Abstract
Human mesenchymal stem cells (hMSCs) can be genetically modified with viral vectors and hold promise as a cell source for regenerative medicine, yet how hMSCs respond to viral vector transduction remains poorly understood, leaving the safety concerns unaddressed. Here, we explored the responses of hMSCs against an emerging DNA viral vector, baculovirus (BV), and discovered that BV transduction perturbed the transcription of 816 genes associated with five signaling pathways. Surprisingly, Toll-like receptor-3 (TLR3), a receptor that generally recognizes double-stranded RNA, was apparently upregulated by BV transduction, as confirmed by microarray, PCR array, flow cytometry, and confocal microscopy. Cytokine array data showed that BV transduction triggered robust secretion of interleukin-6 (IL-6) and IL-8 but not of other inflammatory cytokines and beta interferon (IFN-β). BV transduction activated the signaling molecules (e.g., Toll/interleukin-1 receptor domain-containing adaptor-inducing IFN-β, NF-κB, and IFN regulatory factor 3) downstream of TLR3, while silencing the TLR3 gene with small interfering RNA considerably abolished cytokine expression and promoted cell migration. These data demonstrate, for the first time, that a DNA viral vector can activate the TLR3 pathway in hMSCs and lead to a cytokine expression profile distinct from that in immune cells. These findings underscore the importance of evaluating whether the TLR3 signaling cascade plays roles in the immune response provoked by other DNA vectors (e.g., adenovirus). Nonetheless, BV transduction barely disturbed surface marker expression and induced only transient and mild cytokine responses, thereby easing the safety concerns of using BV for hMSCs engineering.
Toll-like receptors (TLRs) are pattern recognition receptors that recognize a variety of pathogen-associated molecular patterns and are essential for activating innate immunity and potentiating adaptive immunity against pathogens (for a review, see references 2, 15, and 23). To date, 11 TLRs have been identified in humans (2). For example, TLR2 recognizes bacterial lipoproteins and peptidoglycans, TLR3 recognizes virus-derived double-stranded RNA (dsRNA) and a synthetic dsRNA analogue poly(I:C) (polyriboinosinic-polyribocytidylic acid), TLR4 recognizes lipopolysaccharides, and TLR9 recognizes the unmethylated CpG DNA motifs. Upon the engagement of cognate ligands, TLRs are activated and recruit Toll/IL-1 receptor-containing adaptor molecules such as myeloid differentiating factor 88 (MyD88) and Toll/interleukin-1 receptor domain-containing adaptor-inducing beta interferon (TRIF). Among the TLRs, the TLR3 pathway is unique in that its signaling cascade begins by recruiting TRIF (2, 15, 33). TRIF can signal through interferon regulatory factor 3 (IRF-3) phosphorylation, leading to downstream beta interferon (IFN-β) expression. TRIF also can orchestrate with TRAF6 and RIP1, leading to NF-κB activation and subsequent expression of cytokines and chemokines such as interleukin-1 (IL-1), IL-6, IL-8, IL-12, MCP-1 (CCL2), RANTES (CCL5), and MIP-2 (CXCL2).
The baculovirus (BV) Autographa californica multiple nucleopolyhedrovirus is a DNA virus that infects insects as its natural hosts and that has been developed as a biological insecticide. However, BV also efficiently transduces a broad range of mammalian cells in which BV neither replicates nor is toxic. Also, recombinant virus construction, propagation, and handling can be performed readily in biosafety level 1 facilities. These attributes have inspired the development of BV vectors for in vitro and in vivo gene delivery (6, 28), cartilage tissue engineering (3), development of cell-based assays, delivery of vaccine immunogens, production of viral vectors, and cancer therapy (for a review, see references 14 and 17). Furthermore, BV transduces human mesenchymal stem cells (hMSCs) derived from bone marrow at efficiencies greater than 80% (12) and accelerates osteogenesis of hMSCs in vitro and in vivo when expressing an osteogenic growth factor (4). hMSCs are capable of differentiating into multiple cell types (e.g., chondrocytes, osteoblasts, and endothelial cells) and possess immunosuppressive and immunomodulatory properties (32). Therefore, hMSC-based cell therapy has captured growing attention in regenerative medicine and has advanced to various phases of clinical trials for the treatment of damaged myocardium, knee injuries, graft-versus-host disease, and Crohn's disease (22). hMSCs also serve as a gene delivery carrier for the treatment of cancer, osteogenesis imperfecta (13), and various neurological disorders (27). As such, the efficient BV transduction of hMSCs offers a new, attractive option for hMSC engineering.
