Abstract
MicroRNAs (miRNAs) are expressed in a wide variety of organisms, ranging from plants to animals, and are key posttranscriptional regulators of gene expression. Virally encoded miRNAs are unique in that they could potentially target both viral and host genes. Indeed, we have previously demonstrated that a human cytomegalovirus (HCMV)-encoded miRNA, miR-UL112, downregulates the expression of a host immune gene, MICB. Remarkably, it was shown that the same miRNA also downregulates immediate-early viral genes and that its ectopic expression resulted in reduced viral replication and viral titers. The targets for most of the viral miRNAs, and hence their functions, are still unknown. Here we demonstrate that miR-UL112 also targets the UL114 gene, and we present evidence that the reduction of UL114 by miR-UL112 reduces its activity as uracil DNA glycosylase but only minimally affects virus growth. In addition, we show that two additional HCMV-encoded miRNAs, miR-US25-1 and miR-US25-2, reduce the viral replication and DNA synthesis not only of HCMV but also of other viruses, suggesting that these two miRNAs target cellular genes that are essential for virus growth. Thus, we suggest that in addition to miR-UL112, two additional HCMV miRNAs control the life cycle of the virus.
MicroRNAs (miRNAs) are an abundant class of small noncoding RNA molecules that target mRNAs, generally within their 3′ untranslated regions (3′ UTR). miRNAs suppress gene expression, mainly through inhibition of translation or, rarely, through mRNA degradation (2, 11). miRNAs are abundant among various multicellular organisms, and remarkably, several DNA viruses of the herpesvirus family also express miRNAs (12). Herpesviruses belong to a large family of enveloped, double-stranded DNA viruses that are able to maintain a persistent or latent infection during the lifetime of the virus in its host. They are divided into three groups (alpha-, beta-, and gammaherpesviruses). Members of all three groups have been shown to encode miRNAs, indicating that herpesviruses have utilized the RNA interference machinery throughout their evolution (15).
Thus far, cytomegalovirus (CMV) is the only betaherpesvirus found to express miRNAs. Human CMV (HCMV) miRNAs are unique among human herpesviruses, because unlike alpha- and gammaherpesviruses, in which the miRNA genes are clustered within defined genomic regions and are expressed during latent infection, HCMV miRNAs are spread throughout the viral genome and have been demonstrated to be expressed during acute lytic infection (5, 8, 10, 20). In this regard, 3 of the 11 HCMV miRNAs are transcribed from the complementary strand of known open reading frames, 7 miRNAs are located in intergenic regions, and 1 is located within an intron. Whether HCMV miRNAs are also expressed during latency is still an open question, which, at present, is difficult to tackle due to the lack of an appropriate in vitro system.
Viral miRNAs may directly regulate viral genes or, alternatively, they could target host genes. Interestingly, of the 11 HCMV-encoded miRNAs that have been discovered, the function of only 1 miRNA, miR-UL112, has been validated experimentally. Even more remarkable are the observations that this particular viral miRNA is capable of regulating both cellular and viral transcripts (16, 19, 28). We showed that miR-UL112 specifically downregulates a cellular immune gene, MICB, during viral infection in order to escape immune recognition and destruction (28). Since the expression of MICB protein is also inhibited by a viral protein, UL16, a dual mechanism is operating in HCMV in which both a viral miRNA (miR-UL112) and a viral protein (HCMV UL16 [6]) target the host MICB protein.
Remarkably, two additional studies demonstrated that several of the HCMV immediate-early (IE) genes (including the major IE gene, IE72) are also regulated by miR-UL112 (16, 19). Since miR-UL112 is expressed early after infection and accumulates during viral infection (14), it has been suggested that miR-UL112 might inhibit IE72 expression during the late stages of viral replication to promote the transition from productive replication to latent infection. In agreement with this hypothesis, ectopic expression of miR-UL112 early during infection resulted in reduced expression of IE proteins (direct and indirect target genes) and also led to a decrease in viral DNA levels. These results, together with computational data (19) and findings of additional viral targets for other herpesvirus miRNAs (29), led to the hypothesis that virally encoded miRNAs, in general, might inhibit viral replication to establish and maintain latency.
Here we initially show that all HCMV miRNAs identified are expressed by low-passage-number HCMV clinical isolates. We identified an additional target for miR-UL112: the viral uracil DNA glycosylase UL114, which is encoded on the strand antisense to miR-UL112, and we demonstrate that the reduction in UL114 protein levels by miR-UL112 reduces the ability of the virus to properly excise uracil residues from viral DNA. Finally, we show that ectopic expression of two additional HCMV miRNAs, miR-US25-1 and miR-US25-2, resulted in significant reductions in viral DNA synthesis and viral yield.
MATERIALS AND METHODS
Lentiviral constructs and transduction.
To express HCMV miRNAs, we used the pTER vector (30) to generate artificial short hairpin RNAs that function as orthologs of pre-miRNA hairpins. Specific oligonucleotides (see the supplemental material) were annealed and ligated into the pTER vector. When two miRNAs are expressed from one precursor, we expressed the 5′ arm only. The artificial hairpins were then excised from the vector together with the H1 promoter and were ligated into the lentiviral vector SIN18-pRLL-hEFIαp-EGFP-WRPE (32) as previously described (28). For testing the effect of miR-UL112 on the UL114-GFP protein, the green fluorescent protein (GFP) was excised from lentiviral vectors containing miR-UL112 or an empty vector by using the BamHI and SalI sites.
Lentiviruses were produced by transient three-plasmid transfection as described previously (17). The pMD.G envelope expression cassette (3.5 μg), the Gag-Pol packaging construct (6.5 μg), and the relevant vector construct (10 μg) were transfected using the FuGene 6 reagent (Roche). Forty-eight hours after transfection, the supernatants containing the viruses were collected and filtered. These viruses were used to transduce human foreskin fibroblasts (HFF) or 293T cells in the presence of Polybrene (5 μg/ml).
Real-time PCR analysis of viral miRNAs.
