Abstract
A wide variety of commercial products can be potentially made from monomeric sugars produced by the dilute acid hydrolysis of lignocellulosic biomass. However, this process is accompanied by side products such as furfural that hinder microbial growth and fermentation. To investigate the mechanism of furfural inhibition, mRNA microarrays of an ethanologenic strain of Escherichia coli (LY180) were compared immediately prior to and 15 min after a moderate furfural challenge. Expression of genes and regulators associated with the biosynthesis of cysteine and methionine was increased by furfural, consistent with a limitation of these critical metabolites. This was in contrast to a general stringent response and decreased expression of many other biosynthetic genes. Of the 20 amino acids individually tested as supplements (100 μM each), cysteine and methionine were the most effective in increasing furfural tolerance with serine (precursor of cysteine), histidine, and arginine of lesser benefit. Supplementation with other reduced sulfur sources such as d-cysteine and thiosulfate also increased furfural tolerance. In contrast, supplementation with taurine, a sulfur source that requires 3 molecules of NADPH for sulfur assimilation, was of no benefit. Furfural tolerance was also increased by inserting a plasmid encoding pntAB, a cytoplasmic NADH/NADPH transhydrogenase. Based on these results, a model is proposed for the inhibition of growth in which the reduction of furfural by YqhD, an enzyme with a low Km for NADPH, depletes NADPH sufficiently to limit the assimilation of sulfur into amino acids (cysteine and methionine) by CysIJ (sulfite reductase).
Lignocellulose contains up to 70% carbohydrate by weight (35 to 45% cellulose and 20 to 35% hemicellulose) and represents an excellent potential source of sugars for microbial conversion into renewable fuels, plastics, and other chemicals (9, 13, 15, 38). Prior to fermentation, these carbohydrate polymers must be converted to soluble sugars. Hemicellulose can be conveniently hydrolyzed to sugar monomers using dilute mineral acids. However, this process is accompanied by side products that inhibit microbial growth (20, 21, 29, 30, 44-46). Furfural, the dehydration product of pentose sugars, is one of the most important such inhibitors (1). Previous studies have shown that furfural levels directly correlate with toxicity (20, 21). Overliming treatments that render hemicellulose hydrolysates fermentable also reduce the levels of furfural. Full toxicity in overlimed hydrolysates is restored by the addition of furfural. Of the many components of hydrolysates that have been tested for toxicity, only furfural was found to potentiate the toxicity of other agents in binary combinations (45).
A number of approaches have been used to investigate the mechanism of furfural action. Furfural and 5-hydroxymethyl furfural (dehydration product from hexose sugars) have been previously proposed to inhibit growth by damaging DNA (3, 16, 36), inhibiting glycolysis and glycolytic enzymes (5, 11, 25), and chemically reacting with cellular constituents (10, 30). An Escherichia coli enzyme has been partially purified and characterized that catalyzes the NADPH-dependent reduction of furfural to the less toxic alcohol furfuryl alcohol (7, 45, 46). Global transcript analysis was used to identify ADH6 in Saccharomyces cerevisiae as an important enzyme for the reduction of 5-hydroxymethyl furfural (dehydration production of hexose sugars) to a less toxic product, 5-hydroxymethyl furfuryl alcohol (32).
We previously described the isolation of a furfural-tolerant ethanologenic E. coli strain, EMFR9 (23). This mutant strain was isolated by selecting for improved growth in the presence of furfural. Surprisingly, increased furfural tolerance in EMFR9 was accompanied by a decrease in the rate of furfural reduction in vivo and a decrease in the NADPH-dependent furfural reductase activity in vivo. Increased furfural tolerance was found to result in part from the silencing of two genes encoding NADPH-dependent oxidoreductase activity, yqhD and dkgA. Silencing of these two genes was proposed to permit increased growth in the presence of furfural by slowing depletion of the NADPH that is required for biosynthesis. In this study, we have used global transcript analysis to expand our investigations to include the broader cellular response to added furfural in the parent organism, E. coli LY180.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Ethanologenic strains (Table 1) were maintained in AM1 mineral salts medium (22) supplemented with 20 g liter−1 xylose for solid medium and 50 g liter−1 xylose or higher for fermentation experiments. Strain E. coli LY180 (23, 43) is a derivative of KO11 and served as the starting point for this investigation. Note that E. coli W (ATCC 9637) is the parent of strain KO11, initially reported to be a derivative of E. coli B (28).
TABLE 1.
Bacterial strains, plasmids, and primers
| Strain, plasmid, or primer | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| LY180 | ΔfrdBC::(frgZmcelYEc) ΔldhA::(frgZmcasABKo) adhE::(frgZmestZPpFRT) ΔackA::FRT rrlE::(pdc adhA adhB FRT) ΔmgsA::FRT | 23 |
| EMFR9 | LY180 improved for furfural tolerance | 23 |
| TOP10F′ | F′ [lacIq Tn10 (Tetr)] mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen (Carlsbad, CA) |
| Plasmids | ||
| pTrc99a | Ptrc bla oriR rrnB lacIq | 2 |
| pLOI4315 | sthA gene in pTrc99a | This study |
| pLOI4316 | pntAB gene in pTrc99a | This study |
| Primers | ||
| pntAB cloning | Forward: CTCTCTAAGCTTGCTTGTGTGGCTCCTGACAC | This study |
| Reverse: CTCTCTAAGCTTGTTCAGTCCTCGCGGCAATC | ||
| sthA cloning | Forward: CTCTCTAAGCTTATGTTACCATTCTGTTGCTT | This study |
| Reverse: CTCTCTAAGCTTGATGCTGGAAGATGGTCACT |
Zm, Zymomonas mobilis; Ec, Erwinia chrysanthemi; Ko, Klebsiella oxytoca; Pp, Pseudomonas putida; FRT, FLP recombination target.
Fermentations were carried out as previously described (100 g liter−1 xylose, 37°C, 150 rpm, pH 6.5) with the automatic addition of 2 N KOH (22).
Furfural tolerance was examined by measuring growth in standing tubes with a 4-ml total volume of AM1 and 50 g liter−1 filter-sterilized xylose as described previously (23). Tubes were incubated at 37°C and measured after 24 and 48 h. The values reported are an average of four measurements.
Strain constructions.
E. coli transhydrogenase genes were amplified (ribosomal-binding sites, coding regions, and a 200-bp terminator region) from strain LY180 genomic DNA using a Bio-Rad iCycler (Hercules, CA) with primers that provided flanking HindIII sites (23). After digestion with HindIII, the product was ligated into HindIII-digested pTrc99a (vector) and transformed into E. coli TOP10F′ (Carlsbad, CA). Plasmids were purified using a QiaPrep spin miniprep kit (Valencia, CA). Gene orientation was established by digestion with restriction enzymes and by PCR (Table 1). Antibiotics were included as appropriate (24).
Microarray analysis.
