Abstract
Campylobacter jejuni is widely distributed in the environment, and river water has been shown to carry high levels of the organism. In this study, 244 C. jejuni isolates from three river catchment areas in New Zealand were characterized using multilocus sequence typing. Forty-nine of the 88 sequence types identified were new. The most common sequence types identified were ST-2381 (30 isolates), ST-45 (25 isolates), and ST-1225 (23 isolates). The majority of the sequence types identified in the river water could be attributed to wild bird fecal contamination. Two novel clonal complexes (CC) were identified, namely, CC ST-2381 (11 sequence types, 46 isolates) and CC ST-3640 (6 sequence types, 12 isolates), in which all of the sequence types were new. CC ST-2381 was the largest complex identified among the isolates and was present in two of the three rivers. None of the sequence types associated with the novel complexes has been identified among human isolates. The ST-2381 complex is not related to complexes associated with cattle, sheep, or poultry. The source of the novel complexes has yet to be identified.
Contamination of the environment by bacterial pathogens is a significant health concern, as it provides a continuous source of organisms for the infection and reinfection of humans and animals. Enteric pathogens gain entry into the environment through the discharge of sewage into water and via contamination from animal feces (22). Fecal contamination is responsible for the continued presence and spread of a range of pathogenic organisms, including Campylobacter, norovirus, and Escherichia coli O157. Determining the roles of various environmental sources in human enteric disease requires an understanding of the distribution, survival, population structure, and pathogenic potential of the pathogens in the environment.
Campylobacter is the most common cause of gastrointestinal illness in the industrialized world (17), imposing significant economic costs on health systems, and is associated with a number of neurological sequelae (32, 33). The majority of human campylobacter infections are caused by Campylobacter jejuni (90%), with Campylobacter coli mostly responsible for the remainder. Although Campylobacter has been isolated from a wide range of animals (41) and birds (47, 48), contaminated poultry and poultry products remain the most significant sources of human infections (10, 38, 50, 51). Campylobacter is a spiral gram-negative organism that grows best under low-oxygen conditions at 42°C. The organism is unable to grow outside an animal host, and survival in the environment is dependent on ambient temperature, oxygen levels, and sunlight.
Studies worldwide examining rivers and waterways show that there is significant contamination by Campylobacter, with the sources being sewage outflow, direct fecal deposition, and pasture runoff (12, 22, 34, 37, 39). Similarly, coastal waters and estuaries can be contaminated by either sewage or bird fecal deposition (23, 35). The inability of Campylobacter to grow in the environment and its sensitivity to sunlight are thought to ensure that the organism is eventually purged from the system. However, the high levels of the organism identified in water systems have been highlighted as a risk for human infection.
The characterization of campylobacter populations by multilocus sequence typing (MLST) has shown that the organism is weakly clonal and that certain clonal complexes are associated with particular animals (5, 9, 26). Isolates from human cases of infection show a wide variety of sequence types and many clonal complexes. Source attribution studies using MLST have identified poultry as causing approximately 60% of human infections (14, 38, 50). Cattle have been identified as a potential source of infection due to the high level of similarity between bovine and human strains (18, 19). There remains, however, a significant number of infections for which the source is not certain.
New Zealand has one of the highest rates of campylobacteriosis in the developed world. This is due to the significant quantity of fresh chicken consumed coupled with high levels of contamination found in poultry products (1, 10, 51, 52). Campylobacter has been isolated from a range of environmental sources within New Zealand, including its rivers and streams (12, 37). Isolation rates for rivers in New Zealand range from 55 to 90%, comparable to results of studies overseas, and show the same seasonal variation as that seen elsewhere in the world (20). Pulsed-field gel electrophoresis (PFGE) analysis identified indistinguishable macrorestriction profiles for cattle, human, and river isolates, suggesting river water as a potential source of infection (8). In this study, C. jejuni isolates from three rivers in New Zealand, two on the South Island and one on the North Island, were characterized using MLST.
MATERIALS AND METHODS
Sampling of rivers and isolation of Campylobacter.
The Taieri and Ashburton rivers (South Island) and the Manawatu River (North Island) were sampled. Multiple sampling sites were used on the rivers and their tributaries, and these have been described previously (8, 11, 12, 14). The Taieri River catchment area is predominantly rural and is located near Dunedin on the South Island of New Zealand. The surrounding land is used for farming (cattle, sheep, and deer) and forestry. The Ashburton River is also located on the South Island, and its catchment area is used for dairy, beef, and sheep farming. The Taieri and Ashburton rivers are approximately 240 km apart. The Manawatu catchment area is located toward the lower central part of the North Island and is also rural, used for both dairy and sheep farming. Sampling sites on the Manawatu River were principally high-use recreational swimming spots identified by the Regional Council.
The sampling protocol for the Taieri River has been described previously (11). The river water sample was filtered through a 0.45-μm-pore-size membrane, and the filters were incubated in Preston enrichment broth and subcultured onto modified cefoperazone charcoal deoxycholate agar-Preston agar plates. All Taieri River samples were collected between June 2000 and June 2001. Sampling of the Ashburton River has also been described previously (8). In brief, primary and secondary enrichments of filtered water samples were carried out in M-Exeter broth, followed by culture on Exeter agar medium. All Ashburton River samples were collected between January and December 2001. The Manawatu River sampling was performed as described previously (14). Samples were filtered and enriched using Bolton broth and subcultured onto modified cefoperazone charcoal deoxycholate agar plates. All Manawatu River samples were collected between March 2005 and February 2008. Isolates from the Ashburton and Taieri rivers were stored as glycerol stocks at −80°C and resuscitated on 5% Columbia blood agar plates under microaerophilic conditions at 42°C. The Manawatu River isolates were characterized prior to storage in glycerol.
Isolates were identified as C. jejuni based on phenotypic characteristics (Gram stain result and hippurate and oxidase tests) and PCR (7, 42, 49).
MLST.
MLST was carried out as described previously (9). Chromosomal DNA was purified using a Roche PCR template purification kit (Roche, Auckland, New Zealand). Amplifications were performed in a final volume of 25 μl, using Applied Biosystems AmpliTaq Gold master mix (Applied Biosystems, Auckland, New Zealand) and 5 pmol of each primer. Products were sequenced on an ABI 3130XL automated DNA sequencer using ABI BigDye v3.1 technology (Applied Biosystems). Alleles, sequence types, and clonal complexes were assigned using the Campylobacter PubMLST database (http://pubmlst.org/campylobacter/). Novel sequence types not assigned to a clonal complex within the Campylobacter PubMLST database were examined for clonal complexes by use of eBURST3 (13, 40). A stringent requirement of six of seven common alleles for group definition was used to define a clonal complex. Minimum spanning trees were constructed within the BioNumerics program v5.1, using the allelic data set. Complexes in the minimum spanning trees were constructed using a maximum neighbor distance of two changes and a minimum size of two types. Minimum evolution and neighbor-joining trees were generated using the MEGA4 program (43). In total, 244 isolates were characterized from the three rivers, with 62 from the Taieri River, 56 from the Ashburton River, and 126 from the Manawatu River.