Despite the potential of hMSCs for cell and gene therapy, whether the genetic modification provokes undesired cellular responses has yet to be explored. The lack of safety evaluation will hamper future clinical applications of genetically modified hMSCs. Therefore, the overriding objective of this study was to assess how hMSCs respond to BV transduction.
MATERIALS AND METHODS
BV and hMSCs.
A recombinant BV harboring no mammalian transgene cassette was used for transduction. The virus was amplified and harvested, and titers were determined by an end point dilution assay based on virus infectivity in insect cells (25). Bone marrow-derived hMSCs were obtained from Cambrex Co., selected, enriched, cultured in alpha minimal essential medium (α-MEM) containing 10% fetal bovine serum (HyClone) as described previously (11), and expanded to passage 11 for all experiments.
The virus transduction was performed on six-well plates as described previously (21) with minor modifications. Depending on the multiplicity of infection (MOI), a certain volume of virus supernatant was mixed with NaHCO3-deficient α-MEM to adjust the final volume to 500 μl (per well). The transduction was initiated by adding the virus mixture to the cells and was continued by gently shaking the plates on a rocking plate for 4 h at 25 to 27°C. For mock transduction, the cells were incubated under the same conditions with a solution consisting of 400 μl of NaHCO3-deficient α-MEM and 100 μl of fresh TNM-FH medium. After being washed, the cells were replenished with 2 ml of α-MEM containing 10% fetal bovine serum for culture.
For UV inactivation, the virus was exposed to short-wavelength UV radiation at a distance of 5 cm for 30 min on ice (1.6 × 104 mJ/cm2) as described previously (1). For RNase treatment, the virus (per 100 μl) was incubated with 2 μl of RNase A (>70 Kunitz units/mg of protein; Sigma) (35) or 2 μl of TNM-FH medium for 1 h at 37°C as described previously.
Microarray and PCR array.
Total RNA was extracted with an RNeasy Mini Kit (Qiagen) for cDNA synthesis using a Reverse Transcriptase 1st-Strand cDNA Synthesis Kit (Epicentre Biotechnologies). Labeled cRNA was prepared by using an Amino Allyl MessageAmpII aRNA amplification kit (Ambion). The cRNA (10 μg) was fragmented and then hybridized on a Phalanx Human One Array (HOA 4.3; Phalanx Biotech Group, Inc.) as described previously (9). Each microarray contains 32,050 oligonucleotide probes that include 30,968 human gene probes for transcription expression profiling and 1,082 experimental control probes. Detailed descriptions of the gene array list are available from http://www.phalanx.com.tw/tech_support/gene_lists.html. Arrays were scanned by a DNA Microarray Scanner (Agilent Technologies), and the fluorescence intensities were extracted by GenePix Pro, version 6.0 (Molecular Devices). The raw data were preprocessed by log2 transformation and global LOWESS normalization. The preprocessed data were then analyzed by the Limma package of R software. The significantly changed genes, as determined by one-way analysis of variance, were defined as the genes with relative changes in expression of >2-fold or <0.5-fold and adjusted P values of less than 0.05. To identify which known pathways were affected by BV transduction, the significantly changed genes were analyzed by a web-based tool, Pathway-Express (7), which is freely available as part of Onto-Tools (http://vortex.cs.wayne.edu).
Alternatively, the total RNA was reverse transcribed to cDNA using a Moloney murine leukemia virus Reverse Transcriptase 1st-Strand cDNA Synthesis Kit (Epicentre Biotechnologies). The cDNA was analyzed using the a human TLR pathway-focused RT2 Profiler PCR Array (SABiosciences) following the manufacturer's instructions.
Flow cytometry.