Quantitative real-time PCR (qPCR) was performed on mature miRNAs as described previously (26, 28). Total RNA was isolated using Tri Reagent (Sigma). Total RNA was polyadenylated by poly(A) polymerase (Ambion). RNA was then reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Invitrogen) and 0.5 μg poly(T) adapter. The reaction primers (see the supplemental material) were a 3′ adapter primer and primers corresponding to the miRNA sequences. Real-time PCR was performed by using 2× DyNAmo SYBR green qPCR (Finnzymes) on the ABI Prism 7500 real-time PCR system (Applied Biosciences).
Cells and viruses.
HFF were used to propagate HCMV strain AD169 (American Type Culture Collection), the UL114 deletion mutant (ΔUL114 [22]), the IE72 deletion Towne mutant (ΔIE72; CR208) and its revertant recombinant (CRQ208 [13]), and the recombinant UL114-tag virus (23). Infection with the various virus strains was carried out as described previously (31) at a multiplicity of infection (MOI) of 0.5 to 1. Samples of infected-cell supernatants were removed at designated time points and stored at −80°C before titration by a plaque assay on HFF. The human influenza virus A/Puerto Rico/8/34 H1N1 (abbreviated as A/PR8) was generated, and cells were infected, as described previously (1).
Quantification of viral DNA.
For the quantification of HCMV, adenovirus, and herpes simplex virus type 1 (HSV-1) DNA, total cellular DNA was extracted at specified time points from virus-infected and mock-infected cell monolayers by use of the QIAamp DNA minikit (Qiagen, Hilden, Germany). DNA was then subjected to a real-time PCR assay using specific primers and probes (4).
Antibodies, FACS staining, and Western blotting.
The following monoclonal antibodies (MAbs) were used for fluorescence-activated cell sorter (FACS) staining: anti-MICA, anti-MICB, anti-MICA/B, anti ULBP1, anti ULBP2, and anti ULBP3 (all from R&D), antibody W6/32 against major histocompatibility complex (MHC) class I, anti-interferon receptor (R&D), anti-B2M (clone BBM1), anti-PVR (R&D), and the anti-influenza virus type A (H1) antibody H17-L2 (a kind gift from Jonathan W. Yewdell).
For FACS staining, cells (50,000/well) were diluted in 1× phosphate-buffered saline (PBS)-0.5% bovine serum albumin-0.05% azide and incubated on ice with the appropriate antibody (0.2 μg/well) for 0.5 h. Cells were then centrifuged and washed with FACS medium, followed by incubation with fluorochrome-conjugated secondary antibodies for 30 min on ice in the dark. After further washing, labeled cells were analyzed by FACS using CellQuest software.
For propidium iodide (PI) staining, cells were harvested, washed with PBS, and fixed with 70% ethanol. Then cells were washed, resuspended with a staining solution containing 2.5 μg/ml PI and 0.5 mg/ml RNase A in PBS, and analyzed with the FACS.
For Western blot analysis, HFF were infected at an MOI of 1. Cells were collected at 72 h postinfection, washed three times with PBS plus phosphatase inhibitors, and counted. Cells were lysed with 0.5 ml lysis buffer (0.5% NP-40, 150 mM NaCl, 50 mM Tris, 5 mM EDTA, and protease inhibitors), separated by 10% acrylamide gel electrophoresis, and transferred to a nitrocellulose membrane. Mouse MAbs anti-pp65, anti-IE72 (Virusys Corp., Sykesville, MD), anti-ICP4 (Goodwin Institute for Cancer Research, Plantation, FL), and anti-HSP90 (Sigma) were used for immunoblotting.
DNA constructs.
The UL114 gene was amplified using primers 5′-GGA CTC AGA TCT ATG GCC CTC AAG CAG TGG ATG-3′ and 5′-GTC GAC TGC AGA GAA TCT CCC ACA GAG TCG CCA GTC C-3′. The amplified fragment was cloned into the unique BglII and PstI sites of pEGFPN3 (Clontech, Palo Alto, CA) to yield pEGFPN3/UL114.
Luciferase assay.
For the luciferase assay, we used the PsiCHECK vector (Promega). UL114 was amplified by PCR from viral genomic DNA and was inserted into the XhoI site immediately downstream of the Renilla stop codon. Cells were transfected with the various vectors using the LT1 transfection reagent (Mirus). Firefly and Renilla luciferase activities were measured consecutively using Dual-Luciferase assays (Promega) 48 h after transfection.
Computational predictions.
The online target prediction algorithm RNAhybrid (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/submission.html) was used to identify potential target sites in the 3′ UTR of immune system-related genes. We searched for binding sites in the 3′ UTRs of the following genes: MICA, MICB, ULBP1 to ULBP4, HLA-A, HLA-B, HLA-C, HLA-DM, HLA-DR, TAP1, TAP2, Tapasine, PVR, CD48, IFN3, IFN7, alpha interferon (IFN-α), and IFN-α receptor. The algorithm was run using default settings with the additional constraints of full Watson-Crick base pairing in nucleotides 2 to 7 and a free energy score of ≤20 kcal/mol.
RESULTS
HCMV miRNAs are expressed in clinically isolated viruses.
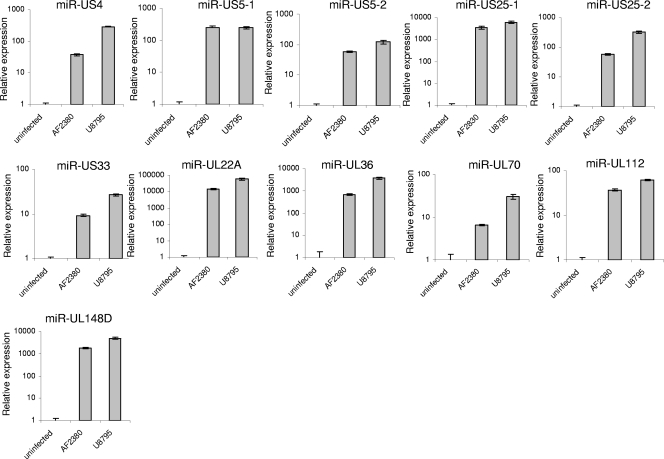
It was recently demonstrated that HCMV encodes 11 miRNAs; however, the functions of only 1 miRNA, miR-UL112, have been revealed (14, 16, 19, 28). Some of these miRNAs were identified in the AD169 (14) and Towne (10) laboratory virus strains, while the rest of the miRNAs were observed in strain VR1814, a virus that was originally isolated from cervical secretions and was extensively propagated in human embryonic lung fibroblasts (20). Thus, it is still not known whether all of these miRNAs are also expressed in clinical HCMV isolates recovered from patients. To address this question, we infected HFF with two low-passage-number clinical isolates, recovered from the amniotic fluid (AF2380) and urine (U8795) of congenitally infected infants, and extracted the total RNA from these cells. As can be seen in Fig. 1, expression of all HCMV miRNAs could be detected in HFF infected with the two low-passage-number clinical isolates, suggesting that the HCMV miRNAs play a role during authentic HCMV infection.