RESULTS
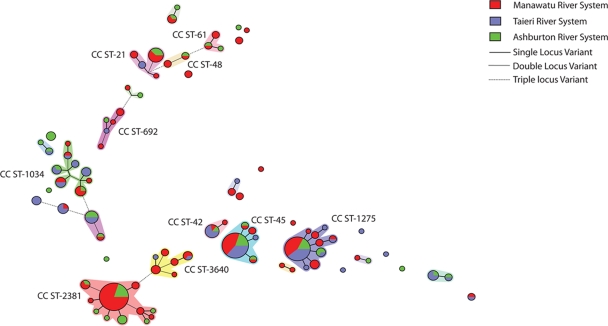
The sequence types identified in each river are given in Table 1. Among the 244 isolates characterized by MLST, 88 sequence types were identified, of which 49 were new and another 7 were unique to New Zealand. Novel sequence types represented less than one-half of the isolates (45%). The major sequence types identified in river water were ST-2381 (30 isolates [12%]), ST-45 (25 isolates [10%]), and ST-1225 (23 isolates [9%]). Seventy sequence types were associated with one or two isolates only (98 isolates [40% of the total]). Thirteen clonal complexes were identified from the Campylobacter PubMLST database, with the major ones being CC ST-1275 (38 isolates [16%]), CC ST-45 (32 isolates [13%]), CC ST-21 (15 isolates [6%]), and CC ST-1034 (13 isolates [5%]). Two additional complexes, the ST-2381 and ST-3640 complexes, were identified using eBURST3, although they have not been assigned in the Campylobacter PubMLST database. A minimum spanning tree for the water complexes is shown in Fig. 1. The ST-2381 complex consisted of 11 sequence types (46 isolates [19%]) and was by far the largest complex among the water isolates. All sequence types within CC ST-2381 are unique to New Zealand. The ST-3640 complex consisted of six sequence types (12 isolates [5%]). One of the six sequence types in CC ST-3640, ST-3673, has subsequently been identified outside New Zealand, as reported in the Campylobacter PubMLST database. It was associated with a C. coli isolate from a chicken sample in the United States. The remaining sequence types within CC ST-3640 are unique to New Zealand. Twenty-six of the sequence types were not assigned to a clonal complex (49 isolates [20%]), and of these, 14 sequence types were identified only in New Zealand (27 isolates [11%]).
TABLE 1.
Sources and sequence types of river water isolates of Campylobacter jejuni from New Zealand
| CC | River | STa | No. of isolates | MLST allele
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| aspA | glnA | gltA | glyA | pgm | tkt | uncA | ||||
| ST-21 | Manawatu | 50 | 2 | 2 | 1 | 12 | 3 | 2 | 1 | 5 |
| Ashburton | 422 | 3 | 2 | 1 | 5 | 3 | 2 | 5 | 5 | |
| Manawatu | 422 | 7 | 2 | 1 | 5 | 3 | 2 | 5 | 5 | |
| Taieri | 520 | 2 | 2 | 1 | 12 | 88 | 2 | 1 | 5 | |
| Manawatu | 3610 | 1 | 2 | 1 | 5 | 88 | 2 | 11 | 5 | |
| ST-42 | Taieri | 42 | 5 | 1 | 2 | 3 | 4 | 5 | 9 | 3 |
| Ashburton | 42 | 1 | 1 | 2 | 3 | 4 | 5 | 9 | 3 | |
| Manawatu | 42 | 1 | 1 | 2 | 3 | 4 | 5 | 9 | 3 | |
| Manawatu | 3676 | 1 | 1 | 307 | 3 | 4 | 5 | 9 | 3 | |
| ST-45 | Ashburton | 25 | 1 | 4 | 7 | 10 | 1 | 1 | 7 | 1 |
| Manawatu | 25 | 1 | 4 | 7 | 10 | 1 | 1 | 7 | 1 | |
| Ashburton | 45 | 5 | 4 | 7 | 10 | 4 | 1 | 7 | 1 | |
| Taieri | 45 | 9 | 4 | 7 | 10 | 4 | 1 | 7 | 1 | |
| Manawatu | 45 | 11 | 4 | 7 | 10 | 4 | 1 | 7 | 1 | |
| Ashburton | 137 | 1 | 4 | 7 | 10 | 4 | 42 | 7 | 1 | |
| Manawatu | 137 | 1 | 4 | 7 | 10 | 4 | 42 | 7 | 1 | |
| Taieri | 3641 | 1 | 81 | 7 | 10 | 4 | 1 | 7 | 1 | |
| Manawatu | 3802 | 2 | 4 | 319 | 10 | 4 | 1 | 7 | 1 | |
| ST-48 | Ashburton | 38 | 1 | 2 | 4 | 2 | 2 | 6 | 1 | 5 |
| Manawatu | 38 | 1 | 2 | 4 | 2 | 2 | 6 | 1 | 5 | |
| Manawatu | 474 | 2 | 2 | 4 | 1 | 2 | 2 | 1 | 5 | |
| ST-52 | Ashburton | 2392 | 1 | 9 | 25 | 2 | 283 | 22 | 3 | 6 |
| ST-61 | Ashburton | 61 | 1 | 1 | 4 | 2 | 2 | 6 | 3 | 17 |
| Manawatu | 61 | 1 | 1 | 4 | 2 | 2 | 6 | 3 | 17 | |
| Ashburton | 2341 | 1 | 1 | 1 | 2 | 2 | 6 | 3 | 17 | |
| Manawatu | 3982 | 2 | 1 | 333 | 2 | 2 | 6 | 3 | 17 | |
| ST-177 | Manawatu | 177 | 2 | 17 | 2 | 8 | 5 | 8 | 2 | 4 |