To characterize surface marker expression, hMSCs were labeled with different antibodies. Anti-CD14-fluorescein isothiocyanate (FITC), anti-CD19-FITC, and anti-CD105-FITC were purchased from Miltenyi Biotec (MACS); anti-CD29-phycoerythrin (PE), anti-CD73-PE, anti-CD44-FITC, anti-CD90-FITC, anti-human leukocyte antigen class I (HLA-I)-PE, and anti-HLA-II-PE were purchased from BD Biosciences; anti-CD45-FITC and anti-CD34-PE were purchased from Chemicon. After being labeled, hMSCs were detached and analyzed with a flow cytometer (FACSCalibur; BD Biosciences). To characterize TLR expression, transduced and mock-transduced cells were fixed and permeabilized with 4% formaldehyde and 0.5% Tween-20. After a washing step, the cells were incubated with different primary antibodies (1:150 dilution) for 1 h at 4°C in the dark. Mouse immunoglobulin G (IgG) primary antibodies were purchased from Abcam and were specific for human TLR2 (ab9100), TLR3 (ab12085), TLR4 (ab30667), and TLR9 (ab17236). After a washing step, the cells were incubated with Alexa Fluor 488-conjugated, goat anti-mouse IgG (Invitrogen) for 1 h at 4°C in the dark. After a washing step, the cells were collected for flow cytometry analyses. As controls, the cells were incubated for 30 min with 500 μl of NaHCO3-deficient α-MEM containing 10 μg/ml poly(I:C) (Sigma), 10 ng/ml lipopolysaccharide (Sigma), or 1 μM CpG oligonucleotide (ODN 2216 and ODN 2006; Invitrogen) and then analyzed for TLR expression in a similar fashion.
Cytokine measurement.
Conditioned medium from the hMSC cultures was analyzed using a fluorescence bead immunoassay (Bender MedSystems) that detects 11 cytokines simultaneously. IL-6 and IL-8 were also measured using Module Sets of enzyme-linked immunosorbent assays (ELISAs) (Bender Medsystems).
Immunofluorescence labeling/confocal microscopy.
The cells were fixed and permeabilized as described above, followed by extensive washing and primary antibody staining (1:150 dilution) for 1 h at 4°C in the dark. The primary antibodies were specific for human TLR3, IRF-3 (ab50772; Abcam), or phosphorylated NF-κB (3033; Cell Signaling Technology). After a washing step, the cells were incubated with the goat anti-mouse (for TLR3 and IRF-3) or goat anti-rabbit (for pNF-κB) antibody conjugated with Alexa Fluor 488 (Invitrogen) for 1 h at 4°C in the dark. After a washing step, the cells were stained with 4,6-diamidino-2-phenylindole (DAPI; Vector Labs) and visualized with a confocal microscope (Nikon TE2000 equipped with the confocal upgrade laser kit).
Western blotting.
The cytoplasmic and nuclear proteins were separately extracted using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce) supplemented with the Halt protease and phosphatase inhibitor cocktails (Pierce), followed by Western blotting. The primary antibodies (1:1,000 dilution) were specific for TRIF (4596; Cell Signaling Technology), phosphorylated IRF-3 (4947; Cell Signaling Technology), and phosphorylated NF-κB or β-actin (A-2066; Sigma). The secondary antibody was species-specific horseradish peroxidase-conjugated IgG (1:5,000 dilution; Amersham Biosciences). The images were developed using an enhanced chemiluminescence kit (Amersham Biosciences).
siRNA knockdown of TLR3.
To knock down TLR3, hMSCs were nucleofected with 5 μg of a control small interfering RNA (siRNA) plasmid (InvivoGen) or a plasmid expressing the TLR3 siRNA (psiTLR3; InvivoGen) using a Human MSC Nucleofector Kit (Amaxa Biosystems). At 48 h posttransfection, hMSCs were transduced with BV or treated with poly(I:C) as described above. The spent medium was collected at 24 h posttransduction (hpt) for ELISA, and the cells were trypsinized for migration assays.
Transwell migration assay.
