Abstract
Nicotiana benthamiana plants were agroinoculated with an infectious cDNA clone of Turnip mosaic virus (TuMV) that was engineered to express a fluorescent protein (green fluorescent protein [GFP] or mCherry) fused to the viral 6K2 protein known to induce vesicle formation. Cytoplasmic fluorescent discrete protein structures were observed in infected cells, corresponding to the vesicles containing the viral RNA replication complex. The vesicles were motile and aligned with microfilaments. Intracellular movement of the vesicles was inhibited when cells were infiltrated with latrunculin B, an inhibitor of microfilament polymerization. It was also observed that viral accumulation in the presence of this drug was reduced. These data indicate that microfilaments are used for vesicle movement and are necessary for virus production. Biogenesis of the vesicles was further investigated by infecting cells with two recombinant TuMV strains: one expressed 6K2GFP and the other expressed 6K2mCherry. Green- and red-only vesicles were observed within the same cell, suggesting that each vesicle originated from a single viral genome. There were also vesicles that exhibited sectors of green, red, or yellow fluorescence, an indication that fusion among individual vesicles is possible. Protoplasts derived from TuMV-infected N. benthamiana leaves were isolated. Using immunofluorescence staining and confocal microscopy, viral RNA synthesis sites were visualized as punctate structures distributed throughout the cytoplasm. The viral proteins VPg-Pro, RNA-dependent RNA polymerase, and cytoplasmic inclusion protein (helicase) and host translation factors were found to be associated with these structures. A single-genome origin and presence of protein synthetic machinery components suggest that translation of viral RNA is taking place within the vesicle.
Positive-strand RNA viruses replicate their genomes on intracellular membranes. Extensive membrane rearrangements leading to cytoplasmic membranous structure production are observed during the infection cycle of many of these viruses (for a review, see reference 32). These virus-induced membrane structures vary greatly in origin, size, and shape. For instance, Flock House virus induces the formation of 50-nm vesicles (spherules), which are outer mitochondrial membrane invaginations with interiors connected to the cytoplasm by a necked channel of approximately 10-nm diameter (24). On the other hand, poxviruses replicate in 1- to 2-μm cytoplasmic foci known as DNA factories (43), which are bounded by rough endoplasmic reticulum (ER). These factories are not only the site of DNA synthesis but also of DNA transcription and RNA translation (21). Similarly, mimiviruses are huge double-stranded DNA viruses that replicate in giant cytoplasmic virus factories (45). Three-dimensional electron microscopic imaging has shown that coronavirus-induced membrane alterations define a reticulovesicular network of modified ER that integrates convoluted membranes, numerous interconnected double-membrane vesicles, and vesicle packets (23), similar to what was observed for dengue viruses (52). These virus-induced structures are known to shelter the virus replication complex, which carries out viral RNA synthesis. The replication complex contains the viral RNA-dependent RNA polymerase (RdRp), positive- and negative-strand viral RNAs, accessory nonstructural viral proteins, and host cell factors. The role of these virus-induced membrane vesicles in regard to viral RNA synthesis is not well understood. They have been proposed to increase the local concentration of components required for replication, to provide a scaffold for anchoring the replication complex, to confine the process of RNA replication to specific cytoplasmic locations, and to aid in preventing the activation of certain host defense functions. The mechanisms that are responsible for the formation of these structures have begun to be deciphered. Several studies have shown that the specific viral proteins are responsible for the formation of the membrane vesicles (3, 42). However, how individual proteins promote their formation is still unexplained. The full role of cellular factors also remains to be investigated in terms of both membrane vesicle formation and viral RNA synthesis. Finally, intracellular trafficking of these vesicles has been reported (15, 25, 29, 54).
Turnip mosaic virus (TuMV) belongs to the genus Potyvirus in the family Potyviridae (44). The TuMV genome is composed of a positive-sense single-stranded RNA molecule of about 10 kb in length (36). The 5′ terminus of the viral RNA is linked covalently to a viral protein known as VPg and the 3′ terminus is polyadenylated. The TuMV RNA is translated into a long polyprotein of 358 kDa and is processed into at least 10 mature proteins by three different virus-encoded proteases. It was demonstrated for Tobacco etch virus (TEV) and Plum pox virus, also members of the Potyvirus genus, that viral RNA synthesis is associated with membranes of the ER (30, 42). In the case of TuMV, the 6K2-VPg-Pro polyprotein, through its hydrophobic 6K2 domain, was shown to be responsible for the formation of cytoplasmic vesicles derived from the ER (4), similar in structure to those observed during TEV infection (42, 51). Besides being involved in vesicle formation, 6K2-VPg-Pro binds a number of proteins of viral and host origin. Interaction with the viral RdRp and the host translation eukaryotic initiation factor (iso) 4E [eIF(iso)4E], poly(A)-binding protein (PABP), heat shock cognate 70-3 (Hsc70-3), and the eukaryotic elongation factor 1A (eEF1A) has been shown to take place within the 6K2-VPg-Pro-induced vesicles (4, 5, 9, 48). Although these vesicles have been referred to as sites for TuMV replication (48), the presence of viral RNA in these vesicles has not been reported. The presence of translation factors with virus replication proteins also brings the question of the physical relationship between viral RNA translation and replication.
In the present study, the biogenesis of the TuMV-induced vesicles was investigated. It was observed that the TuMV-induced vesicles, tagged with a fluorescent protein fused to 6K2, were mobile and aligned with microfilaments. In addition, results using depolymerizing compounds supported the contention that vesicles trafficked on microfilaments and that microtubules were not involved. Evidence is also provided indicating that each vesicle is derived from a single viral genome. Finally, the association of host translation factors with viral RNA synthesis sites was confirmed by double immunofluorescence staining in TuMV-infected protoplasts. The single-genome origin and the presence of translation factors within the replication complex vesicles suggest that viral translation is taking place within the vesicles.
MATERIALS AND METHODS
Plasmid constructions.
pCambiaTunos was obtained by digesting p35Tunos (41) with KpnI and StuI, followed by ligation with pCambiaTunos/6KGFP (48), also digested with KpnI and StuI. Kanamycin-resistant Escherichia coli colonies were screened for plasmids deleted in the region encoding 6K2GFP (i.e., pCambiaTunos/6KGFP). pUC19/6KGFP was obtained by digestion of p35Tunos/ΔNsil-6KGFP (48) with BamHI and KpnI and the 1458-bp fragment was ligated in pUC19 digested with the same enzymes and in which the HindIII restriction site had previously been destroyed. The mCherry coding region was amplified by PCR from pCambia/mCherry (9) by using the forward primer 5′-ATTCGGATCCGTGAGCAAGGGCGAGGAG-3′ and the reverse primer 5′-ATTCAAGCTTCCTTGTACAGCTCGTCCATG-3′ (the restriction sites are underlined). The PCR product was digested with BamHI and HindIII and ligated in pUC19/6KGFP (48), which was also digested with the same enzymes. Ampicillin-resistant E. coli colonies were screened for plasmids containing the fragment encoding 6K2mCherry (i.e., pUC19/6KmCherry). This plasmid was then cut with BamHI and KpnI and the 1451-bp fragment was ligated with p35Tunos/ΔNsil-6KGFP (48) cut with the same enzymes. Ampicillin-resistant E. coli colonies were screened for plasmids containing the fragment encoding 6K2mCherry (i.e., p35Tunos/ΔNsil-6KmCherry). p35Tunos/ΔNsil-6KmCherry was digested with SmaI and KpnI and was ligated with pCambiaTunos/6KGFP, also digested with the same enzymes. Kanamycin-resistant E. coli colonies were screened for plasmids containing the fragment encoding 6K2mCherry (i.e., pCambiaTunos/ 6KmCherry). All plasmid constructs were verified by sequencing.