| Taieri | 2537 | 1 | 17 | 2 | 8 | 5 | 8 | 2 | 143 | |
| ST-354 | Manawatu | 1517 | 1 | 8 | 10 | 149 | 2 | 11 | 12 | 6 |
| ST-403 | Ashburton | 2026 | 1 | 10 | 1 | 16 | 19 | 10 | 5 | 7 |
| Manawatu | 2026 | 2 | 10 | 1 | 16 | 19 | 10 | 5 | 7 | |
| Ashburton | Newb | 1 | Newb | 1 | 16 | 19 | 10 | 5 | 7 | |
| ST-677 | Taieri | 677 | 1 | 10 | 81 | 50 | 99 | 120 | 76 | 52 |
| Manawatu | 677 | 1 | 10 | 81 | 50 | 99 | 120 | 76 | 52 | |
| ST-692 | Taieri | 699 | 1 | 37 | 52 | 57 | 26 | 129 | 29 | 23 |
| Ashburton | 991 | 1 | 37 | 52 | 57 | 26 | 107 | 29 | 23 | |
| Manawatu | 2584 | 1 | 2 | 1 | 57 | 26 | 127 | 29 | 35 | |
| Ashburton | 3235 | 1 | 213 | 1 | 57 | 26 | 127 | 29 | 17 | |
| Manawatu | 3659 | 2 | 37 | 52 | 57 | 26 | 127 | 29 | 1 | |
| Manawatu | 3664 | 1 | 37 | 52 | 4 | 26 | 127 | 29 | 23 | |
| Manawatu | 3804 | 1 | 37 | 52 | 57 | 26 | 129 | 29 | 1 | |
| ST-1034 | Ashburton | 694 | 1 | 2 | 59 | 4 | 105 | 126 | 25 | 23 |
| Manawatu | 694 | 3 | 2 | 59 | 4 | 105 | 126 | 25 | 23 | |
| Ashburton | 977 | 1 | 22 | 61 | 4 | 64 | 74 | 25 | 23 | |
| Taieri | 1255 | 1 | 22 | 146 | 4 | 64 | 74 | 25 | 23 | |
| Ashburton | 1255 | 1 | 22 | 146 | 4 | 64 | 74 | 25 | 23 | |
| Manawatu | 1956 | 1 | 2 | 59 | 4 | 27 | 128 | 25 | 23 | |
| Taieri | 2378 | 2 | 2 | 15 | 4 | 48 | 356 | 25 | 23 | |
| Taieri | 2391 | 2 | 2 | 15 | 4 | 48 | 360 | 25 | 23 | |
| Ashburton | 2391 | 1 | 2 | 15 | 4 | 48 | 360 | 25 | 23 | |
| ST-1275 | Manawatu | 1223 | 2 | 27 | 33 | 22 | 49 | 43 | 9 | 31 |
| Taieri | 1225 | 9 | 27 | 33 | 22 | 49 | 43 | 7 | 31 | |
| Ashburton | 1225 | 4 | 27 | 33 | 22 | 49 | 43 | 7 | 31 | |
| Manawatu | 1225 | 10 | 27 | 33 | 22 | 49 | 43 | 7 | 31 | |
| Taieri | 2348 | 1 | 27 | 8 | 22 | 49 | 43 | 7 | 31 | |
| Ashburton | 2348 | 2 | 27 | 8 | 22 | 49 | 43 | 7 | 31 | |
| Taieri | 3639 | 1 | 27 | 305 | 22 | 49 | 43 | 9 | 31 | |
| Taieri | 3657 | 1 | 27 | 33 | 22 | 104 | 134 | 7 | 31 | |
| Manawatu | 3657 | 1 | 27 | 33 | 22 | 104 | 134 | 7 | 31 | |
| Manawatu | 3661 | 2 | 27 | 33 | 22 | 49 | 134 | 7 | 31 | |
| Manawatu | 3662 | 3 | 27 | 33 | 22 | 49 | 43 | 110 | 31 | |
| Manawatu | 3674 | 1 | 27 | 33 | 22 | 49 | 43 | 350 | 31 | |
| Taieri | 3677 | 1 | 27 | 33 | 22 | 49 | 43 | 351 | 31 | |
| ST-2381 | Ashburton | 2381 | 6 | 175 | 251 | 216 | 282 | 359 | 293 | 102 |
| Manawatu | 2381 | 24 | 175 | 251 | 216 | 282 | 359 | 293 | 102 | |
| Ashburton | 2386 | 1 | 177 | 251 | 216 | 282 | 359 | 293 | 102 | |
| Ashburton | 2618 | 3 | 175 | 251 | 216 | 282 | 370 | 293 | 102 | |
| Ashburton | 2619 | 1 | 191 | 251 | 216 | 282 | 359 | 293 | 214 | |
| Manawatu | 2619 | 2 | 191 | 251 | 216 | 282 | 359 | 293 | 214 | |
| Ashburton | 2620 | 1 | 192 | 259 | 216 | 282 | 359 | 293 | 102 | |
| Ashburton | 3580 | 1 | 223 | 251 | 216 | 282 | 359 | 293 | 102 | |
| Ashburton | 3581 | 1 | 192 | 295 | 216 | 332 | 359 | 293 | 102 | |
| Manawatu | 3656 | 2 | 175 | 251 | 216 | 282 | 359 | 293 | 3 | |
| Manawatu | 3658 | 1 | 1 | 295 | 216 | 282 | 359 | 293 | 102 | |
| Manawatu | 3660 | 2 | 192 | 295 | 216 | 282 | 359 | 293 | 102 | |
| Manawatu | 3801 | 1 | 175 | 318 | 216 | 282 | 359 | 293 | 102 | |
| ST-3640 | Taieri | 3638 | 1 | 175 | 6 | 216 | 282 | 261 | 346 | 3 |
| Manawatu | 3640 | 2 | 1 | 6 | 5 | 4 | 261 | 7 | 3 | |
| Taieri | 3640 | 1 | 1 | 6 | 5 | 4 | 261 | 7 | 3 | |
| Manawatu | 3655 | 2 | 1 | 6 | 5 | 282 | 261 | 7 | 3 | |
| Manawatu | 3663 | 3 | 175 | 6 | 216 | 282 | 261 | 7 | 3 | |
| Manawatu | 3673 | 1 | 175 | 6 | 216 | 4 | 434 | 7 | 3 | |
| Manawatu | 3800 | 2 | 175 | 6 | 5 | 282 | 261 | 7 | 262 | |