The trypsinized hMSCs (5 × 104) were loaded onto the upper chamber of the transwell inserts with 8-μm-pore-size membrane filters (BD Biosciences), while 500 μl of α-MEM was loaded in the bottom chamber. After 4 h of incubation, the upper sides of the filters were carefully washed, and nonmigrated cells were removed with a cotton-tipped applicator. The cells migrating to the lower sides were labeled with DAPI-containing mounting medium and counted under a fluorescence microscope. At least 10 fields (magnification, ×200) in two filters were counted for each sample, and the data are expressed as the average number of migrated cells/field.
Statistical analysis.
All data represent the mean ± standard deviation or mean values of at least three independent culture experiments. The data were statistically analyzed by one-way analysis of variance. A P value of <0.05 was considered significant.
Microarray data accession number.
Array data sets were deposited in the NCBI Gene Omnibus Express database under the accession number GSE15810.
RESULTS
Expression of hMSC surface markers.
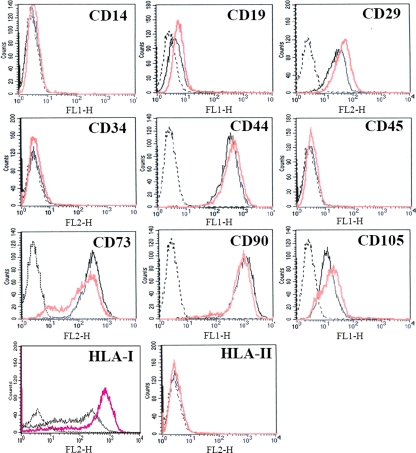
To examine whether BV transduction altered surface characteristics, hMSCs were mock transduced or transduced with a BV carrying no mammalian gene cassette at an MOI of 100, followed by immunofluorescence labeling and flow cytometry analyses at 24 h hpt. In agreement with the surface marker profiles in normal hMSCs (18, 31), the mock-transduced hMSCs expressed CD29, CD44, CD73, CD90, CD105, and HLA-I but were negative for CD14, CD19, CD34, CD45, and HLA-II (Fig. 1). BV transduction did not apparently alter the surface expression profile, except that CD73 expression was slightly diminished while HLA-I expression was elevated.
FIG. 1.
Expression of hMSC surface markers. Cells were mock transduced (black solid lines) or transduced with BV at an MOI of 100 (pink lines) and subjected to immunofluorescence labeling/flow cytometry analyses at 24 hpt.
BV transduction-upregulated genes associated with the TLR signaling pathway.
To explore the global responses of hMSCs to BV transduction, hMSCs were treated as described in the legend of Fig. 1 and subjected to microarray analysis at 24 hpt. Of the 30,968 human genes on the microarray, we identified 548 upregulated (>2-fold) and 268 downregulated (<0.5-fold) known genes after BV transduction compared with the mock transduction control (see Tables S1 and S2 in the supplemental material). Pathway analysis using the Pathway-Express tool demonstrated five signaling pathways that were profoundly disturbed: cell adhesion molecules, TLR, Jak-STAT, apoptosis, and antigen processing and presentation (see Tables S3 to S8 in the supplemental material). Since the activation of the TLR pathway is essential for initiating innate immunity and can trigger the other four pathways, we focused on the TLR pathway in subsequent experiments and analyses.
The microarray data revealed significant upregulation of TLR1, TLR2, and TLR3 but not of other TLR genes (Table 1). Certain genes encoding the TLR signaling molecules (e.g., MyD88 and IRAK2), downstream cytokines (e.g., IL-6 and IL-8), and other genes downstream of the TLR3 pathway (e.g., RSAD2, INDO, and PTGS2) were also significantly upregulated. To confirm the data, transcription was also quantified by using the RT2 Profiler PCR Array, which detects 84 genes involved in the TLR pathway (including TLR1 to TLR10). In accord with the microarray data, the PCR array revealed the upregulation of such genes as TLR3, MyD88, IRAK2, IL-6, IL-8, and PTGS2 (Table 1). In contrast, the PCR array detected upregulation of neither TLR1 nor TLR2 but revealed the upregulation of other genes involved in the TLR pathway (e.g., NFKBIA, TRIF, and TRAF6). The discrepancy between the microarray and PCR array data sets probably arose from the relatively weak stimulation of these genes by BV transduction.
TABLE 1.
Significantly changed genes associated with the TLR signaling pathway by BV transduction
| Gene group and name | Relative change in expression (n-fold) as determined by:a
|
|
|---|---|---|
| Microarray | PCR array | |
| TLR genes | ||
| TLR1 | 2.6 | ND |
| TLR2 | 6.6 | ND |
| TLR3 | 33.4 | 1157.1 |
| TLR signaling-associated molecules | ||
| MYD88 | 8.9 | 5.8 |
| IRAK2 | 5.7 | 10.2 |
| NFKBIA | ND | 9.4 |
| TRIF | ND | 3.4 |
| TRAF6 | ND | 1.8 |
| Cytokines and chemokines | ||
| CXCL10 | 814.0 | 465.1 |
| CCL5 | 231.8 | NA |
| IL1B | 34.6 | 95.4 |
| IL1A | 27.7 | 26.0 |
| IL-6 | 13.7 | 27.0 |
| IL12A | 12.6 | 12.0 |
| CXCL2 | 10.8 | NA |
| IL-8 | 6.2 | 26.4 |
| CCL2 | 4.4 | NA |
| Other genes associated with TLR3 | ||
| RSAD2 | 1220.6 | NA |
| INDO | 106.9 | NA |
| PTGS2 | 5.2 | 19.3 |
| SIGIRR | 2.9 | 3.4 |
| PELI1 | 2.7 | 3.0 |
The data represent the average values from three to five independent culture experiments. ND, not detectable; NA, not available in the PCR array.
BV transduction of hMSCs triggered IL-6 and IL-8 production.
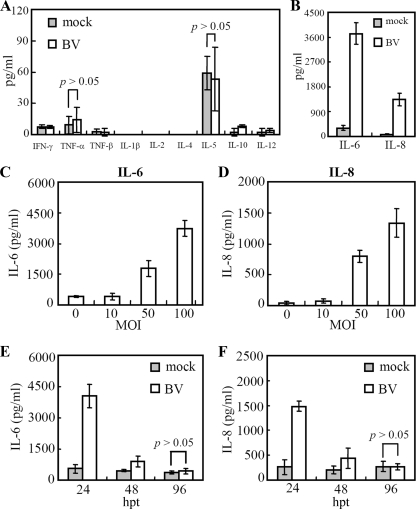
To screen the BV-induced cytokines at the protein level, the conditioned medium collected at 24 hpt was analyzed by a multiplex cytokine array which simultaneously detects 11 cytokines (Fig. 2A and B). Compared with the mock transduction control, BV transduction did not significantly (P > 0.05) elicit the secretion of IFN-γ, tumor necrosis factor alpha (TNF-α), TNF-β, IL-1β, IL-2, IL-4, IL-5, IL-10, and IL-12 but provoked high-level secretion of IL-6 (≈3,722 pg/ml) and IL-8 (≈1,334 pg/ml). Such induction was dose dependent as IL-6 and IL-8 expression increased with elevating MOIs (Fig. 2C and D). To confirm the result and examine the kinetics, the protein concentrations were measured again by ELISA at 24, 48, and 96 hpt. The results shown in Fig. 2E and F demonstrate that the expression of both IL-6 and IL-8 peaked at 24 hpt and fell to background levels at 96 hpt, indicating a transient cytokine response. It is also noteworthy that BV transduction did not provoke the secretion of antiviral IFN-α (5) and IFN-β (see Table S9 in the supplemental material).
FIG. 2.
Cytokine production by BV-transduced hMSCs. (A and B) Cytokine production at an MOI of 100. (C and D) IL-6 and IL-8 production at different MOIs. (E and F) IL-6 and IL-8 production at different times. The mock-transduced hMSCs served as the controls. Cytokine production was measured using a fluorescence bead immunoassay that detects 11 cytokines (A to D) or ELISA kits (E and F).
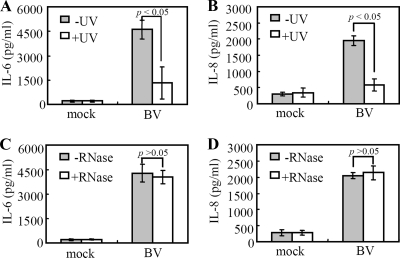
Whether cytokine induction required infectious BV was explored by inactivating the BV with UV prior to transduction. The ELISA data (Fig. 3A and B) showed that UV inactivation significantly (P < 0.05) abolished the BV-induced IL-6 and IL-8 secretion, indicating the essential role of the live virus. Since IL-6 and IL-8 can be elicited by dsRNA as a result of TLR3 activation (18, 31), the virus solutions were treated with RNase or TNM-FH medium prior to transduction. The results shown in Fig. 3C and D show that RNase treatment retarded secretion of neither IL-6 nor IL-8 after BV transduction, thus ruling out a role for RNA. These data collectively confirmed that BV itself provoked the cytokine response.
FIG. 3.
Cytokine production required infectious BV. (A and B) IL-6 and IL-8 production by the hMSCs transduced with virus (MOI of 100) that was untreated (−) or treated (+) with UV light. (C and D) IL-6 and IL-8 production by the hMSCs transduced with virus (MOI of 100) that was pretreated with 2 μl of RNase A (+RNase) or 2 μl of TNM-FH (−RNase) for 1 h at 37°C. In parallel, cells were mock transduced and served as controls. All spent media were collected at 24 hpt for ELISAs.
BV transduction of hMSCs triggered the TLR3 pathway.
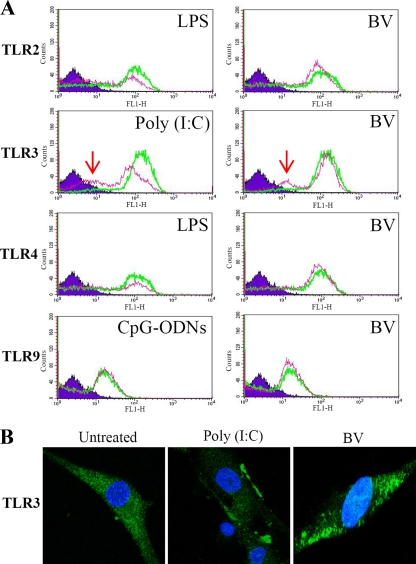
To examine the induction of TLR3 and other TLRs at the protein level, hMSCs were transduced with BV or treated with different TLR agonists and were subjected to immunofluorescence labeling/flow cytometry analyses (Fig. 4A). Compared with the mock transduction control, BV transduction led to the emergence of a peak when cells were labeled with the TLR3 antibody. Such a peak shift was due to receptor activation, internalization, and degradation (31) and was similarly observed for the sample treated with the TLR3 ligand, poly(I:C). However, BV transduction did not apparently provoke TLR2, TLR4, or TLR9.
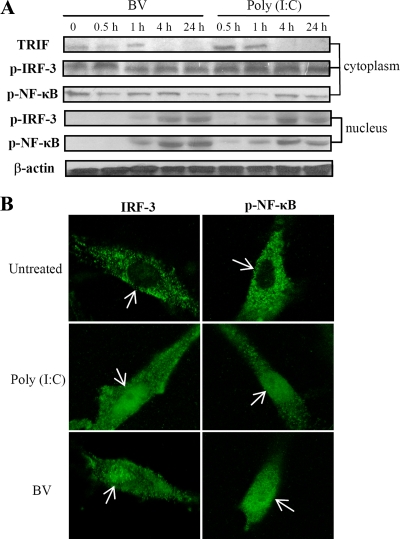
FIG. 4.
BV transduction of hMSCs triggered TLR3 activation. (A) hMSCs were mock transduced or transduced with BV. At 0.5 hpt the cells were subjected to fixation, permeabilization, immunostaining with anti-TLR antibodies, and flow cytometry analyses. The green and pink lines indicate the mock-transduced and transduced cells, respectively. As controls, cells were treated with TLR ligands for 30 min and subjected to the same analyses. The peak shift as indicated by the arrows demonstrated TLR3 activation after BV transduction and poly(I:C) treatment. (B) