4 | Minced meat (pork) | 2 (NT) | ND | − | − | − | − | − | − | − | − | − | A |
| TS09/07 | OR:H19 | Mettwurst | 2e | 2e | − | − | − | − | − | − | − | − | − | A |
| LM22343 | ONT:NM | Wild boar | 2e | ND | − | − | − | − | − | − | − | − | − | D |
| LM22344 | ONT:NM | Wild boar | 2e | ND | − | − | − | − | − | − | − | − | − | D |
| TS03/08 | ONT:NM | Minced meat (pork) | 2e | 2e | − | − | − | − | − | − | − | − | − | A |
| WA (WT) 5555/08 | ONT:NM | Drinking water | 2g, 2EC-1586e | ND | − | − | − | − | − | − | − | − | − | D |
| LM5604/08 | ONT:NM | Lime blossom tea | 2e | ND | − | − | − | − | − | − | − | − | − | D |
| TS29/08 | ONT:NM | Minced meat (mixed) | 2e | 2e | − | − | − | − | − | − | − | − | − | A |
| 03353/1 | O22:H8 | Beef | 2d | ND | − | − | − | − | − | − | − | + | − | E |
| TS04/07 | O113:H4 | Minced meat (beef) | 2NV-206c | 2dact | − | − | − | − | − | − | − | + | − | A |
| TS21/08 | O113:H21 | Minced meat (mixed) | 2EC-1586c | 2dact | − | − | − | − | − | − | − | + | − | A |
| TS23/08 | O113:H21 | Minced meat (beef) | 2EC-1586c | 2dact | − | − | − | − | − | − | − | + | − | A |
| CB11597 | O113:H21 | Minced meat (beef) | 2v-hac | ND | − | − | − | − | − | − | − | + | − | B |
| TS06/07 | O116:NM | Minced meat (mixed) | 2gc | 2g | − | − | − | − | − | − | − | + | − | A |
| 18692/1 | O148:H8 | Minced meat (beef) | 2v-hac | ND | − | − | − | − | − | − | − | + | − | E |
| TS05/07 | OR:H29 | Minced meat (mixed) | 2-O118, 2c | 2g | − | − | − | − | − | − | − | + | − | A |
| TS06/08 | OR:H29 | Minced meat (mixed) | 2-O118, 2v-hbc | 2dact | − | − | − | − | − | − | − | + | − | A |
| TS09/08 | ONT:H4 | Minced meat (mixed) | 2v-hbc | 2dact | − | − | − | − | − | − | − | + | − | A |
| E 917 | O91:NM | Sausage meat | 1 | ND | − | − | − | − | − | − | − | − | − | C |
| 13477/1 | O136:H16 | Raw milk | 1 | ND | − | − | − | − | − | − | − | − | − | E |
| 28504/1 | O136:HNT | Feta cheese | 1c | ND | − | − | − | − | − | − | − | − | − | E |
| TS25/08 | ONT:HNT | Minced meat (mixed) | 1 | ND | − | − | − | − | − | − | − | − | − | A |
| 29087/1 | O8:H19 | Raw milk | 1, 2v-hbd | ND | − | + | − | − | − | − | + | + | − | E |
| TS07/07 | O130:H11 | Minced meat (beef) | 1, 2c | 2g | − | + | − | − | − | − | + | + | − | A |
| E927 | O178:H19 | Minced meat (mixed) | 1, 2NV-206, 2EC-1586e | ND | − | + | − | − | − | − | + | + | − | C |
| TS22/08 | O178:H19 | Minced meat (beef) | 1, 2d | 2g | − | + | − | − | − | − | + | + | − | A |
| TS02/07 | O178:H19 | Minced meat (beef) | 1, 2d | 2g | − | + | − | − | − | − | + | + | − | A |
| K17f | O22:H8 | Raw milk | 2, 2EC-1586, 2NV-206e | ND | − | + | − | + | − | − | + | + | − | D |
| LM5602/08 | O22:H8 | Lime blossom tea | 2v-hac | ND | − | + | − | + | − | − | + | + | − | D |
| CB11588 | O102:NM | Pork | 2v-hbc | ND | − | + | − | + | − | − | + | + | − | B |
| CB11633 | O179:H8 | Mettwurst | 2d | ND | − | + | − | + | − | − | + | + | − | B |
| LM14603/08 | O21:H21 | Deer | 2-O118 | ND | − | − | − | + | − | − | − | + | − | D |
| LM16092/08 | O21:H21 | Deer | 2-O118 | ND | − | − | − | + | − | − | − | + | − | D |
| LM27553 Stx1 | O75:H8 | Deer | 1c, 2-O118 | ND | . | − | − | + | − | − | − | + | − | D |
| TS20/08 | O153:HNT | Minced meat (mixed) | 1, 2e | 2g | − | + | − | + | − | − | + | + | − | A |
| TS26/08 | O179:H8 | Minced meat (mixed) | 1, 2d | 2g | − | + | − | + | − | − | + | + | − | A |
| SF16b | ONT:H11 | Minced meat (mixed) | 1, 2d | ND | − | + | − | + | − | − | + | + | − | C |
| LM27553 Stx2 | O110:H31 | Deer | 2-O118 | ND | − | + | − | + | − | − | − | + | − | D |
| LM 27564 | O113:NM | Deer | 2-O118 | ND | − | + | − | + | − | − | − | + | − | D |
| LM27558 Stx2 | OR:H43 | Deer | 2-O118 | ND | − | + | − | + | − | − | − | + | − | D |
| LM 27555 | OR:NM | Deer | 2-O118 | ND | − | + | − | + | − | − | − | + | − | D |
| 13762/1 | O22:H8 | Mettwurst | 2c | ND | − | + | − | − | − | − | + | + | − | E |
| TS27/08 | OR:NM | Minced meat (beef) | 2v-hbc | 2g | − | − | − | − | − | − | + | + | − | A |
| 17584/1f | O91:H21 | Mettwurst | 1, 2, 2NV-206, 2EC-1586e | ND | − | + | − | − | − | − | − | + | − | E |
| TS28/08 | O113:NM | Minced meat (beef) | 1, 2EC-1586c | 2dact | − | − | − | − | − | − | − | + | − | A |
| TS03/07 | O113:H21 | Minced meat (beef) | 1, 2v-hac | 2dact | − | − | − | − | − | − | + | − | − | A |
| K30 | O55:HNT | Raw milk | 1 | ND | − | − | − | − | − | − | + | + | − | D |
| TS01/08 | O128:NM | Raw milk | 1 | ND | − | − | − | − | − | − | + | + | − | A |