| Unassigned | Manawatu | 436 | 2 | 7 | 21 | 5 | 62 | 4 | 61 | 44 |
| Manawatu | 526 | 2 | 2 | 15 | 4 | 27 | 13 | 80 | 23 | |
| Taieri | 526 | 2 | 2 | 15 | 4 | 27 | 13 | 80 | 23 | |
| Taieri | 992 | 3 | 2 | 59 | 4 | 27 | 126 | 29 | 23 | |
| Ashburton | 995 | 3 | 2 | 4 | 84 | 105 | 126 | 25 | 57 | |
| Taieri | 995 | 3 | 2 | 4 | 84 | 105 | 126 | 25 | 57 | |
| Manawatu | 996 | 1 | 2 | 29 | 84 | 48 | 131 | 25 | 57 | |
| Taieri | 996 | 3 | 2 | 29 | 84 | 48 | 131 | 25 | 57 | |
| Manawatu | 1030 | 1 | 37 | 4 | 4 | 48 | 13 | 25 | 57 | |
| Manawatu | 1243 | 1 | 81 | 155 | 30 | 163 | 231 | 43 | 93 | |
| Ashburton | 1256 | 1 | 10 | 8 | 34 | 6 | 39 | 88 | 3 | |
| Ashburton | 1457 | 3 | 2 | 165 | 73 | 147 | 220 | 190 | 104 | |
| Ashburton | 2347 | 1 | 2 | 4 | 4 | 105 | 10 | 25 | 57 | |
| Manawatu | 2347 | 1 | 2 | 4 | 4 | 105 | 10 | 25 | 57 | |
| Ashburton | 2351 | 1 | 10 | 31 | 63 | 129 | 101 | 45 | 49 | |
| Taieri | 2351 | 3 | 10 | 31 | 63 | 129 | 101 | 45 | 49 | |
| Ashburton | 2352 | 1 | 2 | 29 | 4 | 105 | 131 | 24 | 17 | |
| Ashburton | 2353 | 1 | 10 | 31 | 34 | 47 | 57 | 45 | 1 | |
| Manawatu | 2354 | 1 | 37 | 4 | 4 | 48 | 13 | 25 | 23 | |
| Taieri | 2354 | 1 | 37 | 4 | 4 | 48 | 13 | 25 | 23 | |
| Ashburton | 2385 | 1 | 175 | 251 | 4 | 105 | 10 | 25 | 102 | |
| Manawatu | 3222 | 1 | 33 | 283 | 44 | 82 | 189 | 44 | 17 | |
| Manawatu | 3538 | 2 | 47 | 2 | 4 | 2 | 6 | 5 | 17 | |
| Ashburton | 3642 | 1 | 10 | 31 | 63 | 129 | 101 | 9 | 49 | |
| Taieri | 3642 | 1 | 10 | 31 | 63 | 129 | 101 | 9 | 49 | |
| Taieri | 3643 | 1 | 27 | 22 | 31 | 18 | 178 | 86 | 16 | |
| Taieri | 3670 | 1 | 2 | 33 | 61 | 104 | 134 | 86 | 51 | |
| Taieri | 3671 | 1 | 130 | 155 | 34 | 113 | 39 | 88 | 31 | |
| Manawatu | 3672 | 1 | 236 | 306 | 254 | 339 | 433 | 349 | 255 | |
| Manawatu | 3675 | 2 | 237 | 2 | 254 | 340 | 435 | 349 | 256 | |
| Taieri | 3678 | 3 | 2 | 59 | 4 | 48 | 131 | 352 | 57 | |
| Manawatu | 3803 | 1 | 27 | 8 | 34 | 6 | 39 | 88 | 3 | |
| Manawatu | 3844 | 1 | 33 | 283 | 44 | 82 | 189 | 44 | 1 | |
| Total | 244 | |||||||||
Sequence types shown in bold are new, and those in italics are unique to New Zealand.
The new aspA allele has a deletion, so it was not submitted to the MLST database.
FIG. 1.
Minimum spanning tree of New Zealand water isolates based on sequence types. Clusters are identified by the clonal complex number associated with the sequence types in the cluster.
All of the major clonal complexes (eight isolates or more), except for CC ST-2381 and CC ST-3640, were identified in all three rivers. The ST-2381 complex was the dominant complex of the Ashburton (25% of the Ashburton isolates) and Manawatu (30% of the Manawatu isolates) rivers. The major complex in the Taieri River was CC ST-1275 (19% of the Taieri isolates). No member of CC ST-2381 was found in the Taieri River, and no member of CC ST-3640 was found in the Ashburton River.
Six of the seven alleles in ST-2381 were new, and their sequences were distinct from those of the rest of the C. jejuni alleles in the Campylobacter PubMLST database, differing by up to 10 bases from their nearest allele. A neighbor-joining tree of the individual loci clearly identified them as C. jejuni alleles by comparison to other C. jejuni and C. coli allele sequences (Fig. 2). The ST-2381 complex is distinct from other water sequence types (Fig. 1 and 3) and from all other clonal complexes in the Campylobacter PubMLST database. Of the 19 alleles identified in the complex, only aspA1, aspA175, gltA216, and uncA102 have been reported outside New Zealand.
FIG. 2.
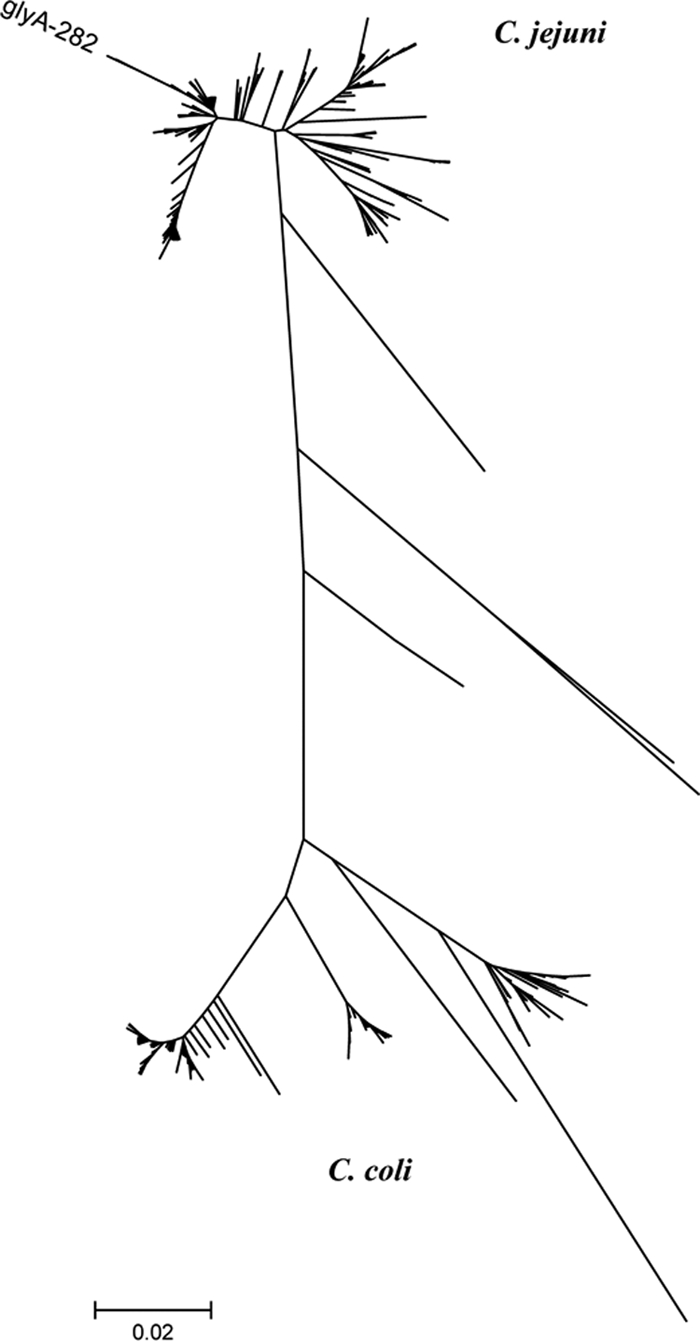
Neighbor-joining tree of the 355 glyA alleles from the Campylobacter PubMLST database (accessed March 2009). The glyA282 allele from the ST-2381 complex is shown.
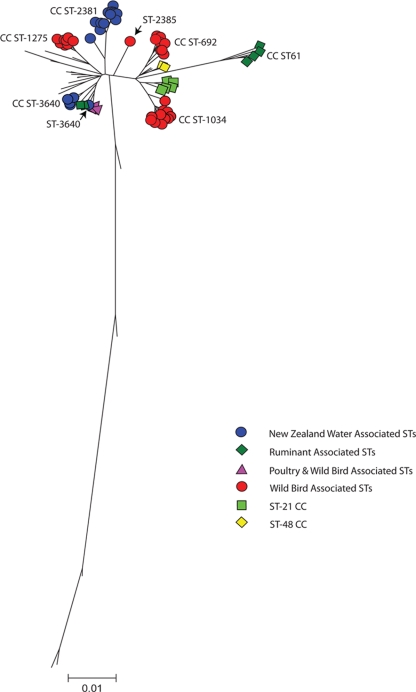
FIG. 3.
A minimum evolution tree was constructed for the concatenated New Zealand river water sequence types (3,309 bp) by use of the Kimura two-parameter method (with 1,000 bootstrap replicates). The tree was drawn to scale, and the units are numbers of base substitutions per site. The sequence type marked “New” in Table 1 was omitted from the analysis. Sequence types from clonal complexes associated with particular environmental niches are shown. The river water-associated sequence types are represented by blue circles, wild bird-associated sequence types are shown with red circles, ruminant-associated sequence types are shown with dark green squares, and poultry/wild bird-associated sequence types are shown with purple triangles.
The ST-2381 and ST-3640 complexes are related, sharing up to three alleles (aspA175, gltA216, and glyA282) (Fig. 1). The majority of the alleles in CC ST-3640 have been identified in isolates from a range of sources worldwide. Based on the minimum evolution tree (Fig. 3), the ST-3640 complex is related to CC ST-42 and CC ST-45.
DISCUSSION
The contamination of drinking water by Campylobacter has resulted in a number of well-characterized outbreaks, highlighting the potential of any water source to act as a transmission route for the spread of Campylobacter infection (3, 25, 36). The recreational use of water has been documented as a risk factor (24, 50), and within New Zealand, it has been estimated to cause 5% of Campylobacter infections (14, 45). The presence and level of contamination of water by Campylobacter have been measured in different water systems, and it is evident that rivers, waterways, and beaches are significantly contaminated and that the levels vary depending on the season and the sampling site (2, 22, 37, 46). Most studies have shown that rivers and waterways have peaks of contamination in spring and early autumn, with reduced numbers during the peak sunshine months. A similar pattern has been identified for rivers in New Zealand, although most human cases of campylobacteriosis are reported during summer.
The source of Campylobacter in rivers and its association with human disease have been investigated using a number of typing methods, including Penner serotyping and PFGE (2, 12, 20). Indistinguishable macrorestriction profiles were identified for water and human samples. Similarly, identical Penner serotypes were found in both sets of isolates. Combining both typing methods suggested that the risk of infection from river water was low, although the same subtypes were identified in both water and human samples (8, 12, 20). The presence of the same subtypes in both sets of isolates could result from human infection from river water or from contamination of river water with human fecal material, and if the latter is true, one need not infer a public health risk from contaminated rivers. More recently, MLST has been used to analyze campylobacter populations in river water (28, 39). The majority of water isolates identified in this study have sequence types not previously associated with isolates from human infections. This finding is in agreement with other studies that have identified novel sequence types associated with environmental water samples (15, 26, 28, 39).
A range of clonal complexes were identified in the river water samples. The major complexes, apart from the two novel ones discussed below, were CC ST-1275 and CC ST-45. The ST-1275 complex was identified only in environmental water samples in Canada and was not found in contemporaneous samples from human infections (28). In the Campylobacter PubMLST database, it is also associated with a range of different wild birds. There have been very few reports of human infections with members of this complex, and it is likely that the major source of CC ST-1275 in New Zealand river water is wild bird feces.
ST-45, the founder sequence type of CC ST-45, was the major type identified in samples taken from river water in northwest England (39). It was also the most prevalent type identified during the early summer peak of campylobacteriosis cases in the same region, especially among rural dwellers and children under 5 years of age. Other studies have also identified ST-45 as a major environmental sequence type (15). It was postulated that the high incidence of ST-45 in river water isolates and in human infections is due to its being an environmentally well-adapted type which can survive under stress better than other sequence types. In this study, ST-45 was the most prominent human-associated sequence type in the river water isolates, although it is not the most prominent type associated with human infection. One sequence type, ST-474, is particularly prevalent among New Zealand human infections and caused over 30% of human infections between 2006 and 2008 (14, 31). It was isolated on only two occasions from the Manawatu River. ST-474 is a member of CC ST-48, for which only four isolates were identified. Similarly, other clonal complexes associated with human infection, such as CC ST-21 and CC ST-42, were not well represented among the water isolates, suggesting that sewerage is not the major source of ST-45 isolates found in rivers, although selection due to increased survival of ST-45 isolates cannot be disregarded. In a previous study, we identified ST-45 as the most common sequence type in the feces of wild birds in New Zealand (16), and it is likely that bird fecal contamination is a significant source of ST-45 in river water.