FIG. 1.
All HCMV miRNAs are expressed in clinical isolates. Total RNA was prepared from HFF infected with two low-passage-number clinical isolates recovered from the amniotic fluid (AF2380) and urine (U8795) of congenitally infected infants. The HCMV miRNAs were detected by real-time PCR analysis. MiR-16 was used as an internal standard.
Several immune surface proteins are not downregulated by HCMV-encoded miRNAs.
We have previously shown that a human HCMV miRNA, miR-UL112, specifically downregulates MICB expression during viral infection and that this downregulation results in reduced killing of infected cells by NK cells (28). Interestingly, the HCMV protein UL16 also targets MICB (among other stress-induced proteins) to prevent its surface expression (6, 9). Since HCMV has developed numerous protein-based mechanisms that interfere with the function of immune proteins (18), and since computer prediction of viral miRNA targets is still inefficient (12), we decided to test if any of the HCMV-encoded miRNAs downregulate additional immune system-related cellular genes. Our criteria for choosing the immune genes to be tested were that these genes should either be shown to be manipulated by the virus or be known to be important for viral detection by the immune system. In addition, we concentrated mainly on proteins that are expressed on the cell surface.
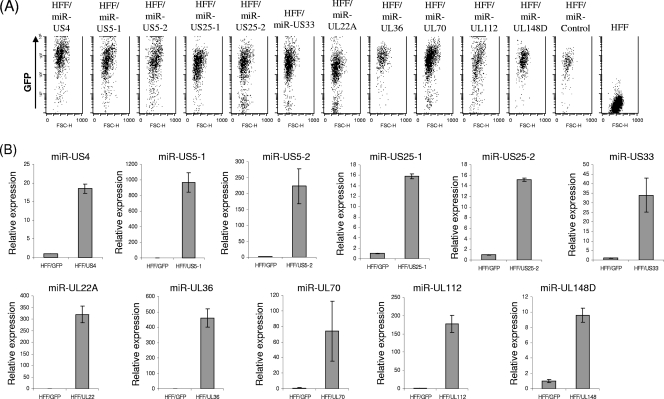
A lentiviral vector system was used to express each of the HCMV miRNAs in primary HFF. The vectors also encoded GFP to allow monitoring of the efficiency of infection (Fig. 2A), and the expression of the different HCMV miRNAs was confirmed by qPCR (Fig. 2B). Analysis of HFF transduced with the various HCMV miRNAs revealed that none of the immune system-related genes tested (MHC class I, MICA, ULBP2, ULBP3, ICAM1, IFN-α receptor, and PVR) were significantly downregulated by any of the HCMV miRNAs (see Fig. S1 in the supplemental material; also data not shown), even though full seed sites (nucleotides 2 to 7) were predicted for some of the targets (see Fig. S2 in the supplemental material). Because there are several important immune molecules that are not expressed on the surfaces of HFF but still might be targeted in vivo by HCMV, we have also used 721.221, an Epstein-Barr virus (EBV)-transformed B-cell line, and expressed in these cells the entire spectrum of HCMV miRNAs. Again, however, none of the immune system-related genes tested (MHC class II, ULBP1, and CD48) were affected by the expression of the viral miRNAs (data not shown).
FIG. 2.
Ectopic expression of HCMV miRNAs in HFF. (A) HFF were transduced with lentiviruses expressing GFP together with the indicated HCMV miRNA or control miRNA (miR-Control). GFP expression was monitored by FACS. (B) qPCR analysis of HCMV miRNA expression in the transduced HFF. MiR-16 was used as an internal standard.
The viral protein UL114 is targeted by miR-UL112.
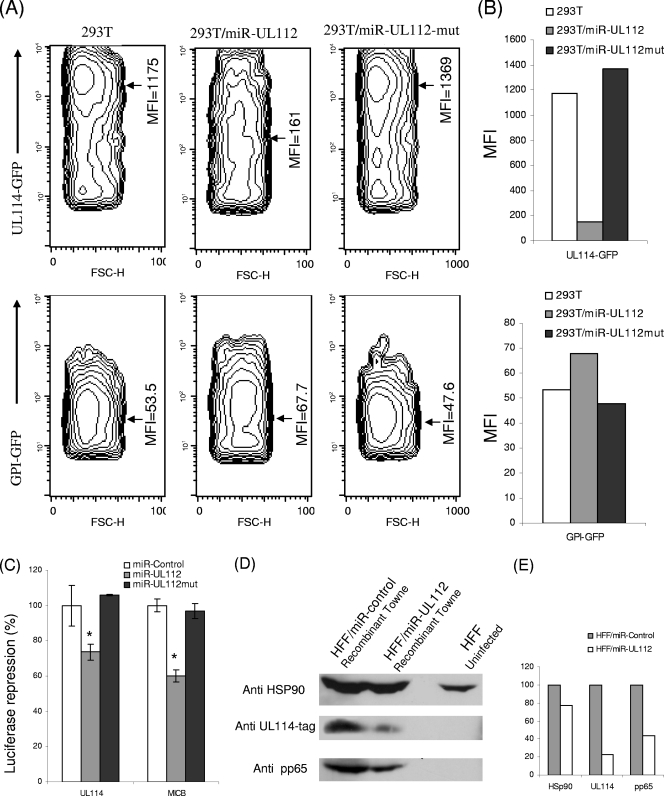
Since we could not detect an obvious repression of cellular immune targets by HCMV-encoded miRNAs, we next examined viral targets. It was previously demonstrated that miR-UL112 targets the IE72 viral protein (16, 19). Interestingly, miR-UL112 resides on a strand complementary to the gene encoding the UL114 protein. UL114 is a uracil DNA glycosylase that has been reported to be important for viral replication (22). To test whether miR-UL112 could downregulate the UL114 protein, we used an in vitro system in which 293T cells were transduced either with a lentivirus expressing miR-UL112 or miR-UL112 mutant (miR-UL112mut) or with an empty vector. The transduction efficiency was monitored through the reduction in MICB expression in 293T cells transduced with miR-UL112 (data not shown). The cells were then transfected either with plasmids encoding UL114 fused to GFP or with a GFP control plasmid in which GFP was processed to allow the addition of a glycosylphosphatidylinositol membrane anchor (GPI-GFP). Importantly, the GFP intensity of UL114-GFP was significantly reduced in 293T cells expressing miR-UL112 compared to that in 293T cells expressing miR-UL112mut or in the control 293T cells (Fig. 3A and B). In Fig. 3A, the GFP intensities in the different cells are presented in contour plots, and the median florescence intensities (MFI) are marked on these plots. The MFI are summarized in Fig. 3B. No differences in the GFP intensity were observed among the various cells when the plasmid encoding only GFP was used (Fig. 3A and B). These results demonstrate that at least in this artificial system, UL114 is targeted by miR-UL112.