Antibodies.
Rabbit antisera were used at the following dilutions: for immunoblot analysis, anti-CP at 1:2,500; and for immunofluorescence labeling, anti-CP at 1:300, anti-RdRp at 1:100 (9), anti-VPg-Pro at 1:200 (27), anti-PABP2 at 1:300 (28), anti-eIF(iso)4E at 1:150 (7), anti-eEF1A at 1:500 (57), and anti-CI at 1:300. Recombinant clones pET-CP and pET-CI in E. coli BL21(DE3) cells were used for anti-CP and anti-CI serum production. Full-length coding sequences of TuMV CP and CI were cloned in frame in the pET11d vector (Novagen). The recombinant proteins were overproduced in E. coli and purified as insoluble inclusion bodies. Inclusion bodies were resuspended in Tris-buffered saline and used for rabbit injection and serum production at McGill University Animal Resources Center. The mouse monoclonal antibody dilutions were as follows: anti-actin8, 1:1000 (Sigma); anti-BrdU, 1:100 (Sigma); and anti-dsRNA, 1:300 (English & Scientific Consulting Bt.). The secondary antibodies were goat anti-mouse conjugated to Alexa Fluor 568 at 1:500 (Molecular Probes) and goat anti-rabbit conjugated to Alexa Fluor 488 at 1:1,000 (Molecular Probes). For immunoblot analysis, the antigen-antibody complexes were visualized by using a horseradish peroxidase-coupled goat anti-rabbit immunoglobulin G (IgG).
Protein expression in plants.
Constructs containing genes for proteins fused to fluorescent tag were introduced by electroporation into Agrobacterium tumefaciens AGL1 and selected on LB ampicillin-kanamycin plates. The pellet of an overnight culture was gently resuspended in water supplemented with 10 mM MgCl2 and 150 μM acetosyringone and left at room temperature for 3 h. The solution was then diluted to an optical density at 600 nm of 0.6. Three-week-old N. benthamiana plants were agroinfiltrated with the fusion construct. Plants were kept for 4 days in growth chamber until observation.
Drug treatments.
Stock solutions of latrunculin B (LatB; 2.5 mM [Calbiochem]), cytochalasin D (CytD; 20 mM [Calbiochem]), trifluralin (20 mM [ChemService]), and oryzalin (10 mM [ChemService]) were prepared in dimethyl sulfoxide (DMSO) and diluted to the desired concentration in water prior to their infiltration into 3-week-old N. benthamiana leaves.
Protoplast isolation and immunofluorescence labeling.
N. benthamiana was agroinfiltrated with pCambiaTunos/6KGFP. Leaves obtained at 4 days postinfection were sliced in 1-mm wide stripes, followed by incubation in an enzyme solution (1.5% cellulose R10, 0.2% macerozyme R10, 0.5 M mannitol, 20 mM KCl, 20 mM MES [pH 5.7], 10 mM CaCl2, 0,1% bovine serum albumin [BSA]) for 3 h in the dark under a vacuum. The protoplast suspension was filtered through a 45-μm-pore-size nylon filter and centrifuged for 4 min at 100 × g. The supernatant was removed. Protoplasts were incubated for 15 min at room temperature with 1 volume of fixing solution (4% paraformaldehyde, 0.25 M mannitol, and 50 mM sodium phosphate in phosphate-buffered saline [PBS]). They were centrifuged and resuspended for 30 min with 2 volumes of fixing solution at room temperature. Protoplasts were washed three times with PBS for 10 min. They were then put on cover slide pretreated with 0.1% poly-l-lysine (Sigma), treated with Triton X-100 0.5% in PBS for 10 min, and incubated for 20 min in a blocking solution of 5% BSA in PBS. The samples were then incubated for 1 h with the primary antibody, washed three times with PBS for 10 min, incubated for another hour with Alexa Fluor 568- or Alexa Fluor 488-conjugated secondary antibody (Molecular Probes), and finally washed three times with PBS for 10 min. Pro-long Gold Antifade (Invitrogen) was used to prepare the slides.
In vivo RNA labeling.
Protoplasts were isolated from plants agroinfiltrated at 4 days postinfection with A. tumefaciens containing pCambiaTunos. They were incubated for 30 min with 10 μg/ml of actinomycin D in order to block RNA transcription from cellular DNA-dependent RNA polymerases. Two mM 5-bromouridine 5′-triphosphate (BrUTP; Sigma) was then added for 3 h. The reaction was stopped by the addition of fixing solution (described above) for 15 min, washed once with PBS, and fixed again for another 30 min. The immunofluorescence labeling was processed as described above using anti-BrdU (1:100), and the detection was done using Alexa Fluor 568-conjugated secondary antibody (Molecular Probes) at a 1:500 dilution.
Immunoblot analysis.
Total proteins were extracted from N. benthamiana agroinfiltrated with pCambiaTunos/6KGFP 4 days before and treated previously with drugs with an extraction buffer (50 mM Tris-HCl [pH 7.6], 50 mM KCl, 0.5 mM EDTA, 20 mM NaCl, 5% glycerol, 0,1% Triton X-100, 0.01% sodium dodecyl sulfate [SDS], and plant protease inhibitor cocktail [Sigma]). Plant tissue (0.5 g) was ground in 500 μl of extraction buffer with a Dounce homogenizer and centrifuged for 1 min at 5,000 × g. The concentration of the supernatant was determined by using a Bradford assay (Bio-Rad) using BSA as a standard. Then, 2 μg of total protein preparation was used to perform an immunoblot analysis after SDS-polyacrylamide gel electrophoresis. The antigen-antibody complexes were visualized using a horseradish peroxidase-coupled goat anti-rabbit IgG or goat anti-mouse IgG according to the manufacturer's recommendations. Immunoreactions were detected with the SuperSignal West Pico chemiluminescent substrate (Pierce).
Confocal microscopy.
Agroinfiltrated leaf sections were placed on a microscope coverslide using immersion oil. Coverslides were inverted on depression slides, aligning the leaf tissue in the well. Cells were observed using a 40X oil immersion objective on a Radiance 2000 confocal microscope (Bio-Rad). Argon-krypton laser was used to excite fluorescent proteins and data from both green and red channels were collected at the same time. A charge-coupled-device camera was used to collect the images. Protoplasts were observed by using a 63X oil immersion objective on a LSM 510 Meta confocal microscope (Zeiss). Argon and HeNe lasers were used to excite fluorescent proteins and data from both green and red channels were collected at sequential time. After acquisition, images were processed by using Metamorph (6.2r6), ImageJ (1.41o), Carl Zeiss LSM Image Browser, and/or Adobe Photoshop software. The Manual Tracking pluging (http://rsbweb.nih.gov/ij/plugins/track/track.html) was used for displaying vesicle movement paths.
RESULTS
Movement of replication complex vesicles.