All three rivers run through areas associated with significant farming activity, and runoff from the land is drained into the rivers. Clonal complexes associated with cattle and sheep, CC ST-61 and CC ST-42 (27), were identified among the water isolates, but not in significant numbers. This was surprising, although similar results were found in the study from northwest England (39). It is possible that adaptation to the ruminant gut has reduced the ability of some types to survive outside their environmental niche.
A surprising result of this study was the identification of two novel complexes, each associated with two of the three rivers tested over different periods. The ST-2381 complex consists of 11 STs, none of which have been identified elsewhere. It was present in the Ashburton (South Island) and Manawatu (North Island) rivers over a period of 7 years, suggesting that the source of the complex is widespread and endemic. Despite its being the most significant complex present in the two rivers, it has not been isolated from any of the commonly reared livestock (cattle and sheep) or poultry in New Zealand. The lack of evidence linking CC ST-2381 to any other niche-associated complex makes it difficult to ascertain the source. Deer are reared in New Zealand, but studies in Norway have shown that deer do not appear to carry significant numbers of C. jejuni organisms (29). Other possibilities include possums, rabbits, or native birds, although no C. jejuni isolates were identified in a New Zealand survey of possums and rabbits (8). C. jejuni isolates have been identified in rabbits in the United Kingdom, but the predominant sequence type was ST-45 (26).
The ST-3640 complex was present in both the Manawatu and Taieri rivers over different periods. Like CC ST-2381, CC ST-3640 is widespread and endemic in New Zealand, and it is unique. One member of the complex has been identified in the United States, isolated from a C. coli chicken meat isolate (Campylobacter PubMLST database [accessed 5 April 2009]). The ST-3640 complex appears to be related to CC ST-42 (Fig. 3), a complex associated with ruminant animals, suggesting a link to cattle and sheep, but no evidence for this has been found. It also appears related to CC ST-45, a complex associated with a range of environmental niches. More interestingly, CC ST-3640 shares a number of rare alleles with CC ST-2381, namely, aspA175, gltA216, and glyA282. The glyA282 allele has not been identified outside New Zealand and has only been found in river water isolates. This suggests that at some stage the two complexes shared a common host, allowing the interchange of alleles. It is likely that the two complexes identified in river water represent host-specific complexes, such as those identified in geese and starlings (4, 6). The large numbers of strains isolated from the different river systems over extended periods also indicate that this may be an environmentally well-adapted complex, as proposed for ST-45 (39).
Despite the presence of Campylobacter in river water and the low infective dose (44), the risk associated with water has been shown to be low (50). This is most likely due to the treatment of drinking water extracted from rivers and the small numbers of potentially pathogenic Campylobacter bacteria present in river water (30). The majority of the isolates identified in river water samples could be linked to wild birds. The clonal complexes CC ST-45, CC ST-177, CC ST-677, CC ST-692, CC ST-1034, and CC ST-1275 (4, 16, 24, 26) have all been identified in isolates from wild birds. Among the isolates not assigned to a clonal complex, seven were identified in wild birds (4), and evidence from microarray studies has linked ST-2381 to avian sequence types (N. French, unpublished results). This suggests that upwards of 60% of isolates found in river water could conceivably be derived from wild birds.
The diversity in the C. jejuni isolates obtained from rivers in New Zealand mirrors the diversity that has been found in environmental samples elsewhere in the world (15, 26, 28). Common sequence types and clonal complexes, such as CC ST-21, CC ST-45, CC ST-48, and CC ST-61, make up approximately half of the isolates, with the rest being rare or new. The new sequence types appear to be local, with few subsequently being identified elsewhere in the world. This pattern of distribution of environmental samples has been seen in Canada (28), the United Kingdom (15, 26), and New Zealand. It reflects the small number of studies that have been performed so far but could also be explained by host-specific sequence types, with the host being a native animal. Our study is the first report of novel complexes specifically associated with water isolates, and we believe that it represents the presence of unique campylobacter populations in New Zealand native fauna. This may be associated with the geographical isolation of New Zealand allowing local campylobacter populations to diverge.
Acknowledgments
We thank Marilyn Grubner and Chris Pope for technical help, members of the ESR Enteric Reference Laboratory for their assistance, Brent Gilpin for help with the PulseNet Aotearoa New Zealand Campylobacter Database, and all contributors to the database. We also thank Ronan O'Toole, School of Biological Sciences Victoria, University of Wellington, for the cosupervision of Sharla McTavish in her master's dissertation.
S.M. is a recipient of an ESR Masters Scholarship. This publication made use of the Campylobacter jejuni Multi Locus Sequence Typing website (http://pubmlst.org/campylobacter/) developed by Keith Jolley and Man-Suen Chan and sited at the University of Oxford (21). The development of this site was funded by the Wellcome Trust. This work was partly funded by the New Zealand Food Safety Authority and the Royal Society of New Zealand Marsden Fund.
Footnotes
Published ahead of print on 31 July 2009.