FIG. 3.
miR-UL112 downregulates the viral protein UL114. (A) 293T cells were transduced either with an empty vector, with miR-UL112, or with miR-UL112mut. These cells were transfected with either the UL114-GFP plasmid or the GPI-GFP plasmid as indicated on the y axis. GFP intensity was assessed by FACS 40 hours after transfection. The contour plots present GFP intensity, with contour lines being drawn to form x and y coordinates that have similar numbers of cells. This gives a 3-dimensional-like display, with the third dimension coming out as the counts. The MFI are specified and marked with arrows. (B) Graphical representation of the MFI of GFP presented in panel A. Data are representative of three independent experiments. (C) Relative luciferase activity after reporter plasmids (indicated on the x axis) were transfected into the indicated cells. Firefly luciferase activity was normalized to Renilla luciferase activity and then normalized to the average activity of the control reporter. Values are means ± standard deviations for triplicate samples. *, P < 0.005 for 293T cells expressing miR-UL112 versus cells expressing miR-UL112mut. (D) HFF were transduced with lentiviruses encoding GFP together with miR-UL112 or a control miRNA. These cells were then infected with the UL114 ICP4-tagged recombinant Towne virus (MOI, 1). Cells were lysed 72 h postinfection and were subjected to Western blot analysis using the indicated antibodies. (E) The amounts of proteins presented in panel D were quantified by densitometry. Protein amounts in the HFF/miR-Control cells were set at 100%, and the amounts of the proteins in HFF/miR-UL112 cells were calculated accordingly.
To further establish that miR-UL112 directly targets the UL114 protein, we used luciferase reporter assays in which UL114 or the 3′ UTR of MICB was appended to the luciferase gene and these constructs were transfected into 293T cells that were transduced with either miR-UL112, miR-UL112mut, or a control miRNA. In agreement with the reduction observed in UL114-GFP expression (Fig. 3A and B), a decrease in the activity of the luciferase reporter gene was observed when it was fused to UL114 (Fig. 3C). The decrease was not observed when miR-UL112mut was used, and it was almost equivalent to the reduction observed with the MICB 3′ UTR, which was used as a positive control (Fig. 3C).
We next wanted to demonstrate that the downregulation of UL114 protein by miR-UL112 could be observed during authentic viral infection. For that purpose, we used a recombinant Towne virus in which the UL114 protein is tagged with an ICP4 epitope (23). HFF were transduced either with miR-UL112 or with a control miRNA and were then infected with the tagged UL114 recombinant virus. Seventy-two hours postinfection, cells were lysed, and the UL114 levels were measured by Western blotting with an anti-ICP4 MAb. As can be seen in Fig. 3D and E, UL114 levels were significantly reduced in infected HFF that ectopically expressed miR-UL112. Since it was previously demonstrated that ectopic expression of miR-UL112 reduces viral titers (16, 19), we could not exclude the possibility that the observed reduction in UL114 protein levels resulted not from direct targeting by miR-UL112 but rather from a general reduction in viral protein expression due to reduced viral replication and titers. Indeed, the level of the viral tegument protein, pp65, which does not contain a potential target site for miR-UL112 (data not shown), was also reduced when the infected cells ectopically expressed miR-UL112. However, this reduction was less significant than the reduction observed in the levels of UL114 protein (Fig. 3D and E), and taking these results together with those discussed above (Fig. 3A to C), we suggest that UL114 is a direct target of miR-UL112.