Cultures were grown in small fermentors to a density of 670 mg dry cell weight liter−1. An initial sample was removed that served as a control. Furfural was immediately added from a 50-g liter−1 aqueous stock (0.5 g liter−1 final concentration) and incubation continued for 15 min prior to a second sampling. Samples were rapidly cooled in an ethanol-dry ice bath, harvested by centrifugation at 4°C, resuspended in Qiagen RNA Later, and stored at −80°C. RNA was extracted using a Qiagen RNeasy minikit, treated with DNase I, and purified by phenol-chloroform extraction and ethanol precipitation. RNA was sent to NimbleGen (Madison, WI) for microarray comparisons using probes designed for E. coli K12. Each sample consisted of pooled material from four fermentors. The complete experiment was performed twice, and the data were averaged (eight fermentors). Data were analyzed using ArrayStar (DNA Star, Madison, WI) and SimPheny software (Genomatica, Inc., San Diego, CA). Expression ratios are presented as the average of the two pooled data sets. In an experiment adding only water as a control, 54 genes (>2% of transcriptome) were found to change by more than twofold after water addition. Only eight of these were also affected by furfural addition, indicating that the transcriptional impact of the furfural delivery procedure was negligible relative to the effect of furfural.
NCA.
Network component analysis (NCA) calculates transcription factor activity ratios from expression ratios and known regulatory connections and was performed as previously described (12, 17, 39). The connectivity file was updated according to Regulon DB and Ecocyc (4, 14). The regulon of the “stringent factor” was defined as previously described, by analysis of the 5-min response to serine starvation via serine hydroxamate treatment during mid-log growth of BW25113 in MOPS (morpholinepropanesulfonic acid)-glucose (12). Regulators with significantly altered regulatory activity were identified by comparison to a null distribution and using a P value cutoff of 0.05.
Microarray data accession number.
The microarray data have been deposited in NCBI's Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) with GEO series accession number GSE17786.
RESULTS
Transcriptomic effect of furfural.
Message levels were compared in actively growing cells before and 15 min after the addition of 0.5 g liter−1 furfural. A water control was also included for comparison. Expression of a small group of genes was found to be altered by more than fivefold in response to furfural (see Table 4). Using a less stringent metric (twofold or greater), expression levels for ∼400 genes (10% of the transcriptome) were altered in response to furfural addition as compared to either the water control or the culture prior to furfural addition. The distribution of these altered genes varied widely among functional groups, providing useful insight into the mechanism of furfural's action (Table 2). In most functional groups, expression levels of less than 10% of the gene members were altered by twofold or greater. Groups with this low frequency of change included cofactors, carbon compounds, regulatory genes, macromolecular synthesis genes (cell structure, DNA, lipids, transcription, and translation), and others (phage, putative/insertion sequence, regulatory, and unclassified/unknown). Expression levels for 10% to 20% of the member genes were altered in four groups (cell processes, central metabolism, energy, and transport). Most of the affected genes associated with central metabolism, energy, and transport increased in expression upon furfural addition. These changes could provide an opportunity to scavenge and metabolize additional compounds that may be available and to increase carbon flow for energy production. Although many of the altered genes concerned with cell processes are involved in motility and chemotaxis, strain LY180 is nonmotile (data not shown), and these genes were not investigated further.
TABLE 4.
Genes with changes in expression ratios of fivefold or greater in response to 0.5 g liter−1 added furfural
| Locus tag designation | Genea | Fold change in gene expression
|
Function | |
|---|---|---|---|---|
| + Furfural | + H2O | |||
| b0365 | tauA* | 9.70 | 2.78 | Periplasmic sulfate-binding component of the taurine ABC transporter |
| b0366 | tauB* | 18.7 | 2.15 | ATP-binding component of the taurine ABC transporter |
| b0367 | tauC | 22.8 | 1.80 | Integral membrane component of the taurine ABC transporter |
| b0368 | tauD | 15.9 | 1.39 | Taurine dioxygenase |
| b0598 | cstA | 7.40 | −1.06 | Peptide transporter, induced by carbon starvation |
| b1188 | ycgB | 6.03 | −1.24 | Putative sporulation protein |
| b1375 | ynaE* | −5.37 | −3.20 | Rac prophage |
| b1544 | ydfK* | −5.58 | −3.38 | Qin prophage |
| b1649 | nemR | 9.95 | 1.40 | DNA-binding transcriptional repressor of nemA |
| b1650 | nemA | 6.90 | 1.14 | N-Ethylmaleimide reductase |
| b1783 | yeaG | 5.25 | −1.05 | Protein kinase |
| b1987 | cbl | 7.42 | −1.06 | Dual regulator of cysteine biosynthesis |
| b2148 | mglC | 5.96 | −1.61 | Integral membrane component of the galactose ABC transporter |
| b2149 | mglA | 6.39 | −1.25 | ATP-binding component of the galactose ABC transporter |
| b2150 | mglB | 7.99 | −1.13 | Periplasmic component of the galactose transport protein |
| b3011 | yqhD | 5.20 | 1.16 | NADP-dependent aldehyde dehydrogenase with furfural-reducing activity |
| b3012 | dkgA | 6.40 | −1.01 | Methylglyoxal reductase |
| b3917 | sbp | 7.45 | 1.06 | Subunit of sulfate-binding protein |
| b4116 | adiY | −6.24 | 1.15 | DNA-binding transcriptional regulator of arginine decarboxylase system |
| b4227 | ytfQ | 8.15 | 1.03 | Periplasmic component of a predicted sugar ABC transporter |
| b4354 | yjiY | 23.2 | −1.13 | Predicted inner membrane protein |
| b4485 | ytfR | 5.25 | 1.04 | ATP-binding component of a predicted sugar ABC transporter |
Some of these genes also changed more than twofold in response to the addition of water in a control experiment (marked with an asterisk).
TABLE 2.