REFERENCES
- 1.Baker, M., N. Wilson, and R. Edwards. 2007. Campylobacter infection and chicken: an update on New Zealand's largest ‘common source outbreak.’ N. Z. Med. J. 120:U2717. [PubMed] [Google Scholar]
- 2.Bolton, F. J., D. Coates, D. N. Hutchinson, and A. F. Godfree. 1987. A study of thermophilic campylobacters in a river system. J. Appl. Bacteriol. 62:167-176. [DOI] [PubMed] [Google Scholar]
- 3.Clark, C. G., L. Price, R. Ahmed, D. L. Woodward, P. L. Melito, F. G. Rodgers, F. Jamieson, B. Ciebin, A. Li, and A. Ellis. 2003. Characterization of waterborne outbreak-associated Campylobacter jejuni, Walkerton, Ontario. Emerg. Infect. Dis. 9:1232-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colles, F. M., K. E. Dingle, A. J. Cody, and M. C. Maiden. 2008. Comparison of Campylobacter populations in wild geese with those in starlings and free-range poultry on the same farm. Appl. Environ. Microbiol. 74:3583-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colles, F. M., K. Jones, R. M. Harding, and M. C. Maiden. 2003. Genetic diversity of Campylobacter jejuni isolates from farm animals and the farm environment. Appl. Environ. Microbiol. 69:7409-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colles, F. M., N. D. McCarthy, J. C. Howe, C. L. Devereux, A. G. Gosler, and M. C. Maiden. 2009. Dynamics of Campylobacter colonization of a natural host, Sturnus vulgaris (European starling). Environ. Microbiol. 11:258-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denis, M., J. Refregier-Petton, M. J. Laisney, G. Ermel, and G. Salvat. 2001. Campylobacter contamination in French chicken production from farm to consumers. Use of a PCR assay for detection and identification of Campylobacter jejuni and Camp. coli. J. Appl. Microbiol. 91:255-267. [DOI] [PubMed] [Google Scholar]
- 8.Devane, M. L., C. Nicol, A. Ball, J. D. Klena, P. Scholes, J. A. Hudson, M. G. Baker, B. J. Gilpin, N. Garrett, and M. G. Savill. 2005. The occurrence of Campylobacter subtypes in environmental reservoirs and potential transmission routes. J. Appl. Microbiol. 98:980-990. [DOI] [PubMed] [Google Scholar]
- 9.Dingle, K. E., F. M. Colles, D. R. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. Willems, R. Urwin, and M. C. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberhart-Phillip, J., N. Walker, N. Garrett, D. Bell, D. Sinclair, W. Rainger, and M. Bates. 1997. Campylobacteriosis in New Zealand: results of a case control study. J. Epidemiol. Community Health 51:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eyles, R., D. Niyogi, C. Townsend, G. Benwell, and P. Weinstein. 2003. Spatial and temporal patterns of Campylobacter contamination underlying public health risk in the Taieri River, New Zealand. J. Environ. Qual. 32:1820-1828. [DOI] [PubMed] [Google Scholar]
- 12.Eyles, R. F., H. J. Brooks, C. R. Townsend, G. A. Burtenshaw, N. C. Heng, R. W. Jack, and P. Weinstein. 2006. Comparison of Campylobacter jejuni PFGE and Penner subtypes in human infections and in water samples from the Taieri River catchment of New Zealand. J. Appl. Microbiol. 101:18-25. [DOI] [PubMed] [Google Scholar]
- 13.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.French, N. 2008. Enhancing surveillance of potentially foodborne enteric diseases in New Zealand: human campylobacteriosis in the Manawatu. New Zealand Food Safety Authority, Wellington, New Zealand. http://www.nzfsa.govt.nz/science/research-projects/Campy_Attribution_Manawatu.pdf.
- 15.French, N., M. Barrigas, P. Brown, P. Ribiero, N. Williams, H. Leatherbarrow, R. Birtles, E. Bolton, P. Fearnhead, and A. Fox. 2005. Spatial epidemiology and natural population structure of Campylobacter jejuni colonizing a farmland ecosystem. Environ. Microbiol. 7:1116-1126. [DOI] [PubMed] [Google Scholar]
- 16.French, N. P., A. Midwinter, B. Holland, J. Collins-Emerson, R. Pattison, F. Colles, and P. Carter. 2009. Molecular epidemiology of Campylobacter jejuni isolates from wild-bird fecal material in children's playgrounds. Appl. Environ. Microbiol. 75:779-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman, C. R., J. Neimann, H. C. Wegener, and R. V. Tauxe. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations, p. 121-138. In I. Nachamkin and M. J. Blase (ed.), Campylobacter. American Society for Microbiology, Washington, DC.
- 18.Gilpin, B. J., B. Thorrold, P. Scholes, R. D. Longhurst, M. Devane, C. Nicol, S. Walker, B. Robson, and M. Savill. 2008. Comparison of Campylobacter jejuni genotypes from dairy cattle and human sources from the Matamata-Piako District of New Zealand. J. Appl. Microbiol. 105:1354-1360. [DOI] [PubMed] [Google Scholar]
- 19.Hannon, S. J., E. N. Taboada, M. L. Russell, B. Allan, C. Waldner, H. L. Wilson, A. Potter, L. Babiuk, and H. G. Townsend. 2009. Genomics-based molecular epidemiology of Campylobacter jejuni isolates from feedlot cattle and from people in Alberta, Canada. J. Clin. Microbiol. 47:410-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hudson, J. A., C. Nicol, J. Wright, R. Whyte, and S. K. Hasell. 1999. Seasonal variation of Campylobacter types from human cases, veterinary cases, raw chicken, milk and water. J. Appl. Microbiol. 87:115-124. [DOI] [PubMed] [Google Scholar]
- 21.Jolley, K. A., M. S. Chan, and M. C. Maiden. 2004. mlstdbNet-distributed multi-locus sequence typing (MLST) databases. BMC Bioinform. 5:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, K. 2001. Campylobacters in water, sewage and the environment. J. Appl. Microbiol. 90:68S-79S. [DOI] [PubMed] [Google Scholar]
- 23.Jones, K., M. Betaieb, and D. R. Telford. 1990. Thermophilic campylobacters in surface waters around Lancaster, UK: negative correlation with Campylobacter infections in the community. J. Appl. Bacteriol. 69:758-764. [DOI] [PubMed] [Google Scholar]
- 24.Karenlampi, R., H. Rautelin, D. Schonberg-Norio, L. Paulin, and M. L. Hanninen. 2007. Longitudinal study of Finnish Campylobacter jejuni and C. coli isolates from humans, using multilocus sequence typing, including comparison with epidemiological data and isolates from poultry and cattle. Appl. Environ. Microbiol. 73:148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuusi, M., J. P. Nuorti, M. L. Hanninen, M. Koskela, V. Jussila, E. Kela, I. Miettinen, and P. Ruutu. 2005. A large outbreak of campylobacteriosis associated with a municipal water supply in Finland. Epidemiol. Infect. 133:593-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwan, P. S., M. Barrigas, F. J. Bolton, N. P. French, P. Gowland, R. Kemp, H. Leatherbarrow, M. Upton, and A. J. Fox. 2008. Molecular epidemiology of Campylobacter jejuni populations in dairy cattle, wildlife, and the environment in a farmland area. Appl. Environ. Microbiol. 74:5130-5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwan, P. S., A. Birtles, F. J. Bolton, N. P. French, S. E. Robinson, L. S. Newbold, M. Upton, and A. J. Fox. 2008. Longitudinal study of the molecular epidemiology of Campylobacter jejuni in cattle on dairy farms. Appl. Environ. Microbiol. 74:3626-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levesque, S., E. Frost, R. D. Arbeit, and S. Michaud. 2008. Multilocus sequence typing of Campylobacter jejuni isolates from humans, chickens, raw milk, and environmental water in Quebec, Canada. J. Clin. Microbiol. 46:3404-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lillehaug, A., B. Bergsjo, J. Schau, T. Bruheim, T. Vikoren, and K. Handeland. 2005. Campylobacter spp., Salmonella spp., verocytotoxic Escherichia coli, and antibiotic resistance in indicator organisms in wild cervids. Acta Vet. Scand. 46:23-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mawer, S. L. 1988. The pathogenicity of environmental campylobacters—a human volunteer experiment. Epidemiol. Infect. 101:295-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McTavish, S. M., C. E. Pope, C. Nicol, K. Sexton, N. French, and P. E. Carter. 2008. Wide geographical distribution of internationally rare Campylobacter clones within New Zealand. Epidemiol. Infect. 136:1244-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mishu, B., and M. J. Blaser. 1993. Role of infection due to Campylobacter jejuni in the initiation of Guillain-Barre syndrome. Clin. Infect. Dis. 17:104-108. [DOI] [PubMed] [Google Scholar]
- 33.Nachamkin, I., B. M. Allos, and T. Ho. 1998. Campylobacter species and Guillain-Barre syndrome. Clin. Microbiol. Rev. 11:555-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obiri-Danso, K., and K. Jones. 1999. Distribution and seasonality of microbial indicators and thermophilic campylobacters in two freshwater bathing sites on the River Lune in northwest England. J. Appl. Microbiol. 87:822-832. [DOI] [PubMed] [Google Scholar]
- 35.Obiri-Danso, K., and K. Jones. 1999. The effect of a new sewage treatment plant on faecal indicator numbers, campylobacters and bathing water compliance in Morecambe Bay. J. Appl. Microbiol. 86:603-614. [DOI] [PubMed] [Google Scholar]
- 36.Richardson, G., D. R. Thomas, R. M. Smith, L. Nehaul, C. D. Ribeiro, A. G. Brown, and R. L. Salmon. 2007. A community outbreak of Campylobacter jejuni infection from a chlorinated public water supply. Epidemiol. Infect. 135:1151-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Savill, M. G., J. A. Hudson, A. Ball, J. D. Klena, P. Scholes, R. J. Whyte, R. E. McCormick, and D. Jankovic. 2001. Enumeration of Campylobacter in New Zealand recreational and drinking waters. J. Appl. Microbiol. 91:38-46. [DOI] [PubMed] [Google Scholar]
- 38.Sheppard, S. K., J. F. Dallas, M. Macrae, N. D. McCarthy, E. L. Sproston, F. J. Gormley, N. J. Strachan, I. D. Ogden, M. C. Maiden, and J. F. Ken. Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland 2005/6. Int. J. Food Microbiol., in press. doi: 10.1016/j.ijfoodmicro.2009.02.010. [DOI] [PMC free article] [PubMed]
- 39.Sopwith, W., A. Birtles, M. Matthews, A. Fox, S. Gee, M. Painter, M. Regan, Q. Syed, and E. Bolton. 2008. Identification of potential environmentally adapted Campylobacter jejuni strains, United Kingdom. Emerg. Infect. Dis. 14:1769-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spratt, B. G., W. P. Hanage, B. C. Li, D. M. Aanensen, and E. J. Feil. 2004. Displaying the relatedness among isolates of bacterial species—the eBURST approach. FEMS Microbiol. Lett. 241:129-134. [DOI] [PubMed] [Google Scholar]
- 41.Stanley, K. N., J. S. Wallace, J. E. Currie, P. J. Diggle, and K. Jones. 1998. The seasonal variation of thermophilic campylobacters in beef cattle, dairy cattle and calves. J. Appl. Microbiol. 85:472-480. [DOI] [PubMed] [Google Scholar]
- 42.Stucki, U., J. Frey, J. Nicolet, and A. P. Burnens. 1995. Identification of Campylobacter jejuni on the basis of a species-specific gene that encodes a membrane protein. J. Clin. Microbiol. 33:855-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 44.Teunis, P., W. Van den Brandhof, M. Nauta, J. Wagenaar, H. Van den Kerkhof, and W. Van Pelt. 2005. A reconsideration of the Campylobacter dose-response relation. Epidemiol. Infect. 133:583-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Till, D., G. McBride, A. Ball, K. Taylor, and E. Pyle. 2008. Large-scale freshwater microbiological study: rationale, results and risks. J. Water Health 6:443-460. [DOI] [PubMed] [Google Scholar]
- 46.Vereen, E., Jr., R. R. Lowrance, D. J. Cole, and E. K. Lipp. 2007. Distribution and ecology of campylobacters in coastal plain streams (Georgia, United States of America). Appl. Environ. Microbiol. 73:1395-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waldenstrom, J., T. Broman, I. Carlsson, D. Hasselquist, R. P. Achterberg, J. A. Wagenaar, and B. Olsen. 2002. Prevalence of Campylobacter jejuni, Campylobacter lari, and Campylobacter coli in different ecological guilds and taxa of migrating birds. Appl. Environ. Microbiol. 68:5911-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wallace, J. S., K. N. Stanley, J. E. Currie, P. J. Diggle, and K. Jones. 1997. Seasonality of thermophilic Campylobacter populations in chickens. J. Appl. Microbiol. 82:219-224. [PubMed] [Google Scholar]
- 49.Wang, G., C. G. Clark, T. M. Taylor, C. Pucknell, C. Barton, L. Price, D. L. Woodward, and F. G. Rodgers. 2002. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J. Clin. Microbiol. 40:4744-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson, D. J., E. Gabriel, A. J. Leatherbarrow, J. Cheesbrough, S. Gee, E. Bolton, A. Fox, P. Fearnhead, C. A. Hart, and P. J. Diggle. 2008. Tracing the source of campylobacteriosis. PLoS Genet. 4:e1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wingstrand, A., J. Neimann, J. Engberg, E. M. Nielsen, P. Gerner-Smidt, H. C. Wegener, and K. Molback. 2006. Fresh chicken as main risk factor for campylobacteriosis, Denmark. Emerg. Infect. Dis. 12:280-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong, T. L., L. Hollis, A. Cornelius, C. Nicol, R. Cook, and J. A. Hudson. 2007. Prevalence, numbers, and subtypes of Campylobacter jejuni and Campylobacter coli in uncooked retail meat samples. J. Food Prot. 70:566-573. [DOI] [PubMed] [Google Scholar]