| LM14960/08 | O23:NM | Deer | 2-O118 | ND | − | − | − | + | − | − | − | − | − | D |
| LM14957/08 | O26:H11 | Wild boar | 2d | ND | + | + | + | − | − | + | − | + | + | D |
| LM15814/08 | O103:H2 | Deer | 1 | ND | + | + | − | − | − | + | − | − | + | D |
| TS17/08 | O113:H21 | Minced meat (pork) | 2c | 2g | − | + | − | − | + | − | + | + | − | A |
| TS18/08 | O113:H21 | Minced meat (mixed) | 2c | 2g | − | + | − | + | + | − | − | + | − | A |
| TS19/08 | O113:H21 | Minced meat (mixed) | 2c | 2g | − | + | − | − | + | − | − | + | − | A |
| LM27558 Stx1 | O128:HNT | Deer | 1, 2-O118 | ND | − | + | − | + | − | − | − | + | − | D |
| LM 3034/08 | O146:H21 | Meat | 1c, 2-O118 | ND | − | + | − | + | − | − | − | − | − | D |
| WA (WT) 4913/08 | O156:NM | Drinking water | 2d | ND | + | + | + | − | − | − | + | − | − | D |
| TS10/08 | O157:H7 | Minced meat mixed | 2 (lost) | ND | + | + | + | − | − | − | + | + | + | A |
| TS24/08 | O178:H19 | Minced meat (beef) | 1, 2d | 2g | − | + | − | − | − | − | − | + | − | A |
| LM22346 | ONT:H28 | Deer | 2-O118 | ND | − | + | − | − | − | − | − | − | − | D |
| TS30/08 | O113:H21 | Minced meat (mixed) | 2c | ND | − | + | − | + | + | − | + | + | − | A |
Results obtained by PCR and RFLP analysis. NT, stx2 could not be subtyped; lost, the stx gene was lost upon subcultivation.
Results obtained by sequencing and database analysis. ND, not determined.
No PstI restriction site is present; mucus activatable according to RFLP analysis.
PstI restriction site is present; not activatable.
PstI RFLP analysis revealed genes both with and without the PstI site in one sample.
RFLP analysis cannot clearly differentiate whether two or three stx2 genes are present. stx2 variants are not completely differentiated by RFLP.
stx2 variant that is highly similar to classical stx2.
A, this study; B, BfR; C, MRI; D, LGL; E, CVUA.
+, present; −, absent.
Characterization and phylogenetic analysis of stx genes.
The stx genes of 72 of the 75 STEC isolates were subtyped by PCR with stx type-specific primers and RFLP analysis as described above. Twenty strains were positive only after amplification with primer pair GK5/GK6, indicating the presence of stx2 and/or particular stx2 variants (see above). Eighteen strains carried the stx2e gene only. Most of the latter strains were derived from minced meat (pork and mixed), mettwurst, and wild boar. Eight strains harbored the stx2-O118 allele, and 12 strains carried stx1 in combination with stx2 and/or stx2 variants. Six strains carried only the stx1 gene. The remaining stx2 alleles occurred rarely. A summary of all stx subtypes and stx combinations detected by PCR is shown in Table 3.
TABLE 3.
Frequencies of stx types and stx combinations in STEC strains analyzed in this study
| stx gene(s)a | No. (%) of strains with indicated gene(s)b |
|---|---|
| stx1 | 6 (8.0) |
| stx1c | 1 (1.4) |
| stx1, stx2, and/or stx2d | 12 (16.7) |
| stx1c, stx2-O118 | 2 (2.8) |
| stx1, stx2-O118 | 1 (1.4) |
| stx2 and/or stx2d | 20 (27.8) |
| stx2 and/or stx2d, stx2-O118 | 2 (2.8) |
| stx2 and/or stx2d, stx2g | 2 (2.8) |
| stx2-O118 | 8 (11.1) |
| stx2e | 18 (25.0) |
“stx2 and/or stx2d” indicates the results of PCR with primers GK5/GK6.
Results for only 72 of 75 strains are included in this table, because TS10/08 has lost its stx gene, and the stx genes of TS14/08 and TS15/08 could not be subtyped. The percentages are rounded.
All GK5/GK6-positive strains were subjected to further analysis to identify particular stx2 subtypes. The results of this stx analysis are shown in Table 4 (left column). In total, 36 GK5/GK6-positive strains were found. Subsequent RFLP analysis of the respective PCR products indicated that 18 strains carry stx2. The remaining 18 strains carried stx2 variants as single toxin genes or in combination (Table 4).
TABLE 4.
Distribution of stx2, stx2 subtypes, and activatable stx2d in 35 food-borne STEC strains
| stx2 and/or stx2d subunit genotype (no. of strains with genotype) | No. of strains with indicated gene(s)a
|
||
|---|---|---|---|
| stx2 | stx2d | stx2 and stx2d | |
| stx2 (18) | 10 | 7 | 1 |
| stx2v-ha (4) | 4 | ||
| stx2v-hb (5) | 1 | 4 | |
| stx2NV-206 (1) | 1 | ||
| stx2EC-1586 (3) | 3 | ||
| stx2g (1) | 1 | ||
| stx2g, stx2EC-1586 (1) | 1 | ||
| stx2EC-1586, stx2NV-206 (1) | 1 | ||
| stx2, stx2EC-1586, stx2NV-206b (2) | 2 | ||
stx2d is used as a synonym for stx2dact.