Genes perturbed more than twofold in the response of LY180 to treatment with furfurala
| Functional group | Total no. of genes | No. (%) of genes differentially regulated | Downregulated genes | Upregulated gene(s) |
|---|---|---|---|---|
| Amino acid biosynthesis and metabolism | 123 | 28 (22.7) | argC, aroL, argA, tyrB, asnA, argD, thrA, ilvD, trpD, argB, trpE, thrC, ilvA, argG, aroH, thrB, sdaB, dapB, ilvM, ilvC | cysM, metC, idcC, dadX, metA, metB, metL, dadA |
| Biosynthesis of cofactors, prosthetic groups, and carriers | 120 | 8 (6.7) | pdxA, ubiX, folC, bioD | idi, trxC, pabC, ybdK |
| Carbon compound metabolism | 133 | 12 (9) | xdhB, acnB, gcd, xdhA, amyA, dhaK, aldB, dhaL, ygjG, fucO, treF, tauD | |
| Cell processes (including adaptation and protection) | 198 | 29 (14.6) | fliQ, fliJ, fliE, fliP, fliL, fliK, fliF, fliG, fliH, cspA, fliN, flgJ, ymcE, lpxP, flgK, fliO, flgH, flgL | ibpA, yfiA, otsA, osmC, sodC, hchA, fic, ecnB, osmY, yqhD, nemA |
| Cell structure | 114 | 8 (7.0) | mreC, mreD, rfaC, fliS, etk, yeiU, lpxB | ybhO |
| Central intermediary metabolism | 162 | 27 (16.7) | pykF, fumB, tktA, pyrH, ppa | poxB, cysQ, gabD, cysN, mqo, acnA, cysC, aceB, aceA, cysI, cysH, dcyD, aldA, gloA, glpK, fumA, glpD, sdhB, sdhC, sdhA, sdhD, dkgA |
| DNA replication, recombination, modification, and repair | 105 | 5 (4.8) | rnhB, fliA, recG | yjiD, aidB |
| Energy metabolism | 139 | 14 (10.1) | hypC, ackA, hypB, atpC | cyoE, frmA, aceE, aceF, fdoH, fdoG, cyoD, cyoB, cyoA, cyoC |
| Fatty acid and phospholipid metabolism | 42 | 3 (7.1) | accC, accB | fadI |
| Nucleotide biosynthesis and metabolism | 62 | 13 (21) | pyrB, purE, carA, pyrD, purH, guaB, purF, purN, purK, purD, carB, pyrE | xdhC |
| Phage, insertion sequence | 295 | 6 (2) | ydfK, ynaE, cspB, cvpA, cspI, ynfN | |
| Putative | 1,167 | 102 (8.7) | mltD, ydjH, fliR, yqeI, yijP, ydjI, bioC, ynjE, yibK, ydjJ, ydjZ, ynjC, ykgK, flhA, dctR, yhgF, yejM, ybjE, yfcC, ydgR, flgI, sdaC, yhiD, ybhA, ecfG, yibQ, ynjI, yliF, yliE, ydjK, yjjB, yibA, yedV | ycbB, paaY, yjgH, ybiC, yeaQ, yecC, yqgD, ydjN, yhbW, ybaT, yagT, yehZ, yfcG, ymgE, yhbO, ygcE, yedY, sfsA, ydgD, nanK, yjiA, yohJ, yhiP, ydcO, yiaG, yigM, yhjG, ydcN, yqfA, dhaM, ygeV, ybaS, ydcT, ynfM, ygiV, nanE, yqaE, yqeF, yfdY, yniA, ydcS, yncG, maeB, ybhP, ygaW, ybdH, yohF, yhcO, ydcK, yddV, yciW, yeiA, sufB, ybeM, yohC, ychH, yeiT, yeeE, yhdW, yjfF, uspB, ytfT, glgS, yqhC, b4485, cstA, ytfQ, ydhM, yjiX |
| Regulatory function | 253 | 18 (7.1) | adiY, evgS, cspG, suhB, cadC, flhC, fis | rssB, sbmC, sdiA, pdhR, crl, bolA, hcaR, metR, phoU, galS, cbl |
| Transcription, RNA processing, and degradation | 61 | 2 (3.3) | trmH, xseB | |
| Translation and posttranslational modification | 184 | 8 (4.3) | truC, rpsT, etp, rpsA | msrB, msrA, clpA, pphA |
| Transport | 353 | 69 (19.5) | artM, nikB, nikA, lysP, proV, artP, proW, nikD, nikC, thiP, tyrP, proX, hisP, aroP, artI, hisJ, nikE, artQ, cusB, btuF, artJ, ampG, cusA, mtr, dcuC, narK, pitA, hisM, emrA, thiQ | xylE, gabP, manX, nagE, araF, sufD, sufC, mntH, livH, kgtP, cysA, ssuC, blc, gltJ, cycA, yahN, pstA, pstC, glpT, glpF, argT, ssuA, pstB, gltK, gltI, b4460, mtlA, pstS, narU, mdtM, sufA, dctA, mglC, mglA, sbp, mglB, tauA, tauB, tauC |
| Unclassified | 14 | 3 (21.4) | ssuD, ssuE, ybdL | |
| Unknown | 681 | 57 (8.5) | yiiQ, intG, flhE, ymdA, ynjB, ydjY, yeeN, ymcA, yibL, yghG, b1172, yjaH | yccT, ybiJ, yjdI, yahO, yjdN, yhcN, ybaA, yedK, yqjD, eutQ, ybgS, yhhA, ompW, yhjY, yghX, yqeB, rtcB, ygaU, erfK, yegS, yeeD, yhcH, ydhS, yegP, yebV, yjfN, yehE, ydcJ, ygaM, yqeC, ybiL, psiF, yhfG, yjfO, nlpA, ybeH, ynhG, ycfR, yjiY, yodD, csiD, yeaH, yedP, yeaG, ycgB |
| Total | 4,206 | 412 (9.8) |
Shown are the genes perturbed more than twofold in response to treatment with 0.5 g liter−1 furfural, sorted by functional group.
Consistent with a previous report (23), two oxidoreductases capable of catalyzing the NADPH-specific reduction of furfural, YqhD and DkgA, were upregulated over fivefold (mRNA) in the parent by the addition of furfural, as previously reported. These two genes were silenced in a furfural-resistant mutant (EMFR9), resulting in an increase in furfural tolerance.
Expression levels for over 20% of the member genes in two functional groups were altered by the addition of furfural: amino acids and nucleotides. In these groups, over 2/3 of the altered genes were reduced by twofold or greater upon the addition of furfural. Expression levels for individual genes affecting the biosynthesis of purines, pyrimidines, and every family of amino acids were reduced by twofold or greater upon the addition of furfural. A single gene involved in nucleotide metabolism and only eight genes involved in amino acid metabolism exhibited a furfural-dependent increase in expression of at least twofold. Together, these changes agree with the general decrease in biosynthesis and growth observed upon the addition of furfural.
Effect of furfural on regulatory activity.
NCA was used to provide a global view of the cellular response to furfural. This analysis uses known regulatory network structure to identify regulators with perturbed activity from transcriptome data (17, 39). Of the 60 regulators included in this analysis, 22 were identified as having significantly altered expression after furfural challenge, where significance was assessed relative to a random network (Fig. 1A; Table 3). Regulators of cysteine and methionine biosynthesis (CysB and MetJ) as well as repressors of amino acid (ArgR) and nucleotide biosynthesis (PurR) were significantly affected by furfural addition. The stringent factor, a collective indicator of the stringent response (diversion of resources away from growth during amino acid and carbon starvation) also shows activation consistent with stalled biosynthesis and an excess of many intermediates. Together, these results indicate that the pools of many amino acids and biosynthetic intermediates have been altered by furfural addition. The fact that expression of genes concerned with cysteine and methionine biosynthesis increased while expression of most other biosynthesis pathways declined is consistent with a depletion of cysteine and methionine pools as an early event resulting from a furfural challenge.
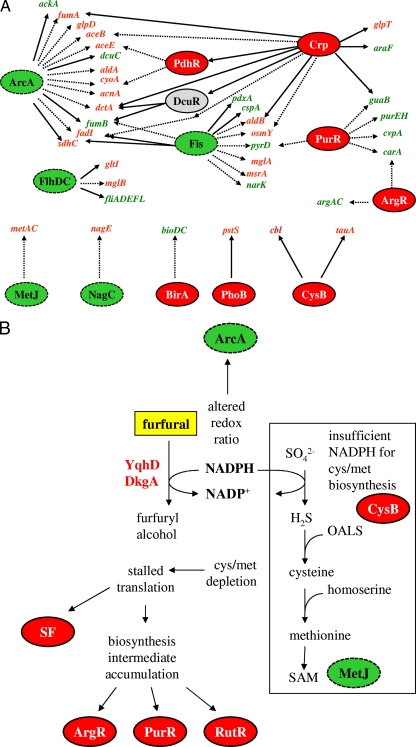
FIG. 1.
Transcriptional and regulatory changes in LY180 following challenge with 0.5 g liter−1 furfural. Regulatory genes that were significantly perturbed were identified by NCA with a P value cutoff of 0.05 relative to a null distribution. Regulators with increased activity are shown in red with a solid border, regulators with decreased activity are shown in green with a dashed border. Regulators that showed a mixed activity are shown in gray. Table 3 gives a more comprehensive listing of all perturbed regulators. (A) Partial regulator gene response map. Representative genes that were perturbed more than twofold are shown, with red (green) indicating genes with increased (decreased) expression. Solid lines indicate activation by the connected regulator, and dashed lines indicate repression. Because the direction of perturbation for DcuR is unclear, this regulator is shown in gray. (B) Model illustrating the mechanism of growth inhibition by furfural. The addition of furfural induces two NADPH-dependent oxidoreductases (YqhD and DkgA) that compete with essential biosynthetic reactions for NADPH. Assimilation of sulfate requires 4 NADPH molecules per cysteine and appears to be the most vulnerable of these biosynthetic reactions, as evidenced by perturbation of CysB and MetJ and by the direct benefit of supplementation with alternative sulfur sources. Secondary consequences include depletion of sulfur amino acids and a cascade of events from stalled translation and accumulation of many non-sulfur building block intermediates to a more general stress response, as evidenced by perturbation of the stringent factor (SF) and biosynthesis regulators ArgR, PurR, and RutR. OALS, O-acetyl-l-serine; SAM, S-adenosylmethionine.
TABLE 3.
Regulators significantlya perturbed in the LY180 furfural response relative to a null distribution, as determined by NCA
| Regulator | Perturbation direction | Description | Activation mechanism |
|---|---|---|---|
| ArcA | Down | Aerobic respiration control | Phosphorylation by ArcB |
| ArgR | Up | Repressor of arginine biosynthesis | Binding of l-arginine |
| BirA | Up | Repressor of biotin biosynthesis | Binding of bio-5′-AMP |
| CRP | Up | Global regulator of catabolite-sensitive operons | Binding of cyclic AMP |
| CysB | Up | Regulator of cysteine biosynthesis | Binding of O-acetyl-l-serine |
| DcuR | Unclear | Activator of genes involved in C-4 dicarboxylate metabolism | Phosphorylation by DcuS |
| FIS | Down | Global regulator associated with nutritional upshift | Inherently active |
| FlhDC | Down | Master motility regulator | [FlhD]4[FlhC]2 |
| FliA | Down | Minor sigma factor, regulates motility-associated genes | Inherently active |
| His | Up | Histidine, regulates histidine biosynthesis via transcriptional attenuation | Inherently active |
| MetJ | Down | Repressor of methionine biosynthesis | Binding of S-adenosylmethionine |
| NagC | Down | Coordinates biosynthesis and catabolism of amino sugars | Binding of GlcNAc-6-P |
| PdhR | Down | Repressor of pyruvate dehydrogenase complex | Absence of binding by pyruvate |
| PhoB | Up | Regulator of inorganic phosphate uptake | Phosphorylation by PhoR |
| PhoP | Up | Regulator of divalent cation starvation response | Phosphorylation by PhoQ |
| PurR | Up | Repressor of purine nucleotide biosynthesis | Binding of hypoxanthine |
| RpoH | Up | Heat shock sigma factor | Inherently active |
| RpoN | Up | Nitrogen-related sigma factor | Inherently active |
| RpoS | Up | General stress response sigma factor | Multiple mechanisms |
| RutR | Up | Proposed repressor of pyrimidine degradation | Unknown |
| SF | Up | Lumped “stringent factor” | Amino acid starvation |
P < 0.05.
RpoS, a sigma factor that acts as a signal for general stress response, was also affected by the addition of furfural, indicating that the cell recognizes the presence of a stress-inducing agent. Since up to 10% of E. coli's genome is regulated in some fashion by RpoS (31, 42), it is difficult to determine a specific inhibitory response. Examples of RpoS-regulated genes with increased expression include poxB (conversion of pyruvate into acetate and CO2) (47) and otsA (trehalose production during osmotic stress response) (37).
ArcA appeared to be significantly downregulated by NCA, as evidenced by increased expression of numerous genes in central metabolism (aceB, aceE, sdhC, dctA, cyoA, and fumA) and downregulation of fumB, a gene typically known to be active under anaerobic conditions. The upregulation of several glycerol metabolism genes (glpT and glpD in Fig. 1A, and glpF and glpK in Table 2) led us to investigate the effect of glycerol supplementation on furfural tolerance. However, the addition of glycerol (1.0 to 20 g liter−1) had no effect on furfural tolerance (data not included). Several other regulators, including fis and crp, were found to be significantly altered by NCA.
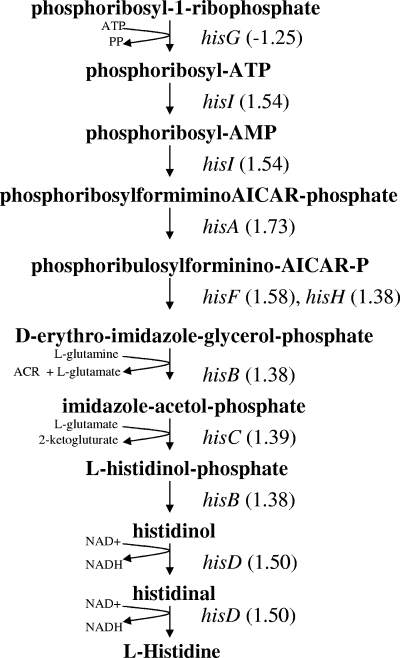
Histidine may also be limited by the addition of furfural. Genes (hisA, hisB, hisC, hisD, hisF, hisH, and hisI) under the control of the His regulator (histidinyl-tRNA) were generally increased after the addition of furfural, although less than twofold (Fig. 2). The two terminal steps in histidine biosynthesis involve the reduction of NAD+ to NADH, a reaction that may be slowed by the high NADH/NAD+ ratio associated with fermentation.
FIG. 2.
Addition of furfural moderately increased expression of most genes in the histidine pathway. With the exception of hisG, all other genes in this pathway were upregulated in response to furfural addition. Ratios for these changes are shown in parentheses. ACR, aminoimidazole carboxamide ribonucleotide.
Effects of furfural on expression of genes concerned with sulfur assimilation into amino acids.