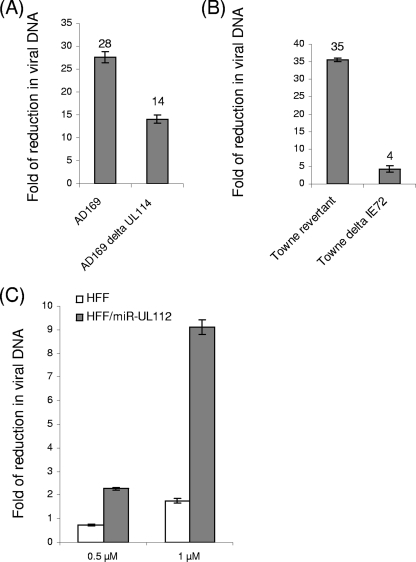
It has been demonstrated that several of the IE genes—IE72, UL112/113, and UL120/121—are targeted by miR-UL112 (16, 19), resulting in a reduction in viral loads. However, the observed effects of ectopic expression of miR-UL112 were different from the effects observed in the IE72 deletion virus, suggesting that miR-UL112 might target other viral genes that are involved in the phenotype of reduced viral titers (12). As demonstrated above, UL114—a uracil DNA glycosylase, which has been suggested to play a role in viral DNA synthesis and viral replication (22, 23)—is also targeted by miR-UL112. To test whether the reduction of UL114 expression by miR-UL112 also affects viral replication, HFF expressing miR-UL112 or a control miRNA were infected with AD169 (wild type [WT]) and with AD169 ΔUL114 (22), and viral DNA levels were measured at 96 h postinfection (Fig. 4A). As expected, significant reductions in the levels of viral DNA were observed when the cells that ectopically expressed miR-UL112 were infected with the WT AD169 virus (mean 28-fold reduction [Fig. 4A]). Importantly, when a mutant virus in which UL114 was deleted was used, a significant reduction in viral DNA was still noticed (14-fold [Fig. 4A]), suggesting that the targeting of UL114 by miR-UL112 is only minimally involved in reducing viral DNA synthesis. To further investigate this point, we also used a Towne virus in which the IE72 gene was deleted and its recombinant revertant (13). Using this system, we could observe only a small reduction in viral titers when the IE72 gene was deleted compared to the reduction in the titers of its revertant (4- versus 35-fold [Fig. 4B]), indicating that the miRNA-dependent reduction in viral DNA levels is mostly due to miR-ULl12-mediated reduction of the IE72 protein levels and is not due to the UL114 reduction.
FIG. 4.
Role of miR-UL112 during infection. (A and B) HFF were transduced with lentiviruses encoding GFP together with miR-UL112 or with miR-Control. These cells were then infected (MOI, 0.5) with WT AD169 or AD169 ΔUL114 (A) or with Towne ΔIE72 or its revertant (B). Ninety-six hours postinfection, viral DNA accumulated in infected cells was quantified by real-time PCR, and the reductions in viral DNA levels in cells expressing miR-UL112 compared to cells expressing miR-Control are presented. Data are representative of three independent experiments. (C) HFF were transduced with lentiviruses encoding GFP together with miR-UL112 or miR-Control and were infected (MOI, 2) with WT AD169 in the presence of different BrdU concentrations. Ninety-six hours postinfection, viral DNA accumulated in infected cells was quantified by real-time PCR. Reductions in viral DNA levels in different concentrations of BrdU (indicated on the y axis), compared to those with no BrdU, are shown for in HFF/miR-control and HFF/miRUL112.
The most established function of the UL114 protein is its function as a uracil DNA glycosylase (22), and thus, if miR-UL112 indeed targets UL114 directly, the ability of this protein to excise uracil residues from DNA should be impaired. It has been shown that uracil nucleoside analogs that are halogenated at the 5 position are also potent inhibitors of herpesvirus replication (23). Furthermore, it has been demonstrated that UL114 is able to remove some of the halogenated uracil analogs from viral DNA and that the virus is very sensitive to these analogs in the absence of UL114 activity (23). To test whether the downregulation of UL114 by miR-UL112 would affect its ability to remove the halogenated uracil analogs, we compared viral DNA replication in HFF and in HFF ectopically expressing miR-UL112. We used concentrations of 5-bromodeoxyuridine (BrdU) that were shown to inhibit virus replication in the absence of UL114 activity (18). As in all of the experiments discussed above, the expression of miR-UL112 in HFF resulted in a significant reduction in viral DNA levels, which was mediated, as shown above, by the combined targeting primarily of IE72 and also of UL114. Importantly, in the presence of a low concentration of BrdU, DNA synthesis was inhibited much more profoundly in cells expressing miR-UL112 than in control HFF (Fig. 4C). This result indicates that expression of miR-UL112 reduces uracil DNA glycosylase activity, and it further supports the observation that UL114 is a direct target of miR-UL112.
Ectopic expression of miR-US25-1 and miR-US25-2 reduces the titers of several viruses.
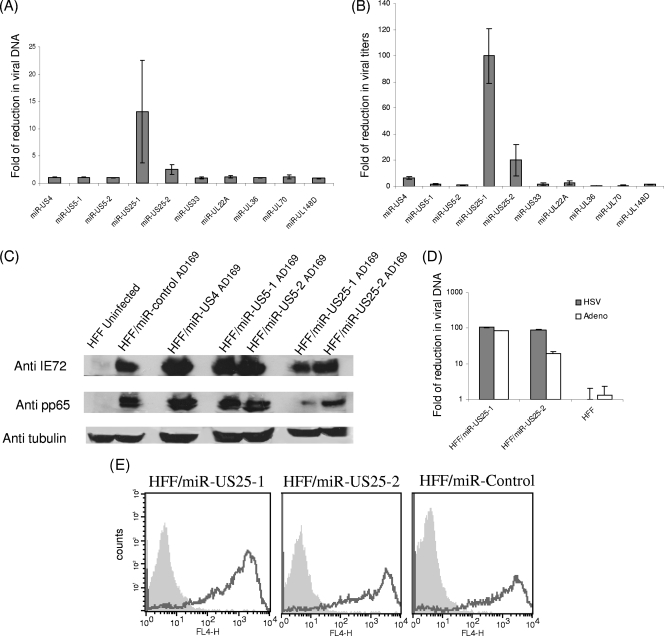
Several recent reports (16, 19) establish a concept in which various viral miRNAs derived from different herpesviruses, such as HCMV, EBV, and HSV, inhibit viral growth. We therefore tested whether additional HCMV-encoded miRNAs would also affect DNA synthesis and viral titers. For this purpose, HFF expressing the various HCMV miRNAs or a control miRNA were infected (MOI, 0.5) either with AD169 or with TB40, and the levels of viral DNA and viral titers were measured 96 h postinfection. miR-UL112 was not included in this assay, because it had already been shown to reduce viral replication (16, 19) (Fig. 4). Interestingly, the expression of miR-US25-1 and miR-US25-2 resulted in significant reductions in viral DNA levels, as detected by qPCR analysis of infected cells (Fig. 5A for AD169; data not shown for TB40). In agreement with these results, ectopic expression of miR-US25-1 and miR-US25-2 also resulted in pronounced reductions in viral titers when cells were infected with either AD169 or TB40 (Fig. 5B; also data not shown). To gain an initial insight into the mechanisms by which miR-US25-1 and miR-US25-2 reduce viral titers and to verify the observed effect, we examined the expression of IE and late proteins (IE72 and pp65, respectively) in the presence if miR-US25-1 and miR-US25-2. Importantly, the levels of both IE72 and pp65 were significantly reduced by miR-US25-1 and miR-US25-2 72 h postinfection, while there was no major alteration in the expression of these proteins in the presence of a control miRNA, miR-US4, miR-US5-1, or miR-US5-2 (Fig. 5C). Immunofluorescence analysis of the expression of IE72 confirmed these results (data not shown). Thus, miR-US25-1 and miR-US25-2 block HCMV infection, probably at early stages of infection.
FIG. 5.
Ectopic expression of miR-US25-1 and HCMV-miR-US25-2 reduces viral titers. (A and B) HFF were transduced with lentiviruses encoding GFP together with the indicated HCMV miRNA or a control miRNA. These cells were then infected (MOI, 0.5) with WT HCMV AD169. At 96 h postinfection, viral DNA was quantified by real-time PCR (A), and viral titers were measured by a plaque assay (B). The reductions in viral DNA levels (A) and viral titers (B) in cells expressing each of the miRNAs the control miRNA compared to those in cells expressing are presented. Data are averages for three independent experiments. (C) HFF were transduced with lentiviruses encoding GFP together with the indicated HCMV miRNA or control miRNA. These cells were then infected (MOI, 1) with WT AD169 virus. Cells were lysed at 72 h postinfection and were then subjected to Western blot analysis using the indicated antibodies. (D) HFF were transduced with lentiviruses encoding GFP together with the indicated HCMV miRNA or control miRNA. These cells were then infected at an MOI of 10−3 for adenovirus (Adeno) and an MOI of 0.1 for HSV-1. Viral DNA was quantified by real time PCR. The reductions in viral DNA levels in cells expressing each of the miRNAs compared to those in cells expressing the control miRNA are presented. (E) FACS staining of uninfected cells (shaded histograms) or of cells infected with A/PR8 virus (open histograms) with an anti-H1 MAb.
To verify that the observed effects did not result from cell apoptosis mediated by miR-US25-1 and miR-US25-2, we performed PI staining of all cells ectopically expressing the HCMV-encoded miRNAs. The PI staining measures DNA content, and thus apoptotic cells and cell cycle progression could be monitored. As we observed no significant changes in the percentage of apoptotic cells or in cell cycle progression in the cells ectopically expressing the various HCMV miRNAs (see Fig. S3 in the supplemental material), we concluded that the miR-US25-1 and miR-US25-2 effect probably did not result from enhanced cell death.
To further examine if the observed phenotype is mediated by direct targeting of HCMV genes or if it is related to the targeting of cellular genes, we tested whether infection with other viruses, such as HSV-1 and adenovirus, would be affected by miR-US25-1 and HCMV-miR-US25-2. Importantly, ectopic expression of miR-US25-1 and miR-US25-2 resulted in significant reductions in the DNA levels of both viruses, as detected by qPCR analysis of infected cells (Fig. 5D). Since all three viruses (HCMV, HSV-1 and adenovirus) contain different genomes, these results suggest that miR-US25-1 and miR-US25-2 reduce viral titers by inhibiting the expression of cellular proteins that are important for viral infection. Finally we tested whether ectopic expression of these miRNAs will also affect infection with RNA viruses. For that purpose, 721.221 cells were transduced with either miR-US25-1, miR-US25-2, or a control miRNA, and the cells were then infected with A/PR8 H1N1 influenza virus. The infection efficiency was monitored either by detection of the hemagglutinin protein on the surfaces of the infected cells or by measuring viral DNA levels, as detected by qPCR analysis. In both assays (Fig. 5E and data not shown), we observed no significant changes in influenza infection with ectopic expression of miR-US25-1 and miR-US25-2.
Thus, in the results presented above, we identify a third target for miR-UL112, which is UL114, and we show that miR-US25-1 and miR-US25-2 probably target cellular genes that are important for DNA virus replication.
DISCUSSION
In 2004, Pfeffer et al. showed that EBV contains several miRNAs in its genome (21). Since then, several DNA viruses, including all herpesviruses examined to date, have been found to express viral miRNAs (12). We show here that all of the HCMV-encoded miRNAs are also expressed in clinical isolates and thus that the identification of the HCMV miRNAs is clinically relevant.
Viral miRNAs are interesting because they could potentially regulate both viral and host genes. However, although more than 100 viral miRNAs have now been identified, validated targets and consequently the functions of the vast majority of viral miRNAs are still unknown. The identification of viral miRNA targets is particularly challenging, because most available target prediction algorithms are based on evolutionary conservation of the miRNA sites (24). However, in the case of viral miRNAs, this approach is problematic because viruses are much less evolutionarily conserved. In addition, even though some current algorithms could be used for viral miRNA target prediction, the false-positive rate of target predictions is still very high (up to ∼66%) (3). Since we demonstrated that miR-UL112 downregulates MICB (28), and since it was demonstrated that a viral protein, UL16 (6), also inhibits MICB expression, we initially tried to determine if any additional immune genes that are manipulated by viral proteins would also be targeted by the HCMV-encoded miRNAs. We observed no effect of the miRNAs on the immune genes that we tested. Although it is still possible that these immune genes are affected by HCMV-encoded miRNAs in vivo, our functional analysis suggests that the HCMV miRNAs do not play a significant role in the targeting of these genes.
Of the 11 HCMV miRNAs, 3 miRNAs; miR-US33, miR-UL112, and miR-UL148, are located on the complementary strands of open reading frames of viral genes. We demonstrate here by several modalities that miR-UL112 downregulates the expression of its complementary gene, UL114, as was suggested when this miRNA was first discovered (20). UL114 has uracil DNA glycosylase activity (an important enzyme in DNA repair that removes incorporated uracil residues from DNA) and has been demonstrated to function as part of the DNA replication machinery (22). The uracil DNA glycosylase activity of UL114 is low compared to that of its cellular homolog (25), and thus, it has likely evolved a second function in DNA replication, as observed for vaccinia virus (27). Supporting this notion, it has been demonstrated that UL114 deletion delays the onset of viral DNA synthesis, resulting in a prolonged replication cycle (23). Interestingly, the function of the UL114 protein was determined by using a mutant virus in which UL114 was deleted, and although it was not known at that time, miR-UL112 was also deleted in this mutant. Thus, it is possible that some of the defects observed in the UL114 deletion virus were due to the deletion of miR-UL112. Indeed, it was demonstrated that immediately after infection, the ΔUL114 mutant and WT viruses generated similar quantities of the IE protein IE72, but the ΔUL114 mutant continued to produce high levels of IE72 late in infection, which could explain the prolonged replication cycle of this virus (22). We know today that the observed upregulation of IE72 in the ΔUL114 virus was probably due to the absence of miR-UL112, which targets this gene (16, 19). Importantly, however, a cell line expressing the UL114 protein complemented some of the observed phenotype of a UL114 deletion virus, confirming that at least some of the observed defects in DNA synthesis were the result of a deficiency in this specific gene (23).
In agreement with previous publications (16, 19), we show here that the ectopic expression of miR-UL112 resulted in significant reductions in viral DNA levels. By using mutated viruses in which either the IE72 or the UL114 gene was deleted, we show that the reduced viral replication is due mainly to miR-UL112-mediated suppression of the IE72 protein and that in this scenario and with these types of assays, the reduction in the level of the UL114 protein contributes minimally to this phenomenon.
We show that the miR-UL112-mediated downregulation of UL114 reduces the ability of the virus to excise uracil residues from its genome and renders it hypersensitive to BrdU. It is tempting to speculate that the increase in uracil content due to UL114 targeting by miR-UL112 would increase the mutation rates in the viral genome, which might be beneficial for the virus at later stages of replication, when miR-UL112 is expressed. However, this option is quite unlikely, since a cellular homolog of this enzyme also exists which could probably compensate for the reduction in the viral enzyme levels.
We therefore suggest that the targeting of UL114 by miR-UL112 might be important for the life cycle of the virus. Indeed, the UL114 protein has been shown to be directly involved in viral DNA replication (through association with the viral DNA polymerase), and this protein has been demonstrated to be required during the transition to late-phase viral DNA replication (7, 23). It is also possible that the suppression of UL114 levels might be used to inhibit DNA replication during the establishment of latency, a mechanism that is currently difficult to test experimentally due to the lack of an appropriate in vitro system.
In light of these results, it is conceivable that miR-US33 and miR-UL148 would also target their complementary genes (the US29 and UL150 genes, respectively). However, the functions of both of these genes are still unknown, and no antibodies are currently available for these proteins.
We also identified here two additional HCMV-encoded miRNAs, miR-US25-1 and miR-US25-2, that reduce viral DNA synthesis and viral titers. Interestingly, these miRNAs follow an expression pattern similar to that of miR-UL112: they are expressed early after infection, with a continuing increase in their levels over time (14). These HCMV-derived miRNAs inhibited the DNA levels not only of HCMV but also of HSV-1 and adenovirus but, importantly, had no effect on influenza virus infection, and thus it is most likely that they target cellular genes that are generally important for the life cycle of DNA viruses. As stated above, the identification of such cellular genes is currently very challenging due to the lack of an appropriate system that would allow accurate and efficient identification of viral miRNA targets.
We still do not understand why different miRNAs are needed to reduce viral replication, and although it was postulated that the reduction in viral titers would drive the virus into latency, experimental validation is still required for this assumption. Nevertheless, the discovery of two additional miRNAs that might affect viral titers is significant, especially since our results suggest that these viral miRNAs probably target cellular genes, a mechanism that would enable the virus to reduce its titers. Finally, the discovery that three different viral miRNAs, miR-UL112, miR-US25-1, and miR-US25-2, might reduce the level of viral replication could lead to the development of an miRNA-based therapeutic approach aimed at reducing the severity of virus infection in immunocompromised individuals.
Supplementary Material
Acknowledgments
We thank E. S. Mocarski for the ΔIE72 virus and for critical reading of our manuscript. We also thank F. Gray and J. Nelson for sharing unpublished results.
This study was supported by grants from the U.S.-Israel Binational Science Foundation, the Israeli Cancer Research Foundation, The Israeli Science Foundation, The Israeli Science Foundation (Morasha), the Association for International Cancer Research (AICR), and an Israel-Croatia research grant (all to O.M.), by the Israeli Ministry of Science and Israel Scientific Foundation (to D.G.W.), and by NO1-AI-30049 from NIAID, NIH (to M.P.). O.M. is a Crown Professor of Molecular Immunology. N.S.-G. is supported by the Adams Fellowship Program of the Israel Academy of Sciences and Humanities.
Footnotes
Published ahead of print on 5 August 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Achdout, H., T. I. Arnon, G. Markel, T. Gonen-Gross, G. Katz, N. Lieberman, R. Gazit, A. Joseph, E. Kedar, and O. Mandelboim. 2003. Enhanced recognition of human NK receptors after influenza virus infection. J. Immunol. 171:915-923. [DOI] [PubMed] [Google Scholar]
- 2.Ambros, V. 2004. The functions of animal microRNAs. Nature 431:350-355. [DOI] [PubMed] [Google Scholar]
- 3.Baek, D., J. Villen, C. Shin, F. D. Camargo, S. P. Gygi, and D. P. Bartel. 2008. The impact of microRNAs on protein output. Nature 455:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeckh, M., M. Huang, J. Ferrenberg, T. Stevens-Ayers, L. Stensland, W. G. Nichols, and L. Corey. 2004. Optimization of quantitative detection of cytomegalovirus DNA in plasma by real-time PCR. J. Clin. Microbiol. 42:1142-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buck, A. H., J. Santoyo-Lopez, K. A. Robertson, D. S. Kumar, M. Reczko, and P. Ghazal. 2007. Discrete clusters of virus-encoded microRNAs are associated with complementary strands of the genome and the 7.2-kilobase stable intron in murine cytomegalovirus. J. Virol. 81:13761-13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cosman, D., J. Mullberg, C. L. Sutherland, W. Chin, R. Armitage, W. Fanslow, M. Kubin, and N. J. Chalupny. 2001. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity 14:123-133. [DOI] [PubMed] [Google Scholar]
- 7.Courcelle, C. T., J. Courcelle, M. N. Prichard, and E. S. Mocarski. 2001. Requirement for uracil-DNA glycosylase during the transition to late-phase cytomegalovirus DNA replication. J. Virol. 75:7592-7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dölken, L., J. Perot, V. Cognat, A. Alioua, M. John, J. Soutschek, Z. Ruzsics, U. Koszinowski, O. Voinnet, and S. Pfeffer. 2007. Mouse cytomegalovirus microRNAs dominate the cellular small RNA profile during lytic infection and show features of posttranscriptional regulation. J. Virol. 81:13771-13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunn, C., N. J. Chalupny, C. L. Sutherland, S. Dosch, P. V. Sivakumar, D. C. Johnson, and D. Cosman. 2003. Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity. J. Exp. Med. 197:1427-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn, W., P. Trang, Q. Zhong, E. Yang, C. van Belle, and F. Liu. 2005. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell. Microbiol. 7:1684-1695. [DOI] [PubMed] [Google Scholar]
- 11.Filipowicz, W. 2005. RNAi: the nuts and bolts of the RISC machine. Cell 122:17-20. [DOI] [PubMed] [Google Scholar]
- 12.Gottwein, E., and B. R. Cullen. 2008. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 3:375-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J. Virol. 72:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grey, F., A. Antoniewicz, E. Allen, J. Saugstad, A. McShea, J. C. Carrington, and J. Nelson. 2005. Identification and characterization of human cytomegalovirus-encoded microRNAs. J. Virol. 79:12095-12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grey, F., L. Hook, and J. Nelson. 2008. The functions of herpesvirus-encoded microRNAs. Med. Microbiol. Immunol. 197:261-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grey, F., H. Meyers, E. A. White, D. H. Spector, and J. Nelson. 2007. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 3:e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kafri, T., U. Blomer, D. A. Peterson, F. H. Gage, and I. M. Verma. 1997. Sustained expression of genes delivered directly into liver and muscle by lentiviral vectors. Nat. Genet. 17:314-317. [DOI] [PubMed] [Google Scholar]
- 18.Mocarski, E. S., Jr. 2004. Immune escape and exploitation strategies of cytomegaloviruses: impact on and imitation of the major histocompatibility system. Cell. Microbiol. 6:707-717. [DOI] [PubMed] [Google Scholar]
- 19.Murphy, E., J. Vanicek, H. Robins, T. Shenk, and A. J. Levine. 2008. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc. Natl. Acad. Sci. USA 105:5453-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grasser, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269-276. [DOI] [PubMed] [Google Scholar]
- 21.Pfeffer, S., M. Zavolan, F. A. Grasser, M. Chien, J. J. Russo, J. Ju, B. John, A. J. Enright, D. Marks, C. Sander, and T. Tuschl. 2004. Identification of virus-encoded microRNAs. Science 304:734-736. [DOI] [PubMed] [Google Scholar]
- 22.Prichard, M. N., G. M. Duke, and E. S. Mocarski. 1996. Human cytomegalovirus uracil DNA glycosylase is required for the normal temporal regulation of both DNA synthesis and viral replication. J. Virol. 70:3018-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prichard, M. N., H. Lawlor, G. M. Duke, C. Mo, Z. Wang, M. Dixon, G. Kemble, and E. R. Kern. 2005. Human cytomegalovirus uracil DNA glycosylase associates with ppUL44 and accelerates the accumulation of viral DNA. Virol. J. 2:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajewsky, N. 2006. microRNA target predictions in animals. Nat. Genet. 38(Suppl.):S8-S13. [DOI] [PubMed] [Google Scholar]
- 25.Ranneberg-Nilsen, T., H. A. Dale, L. Luna, R. Slettebakk, O. Sundheim, H. Rollag, and M. Bjoras. 2008. Characterization of human cytomegalovirus uracil DNA glycosylase (UL114) and its interaction with polymerase processivity factor (UL44). J. Mol. Biol. 381:276-288. [DOI] [PubMed] [Google Scholar]
- 26.Shi, R., and V. L. Chiang. 2005. Facile means for quantifying microRNA expression by real-time PCR. BioTechniques 39:519-525. [DOI] [PubMed] [Google Scholar]
- 27.Stanitsa, E. S., L. Arps, and P. Traktman. 2006. Vaccinia virus uracil DNA glycosylase interacts with the A20 protein to form a heterodimeric processivity factor for the viral DNA polymerase. J. Biol. Chem. 281:3439-3451. [DOI] [PubMed] [Google Scholar]
- 28.Stern-Ginossar, N., N. Elefant, A. Zimmermann, D. G. Wolf, N. Saleh, M. Biton, E. Horwitz, Z. Prokocimer, M. Prichard, G. Hahn, D. Goldman-Wohl, C. Greenfield, S. Yagel, H. Hengel, Y. Altuvia, H. Margalit, and O. Mandelboim. 2007. Host immune system gene targeting by a viral miRNA. Science 317:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Umbach, J. L., M. F. Kramer, I. Jurak, H. W. Karnowski, D. M. Coen, and B. R. Cullen. 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454:780-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van de Wetering, M., I. Oving, V. Muncan, M. T. Pon Fong, H. Brantjes, D. van Leenen, F. C. Holstege, T. R. Brummelkamp, R. Agami, and H. Clevers. 2003. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 4:609-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wolf, D. G., N. S. Lurain, T. Zuckerman, R. Hoffman, J. Satinger, A. Honigman, N. Saleh, E. S. Robert, J. M. Rowe, and Z. Kra-Oz. 2003. Emergence of late cytomegalovirus central nervous system disease in hematopoietic stem cell transplant recipients. Blood 101:463-465. [DOI] [PubMed] [Google Scholar]
- 32.Xu, K., H. Ma, T. J. McCown, I. M. Verma, and T. Kafri. 2001. Generation of a stable cell line producing high-titer self-inactivating lentiviral vectors. Mol. Ther. 3:97-104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.