RFLP analysis cannot clearly differentiate whether two or three stx2 genes are present.
In addition, the 36 strains were used for further PCR with primers CKS2 and SLT-II-vc and subsequent restriction analysis to detect stx sequences with the same RFLP as stx2d (synonym for stx2dact) (Table 4, right columns). With the latter RFLP analysis, stx2 subfragments were restricted in order to define mucus-activatable sequences, according to the protocol of Zheng et al. (73). As an example, from the 18 stx2 genes investigated, 7 appear to be mucus activatable according to the RFLP analysis. Interestingly, stx2g also belongs to the group of genes encoding mucus-activatable toxins (Table 4). All in all, 25 stx2 genes were found that did not contain a PstI restriction site and were therefore considered to be mucus activatable (Table 2).
We decided to analyze stx2 in more detail and to confirm the results obtained by RFLP analysis. Therefore, we determined the nucleotide sequences of 28 stx2 genes from strains which were isolated in our laboratory. The lengths of the sequences varied from 1,236 bp up to 1,241 bp. Due to database comparison, we were able to assign those sequences to already known stx variants (Fig. 1, box 2). Nine of the strains carried stx2e, 11 strains carried a stx2 variant that is closely related to stx2, 7 strains carried the stx2dact sequence, and strain TS06/07 harbored stx2g. We translated the sequences into the putative amino acid sequences of the A and B subunits and performed a phylogenetic analysis (Fig. 1). We used the following sequences as references: stx2 (accession no. X07865), stx2c (accession no. M59432), stx2dact (accession no. AF479828), stx2d (synonym, stx2-O118) (accession no. AF043627), stx2e (accession no. AJ249351), stx2g (accession no. AJ966783), and stx2f (accession no. AJ010730). Due to sequence similarities, we organized the analyzed strains in four sequence groups, encoding Stx2e, Stx2dact, Stx2g, and a group of Stx2 variants (Fig. 1). Slight differences have been found in the analysis of A and B subunit genes. Whereas Stx2 and Stx2c cannot be easily distinguished by comparison of the sequences of the A subunit (Fig. 1A, box 1), they are distinguishable by analysis of the B subunit (Fig. 1B, box 1 and box 2). Moreover, Stx2c and Stx2dact are indistinguishable by B subunit analysis (Fig. 1B, box 2) but can be differentiated by analysis of the sequences of the A subunit (Fig. 1A, boxes 1 and 2).
FIG. 1.
Phylogenetic dendrogram of the deduced amino acid sequences of the A subunit (panel A) and the B subunit (panel B) of stx2 genes, based on the unweighted pair-group method with arithmetic mean algorithm using Mega 3.1 software (42). The shaded boxes show the alignment of the stx2 sequences to reference sequences (see Discussion).
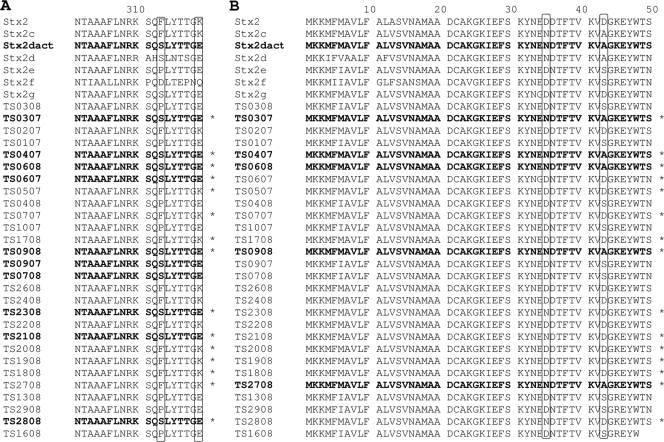
The 28 sequences were also analyzed for amino acid substitutions described to be typical for mucus-activatable toxins as published by Melton-Celsa et al. (47) and Zheng et al. (73). With regard to classical Stx2, Stx2dact carries the two amino acid substitutions S313F and E319K in the A subunit and N35D and A43D in the B subunit (Fig. 2).
FIG. 2.
Alignment of the deduced amino acid sequences of the C-terminal ends of the A subunits (A) and B subunits (B) of 28 Stx2 sequences analyzed in this study. The amino acid positions of signature amino acids which are indicative for activation by mucus are labeled with boxes. Asterisks show stx2 sequences which are activatable according to the RFLP analysis. Amino acid sequences in bold font show the typical activatable signature as analyzed by sequencing.
We found 10 A subunits carrying the amino acid substitutions for Stx2dact and 5 B subunits (Fig. 2) with the respective amino acid substitutions. Only the Stx sequences from strains TS03/07, TS04/07, TS06/08, and TS09/08 carried these substitutions in both subunits and are therefore considered to be activatable (Stx2dact). These results were confirmed by RFLP analysis (Table 2). In addition, 11 further stx genes were activatable according to RFLP analysis (Table 4). These strains are labeled with asterisks in Fig. 2. Two strains carried the Stx2dact-specific amino acid substitutions in the A subunit but appeared not to be activatable according to the RFLP scheme, and seven strains that were activatable according to the RFLP analysis did not contain the respective amino acid substitutions (Fig. 2). In eight cases, both procedures gave the same results for the A subunit. Ten strains carried Stx2dact in the B subunit according to the RFLP pattern but not the respective amino acid substitutions. Concordance of the results of both procedures was revealed for the B subunit of five strains (Fig. 2). Using these procedures, not only Stx2dact but also Stx2g (TS06/07) appears to be activatable; Stx2g of TS06/07 carries the S313F and E319K substitution in the A subunit and is activatable according to the RFLP analysis. The Stx2e-positive strains TS09/07 and TS07/08 also carried the signature amino acids for Stx2dact in the A subunit but were not mucus activatable according to the RFLP analysis (Fig. 2).
Analysis of STEC serotypes and virulence markers.
In 57 of the 75 STEC strains, the O antigens could be determined. Eleven strains were O nontypeable, and six STEC strains were O rough. The H antigen was determined in 46 strains, 22 were nonmotile, and 7 strains were H nontypeable. In 38 strains, the O-H types could be completely determined, and 20 different complete O-H serotypes could be described (Table 2). The most frequent O groups were O8 (n = 10), O113 (n = 8), O22 (n = 4), and O178 (n = 4). We detected one STEC strain each of the typical EHEC serotypes O26:H11, O103:H2, and O157:H7 (Table 2). The E. coli O26:H11 and E. coli O103:H2 strains showed the sorbitol-positive phenotype, whereas E. coli O157:H7 was sorbitol negative (data not shown).
In order to assess the virulence profiles of the isolated STEC strains, PCR investigations of virulence markers were conducted (Table 2). Interestingly, 22 strains carried only stx2 variants as virulence markers and none of the other virulence genes tested. The iha gene could be detected in 43 (57.3%) strains and was the most frequent virulence gene present. The gene for the adherence factor efa1 was present only in the three STEC strains of serogroups O26, O103, and O157 (Table 2). The ehxA gene was found in 30 (40.0%) strains, and espP in 21 (28.0%) strains. Seventeen of the espP-positive strains were also positive for ehxA. Only four strains carried eae, three of which were nleA positive and two of which were cif positive (Table 2). One of these strains harbored both genes, nleA and cif. Analysis of the toxin genes cdt-V and subAB demonstrated that 19 (25.3%) strains were subAB positive, and only 4 O113:H21 strains carried cdt-V, with 2 of these strains carrying both toxin genes. In general, the EHEC O26, O103, and O157 strains carried most of the virulence markers (Table 2).
Food samples contained two or three different STEC isolates.
Two or three different isolates were found in five minced-meat samples from the current study, as described above. Table 5 summarizes the molecular characteristics of these strains. Strains TS05/07 and TS06/07 of food sample A had different serotypes and showed almost identical virulence profiles; they differed only in their stx variants (Table 5). In food sample B, the strains had different serotypes but identical virulence profiles. In food sample C, strains TS18/08, TS19/08, and TS30/08 had the same serotype but differed in the presence of subAB and espP. Strains TS20/08 and TS21/08 of food sample D differed in serotype, stx variant, and accessory virulence markers (Table 5). In food sample E, E. coli O178:H19 strains TS22/08 and TS24/08 were espP positive and espP negative, respectively. The third strain found in sample E differed in both serotype and virulence profile.
TABLE 5.
Molecular characteristics of food-borne STEC strains originating from the same samplesb
| Sample | Strain | Serotype | Origin | stx type(s) | eae | hlyA | nleA | subAB | cdt-V | cif | espP | iha | efa1 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | TS05/07 | OR:H29 | Minced meat (mixed) | 2, 2-O118 | − | − | − | − | − | − | − | + | − |
| TS06/07 | O116:NM | Minced meat (mixed) | 2g | − | − | − | − | − | − | − | + | − | |
| B | TS14/08 | O153:NM | Minced meat (pork) | 2a | − | − | − | − | − | − | − | − | − |
| TS15/08 | ONT:H4 | Minced meat (pork) | 2a | − | − | − | − | − | − | − | − | − | |
| C | TS18/08 | O113:H21 | Minced meat (mixed) | 2 | − | + | − | + | + | − | − | + | − |
| TS19/08 | O113:H21 | Minced meat (mixed) | 2 | − | + | − | − | + | − | − | + | − | |
| TS30/08 | O113:H21 | Minced meat (mixed) | 2 | − | + | − | + | + | − | + | + | − | |
| D | TS20/08 | O153:HNT | Minced meat (mixed) | 1, 2 | − | + | − | + | − | − | + | + | − |
| TS21/08 | O113:H21 | Minced meat (mixed) | 2EC-1586 | − | − | − | − | − | − | − | + | − | |
| E | TS24/08 | O178:H19 | Minced meat (beef) | 1, 2 | − | + | − | − | − | − | − | + | − |
| TS22/08 | O178:H19 | Minced meat (beef) | 1, 2 | − | + | − | − | − | − | + | + | − | |
| TS23/08 | O113:H21 | Minced meat (beef) | 2EC-1586 | − | − | − | − | − | − | − | + | − |
stx2 gene was not subtypeable.
+, present; −, absent.
DISCUSSION
Seventy-five STEC strains originating from foods and water were characterized with molecular methods, and a number of interesting issues arose from the results of this study.
STEC strains TS10/08 (O157:H7), LM15814/08 (O103:H2), and LM14597/08 (O26:H11) can be considered highly pathogenic due to their serotypes and virulence gene composition (Table 2). Moreover, we characterized isolates with serotypes such as O8:H19, O22:H8, O91:H21, and O113:H21. These serotypes have also been associated with severe human disease (33). An example of this is the serotype O113:H21. Paton et al. (53) described an eae-negative O113:H21 isolate from an outbreak of HUS which harbored stx2, ehxA, and subAB. This is comparable to the eae-negative STEC O113:H21 strain TS18/08, which also carries stx2, ehxA, and subAB. In addition, this strain harbors cdt-V and may be considered a putative pathogen for humans.
The O156:NM strain WA (WT) 4913/08 is also noteworthy, because it contains stx2, eae, ehxA, nleA, and espP. It probably harbors the pO157 virulence plasmid. STEC O156 strains have been reported to cause illness, but to our knowledge, they were not isolated from HUS patients (33). STEC O156 strains have been isolated from ruminants, especially from sheep. In our study, strain WA (WT) 4913/08 was found in drinking water.
Strain TS10/08 lost its stx gene upon subcultivation. The loss of stx genes has been described in a number of publications, but the mechanism remains unclear. Karch et al. (38) reported the loss of stx genes upon subcultivation in 15 out of 45 cases. The STEC strains belonged to serotypes O2:H5, O26:H11, O73:H34, and O100:H32. In another study, an E. coli O157:H7 strain was isolated that displayed two morphotypes on agar plates. One of these morphotypes had lost its stx gene (24). Gain and loss of stx2 and Stx2 phages have been described for E. coli O157:H− strains (45). Such strains can pose a problem because they may be missed during diagnostic procedures that target only stx genes. Strains that have lost their stx genes, however, retain their full spectrum of accessory virulence genes and are able to regain infectious Stx phages (45).
A large group of strains contain stx2e but none of the other virulence factors investigated (Tables 2 and 3). Since stx2e is only rarely detected in isolates from patients with severe disease, these strains may be considered to be less pathogenic, as mentioned by Beutin et al. (5). The analysis of the stx2e-carrying strains in our study confirmed these findings. However, a deeper view into the genetic structure is necessary to establish whether these strains can be considered harmless. In this context, integration sites of phages and pathogenicity islands should be investigated to screen for accessory virulence determinants. Moreover, uptake of phages and other mobile genetic elements can occur and may convert a harmless strain into a pathogen. This should be kept in mind when assessing the risk for human infection of stx2e-positive strains.
The results obtained by using different methods for determining genes for mucus-activatable toxins are somewhat conflicting. Although in a number of cases RFLP and sequencing results are in concordance (see above), there are a number of cases where the results of these analytical methods do not agree. In the current study, a number of stx2 genes were sequenced that were highly similar to classical stx2 but did not contain the PstI restriction site, due to single-base-pair exchanges. Since the absence of the PstI restriction site is the only criterion that defines a gene for a mucus-activatable toxin (6, 30), Stx2 could probably be falsely identified as activatable. Inconsistent data from RFLP, DNA sequence, and functional investigations were already described by Tasara et al. (67). Therefore, the genotypic methods described should be used primarily for subtyping of strains rather than for determination of mucus activation of stx2d. To determine this, functional analyses, such as elastase tests, should be performed.
The subAB gene has been detected in a number of STEC serogroups, examples of which are O26, O91, O111, O113, O128, and O157 (52). In another study, subAB was found in 48.4% of strains isolated from human infections, cattle, and ground meat (20). The subAB gene fragment was detected mainly in eae-negative strains, which contained more than one stx gene copy. A further study conducted with STEC strains from the United States determined the percentage of subAB strains to be 21.7% (40). The subAB genes frequently occur together with stx2, stx2c, and ehxA.
In our study, we detected the subAB fragment in 19 strains (25.3%), which were all eae negative (Table 2). These strains frequently contained stx2 and ehxA. Remarkably, most of our deer meat isolates contained subAB. STEC O113:H21 strain TS18/08 contained both subAB and cdt-V. Furthermore, we detected four cdt-V-positive strains in minced meat which were eae negative and of serotype O113:H21. In another study, 340 STEC strains of non-O157 serotypes from patients were screened for cdt alleles; 4 of these were of serotype O113:H21 (7). These findings pose the question of whether such food isolates are potentially pathogenic. Further molecular investigations for the comparison of isolates from food and patients, such as fingerprinting, multilocus sequence analysis, etc., are necessary to substantiate this hypothesis.
Another interesting feature was the detection of five food samples which harbored more than one STEC isolate. STEC strains TS18/08, TS19/08, and TS30/08, as well as TS24/08 and TS22/08, had the same serotype but differed in only one virulence plasmid marker. It is conceivable that those strains are phylogenetically closely related and had lost or gained a single gene. Isolate pairs TS05/07 and TS06/07, TS14/08 and TS15/08, and TS20/08 and TS21/08, as well as strains TS24/08, TS22/08, and TS23/08, had different serotypes and, frequently, different virulence marker combinations, and are therefore considered different strains. The presence of two or three different STEC strains in a given food sample may possibly cause double or triple infections. In a study about the role of STEC in HUS patients, some of the patients had an antibody response against the O157 lipopolysaccharide and, in addition, a STEC strain with a different serotype was isolated from the respective stool sample (29). At this point, it is not clear whether all strains in double or triple infections contribute to the development of disease; further investigations are needed to better define the role of particular strains in multiple infections.
The results of this study show that food, especially from animal sources, may contain potentially pathogenic STEC strains. The characterization of stx variants and detection of additional virulence factors in eae-negative STEC strains may help to better assess the capability of those strains to cause human infections.
Acknowledgments
We thank Grit Fogarassy, Markus Kranz, and Stefanie Selent for skillful technical assistance. Lothar Wieler, Berlin, and Helge Karch, Münster, were always open to helpful discussions.
This work was supported by grant 01Kl 07128 (Food-Borne Zoonotic Infections of Humans) from the German Federal Ministry of Education and Research (BMBF).
Footnotes
Published ahead of print on 14 August 2009.