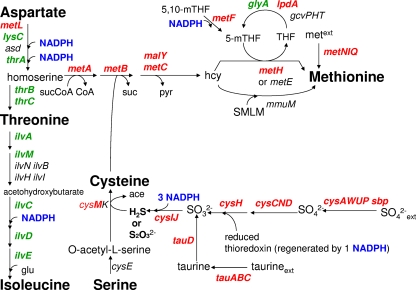
Many genes (cysC, cysH, cysI, cysM, cysN, cysQ, metA, metB, metC, metL, sbp, tauA, tauB, tauC, and tauD) concerned with sulfur assimilation into cysteine and methionine were increased by twofold or greater. These are scattered within several functional groups (Table 2): amino acids, central metabolism, regulation, and transport. Many additional genes involved in sulfur assimilation were also upregulated less than twofold and have been included to demonstrate the broad furfural response (Fig. 3). Sulfur is supplied as sulfate in AM1 medium and must be reduced to the level of hydrogen sulfide for incorporation, an energy-intensive reaction requiring 4 molecules of NADPH. The furfural-induced increase in expression of these genes is in sharp contrast to the decreases observed for many other genes concerned with the biosynthesis of amino acids, purines, and pyrimidines (Table 2). Expression of the taurine transport genes (tauABCD; alternative source of sulfur), the sulfate-binding transport protein (sbp), and the transcriptional activator of many cysteine biosynthetic genes (cbl) were increased by more than fivefold in response to added furfural (Table 4). Together, these results suggest that the addition of furfural results in an intracellular deficit in sulfur-containing amino acids (cysteine and methionine) which may be associated with the high NADPH requirement in this pathway.
FIG. 3.
Furfural increased expression of most genes concerned with sulfur assimilation into cysteine and methionine. Pathways for the synthesis of threonine and isoleucine from aspartate are included for comparison. Genes upregulated by 1.5-fold or greater are shown in red. Genes downregulated by 1.5-fold or greater are shown in green (negative sign assigned). All others are shown in black.
Effect of amino acid supplements on furfural tolerance.
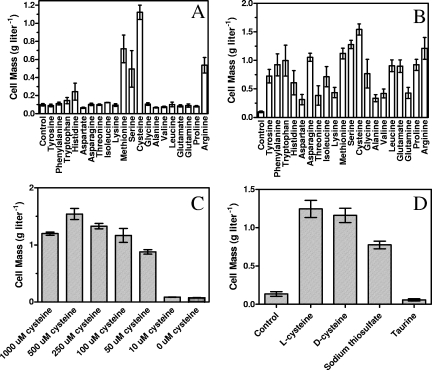
All 20 amino acids were individually tested for their ability to improve the growth of LY180 in AM1 mineral salts medium (Fig. 4A). A concentration of 0.1 mM was selected roughly based on the cellular content of individual amino acids (27). Only five amino acids improved furfural resistance when supplied at this low concentration: cysteine > methionine > serine and arginine > histidine. The two sulfur amino acids were clearly the most beneficial for furfural resistance. When supplied at a fivefold-higher concentration (0.5 mM), all amino acids were beneficial to some degree (Fig. 4B). However, cysteine remained the most effective, followed by serine, methionine, and arginine. A cysteine concentration of 0.05 mM allowed LY180 to grow to a density of 0.8 g liter−1 in the presence of 1 g liter−1 furfural, approximately equal to the total cellular sulfur (Fig. 4C). No measurable improvement in furfural resistance was observed with 0.01 mM cysteine.
FIG. 4.
Effect of medium supplements on growth in the presence of 1 g liter−1 furfural. Cultures were compared after incubation for 48 h in AM1 medium with 50 g liter−1 xylose at 37°C. (A) Addition of individual amino acids at 0.1 mM each; (B) addition of amino acids at 0.5 mM each; (C) addition of cysteine; (D) addition of alternative sulfur sources.
We considered the possibility that the protective effect of l-cysteine could result from a chemical reaction with furfural in AM1 medium. However, protective concentrations of cysteine (0.05 mM) were 200-fold lower than that of 1 g liter−1 furfural (10 mM), making this unlikely. Furfural in mineral salts medium can be readily quantified by its characteristic spectrum (19, 20) and remained unchanged during 48 h of incubation at 37°C, consistent with minimal chemical reactivity.
The beneficial effects of histidine, serine, and arginine for furfural tolerance are not immediately apparent. Most genes concerned with histidine biosynthesis increased in response to furfural addition, although less than twofold (Fig. 2). De novo biosynthesis of histidine during fermentation may be constrained by the high NADH/NAD+ ratio during anaerobic growth and the requirement for further reduction of NAD+ in the two terminal steps of biosynthesis. Similarly, the first committed step in serine biosynthesis also involves the reduction of NAD+ and may be hindered during fermentation. Increasing serine may also increase the efficiency of incorporating reduced sulfur from H2S into cysteine. Genes concerned with arginine biosynthesis (argA, argB, argC, argD, carA, carB, and argG) were generally lowered by more than twofold upon the addition of furfural. The expression level of speA encoding arginine decarboxylase was increased by the addition of furfural. The degradation of arginine may provide useful intermediates and cofactors for biosynthesis.
Effect of alternative sulfur sources on furfural tolerance.
The addition of furfural inhibited growth and increased the transcription of genes concerned with sulfur assimilation. Genes involved in the uptake and incorporation of the alternative sulfur compound taurine (tauABC and tauD) were among the 10 genes with the largest increases in expression (Table 4). The tau genes are typically expressed only during sulfur starvation (40). Since cysteine was effective in relieving furfural inhibition, the increased expression of these genes can be presumed to result from a reduction in the pool of sulfur amino acids by furfural. Furfural could inhibit sulfur amino acid biosynthesis either by limiting the availability of reduced sulfur (H2S) from sulfate or by inhibiting the incorporation of reduced sulfur into cysteine.
These hypotheses were examined during growth in AM1 medium containing 1 g liter−1 furfural by comparing the effects of alternative sulfur sources (l-cysteine, d-cysteine, taurine, and sodium thiosulfate) that enter metabolism at different levels of reduction (Fig. 4D). Note that 4 NADPH molecules and two reductase enzymes (CysH and CysIJ) are required to fully reduce sulfate prior to assimilation into cysteine. l-Cysteine, d-cysteine, and thiosulfate bypass both reductase enzymes, and all were effective at relieving furfural inhibition. d-Cysteine cannot be incorporated directly and is first catabolized to H2S (26). Thiosulfate also serves as a source of reduced sulfur for incorporation by CysM (18). Taurine is catabolized to sulfite in the cytoplasm and must be reduced by sulfite reductase (CysIJ and 3 NADPH molecules) prior to assimilation into cysteine (35). Unlike cysteine and thiosulfate, taurine was not effective in preventing the inhibition of growth by 1 g liter−1 furfural.
These results with alternative sulfur sources indicate that furfural acts to inhibit growth by limiting the production of reduced H2S from sulfate rather than by inhibiting the incorporation of reduced sulfur into cysteine. With taurine as a sulfur source, furfural must act at the level of sulfite reductase (CysIJ), which has a higher Km for NADPH (80 μM) (34) than the furfural reductase YqhD (8 μM) (23). With sulfate as a sulfur source, further effects of furfural at earlier steps in sulfur assimilation cannot be excluded.
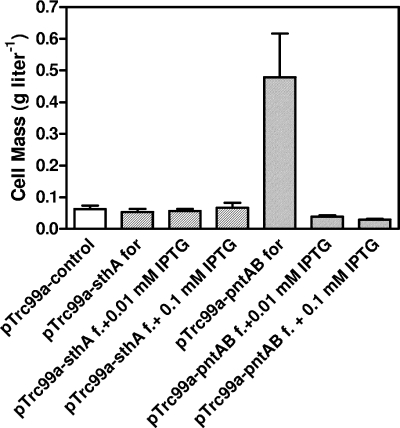
Effect of increasing transhydrogenase expression on furfural tolerance.
Previous studies have shown that growth is inhibited only while furfural is being metabolized and resumes after complete reduction to furfuryl alcohol by NADPH-dependent enzymes (7, 23). Silencing two genes (yqhD and dkgA) encoding low-Km, NADPH-dependent furfural reductases provided resistance to over 1.0 g liter−1 furfural. Although furfural may inhibit sulfur amino acid production by directly affecting enzymes concerned with the conversion of sulfate and sulfite to H2S, it is also possible that the inhibition of this process results from an indirect effect of furfural on the availability of NADPH. To test this hypothesis, the two E. coli transhydrogenases (SthA and PntAB) were cloned into pTrc99a and confirmed by sequencing.
SthA is a cytoplasmic transhydrogenase with kinetic characteristics that promote function primarily in the direction of NADPH oxidation (33). Expression of the sthA gene from a plasmid did not alter furfural tolerance with or without IPTG (isopropyl-β-d-thiogalactopyranoside) induction (Fig. 5). Functionality of the cloned gene was confirmed by in vitro assays. Upon induction with 0.1 mM IPTG, activity was found to increase from ∼1.0 nmol min−1 mg protein−1 to 18 nmol min−1 mg protein−1.
FIG. 5.
Effect of increased expression of transhydrogenases (SthA and PntAB) on furfural tolerance. Cultures were grown for 48 h in AM1 minimal media containing 50 liter−1 xylose and 1.0 g liter−1 furfural (f.). The empty vector served as a control. Inducer was added prior to inoculation.
PntAB is a proton-translocating transhydrogenase that is not known to function during fermentative growth but is potentially capable of increasing the pool of NADPH (33). Leaky expression of pntAB from an uninduced plasmid partially re-stored growth in the presence of 1 g liter−1 furfural (Fig. 5). Addition of IPTG to express higher levels of this enzyme eliminated resistance to furfural and also inhibited the growth of cells in the absence of furfural. Based on these results, furfural appears to inhibit growth by depleting the supply of NADPH needed for biosynthesis. The large requirement of NADPH for sulfate assimilation (4 molecules per cysteine equivalent) and the limited routes for NADPH production from xylose during fermentation appear to have made the production of sulfur amino acids most vulnerable to competition from furfural reductases for NADPH.
DISCUSSION
Based on the transcriptional and regulatory changes that were observed in response to the addition of furfural, a partial response map was assembled (Fig. 1B). This map combined with a summary of perturbed genes (Table 2) allowed the identification of sulfur assimilation into amino acids as an early site of furfural action. Furfural increased the expression of many genes and regulators concerned with sulfur assimilation into cysteine and methionine (Fig. 3), consistent with deficiency in these sulfur amino acids. In contrast, furfural lowered the expression of many other biosynthetic genes for building block molecules, consistent with their excess. Furthermore, the addition of low concentrations (0.1 mM) of cysteine and methionine relieved growth inhibition by 1 g liter−1 furfural (Fig. 4A). The minimum effective level of cysteine (0.05 mM) was similar to the estimated sulfur amino acid content of the cells that grew in the presence of 1 g liter−1 furfural.
Previous studies investigated the transcriptional response of E. coli strain K-12 to sulfur limitation during growth in minimal medium. Sulfur limitation was induced by replacing sulfate with either 0.25 mM taurine or 0.25 mM glutathione (8). In their study, a sulfur limitation reduced the rate of synthesis of cysteine and methionine and induced oxidative stress. Interestingly, the sulfur limitation also increased the transcription of cbl and the taurine transport genes (tauABC), two effects also observed in our furfural response data (Table 2).
At low concentrations, serine (a precursor of cysteine), histidine, and arginine were also effective at reducing furfural inhibition of growth. All amino acids were effective to some extent when added at a fivefold-higher concentration (0.5 mM). Transcript levels for most genes concerned with histidine biosynthesis were elevated, indicating a shortage of histidine (Fig. 2). Intracellular pools of histidine and serine may be limited to some extent during anaerobic growth since both biosynthetic pathways include reactions that reduced NAD+ to NADH. These reactions may be hindered by the high NADH/NAD+ ratios typical of fermentation, reducing pool sizes. The beneficial action of arginine was surprising because expression levels for genes concerned with arginine biosynthesis (argA, argB, argC, argD, carA, carB, and argG) were lowered by the addition of furfural, consistent with an excess of this amino acid. Furfural also increased the expression of arginine decarboxylase (speA), an enzyme concerned with arginine increasing intracellular pH and degradation. It is possible that degradation products of arginine increase furfural tolerance.
The inhibition of sulfur amino acid synthesis by furfural was localized to the steps prior to sulfur assimilation into cysteine (Fig. 3). Alternative sulfur sources at different degrees of reduction were tested as supplements during growth with furfural (1 g liter−1) (Fig. 4D). The cytoplasmic degradation of d-cysteine and thiosulfate both provided a source of reduced sulfur for direct incorporation into cysteine and relieved the inhibition of growth by furfural. Taurine is degraded intracellularly to sulfite, a partially reduced sulfur source that needs further reduction by CysIJ prior to incorporation into cysteine. Unlike thiosulfate, the addition of taurine (sulfite) had no effect on furfural tolerance despite the high expression levels of genes encoding taurine transport (tauABC) and degradation (tauD). Together, these results indicate that furfural inhibits sulfate assimilation by interfering with the reduction of sulfite by CysIJ. Additional inhibitory effects may also be present at earlier steps and cannot be excluded.
The inhibition of sulfate reduction is unlikely to represent the initial action of furfural that inhibits growth (Fig. 1B). Prior studies have reported the isolation of a furfural-resistant mutant of E. coli (23). Resistance was demonstrated to result partially from the silencing of two NADPH-dependent enzymes (YqhD and DkgA) that reduce furfural to furfuryl alcohol, a less toxic compound (45, 46). Based on these results, furfural was proposed to inhibit growth by limiting the available NADPH for biosynthesis. Results from our gene array studies provide further support for this hypothesis. Sulfate assimilation, the most NADPH-intensive pathway in metabolism, was found to be the most vulnerable site for furfural action. Growth inhibition by 1 g liter−1 furfural was relieved by supplying reduced sulfur for amino acid biosynthesis. Many other NADPH-dependent biosynthetic reactions would also be adversely affected by the NADPH-dependent reduction of furfural. Supplying reduced sulfur for biosynthesis as well as other building block metabolites would have a general sparing effect on the NADPH pool, consistent with the growth benefit provided by individual amino acids (Fig. 4A and B). A direct linkage between furfural inhibition of growth and NADPH was further demonstrated by expression of the proton-translocating transhydrogenase genes, pntAB. Low-level expression of these genes without inducer was shown to increase furfural tolerance.
Depletion of cysteine and methionine levels by furfural would be expected to initiate a cascade of cellular events (Fig. 1B), including stalled ribosomes that trigger a stringent response (6). The accumulation of other amino acids and nucleotides would activate repressors of biosynthesis such as ArgR and PurR and decrease expression of many biosynthetic pathways. NADPH depletion can also explain the altered activity of ArcA and the resulting increase in expression of tricarboxylic acid cycle-related genes, as ArcAB activity is known to be sensitive to the cellular redox ratio (41).
Nearly half of the genes perturbed more than twofold by furfural addition have either putative function (102 genes) or are unclassified (3 genes) or unknown (57 genes). With so little information available about these, it is difficult to classify their role in the furfural response. However, it is clear from the available data that inhibition of sulfate assimilation is a major component of the furfural-mediated response.
Acknowledgments
We gratefully acknowledge research support by grants from the U.S. Department of Energy (DE-FG02-96ER20222, DE-FG36-08GO88142, and DE-FC36-GO17058) and by the Verenium Corporation.
This work was facilitated by the BioCyc databases (14).
Footnotes
Published ahead of print on 14 August 2009.
REFERENCES
- 1.Almeida, J. R. M., M. Betilsson, M. F. Gorwa-Grauslund, S. Gorsich, and G. Liden. 2009. Metabolic effects of furaldehydes and impacts on biotechnological processes. Appl. Microbiol. Biotechnol. 82:625-638. [DOI] [PubMed] [Google Scholar]
- 2.Amann, E., B. Ochs, and K. J. Abel. 1988. Tightly regulated tac promoter vectors for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. [DOI] [PubMed] [Google Scholar]
- 3.Barciszewski, J., G. E. Siboska, B. O. Pedersen, B. F. C. Clark, and S. I. S. Ratten. 1997. A mechanism for the in vivo formation of N-6-furfuryladenine, kinetin, as a secondary oxidative damage product of DNA. FEBS Lett. 414:457-460. [DOI] [PubMed] [Google Scholar]
- 4.Gama-Castro, S., J. J. Veronica, M. Peralta-Gil, A. Santos-Zavaleta, M. I. Peñaloza-Spindola, B. Contreras-Moreira, J. Segura-Salazar, L. Muñiz-Rascado, I. Martinez-Flores, H. Salgado, C. Bonavides-Martinez, C. Abreu-Goodger, C. Rodriguez-Penagos, J. Miranda-Rios, E. Morett, E. Merino, A. M. Huerta, and J. Collado-Vides. 2008. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 36:D120-D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorsich, S. W., B. S. Dien, N. N. Nichols, P. J. Slinger, Z. L. Liu, and C. D. Skory. 2006. Tolerance to furfural-induced stress is associated with pentose phosphate pathway genes ZWF1, GND1, RPE1, and TKL1 in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 71:339-349. [DOI] [PubMed] [Google Scholar]
- 6.Gralla, J. D. 2005. Escherichia coli ribosomal RNA transcription: regulatory roles for ppGpp, NTPs, architectural proteins and a polymerase-binding protein. Mol. Microbiol. 55:973-977. [DOI] [PubMed] [Google Scholar]
- 7.Gutiérrez, T., L. O. Ingram, and J. F. Preston. 2006. Purification and characterization of a furfural reductase (FFR) from Escherichia coli strain LY01—an enzyme important in the detoxification of furfural during ethanol production. J. Biotechnol. 121:154-164. [DOI] [PubMed] [Google Scholar]
- 8.Gyaneshwar, P., O. Paliy, J. McAuliffe, D. L. Papham, M. I. Jordan, and S. Kustu. 2005. Sulfur and nitrogen limitation in Escherichia coli K-12: specific homeostatic responses. J. Bacteriol. 187:1074-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn-Hägerdal, B., M. Galbe, M. F. Gorwa-Grauslund, G. Liden, and G. Zacchi. 2006. Bio-ethanol—the fuel of tomorrow from the residues of today. Trends Biotechnol. 24:549-556. [DOI] [PubMed] [Google Scholar]
- 10.Horvath, I. S., M. J. Taherzadeh, C. Niklasson, and G. Liden. 2001. Effects of furfural on anaerobic continuous cultivation of Saccharomyces cerevisiae. Biotechnol. Bioeng. 75:540-549. [DOI] [PubMed] [Google Scholar]
- 11.Hristozova, T., A. Angelov, B. Tzvetkova, D. Paskaleva, V. Gotcheva, S. Gargova, and K. Pavlova. 2006. Effect of furfural on carbon metabolism key enzymes of lactose-assimilating yeasts. Enzyme Microbiol. Technol. 39:1108-1112. [Google Scholar]
- 12.Hyduke, D. H., L. R. Jarboe, L. M. Tran, K. J. Y. Chou, and J. C. Liao. 2007. Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc. Natl. Acad. Sci. USA 104:8484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarboe, L. R., T. B. Grabar, L. P. Yomano, K. T. Shanmugan, and L. O. Ingram. 2007. Development of ethanologenic bacteria. Biofuels 108:237-261. [DOI] [PubMed] [Google Scholar]
- 14.Karp, P. D., I. M. Keseler, A. Shearer, M. Latendresse, M. Krummenacker, S. M. Paley, I. Paulsen, J. Collado-Vides, S. Gama-Castro, M. Peralta-Gil, A. Santos-Zavaleta, M. I. Penaloza-Spinola, C. Bonavides-Martinez, and J. Ingraham. 2007. Multidimensional annotation of the Escherichia coli K-12 genome. Nucleic Acids Res. 35:7577-7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katahira, S., A. Mizuike, H. Fukuda, and A. Kondo. 2006. Ethanol fermentation from lignocellulosic hydrolysate by a recombinant xylose and cellooligosaccharide-assimilating yeast strain. Appl. Microbiol. Biotechnol. 72:1136-1143. [DOI] [PubMed] [Google Scholar]
- 16.Khan, Q. A., F. A. Shamsi, and S. M. Hadi. 1995. Mutagenicity of furfural in plasmid DNA. Cancer Lett. 89:95-99. [DOI] [PubMed] [Google Scholar]
- 17.Liao, J. C., R. Boscolo, Y. L. Yang, L. M. Tran, C. Sabatti, and V. P. Roychowdhury. 2003. Network component analysis: reconstruction of regulatory signals in biological systems. Proc. Natl. Acad. Sci. USA 100:15522-15527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maier, T. H. 2003. Semisynthetic production of L-alpha-amino acids by metabolic engineering of the cysteine-biosynthetic pathway. Nat. Biotechnol. 4:422-427. [DOI] [PubMed] [Google Scholar]
- 19.Martinez, A., M. E. Rodriguez, S. W. York, J. F. Preston, and L. O. Ingram. 2000. Use of UV absorbance to monitor furans in dilute acid hydrolysates of biomass. Biotechnol. Prog. 16:637-641. [DOI] [PubMed] [Google Scholar]
- 20.Martinez, A., M. E. Rodriguez, S. W. York, J. F. Preston, and L. O. Ingram. 2000. Effects of Ca(OH)2 treatments (“overliming”) on the composition and toxicity of bagasse hemicellulose hydrolysates. Biotechnol. Bioeng. 69:526-536. [DOI] [PubMed] [Google Scholar]
- 21.Martinez, A., M. E. Rodriguez, M. L. Wells, S. W. York, J. F. Preston, and L. O. Ingram. 2001. Detoxification of dilute acid hydrolysates of lignocellulose with lime. Biotechnol. Prog. 17:287-293. [DOI] [PubMed] [Google Scholar]
- 22.Martinez, A., T. B. Grabar, K. T. Shanmugam, L. P. Yomano, S. W. York, and L. O. Ingram. 2007. Low salt medium for lactate and ethanol production by recombinant Escherichia coli B. Biotechnol. Lett. 29:397-404. [DOI] [PubMed] [Google Scholar]
- 23.Miller, E. M., L. R. Jarboe, L. P. Yomano, S. W. York, K. T. Shanmugam, and L. O. Ingram. 2009. Silencing of NADPH-dependent oxidoreductases (yqhD and dkgA) in furfural-resistant ethanologenic Escherichia coli. Appl. Environ. Microbiol. 75:4315-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, J. H. 1992. A short course in bacterial genetics. CSHL Press, Plainview, NY.