There are reports indicating that virus replication complexes are motile (15, 25, 29, 54). Trafficking of TuMV-induced vesicles was thus investigated with an infectious clone of TuMV (pCambiaTunos/6KGFP) (Fig. 1) that was previously engineered for expressly tagging replication complexes with green fluorescent protein (GFP) (48). The coding sequence of GFP was fused with the gene encoding 6K2 and inserted between the P1 and HCPro coding genes as an in-frame translational fusion containing flanking P1 and VPg-Pro cleavage site coding sequences: 6K2GFP is thus released when the polyprotein is processed during infection. The resulting recombinant TuMV induced the formation of fluorescent discrete structures in N. benthamiana (Fig. 2). They were irregular in shape and heterogeneous in size, ranging from 0.6 to 4.3 μm in diameter, the mean being 1.68 μm ± 0.91 μm (n = 52). Optical cross-sections show that generally GFP fluorescence fill the whole vesicle structure, with occasional observation of ringlike vesicles (described in reference 51) (Fig. 2C). These 6K2GFP-tagged structures were previously shown to contain 6K2-VPg-Pro, RdRP, eIF(iso)4E, PABP, and eEF1a (9, 48) and were reminiscent of those induced by 6K2-VPg-Pro alone (4). To confirm that the 6K2GFP fluorescence matched with viral replication sites, these were visualized by staining with antibodies directed against double-stranded RNA (dsRNA) (1, 10). N. benthamiana leaves were agroinfiltrated with A. tumefaciens Agl1 strain containing pCambiaTunos/6KGFP, and protoplasts were isolated from infiltrated leaves 4 days later. The fixed and permeabilized protoplasts were then reacted with a mouse antibody recognizing dsRNA and Alexa Fluor 586 (red)-conjugated anti-mouse antibodies. Protoplasts were subsequently observed by confocal microscopy. dsRNA was associated with punctate structures of 2.1 μm ± 0.5 μm in diameter distributed throughout the cytoplasm in TuMV-infected protoplasts (see Fig. 8I). No immunofluorescence was observed in uninfected protoplasts (data not shown). The presence of dsRNA coincided with fluorescence emitted by GFP, confirming that 6K2GFP tagging constitutes a good marker for the visualization of TuMV replication complexes.
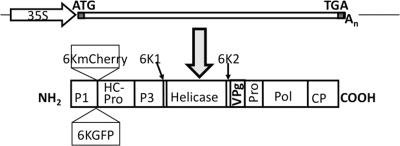
FIG. 1.
Schematic representation of recombinant TuMV expressing 6K2GFP or 6K2mCherry. At the top, the TuMV genome is shown as a rectangle, and an arrow represents the CaMV 35S promoter. Start and stop codons are represented, and An indicates the position of the polyadenylated tail. At the bottom, a TuMV polyprotein is shown as a rectangle in which individual proteins are delineated by vertical lines. The position within the polyprotein of 6K2GFP and 6K2mCherry is indicated.
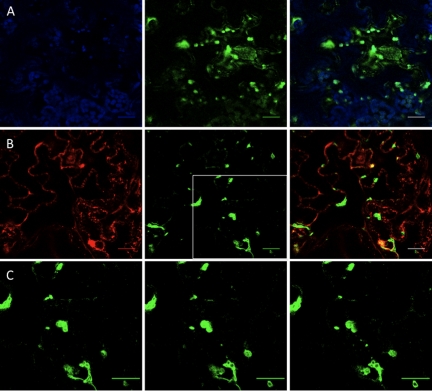
FIG. 2.
6K2GFP-tagged TuMV-induced vesicles. N. benthamiana cells expressing 6K2GFP-tagged TuMV-induced vesicles observed by confocal microscopy at 4 days after agroinfiltration. (A) Optical images (1-μm thick) showing 6K2GFP-tagged vesicles (green) with chloroplasts (in blue), with merge colors in the right panel. (B) Optical images (1-μm thick) showing 6K2GFP-tagged vesicles (green) and the Golgi marker fused to mCherry (red), with merge colors in the right panel. (C) Consecutive 1-μm thick single plane images (from left to right) of the depicted square in B that show GFP distribution within the vesicles. Scale bar, 10 μm.
FIG. 8.
Localization of dsRNA and BrUTP-labeled RNA with plant and viral proteins in TuMV-infected protoplasts. N. benthamiana leaves agroinfiltrated with A. tumefaciens Agl1 containing pCambiaTunos were collected for protoplast isolation. TuMV-infected protoplasts were processed for double-label immunofluorescence with antisera raised against dsRNA and VPg-Pro (A), BrdU and VPg-Pro (B), dsRNA and RdRp (C), dsRNA and CI (helicase) (D), dsRNA and CP (E), dsRNA and eIF(iso)4E (F), dsRNA and PABP2 (G), and dsRNA and eEF1A (H). Goat anti-mouse conjugated to Alexa Fluor 568 (red, for visualization of dsRNA and BrdU) and goat anti-rabbit conjugated to Alexa Fluor 488 (green, for visualization of viral or host proteins) were used as secondary antibodies. (I) N. benthamiana leaves agroinfiltrated with A. tumefaciens Agl1 containing pCambiaTunos/6KGFP and protoplasts were processed for dsRNA immunofluorescence detection as described above. The left panels show fluorescence emitted by the green channel only, the middle panels show fluorescence emitted by the red channel only, and the right panels show the merging of the red and green channels. The inset in panel E (left panel) is a close-up view of depicted square. Scale bar, 10 μm.
These discrete structures are likely to be membrane-bound vesicles and not protein aggregates. First, 6K2-VPg-Pro, which has been shown to colocalize with 6K2GFP (48), and RdRp “float” in membrane flotation experiments (5, 9), indicating that these replication-associated proteins are membrane associated and not aggregates or inclusions. Furthermore, Schaad et al. (42) have shown that the 6K2 protein of the related tobacco etch virus fused to GFP and expressed using the same approach as here was associated with large vesicular compartments derived from the ER. Although some are relatively large and akin to mini-organelles, these TuMV-induced structures will be designated as vesicles from now on.
The large size of the 6K2GFP-tagged TuMV-induced vesicles suggests that they are linked to chloroplasts or Golgi bodies. The Bio-Rad confocal microscope used in the present study possesses three detectors with filter sets calibrated to absorb light at wavelengths of 515 nm ± 15 nm (GFP fluorescence [green channel]), of 600 nm ± 20 nm (mCherry fluorescence [red channel]), and of >660 nm (chlorophyll autofluorescence [blue channel]). Such setup allows chlorophyll fluorescence to be separated from the fluorescence emitted by GFP and mCherry. Observation of serial optical images showed that the bulk of the chloroplasts were generally found near the surface of the plasma membrane, whereas the virus-induced vesicles were located more in the interior of the cell, often near the nucleus. Figure 2A shows a single 1-μm optical slice where 6K2GFP-tagged vesicles can be seen concomitantly with chloroplasts. Some vesicles were found in close association with chloroplasts, which possibly contribute along the ER to membrane structures at least in the case of plum pox virus replication (30). However, more often, vesicles and chloroplasts were seen as different entities. Golgi localization was based on the cytoplasmic tail and transmembrane domain (first 49 amino acids) of the soybean α-1,2-mannosidase I (40) fused to mCherry (34). This Golgi marker consists of a large number of small (<1 μm) independent stacks (34) that appeared as round discs of uniform size (Fig. 2B). The Golgi marker also showed ER labeling, resulting from the continuous recycling of Golgi resident proteins through the ER round discs (6). The 6K2GFP-tagged vesicles were found to align with the ER (as expected) but generally did not colocalize with the Golgi bodies. Occasional association with Golgi is explained by the biogenesis of potyvirus replication vesicles occurring at ER exit sites in a COPI- and COPII-dependent manner (51). The 6K2GFP-tagged TuMV-induced vesicles are thus distinct from Golgi bodies.