REFERENCES
- 1.Aldick, T., M. Bielaszewska, B. E. Uhlin, H. U. Humpf, S. N. Wai, and H. Karch. 2009. Vesicular stabilization and activity augmentation of enterohaemorrhagic Escherichia coli haemolysin. Mol. Microbiol. 71:1496-1508. [DOI] [PubMed] [Google Scholar]
- 2.Badea, L., S. Doughty, L. Nicholls, J. Sloan, R. M. Robins-Browne, and E. L. Hartland. 2003. Contribution of Efa1/LifA to the adherence of enteropathogenic Escherichia coli to epithelial cells. Microb. Pathog. 34:205-215. [DOI] [PubMed] [Google Scholar]
- 3.Beutin, L., S. Kaulfuss, S. Herold, E. Oswald, and H. Schmidt. 2005. Genetic analysis of enteropathogenic and enterohemorrhagic Escherichia coli serogroup O103 strains by molecular typing of virulence and housekeeping genes and pulsed-field gel electrophoresis. J. Clin. Microbiol. 43:1552-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beutin, L., G. Krause, S. Zimmermann, S. Kaulfuss, and K. Gleier. 2004. Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. J. Clin. Microbiol. 42:1099-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beutin, L., U. Kruger, G. Krause, A. Miko, A. Martin, and E. Strauch. 2008. Evaluation of major types of Shiga toxin 2E-producing Escherichia coli bacteria present in food, pigs, and the environment as potential pathogens for humans. Appl. Environ. Microbiol. 74:4806-4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beutin, L., A. Miko, G. Krause, K. Pries, S. Haby, K. Steege, and N. Albrecht. 2007. Identification of human-pathogenic strains of Shiga toxin-producing Escherichia coli from food by a combination of serotyping and molecular typing of Shiga toxin genes. Appl. Environ. Microbiol. 73:4769-4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bielaszewska, M., M. Fell, L. Greune, R. Prager, A. Fruth, H. Tschäpe, M. A. Schmidt, and H. Karch. 2004. Characterization of cytolethal distending toxin genes and expression in shiga toxin-producing Escherichia coli strains of non-O157 serogroups. Infect. Immun. 72:1812-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bielaszewska, M., A. W. Friedrich, T. Aldick, R. Schurk-Bulgrin, and H. Karch. 2006. Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin. Infect. Dis. 43:1160-1167. [DOI] [PubMed] [Google Scholar]
- 9.Bielaszewska, M., B. Sinha, T. Kuczius, and H. Karch. 2005. Cytolethal distending toxin from Shiga toxin-producing Escherichia coli O157 causes irreversible G2/M arrest, inhibition of proliferation, and death of human endothelial cells. Infect. Immun. 73:552-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bielaszewska, M., W. Zhang, P. I. Tarr, A. K. Sonntag, and H. Karch. 2005. Molecular profiling and phenotype analysis of Escherichia coli O26:H11 and O26:NM: secular and geographic consistency of enterohemorrhagic and enteropathogenic isolates. J. Clin. Microbiol. 43:4225-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brockmeyer, J., M. Bielaszewska, A. Fruth, M. L. Bonn, A. Mellmann, H. U. Humpf, and H. Karch. 2007. Subtypes of the plasmid-encoded serine protease EspP in Shiga toxin-producing Escherichia coli: distribution, secretion, and proteolytic activity. Appl. Environ. Microbiol. 73:6351-6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brooks, J. T., E. G. Sowers, J. G. Wells, K. D. Greene, P. M. Griffin, R. M. Hoekstra, and N. A. Strockbine. 2005. Non-O157 Shiga toxin-producing Escherichia coli infections in the United States, 1983-2002. J. Infect. Dis. 192:1422-1429. [DOI] [PubMed] [Google Scholar]
- 13.Brunder, W., H. Karch, and H. Schmidt. 2006. Complete sequence of the large virulence plasmid pSFO157 of the sorbitol-fermenting enterohemorrhagic Escherichia coli O157:H- strain 3072/96. Int. J. Med. Microbiol. 296:467-474. [DOI] [PubMed] [Google Scholar]
- 14.Brunder, W., H. Schmidt, M. Frosch, and H. Karch. 1999. The large plasmids of Shiga-toxin-producing Escherichia coli (STEC) are highly variable genetic elements. Microbiology 145(Pt. 5):1005-1014. [DOI] [PubMed] [Google Scholar]
- 15.Brunder, W., H. Schmidt, and H. Karch. 1997. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol. Microbiol. 24:767-778. [DOI] [PubMed] [Google Scholar]
- 16.Burk, C., R. Dietrich, G. Acar, M. Moravek, M. Bulte, and E. Märtlbauer. 2003. Identification and characterization of a new variant of Shiga toxin 1 in Escherichia coli ONT:H19 of bovine origin. J. Clin. Microbiol. 41:2106-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burland, V., Y. Shao, N. T. Perna, G. Plunkett, H. J. Sofia, and F. R. Blattner. 1998. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 26:4196-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camguilhem, R., and A. Milon. 1989. Biotypes and O serogroups of Escherichia coli involved in intestinal infections of weaned rabbits: clues to diagnosis of pathogenic strains. J. Clin. Microbiol. 27:743-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cebula, T. A., W. L. Payne, and P. Feng. 1995. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J. Clin. Microbiol. 33:248-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cergole-Novella, M. C., L. S. Nishimura, L. F. Dos Santos, K. Irino, T. M. Vaz, A. M. Bergamini, and B. E. Guth. 2007. Distribution of virulence profiles related to new toxins and putative adhesins in Shiga toxin-producing Escherichia coli isolated from diverse sources in Brazil. FEMS Microbiol. Lett. 274:329-334. [DOI] [PubMed] [Google Scholar]
- 21.Creuzburg, K., J. Recktenwald, V. Kuhle, S. Herold, M. Hensel, and H. Schmidt. 2005. The Shiga toxin 1-converting bacteriophage BP-4795 encodes an NleA-like type III effector protein. J. Bacteriol. 187:8494-8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Creuzburg, K., and H. Schmidt. 2007. Molecular characterization and distribution of genes encoding members of the type III effector nleA family among pathogenic Escherichia coli strains. J. Clin. Microbiol. 45:2498-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 24.Feng, P., M. Dey, A. Abe, and T. Takeda. 2001. Isogenic strain of Escherichia coli O157:H7 that has lost both Shiga toxin 1 and 2 genes. Clin. Diagn. Lab. Immunol. 8:711-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 26.Friedrich, A. W., M. Bielaszewska, W. L. Zhang, M. Pulz, T. Kuczius, A. Ammon, and H. Karch. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74-84. [DOI] [PubMed] [Google Scholar]
- 27.Friedrich, A. W., S. Lu, M. Bielaszewska, R. Prager, P. Bruns, J. G. Xu, H. Tschäpe, and H. Karch. 2006. Cytolethal distending toxin in Escherichia coli O157:H7: spectrum of conservation, structure, and endothelial toxicity. J. Clin. Microbiol. 44:1844-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garmendia, J., G. Frankel, and V. F. Crepin. 2005. Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect. Immun. 73:2573-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerber, A., H. Karch, F. Allerberger, H. M. Verweyen, and L. B. Zimmerhackl. 2002. Clinical course and the role of shiga toxin-producing Escherichia coli infection in the hemolytic-uremic syndrome in pediatric patients, 1997-2000, in Germany and Austria: a prospective study. J. Infect. Dis. 186:493-500. [DOI] [PubMed] [Google Scholar]
- 30.Gobius, K. S., G. M. Higgs, and P. M. Desmarchelier. 2003. Presence of activatable Shiga toxin genotype (stx2d) in Shiga toxigenic Escherichia coli from livestock sources. J. Clin. Microbiol. 41:3777-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gruenheid, S., I. Sekirov, N. A. Thomas, W. Deng, P. O'Donnell, D. Goode, Y. Li, E. A. Frey, N. F. Brown, P. Metalnikov, T. Pawson, K. Ashman, and B. B. Finlay. 2004. Identification and characterization of NleA, a non-LEE-encoded type III translocated virulence factor of enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 51:1233-1249. [DOI] [PubMed] [Google Scholar]
- 32.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 33.Hussein, H. S. 2007. Prevalence and pathogenicity of Shiga toxin-producing Escherichia coli in beef cattle and their products. J. Anim. Sci. 85:E63-E72. [DOI] [PubMed] [Google Scholar]
- 34.Janka, A., M. Bielaszewska, U. Dobrindt, L. Greune, M. A. Schmidt, and H. Karch. 2003. Cytolethal distending toxin gene cluster in enterohemorrhagic Escherichia coli O157:H− and O157:H7: characterization and evolutionary considerations. Infect. Immun. 71:3634-3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janka, A., M. Bielaszewska, U. Dobrindt, and H. Karch. 2002. Identification and distribution of the enterohemorrhagic Escherichia coli factor for adherence (efa1) gene in sorbitol-fermenting Escherichia coli O157:H−. Int. J. Med. Microbiol. 292:207-214. [DOI] [PubMed] [Google Scholar]
- 36.Johnson, K. E., C. M. Thorpe, and C. L. Sears. 2006. The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli. Clin. Infect. Dis. 43:1587-1595. [DOI] [PubMed] [Google Scholar]
- 37.Kaper, J. B. 1998. The locus of enterocyte effacement pathogenicity island of Shiga toxin-producing Escherichia coli O157:H7 and other attaching and effacing E. coli. Jpn. J. Med. Sci. Biol. 51(Suppl.):S101-S107. [DOI] [PubMed] [Google Scholar]
- 38.Karch, H., T. Meyer, H. Rüssmann, and J. Heesemann. 1992. Frequent loss of Shiga-like toxin genes in clinical isolates of Escherichia coli upon subcultivation. Infect. Immun. 60:3464-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kenny, B. 2002. Mechanism of action of EPEC type III effector molecules. Int. J. Med. Microbiol. 291:469-477. [DOI] [PubMed] [Google Scholar]
- 40.Khaitan, A., D. M. Jandhyala, C. M. Thorpe, J. M. Ritchie, and A. W. Paton. 2007. The operon encoding SubAB, a novel cytotoxin, is present in Shiga toxin-producing Escherichia coli isolates from the United States. J. Clin. Microbiol. 45:1374-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim, J., A. Thanabalasuriar, T. Chaworth-Musters, J. C. Fromme, E. A. Frey, P. I. Lario, P. Metalnikov, K. Rizg, N. A. Thomas, S. F. Lee, E. L. Hartland, P. R. Hardwidge, T. Pawson, N. C. Strynadka, B. B. Finlay, R. Schekman, and S. Gruenheid. 2007. The bacterial virulence factor NleA inhibits cellular protein secretion by disrupting mammalian COPII function. Cell Host Microbe 2:160-171. [DOI] [PubMed] [Google Scholar]
- 42.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 43.Leung, P. H., J. S. Peiris, W. W. Ng, R. M. Robins-Browne, K. A. Bettelheim, and W. C. Yam. 2003. A newly discovered verotoxin variant, VT2g, produced by bovine verocytotoxigenic Escherichia coli. Appl. Environ. Microbiol. 69:7549-7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marches, O., T. N. Ledger, M. Boury, M. Ohara, X. Tu, F. Goffaux, J. Mainil, I. Rosenshine, M. Sugai, R. J. De, and E. Oswald. 2003. Enteropathogenic and enterohaemorrhagic Escherichia coli deliver a novel effector called Cif, which blocks cell cycle G2/M transition. Mol. Microbiol. 50:1553-1567. [DOI] [PubMed] [Google Scholar]
- 45.Mellmann, A., S. Lu, H. Karch, J. G. Xu, D. Harmsen, M. A. Schmidt, and M. Bielaszewska. 2008. Recycling of Shiga toxin 2 genes in sorbitol-fermenting enterohemorrhagic Escherichia coli O157:NM. Appl. Environ. Microbiol. 74:67-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melton-Celsa, A. R., S. C. Darnell, and A. D. O'Brien. 1996. Activation of Shiga-like toxins by mouse and human intestinal mucus correlates with virulence of enterohemorrhagic Escherichia coli O91:H21 isolates in orally infected, streptomycin-treated mice. Infect. Immun. 64:1569-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Melton-Celsa, A. R., J. F. Kokai-Kun, and A. D. O'Brien. 2002. Activation of Shiga toxin type 2d (Stx2d) by elastase involves cleavage of the C-terminal two amino acids of the A2 peptide in the context of the appropriate B pentamer. Mol. Microbiol. 43:207-215. [DOI] [PubMed] [Google Scholar]
- 48.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newton, H. J., J. Sloan, D. M. Bulach, T. Seemann, C. C. Allison, M. Tauschek, R. M. Robins-Browne, J. C. Paton, T. S. Whittam, A. W. Paton, and E. L. Hartland. 2009. Shiga toxin-producing Escherichia coli strains negative for locus of enterocyte effacement. Emerg. Infect. Dis. 15:372-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicholls, L., T. H. Grant, and R. M. Robins-Browne. 2000. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol. Microbiol. 35:275-288. [DOI] [PubMed] [Google Scholar]
- 51.O'Brien, A. D., V. L. Tesh, A. Donohue-Rolfe, M. P. Jackson, S. Olsnes, K. Sandvig, A. A. Lindberg, and G. T. Keusch. 1992. Shiga toxin: biochemistry, genetics, mode of action, and role in pathogenesis. Curr. Top. Microbiol. Immunol. 180:65-94. [DOI] [PubMed] [Google Scholar]
- 52.Paton, A. W., P. Srimanote, U. M. Talbot, H. Wang, and J. C. Paton. 2004. A new family of potent AB(5) cytotoxins produced by Shiga toxigenic Escherichia coli. J. Exp. Med. 200:35-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paton, A. W., M. C. Woodrow, R. M. Doyle, J. A. Lanser, and J. C. Paton. 1999. Molecular characterization of a Shiga toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J. Clin. Microbiol. 37:3357-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Persson, S., K. E. Olsen, S. Ethelberg, and F. Scheutz. 2007. Subtyping method for Escherichia coli Shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J. Clin. Microbiol. 45:2020-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pierard, D., G. Muyldermans, L. Moriau, D. Stevens, and S. Lauwers. 1998. Identification of new verocytotoxin type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J. Clin. Microbiol. 36:3317-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prager, R., A. Fruth, U. Siewert, U. Strutz, and H. Tschäpe. 2009. Escherichia coli encoding Shiga toxin 2f as an emerging human pathogen. Int. J. Med. Microbiol. 299:343-353. [DOI] [PubMed] [Google Scholar]
- 57.Prager, R., U. Strutz, A. Fruth, and H. Tschäpe. 2003. Subtyping of pathogenic Escherichia coli strains using flagellar (H)-antigens: serotyping versus fliC polymorphisms. Int. J. Med. Microbiol. 292:477-486. [DOI] [PubMed] [Google Scholar]
- 58.Rangel, J. M., P. H. Sparling, C. Crowe, P. M. Griffin, and D. L. Swerdlow. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg. Infect. Dis. 11:603-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmidt, H., L. Beutin, and H. Karch. 1995. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect. Immun. 63:1055-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmidt, H., E. Maier, H. Karch, and R. Benz. 1996. Pore-forming properties of the plasmid-encoded hemolysin of enterohemorrhagic Escherichia coli O157:H7. Eur. J. Biochem. 241:594-601. [DOI] [PubMed] [Google Scholar]
- 61.Schmidt, H., B. Plaschke, S. Franke, H. Rüssmann, A. Schwarzkopf, J. Heesemann, and H. Karch. 1994. Differentiation in virulence patterns of Escherichia coli possessing eae genes. Med. Microbiol. Immunol. 183:23-31. [DOI] [PubMed] [Google Scholar]
- 62.Schmidt, H., H. Rüssmann, A. Schwarzkopf, S. Aleksic, J. Heesemann, and H. Karch. 1994. Prevalence of attaching and effacing Escherichia coli in stool samples from patients and controls. Zentralbl. Bakteriol. 281:201-213. [DOI] [PubMed] [Google Scholar]
- 63.Schmidt, H., J. Scheef, S. Morabito, A. Caprioli, L. H. Wieler, and H. Karch. 2000. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Appl. Environ. Microbiol. 66:1205-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidt, H., W. L. Zhang, U. Hemmrich, S. Jelacic, W. Brunder, P. I. Tarr, U. Dobrindt, J. Hacker, and H. Karch. 2001. Identification and characterization of a novel genomic island integrated at selC in locus of enterocyte effacement-negative, Shiga toxin-producing Escherichia coli. Infect. Immun. 69:6863-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schmitt, C. K., M. L. McKee, and A. D. O'Brien. 1991. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H− strain E32511. Infect. Immun. 59:1065-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tasara, T., M. Bielaszewska, S. Nitzsche, H. Karch, C. Zweifel, and R. Stephan. 2008. Activatable Shiga toxin 2d (Stx2d) in STEC strains isolated from cattle and sheep at slaughter. Vet. Microbiol. 131:199-204. [DOI] [PubMed] [Google Scholar]
- 68.Teel, L. D., A. R. Melton-Celsa, C. K. Schmitt, and A. D. O'Brien. 2002. One of two copies of the gene for the activatable Shiga toxin type 2d in Escherichia coli O91:H21 strain B2F1 is associated with an inducible bacteriophage. Infect. Immun. 70:4282-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tobe, T., S. A. Beatson, H. Taniguchi, H. Abe, C. M. Bailey, A. Fivian, R. Younis, S. Matthews, O. Marches, G. Frankel, T. Hayashi, and M. J. Pallen. 2006. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc. Natl. Acad. Sci. USA 103:14941-14946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weinstein, D. L., M. P. Jackson, J. E. Samuel, R. K. Holmes, and A. D. O'Brien. 1988. Cloning and sequencing of a Shiga-like toxin type II variant from Escherichia coli strain responsible for edema disease of swine. J. Bacteriol. 170:4223-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Werber, D., L. Beutin, R. Pichner, K. Stark, and A. Fruth. 2008. Shiga toxin-producing Escherichia coli serogroups in food and patients, Germany. Emerg. Infect. Dis. 14:1803-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, W., M. Bielaszewska, T. Kuczius, and H. Karch. 2002. Identification, characterization, and distribution of a Shiga toxin 1 gene variant (stx1c) in Escherichia coli strains isolated from humans. J. Clin. Microbiol. 40:1441-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng, J., S. Cui, L. D. Teel, S. Zhao, R. Singh, A. D. O'Brien, and J. Meng. 2008. Identification and characterization of Shiga toxin type 2 variants in Escherichia coli isolates from animals, food, and humans. Appl. Environ. Microbiol. 74:5645-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]