- 25.Modig, T., G. Liden, and M. J. Taherzadeh. 2002. Inhibition effects of furfural on alcohol dehydrogenase, aldehyde dehydrogenase, and pyruvate dehydrogenase. Biochem. J. 363:769-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagasawa, T., T. Ishi, H. Kumagai, and H. Yamada. 1985. d-Cysteine desulfhydrase of Escherichia coli—purification and characterization. Eur. J. Biochem. 153:541-551. [DOI] [PubMed] [Google Scholar]
- 27.Neidhardt, F. C., J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.). 1987. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, DC.
- 28.Ohta, K., D. S. Beall, J. P. Meijia, K. T. Shanmugam, and L. O. Ingram. 1991. Genetic improvement of Escherichia coli for ethanol production: chromosomal integration of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Appl. Environ. Microbiol. 57:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palmqvist, E., and B. Hahn-Hagerdal. 2000. Fermentation of lignocellulosic hydrolysates. I. Inhibition and detoxification. Bioresour. Technol. 74:17-24. [Google Scholar]
- 30.Palmqvist, E., and B. Hahn-Hagerdal. 2000. Fermentation of lignocellulosic hydrolysates. II. Inhibitors and mechanisms of inhibition. Bioresour. Technol. 74:25-33. [Google Scholar]
- 31.Patten, C. L., M. G. Kirchhof, M. R. Schertzberg, R. A. Morton, and H. E. Schellhorn. 2004. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genomics 272:580-591. [DOI] [PubMed] [Google Scholar]
- 32.Petersson, A., J. R. M. Almeida, T. Modig, K. Karhumaa, B. Hahn-Hagerdal, M. F. Gorwa-Grauslund, and G. Liden. 2006. A 5-hydroxymethyl furfural reducing enzyme encoded by Saccharomyces cerevisiae ADH6 gene conveys HMF tolerance. Yeast 23:455-464. [DOI] [PubMed] [Google Scholar]
- 33.Sauer, U., F. Canonaco, S. Heri, A. Perrenoud, and E. Fisher. 2004. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J. Biol. Chem. 279:6613-6619. [DOI] [PubMed] [Google Scholar]
- 34.Schomburg, I., A. Chang, O. Hofmann, C. Ebeling, F. Ehrentreich, and D. Schomburg. 2002. BRENDA: a resource for enzyme data and metabolic information. Trends Biochem. Sci. 27:54-56. [DOI] [PubMed] [Google Scholar]
- 35.Siegel, L. M., and P. S. Davis. 1974. Reduced nicotinamide adenine dinucleotide phosphate-sulfite reductase of Enterobacteria. J. Biol. Chem. 249:1587-1598. [PubMed] [Google Scholar]
- 36.Singh, N. P., and A. Khan. 1995. Acetaldehyde: genotoxicity and cytotoxicity in human lymphocytes. Mutat. Res. 337:9-17. [DOI] [PubMed] [Google Scholar]
- 37.Strom, A. R., and I. Kaasen. 1993. Trehalose metabolism in Escherichia coli: stress protection and stress regulation of gene expression. Mol. Microbiol. 8:205-210. [DOI] [PubMed] [Google Scholar]
- 38.Tokiwa, Y., and B. P. Calabia. 2008. Biological production of functional chemicals from renewable resources. Can. J. Chem. 86:548-555. [Google Scholar]
- 39.Tran, L. M., M. P. Brynildsen, K. C. Kao, J. K. Suen, and J. C. Liao. 2005. gNCA: a framework for determining transcription factor activity based on transcriptome: identifiability and numerical implementation. Metab. Eng. 7:128-141. [DOI] [PubMed] [Google Scholar]
- 40.van der Ploeg, J. R., M. A. Weiss, E. Saller, H. Nahimoto, N. Saito, M. A. Kertesz, and T. Leisinger. 1996. Identification of sulfate starvation-regulated genes in Escherichia coli: a gene cluster involved in the utilization of taurine as a sulfur source. J. Bacteriol. 178:5438-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vemuri, G. N., E. Altman, D. P. Sangurdekar, A. B. Khodursky, and M. A. Eiteman. 2006. Overflow metabolism in Escherichia coli during steady-state growth: transcriptional regulation and effect of the redox ratio. Appl. Environ. Microbiol. 72:3653-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber, H., T. Polen, J. Heuveling, V. F. Wendish, and R. Hengge. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yomano, L. P., S. W. York, K. T. Shanmugam, and L. O. Ingram. 2009. Deletion of methylglyoxal synthase gene (mgsA) increased sugar co-metabolism in ethanol-producing Escherichia coli. Biotechnol. Lett. 31:1389-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaldivar, J., and L. O. Ingram. 1999. Effect of organic acids on the growth and fermentation of ethanologenic Escherichia coli LY01. Biotechnol. Bioeng. 66:203-210. [DOI] [PubMed] [Google Scholar]
- 45.Zaldivar, J., A. Martinez, and L. O. Ingram. 1999. Effect of selected aldehydes on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol. Bioeng. 65:24-33. [DOI] [PubMed] [Google Scholar]
- 46.Zaldivar, J., A. Martinez, and L. O. Ingram. 2000. Effect of alcohol compounds found in hemicellulose hydrolysate on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol. Bioeng. 68:524-530. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, T. F., and L. P. Hager. 1987. A single-step large scale purification of pyruvate oxidase. Arch. Biochem. Biophys. 257:485-487. [DOI] [PubMed] [Google Scholar]