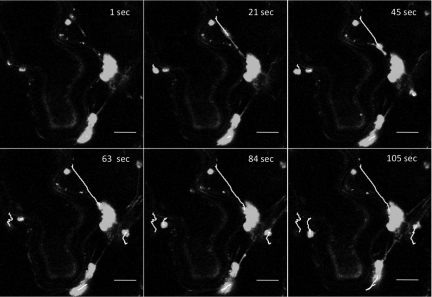
The intracellular trafficking of vesicles induced by TuMV and tagged with either fluorescent protein was then investigated. Three-week-old N. benthamiana leaves were agroinfiltrated with A. tumefaciens Agl1 containing pCambiaTunos/6KGFP, and movement of the fluorescing vesicles was investigated by time-lapse imaging after 4 days. The vesicles were motile, but each vesicle moved at different speed, with average velocities of 0.45 μm/s ± 0.27 μm/s (n = 10) (Fig. 3 and see Movie S2 in the supplemental material). Movement was unidirectional, which was accompanied with stop-and-go activity. Although the exact destination was not known, occasional fusion with perinuclear vesicles was observed, as shown in Fig. 3. Similar data were obtained when vesicles were tagged with mCherry. In comparison, cauliflower mosaic virus (CaMV) P6 inclusion bodies move with an average velocity of 2 μm/s (maximum of approximately 8 μm/s) (15), and the average velocities are ∼1 μm/s for tobacco mosaic virus 126K bodies and viral replication complexes (maximum of 8 μm/s) (29).
FIG. 3.
Movement of TuMV-induced vesicles. N. benthamiana cells expressing 6K2GFP-tagged TuMV-induced vesicles observed 4 days after agroinfiltration by confocal microscopy. Time-lapse images illustrate the movement of the vesicles over the indicated time periods. Lines depict the path taken by individual vesicles. Scale bar, 10 μm.
Since 6K-VPg-Pro alone induces vesicle formation (4), their trafficking was also investigated. N. benthamiana plants were agroinfiltrated with pGreen6KVPgProGFP, and the movement of the green fluorescing vesicles was observed after 3 days. 6K2-VPg-Pro-induced vesicles were also motile (see Movie S3 in the supplemental material), suggesting that the virus molecular determinant for movement lies within 6K2-VPg-Pro.
Stop and go movement is indicative of Golgi and was consequently compared directly to that of TuMV-induced vesicles. The Golgi stacks can be seen moving and extensively along the polygonal cortical ER network (see Movie S4 in the supplemental material) with an estimated speed of 0.35 μm/s. These data indicated that TuMV-induced 6K2GFP-tagged and Golgi vesicles are distinct structures.
Vesicle alignment with microfilaments.
Since the protein content and organized nature of the cytoplasm restrict diffusion of large molecular complexes, movement of virus-induced vesicles is likely to require cytoskeleton elements (17). To determine whether TuMV vesicle movement is associated with microfilaments, an A. tumefaciens Agl1 mixture containing pCambiaTunos/6KmCherry (Fig. 1) and a pCambia vector containing the 35S::GFP-ABD2-GFP construct (50) was agroinfiltrated in N. benthamiana. The latter construct codes for GFP fused to both termini of the actin-binding domain of fimbrin (GFP-ABD2-GFP) and provides improved imaging of actin filaments. Figure 4A shows the positioning of the mCherry-tagged vesicles with respect to the GFP-ADB2-GFP-labeled actin filaments. A dense filament network is seen radiating from the nucleus (denoted by an asterisk in the figure) to the cell periphery, and the TuMV vesicles are randomly distributed in the cell. Alignment of the vesicles with the ABD2 filaments is readily observed, with each vesicle concurrently interacting with several filaments. Vesicles seen not to align with ABD2 filaments are from adjoining cells that were not expressing the latter protein. It must be noted that in cells expressing both proteins, no vesicles were observed in subcompartments of the cell devoid of ABD2 filaments, suggesting that alignment of the vesicles with the filaments is not the indirect consequence of the density of the network trapping any large structures. The red vesicles cannot be interpreted as being chloroplasts, as explained above. Moreover, no mCherry fluorescence is observed in the red channel in mock-inoculated plants or in cells expressing GFP fusions only. Furthermore, experiments using mCherry fusions developed previously (9, 48) showed that the red fluorescence does not result from chlorophyll autofluorescence.
FIG. 4.
Coalignment of vesicles with microfilaments. (A) N. benthamiana cells expressing 6K2GFP-tagged TuMV-induced vesicles and GFP-ABD2-GFP observed by confocal microscopy at 4 days after agroinfiltration. (A) No treatment was applied. (B) Leaves were infiltrated with 5 μM LatB 4 h prior to confocal microscope observation. (C) Leaves were infiltrated with 5 μM LatB 4 h prior to agroinfiltration. Photographs are three-dimensional renderings of 40 1-μm-thick slices that overlap by 0.5 μm. An asterisk indicates the position of the nucleus. The inset image is a close-up view of the depicted square. Arrows indicate the presence of TuMV vesicles “trapped” within microfilament bundles. Scale bar, 10 μm.
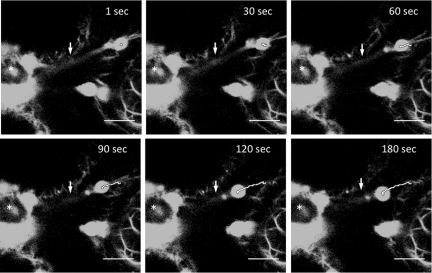
Movement of the vesicles was investigated in the presence of GFP-ABD2-GFP. Trafficking of the vesicles was difficult to discern, a phenomenon observed also for the movement of the CaMV P6 inclusion bodies (15). It has been reported that the use of live markers to visualize actin in plants could affect motility of cellular compounds because of the overexpression of the markers (19). However, despite this inhibition, TuMV vesicles were seen to traffic along GFP-ABD2-GFP filaments (Fig. 5; see also Movie S5 in the supplemental material).
FIG. 5.
Vesicle trafficking along microfilaments. N. benthamiana cell expressing 6K2GFP-tagged TuMV-induced vesicles and GFP-ABD2-GFP observed 4 days after agroinfiltration by confocal microscopy. Time-lapse images illustrate the movement of a vesicle over the indicated time periods. A line depicts the path taken by the vesicle. An asterisk denotes the position of the nucleus, and arrows indicate the position of a single microfilament. Scale bar, 10 μm.
To confirm the implication of microfilaments in viral vesicle movement, LatB was used to disassemble microfilaments. Since LatB's effect on microfilaments is likely to affect several processes, which indirectly might affect vesicle trafficking, movement was first evaluated on preestablished vesicles as soon as possible after drug application. N. benthamiana leaves were agroinfiltrated with A. tumefaciens Agl1 suspensions containing pCambiaTunos/6KGFP and the 35S::GFP-ABD2-GFP construct. At 4 days postinfection and 4 h before confocal observation, the agroinfiltrated leaves were treated with 5 μM LatB or 1% DMSO solvent control. The DMSO infiltration did not affect the microfilament network (data not shown), but after LatB treatment the ABD2 filaments had begun the depolymerization process, as noted by the blurred appearance of the filaments near the nucleus (Fig. 4B). When LatB was infiltrated 4 h prior to TuMV agroinfection, the network, when observed by confocal microscopy 4 days later, was sparser, the filaments were thicker, and several microfilament bundles were observed (Fig. 4C). Within that time period, it is expected that plant cells had metabolized some of the LatB molecules and that the microfilament network is in the process of regaining its normal state. Interestingly, the 6K2mCherry-tagged TuMV-induced vesicles were tightly enclosed within the microfilament bundles (4C).
Movement was assessed with N. benthamiana leaves agroinfiltrated with A. tumefaciens Agl1 suspensions containing pCambiaTunos/6KGFP or the GFP-labeled Golgi marker construct. The latter marker was used as a drug control since Golgi trafficking in dependent on myosin (2). DMSO had no inhibitory effect on Golgi body and TuMV vesicle movement (data not shown). On the other hand, LatB treatment 4 h before confocal microscope observation abolished trafficking of Golgi bodies (see Movie S6 in the supplemental material) and TuMV vesicles (see Movie S7 in the supplemental material).
TuMV infection is inhibited by LatB treatment.
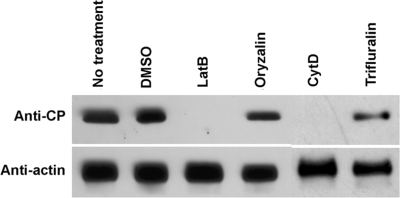
To assess the effect of cytoskeleton-affecting drugs on the initiation of TuMV infection, N. benthamiana leaves were infiltrated either with LatB, CytD, oryzalin, trifluralin, or DMSO 24 h prior to agroinfiltration with A. tumefaciens Agl1 containing pCambiaTunos/6KGFP. CytD depolymerizes microfilaments, while oryzalin and trifluarin are agents that disassemble microtubules. Plant tissues were collected 4 days later, and the total proteins were extracted, separated by SDS-polyacrylamide gel electrophoresis, and subjected to immunoblot analysis. Virus accumulation was assessed using a rabbit serum raised against the TuMV coat protein (CP). Figure 6 shows virus accumulation in plants following different drug treatments. Treatment with 5 μM LatB or 20 μM CytD reduced CP production below the level of detection by the Western immunoblot, whereas treatment with 20 μm oryzalin or trifluralin did not significantly inhibit CP production. Diminished virus production was correlated by confocal observation of agroinoculated leaves. The number of vesicles per cell was reduced after treatment with LatB and CytD compared to untreated plants or plants treated with DMSO, oryzalin, or trifluralin (data not shown). The presence of vesicles (albeit to a decreased level) suggests that translation and initial vesicle formation is possible in the presence of microfilaments depolymerization drugs. However, an intact microfilament network appears to be required for the establishment of a fully productive TuMV infection, whereas microtubules do not appear to be involved.
FIG. 6.
Effect of cytoskeleton-affecting drugs on the initiation of TuMV infection. N. benthamiana leaves were infiltrated with 5 μM LatB or 20 μM CytD, oryzalin, or trifluralin 24 h prior to infiltration with A. tumefaciens Agl1 containing pCambiaTunos/6KGFP. Total proteins were extracted 4 days after agroinfiltration, and 2 μg was analyzed by immunoblot analysis with antibodies raised against TuMV CP. Antibody against actin-8 was used as a loading control.
Vesicles derive from single genome.
Intracellular movement of and likely fusion (see Fig. 3) among the vesicles raise the question on the biogenesis of the vesicles. TuMV-induced vesicles contain viral proteins (and also host proteins), but how these proteins are imported in these vesicles is not known. One possibility is that viral RNA is translated in the surrounding cytoplasm and the viral proteins exported to growing 6K2-VPg-Pro-induced vesicles. Because of the presence of translation factors within the vesicles (4, 5, 9, 48), the other possibility is that viral protein synthesis is taking place within the virus-induced vesicles. To resolve this issue, two recombinant TuMV were used: one that expressed 6K2GFP and the other that expressed 6K2mCherry. The rational was that in a cell that is simultaneously infected with the two recombinant viruses, if the viral RNA is translated in the cytoplasm and the viral proteins are indiscriminately exported to the vesicles, one should observe vesicles that are both green and red fluorescing. On the other hand, if the vesicles are derived from a single genome and translation occurs within the vesicle, one should observe green- and red-only fluorescing vesicles. Leaves were agroinfected with a 1:1 mixture of the two viruses and individual cells that exhibited both green and red fluorescing vesicles were screened for. Among these cells were examples with individual green and red vesicles (Fig. 7A and B), suggesting that each vesicle originated from a single viral genome. Interestingly, there were vesicles that exhibited both green and red fluorescence (Fig. 7C). However, 6K2GFP and 6K2mCherry were not always perfectly colocalizing, resulting in variegated vesicles (i.e., a mixture of red, green, and yellow sectors). This uneven mixing of the fluorescent proteins is an indication that fusion among individual vesicles is also possible, a phenomenon that was observed during vesicle movement (see Fig. 3).
FIG. 7.
Presence of TuMV-induced vesicles tagged with 6K2GFP and 6K2mCherry within the same cell. N. benthamiana leaves were agroinfiltrated with A. tumefaciens Agl1 containing pCambiaTunos/6KGFP and pCambiaTunos/6KmCherry in a 1:1 mixture and observed 4 days after agroinfiltration. (A to C) Three different cells infected by both viruses. Photographs are consecutive 1-μm-thick single optical images (from left to right) taken every 1 μm (A and C) or 2 μm (B). Arrowheads delimit cellular membrane between two cells in panel B. Arrows highlight chimeric vesicles. Scale bar, 10 μm.
Localization of host and viral proteins within virus replication complexes.
Formation of a large vesicle derived from a single viral genome presupposes that a cis mechanism is involved: proteins synthesized from the same viral RNA are incorporated in the same vesicle, with no outside contribution from other genomes. This is possible if viral RNA translation is taking place within the vesicle. It was previously shown that 6K2-VPg-Pro of TuMV induces the formation of vesicles that contain host translation factors such as eIF(iso)4E, PABP, eEF1A, and Hsc70 (4, 5, 9, 48). The question however remains whether these same vesicles are also the sites of viral RNA synthesis. RNA replication sites can be visualized by immunofluorescence staining using antibodies either directed against dsRNA or neosynthesized 5-bromouridine-labeled RNA (1, 10). To confirm that the above-listed host proteins colocalize with viral replication sites, double immunofluorescence staining of TuMV-infected protoplasts was performed as described above. The fixed and permeabilized protoplasts were then reacted with rabbit sera raised against viral or plant proteins and mouse antibodies recognizing dsRNA or bromouridine. For bromouridine labeling, protoplasts were treated with actinomycin D prior to BrUTP incorporation in order to block host DNA transcription without affecting TuMV-directed RNA-dependent RNA synthesis. Rabbit and mouse antibodies were labeled with Alexa Fluor 488 (green)- and Alexa Fluor 586 (red)-conjugated secondary antibodies, respectively, and protoplasts were observed by confocal microscopy. To assess the background fluorescence, protoplasts from mock-inoculated leaves were subjected to the same immunofluorescence labeling conditions, and fluorescent signals were adjusted to set the background threshold level. For virus-specific elements, no significant background was detected in uninfected protoplasts (data not shown). Figure 8 shows the intracellular localization of dsRNA or bromouridine-labeled RNA, along with plant and viral proteins in TuMV-infected protoplasts. As mentioned above, dsRNA was associated with punctate structures distributed throughout the cytoplasm. Similar structures were also observed for bromouridine-labeled RNA, indicating that the presence of dsRNA likely corresponds to area of active RNA synthesis. No immunofluorescence was observed in uninfected protoplasts (data not shown). If the dsRNA punctate structures are true indicators of the virus replication complex, viral proteins known to be involved in virus replication must be associated with these structures. The anti-VPg-Pro antibodies react with VPg-Pro, its precursor (6K-VPg-Pro), as well as processed forms (VPg and Pro) (4). Staining with this serum was observed throughout the cytoplasm, as expected from previous cell fractionation data (4, 28). However, a subpopulation of the proteins recognized by the anti-VPg-Pro antibodies was also associated with the dsRNA or bromouridine-labeled RNA punctate structures (Fig. 8A and B). A similar fluorescent pattern was observed for RdRp (Fig. 8C). Interestingly, the cytoplasmic inclusion protein (CI), which has helicase activity (26), was associated with punctate structures that did not always coincide with the presence of dsRNA. However, dsRNA structures always correlated with the presence of CI (Fig. 8D). This suggests that CI may have additional roles besides being involved with the replication complex. The CP, which is not known to be directly involved in RNA replication, was very rarely associated with dsRNA punctate structures (Fig. 8E), suggesting that RNA synthesis and capsid-related events (e.g., virion assembly) are physically separated.
Host proteins that were shown to be enclosed within 6K-VPg-Pro-induced vesicles were then tested. eIF(iso)4E, PABP, and eEF1A were distributed throughout the cytoplasm (Fig. 8F to H), as expected from the fractionation data (4, 28, 48), but subpopulations of the different proteins were found to colocalize with the dsRNA punctate structures. Taken together, these results indicate that TuMV RNA replication sites not only contained viral proteins expected to be involved in RNA synthesis but also host factors implicated in protein synthesis.
DISCUSSION
A major finding of this investigation is that a TuMV-induced vesicle originates from a single viral genome. Infecting cells simultaneously with two recombinant viruses, one tagging vesicles with 6K2GFP and the other with 6K2mCherry, showed that green- and red-only vesicles were observed within the same cell. Vesicles tagged with both 6K2GFP and 6K2mCherry were also found and were often characterized by uneven mixing of the fluorescent protein (i.e., the presence of green, red, and yellow sectors). This indicates that fusion between vesicles is taking place, a phenomenon that has been regularly observed while looking at vesicle trafficking. A single-genome origin means that there is a cis-acting mechanism that incorporates the proteins resulting from the translation of a viral RNA into the same vesicle and also prevents the importation of viral proteins synthesized from surrounding viral RNAs. The cis-acting mechanism that naturally comes to mind is that viral RNA translation is taking place within the vesicle. As soon as the polyprotein is undergoing synthesis and processing, sufficient copies of 6K2-VPg-Pro are produced after a few rounds of translation to induce the formation of a membrane vesicle. Upon its formation, it encloses the viral RNA and the protein synthetic machinery through protein interaction with viral proteins, notably 6K2-VPg-Pro and RdRp (4, 5, 9, 48). Viral RNA translation continues within the vesicle and the viral proteins produced in situ contribute to the size increase of the vesicle and its maturation.
One requirement for this hypothesis is the presence of translation initiation factors within the replication complex vesicles. PABP, eIF(iso)4E, Hsc70, and eEF1A have been shown to interact with RdRp and/or VPg-Pro within 6K2-VPg-Pro-induced vesicles (4, 5, 9, 48, 53). PABP and Hsc70 undergo intracellular redistribution and a population of the proteins becomes associated with membranes during TuMV infection (5, 9). Here, colocalization of viral RNA with these host proteins has been demonstrated. Viral RNA synthesis sites were visualized as punctate structures distributed throughout the cytoplasm, likely enclosed within membrane vesicles induced by 6K2-VPg-Pro. This dsRNA punctate distribution is commonly observed for plant positive-strand RNA viruses (10, 31). As expected, TuMV proteins suspected to be involved in RNA replication, such as VPg-Pro, RdRp, and CI (helicase), were found to colocalize with these same punctate structures. Thus, the presence of three translation proteins enclosed within vesicles containing actively transcribing RNA was confirmed. Association of PABP or eEF1A with viral replication sites has been reported (8, 18, 20, 55, 57), and this association has generally been linked to replication of the viral genome. Here, at least three factors (and possibly Hsc70) were found with actively replicating RNA. Although a role in viral RNA synthesis cannot be excluded, the presence of so many translation factors is an indication that they are probably involved in viral protein synthesis.
An electron microscopy tomography study showed that coronavirus replication is supported by a reticulovesicular network of modified ER (23). This network integrates convoluted membranes, numerous interconnected double-membrane vesicles and vesicle packets. Although not found within them, the presence of ribosomes on the outer membrane of double-membrane vesicles and vesicle packets was noted. It is possible that the TuMV-induced vesicles at a higher resolution may show a similar assemblage of membrane structures and vesicles. Consequently, instead of translation taking place within the vesicles, viral proteins may be synthesized in the vicinity of the vesicle (i.e., on the cytoplasmic side) and being preferentially imported to the same vesicle.
In the case of positive-sense RNA viruses, virology textbooks generally depict viral RNA translation and synthesis as distinct processes that are physically separated (for example, see Fig. 12 and its legend in the appendix of reference 12). In this model, viral RNA is translated on ribosomes distributed randomly in the cytoplasm and resulting viral proteins necessary for viral RNA replication are exported to vesicle-enclosed replication complexes. In the case of poliovirus, it was even suggested that the viral RNA intended to be translated is structurally different (i.e., it does not have a VPg) from the RNA found associated with the replication complex (37). However, there are increasing reports indicating that viral RNA translation and replication are tightly coupled events. This is the case for picornaviruses (13) or ambisense viruses (35), where viral replication and/or transcription necessitates continuous viral protein synthesis. Egger et al. (11) demonstrated that preformed poliovirus vesicles could not accept viral RNA and proteins involved in replication and concluded that vesicle formation, viral RNA translation, and replication are cis-linked events. Inefficient complementation activity of poliovirus 2C and 3D proteins for the rescue of lethal mutations in the viral genome indicates that poliovirus RNA replication shows marked preference for proteins contributed in cis (47). Efficient Brome mosaic virus RNA1 replication requires 1a synthesis from RNA1 in cis (56), and eIF3 is associated with purified Brome mosaic virus replication complexes (38). Finally, it was found that coupling between translation and replication of RNA2 may occur in cells infected with Red clover necrotic mosaic virus (33). However, no physical mechanism responsible for the coupling of viral RNA translation with replication and/or transcription has been proposed for these studies.
Recently, Katsafanas and Moss showed that the host protein synthetic machinery can be sequestered within virus-induced structures (21). Poxviruses are large DNA viruses that replicate in cytoplasmic foci known as DNA factories. Poxvirus infection redistributes associated components of eIF4F and concentrates them within these factories (21, 49). To demonstrate that the factories were not only sites of DNA replication and transcription but also of viral RNA translation, these authors coinfected cells with two engineered viruses, each of which expressed a different fluorescent protein (yellow or cyan) fused to a late virion structural protein. The factories that stained uniquely with either yellow or cyan were readily observed, thus demonstrating that protein synthesis was taking place within the factories. This observation parallels the one obtained for TuMV. It will be interesting to learn for other viruses whether RNA translation is taking place in replication complex vesicles.
Another hallmark of TuMV-induced vesicles is their motility, which is dependent on microfilaments. Numerous examples of microtubule- or microfilament-mediated transport of viral components have been reported (14, 17). In plant-virus interactions, viruses appears to utilize the plant cytoskeleton for disease spread (46). For instance, viral movement proteins interact with actin microfilaments and microtubules and evidence has accumulated that both cytoskeletal systems may act as conduits not only for viral RNAs and virions but also for plant macromolecules to reach plasmodesmata (16). Recently, movement of RNA virus replication complexes has been documented. Hepatitis C virus replication complexes showed microtubule- and microfilament-dependent transport (25, 54). Tobacco mosaic virus replication complex trafficking is dependent on actin filaments (29) and is able to move in a cell-to-cell manner (22). The CaMV protein P6 forms motile inclusions that traffic along actin microfilaments and stabilize microtubules (15). Furthermore, initiation of viral infection requires an intact microtubule or microfilament network (15, 39). The biological function of TuMV vesicle movement is not clear but appears to be a requirement for the virus replication cycle since disruption of microfilaments inhibited virus production. One possible mechanism for inhibition of virus production appears to be the confinement of the vesicles in microfilament bundles, which is perhaps preventing fusion of vesicles among themselves. Because microfilament disassembly results in a collapse of the ER network, replication defect can also be attributed to the inability of virus to recruit ER membranes for replication or other processes.
Supplementary Material
Acknowledgments
We thank Marcel Desrosiers and Christian Charbonneau for helping with confocal microscopy, Karen S. Browning for the anti-eEF1A serum, Andreas Nebenführ for the Golgi organelle marker, and Elison B. Blancaflor for the GFP-ABD2-GFP construct. We thank Hélène Sanfaçon for critical reading of the manuscript and the anonymous reviewers of a previous submission.
This study was supported the National Science and Engineering Research Council of Canada and from Le Fonds de la Recherche sur la Nature et les Technologies from the Government of Québec.
Footnotes
Published ahead of print on 5 August 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Aizaki, H., K. S. Choi, M. Liu, Y. J. Li, and M. M. Lai. 2006. Polypyrimidine-tract-binding protein is a component of the HCV RNA replication complex and necessary for RNA synthesis. J. Biomed. Sci. 13:469-480. [DOI] [PubMed] [Google Scholar]
- 2.Avisar, D., M. Abu-Abied, E. Belausov, E. Sadot, C. Hawes, and I. A. Sparkes. 2009. A comparative study of the involvement of 17 Arabidopsis myosin family members on the motility of Golgi and other organelles. Plant Physiol. 150:700-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barco, A., and L. Carrasco. 1995. A human virus protein, poliovirus protein 2BC, induces membrane proliferation and blocks the exocytic pathway in the yeast Saccharomyces cerevisiae. EMBO J. 14:3349-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beauchemin, C., N. Boutet, and J.-F. Laliberté. 2007. Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in planta. J. Virol. 81:775-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beauchemin, C., and J.-F. Laliberté. 2007. The poly(A) binding protein is internalized in virus-induced vesicles or redistributed to the nucleolus during turnip mosaic virus infection. J. Virol. 81:10905-10913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandizzi, F., E. L. Snapp, A. G. Roberts, J. Lippincott-Schwartz, and C. Hawes. 2002. Membrane protein transport between the endoplasmic reticulum and the Golgi in tobacco leaves is energy dependent but cytoskeleton independent: evidence from selective photobleaching. Plant Cell 14:1293-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browning, K. S., J. Humphreys, W. Hobbs, G. B. Smith, and J. M. Ravel. 1990. Determination of the amounts of the protein synthesis initiation and elongation factors in wheat germ. J. Biol. Chem. 265:17967-17973. [PubMed] [Google Scholar]
- 8.Davis, W. G., J. L. Blackwell, P. Y. Shi, and M. A. Brinton. 2007. Interaction between the cellular protein eEF1A and the 3′-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J. Virol. 81:10172-10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dufresne, P. J., K. Thivierge, S. Cotton, C. Beauchemin, C. Ide, E. Ubalijoro, J.-F. Laliberté, and M. G. Fortin. 2008. Heat shock 70 protein interaction with turnip mosaic virus RNA-dependent RNA polymerase within virus-induced membrane vesicles. Virology 374:217-227. [DOI] [PubMed] [Google Scholar]
- 10.Dunoyer, P., C. Ritzenthaler, O. Hemmer, P. Michler, and C. Fritsch. 2002. Intracellular localization of the Peanut clump virus replication complex in tobacco BY-2 protoplasts containing green fluorescent protein-labeled endoplasmic reticulum or Golgi apparatus. J. Virol. 76:865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egger, D., N. Teterina, E. Ehrenfeld, and K. Bienz. 2000. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 74:6570-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flint, S. J., L. W. Enquist, V. R. Racaniello, and A. M. Skalka. 2009. Principles of virology: molecular biology, pathogenesis, and control of animal viruses, 3rd ed. ASM Press, Washington, DC.
- 13.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greber, U. F., and M. Way. 2006. A superhighway to virus infection. Cell 124:741-754. [DOI] [PubMed] [Google Scholar]
- 15.Harries, P. A., K. Palanichelvam, W. Yu, J. E. Schoelz, and R. S. Nelson. 2009. The cauliflower mosaic virus protein P6 forms motile inclusions that traffic along actin microfilaments and stabilize microtubules. Plant Physiol. 149:1005-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinlein, M. 2002. The spread of tobacco mosaic virus infection: insights into the cellular mechanism of RNA transport. Cell Mol. Life Sci. 59:58-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry, T., J.-P. Gorvel, and S. Méresse. 2006. Molecular motors hijacking by intracellular pathogens. Cell. Microbiol. 8:23-32. [DOI] [PubMed] [Google Scholar]
- 18.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holweg, C. L. 2007. Living markers for actin block myosin-dependent motility of plant organelles and auxin. Cell Motil. Cytoskelet. 64:69-81. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, C. M., D. R. Perez, R. French, W. C. Merrick, and R. O. Donis. 2001. The NS5A protein of bovine viral diarrhoea virus interacts with the {alpha} subunit of translation elongation factor-1. J. Gen. Virol. 82:2935-2943. [DOI] [PubMed] [Google Scholar]
- 21.Katsafanas, G. C., and B. Moss. 2007. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2:221-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawakami, S., Y. Watanabe, and R. N. Beachy. 2004. Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc. Natl. Acad. Sci. USA 101:6291-6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knoops, K., M. Kikkert, S. H. Worm, J. C. Zevenhoven-Dobbe, Y. van der Meer, A. J. Koster, A. M. Mommaas, and E. J. Snijder. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopek, B. G., G. Perkins, D. J. Miller, M. H. Ellisman, and P. Ahlquist. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5:e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai, C.-K., K.-S. Jeng, K. Machida, and M. M. C. Lai. 2008. Association of hepatitis C virus replication complexes with microtubules and actin filaments is dependent on the interaction of NS3 and NS5A. J. Virol. 82:8838-8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lain, S., M. T. Martin, J. L. Riechmann, and J. A. Garcia. 1991. Novel catalytic activity associated with positive-strand RNA virus infection: nucleic acid-stimulated ATPase activity of the plum pox potyvirus helicaselike protein. J. Virol. 65:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laliberté, J.-F., O. Nicolas, H. Chatel, C. Lazure, and R. Morosoli. 1992. Release of a 22-kDa protein derived from the amino-terminal domain of the 49-kDa NIa of turnip mosaic potyvirus in Escherichia coli. Virology 190:510-514. [DOI] [PubMed] [Google Scholar]
- 28.Léonard, S., C. Viel, C. Beauchemin, N. Daigneault, M. G. Fortin, and J.-F. Laliberté. 2004. Interaction of VPg-Pro of Turnip mosaic virus with the translation initiation factor 4E and the poly(A)-binding protein in planta. J. Gen. Virol. 85:1055-1063. [DOI] [PubMed] [Google Scholar]
- 29.Liu, J.-Z., E. B. Blancaflor, and R. S. Nelson. 2005. The Tobacco mosaic virus 126-kilodalton protein, a constituent of the virus replication complex, alone of within the complex aligns with and traffics along microfilaments. Plant Physiol. 138:1853-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin, M. T., M. T. Cervera, and J. A. Garcia. 1995. Properties of the active plum pox potyvirus RNA polymerase complex in defined glycerol gradient fractions. Virus Res. 37:127-137. [DOI] [PubMed] [Google Scholar]
- 31.McCartney, A. W., J. S. Greenwood, M. R. Fabian, K. A. White, and R. T. Mullen. 2005. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17:3513-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, S., and J. Krijnse-Locker. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizumoto, H., H.-O. Iwakawa, M. Kaido, K. Mise, and T. Okuno. 2006. Cap-independent translation mechanism of Red clover necrotic mosaic virus RNA2 differs from that of RNA1 and is linked to RNA replication. J. Virol. 80:3781-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson, B. K., X. Cai, and A. Nebenfuhr. 2007. A multicolored set of in vivo organelle markers for colocalization studies in Arabidopsis and other plants. Plant J. 51:1126-1136. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen, M., and A.-L. Haenni. 2003. Expression strategies of ambisense viruses. Virus Res. 93:141-150. [DOI] [PubMed] [Google Scholar]
- 36.Nicolas, O., and J.-F. Laliberté. 1992. The complete nucleotide sequence of Turnip mosaic potyvirus RNA. J. Gen. Virol. 73:2785-2793. [DOI] [PubMed] [Google Scholar]
- 37.Nomoto, A., N. Kitamura, F. Golini, and E. Wimmer. 1977. The 5′-terminal structures of poliovirion RNA and poliovirus mRNA differ only in the genome-linked protein VPg. Proc. Natl. Acad. Sci. USA 74:5345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quadt, R., C. C. Kao, K. S. Browning, R. P. Hershberger, and P. Ahlquist. 1993. Characterization of a host protein associated with brome mosaic virus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 90:1498-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roohvand, F., P. Maillard, J.-P. Lavergne, S. Boulant, M. Walic, U. Andreo, L. Goueslain, F. Helle, A. Mallet, J. McLauchlan, and A. Budkowska. 2009. Initiation of hepatitis C virus infection requires the dynamic microtubule network: role of the viral nucleocapsid protein. J. Biol. Chem. 284:13778-13791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saint-Jore-Dupas, C., A. Nebenführ, A. Boulaflous, M.-L. Follet-Gueye, C. Plasson, C. Hawes, A. Driouich, L. Faye, and V. Gomord. 2006. Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell 18:3182-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanchez, F., D. Martinez-Herrera, I. Aguilar, and F. Ponz. 1998. Infectivity of Turnip mosaic potyvirus cDNA clones and transcripts on the systemic host Arabidopsis thaliana and local lesion hosts. Virus Res. 55:207-219. [DOI] [PubMed] [Google Scholar]
- 42.Schaad, M. C., P. E. Jensen, and J. C. Carrington. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16:4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schramm, B., and J. K. Locker. 2005. Cytoplasmic organization of POXvirus DNA replication. Traffic 6:839-846. [DOI] [PubMed] [Google Scholar]
- 44.Shukla, D. D., C. W. Ward, and A. A. Brunt. 1994. The Potyviridae. CAB International, Wallingford, United Kingdom.
- 45.Suzan-Monti, M., B. L. Scola, L. Barrassi, L. Espinosa, and D. Raoult. 2007. Ultrastructural characterization of the giant volcano-like virus factory of Acanthamoeba polyphaga mimivirus. PLoS ONE 2:e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takemoto, D., and A. R. Hardham. 2004. The cytoskeleton as a regulator and target of biotic interactions in plants. Plant Physiol. 136:3864-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teterina, N., W. Zhou, M. Cho, and E. Ehrenfeld. 1995. Inefficient complementation activity of poliovirus 2C and 3D proteins for rescue of lethal mutations. J. Virol. 69:4245-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thivierge, K., S. Cotton, P. J. Dufresne, I. Mathieu, C. Beauchemin, C. Ide, M. G. Fortin, and J.-F. Laliberté. 2008. Eukaryotic elongation factor 1A interacts with turnip mosaic virus RNA-dependent RNA polymerase and VPg-Pro in virus-induced vesicles. Virology 377:216-225. [DOI] [PubMed] [Google Scholar]
- 49.Walsh, D., C. Arias, C. Perez, D. Halladin, M. Escandon, T. Ueda, R. Watanabe-Fukunaga, R. Fukunaga, and I. Mohr. 2008. Eukaryotic translation initiation factor 4F architectural alterations accompany translation initiation factor redistribution in poxvirus-infected cells. Mol. Cell. Biol. 28:2648-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, Y.-S., C.-M. Yoo, and E. B. Blancaflor. 2008. Improved imaging of actin filaments in transgenic Arabidopsis plants expressing a green fluorescent protein fusion to the C and N termini of the fimbrin actin-binding domain 2. New Phytologist 177:525-536. [DOI] [PubMed] [Google Scholar]
- 51.Wei, T., and A. Wang. 2008. Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit Sites in a COPI- and COPII-dependent manner. J. Virol. 82:12252-12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welsch, S., S. Miller, I. Romero-Brey, A. Merz, C. K. Bleck, P. Walther, S. D. Fuller, C. Antony, J. Krijnse-Locker, and R. Bartenschlager. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wittmann, S., H. Chatel, M. G. Fortin, and J.-F. Laliberté. 1997. Interaction of the viral protein genome linked of Turnip mosaic potyvirus with the translational eukaryotic initiation factor (iso) 4E of Arabidopsis thaliana using the yeast two-hybrid system. Virology 234:84-92. [DOI] [PubMed] [Google Scholar]
- 54.Wolk, B., B. Buchele, D. Moradpour, and C. M. Rice. 2008. A dynamic view of hepatitis C virus replication complexes. J. Virol. 82:10519-10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamaji, Y., T. Kobayashi, K. Hamada, K. Sakurai, A. Yoshii, M. Suzuki, S. Namba, and T. Hibi. 2006. In vivo interaction between Tobacco mosaic virus RNA-dependent RNA polymerase and host translation elongation factor 1A. Virology 347:100-108. [DOI] [PubMed] [Google Scholar]
- 56.Yi, G., and C. Kao. 2008. cis- and trans-acting functions of brome mosaic virus protein 1a in genomic RNA1 replication. J. Virol. 82:3045-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeenko, V. V., L. A. Ryabova, A. S. Spirin, H. M. Rothnie, D. Hess, K. S. Browning, and T. Hohn. 2002. Eukaryotic elongation factor 1A interacts with the upstream pseudoknot domain in the 3′ untranslated region of tobacco mosaic virus RNA. J. Virol. 76:5678-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.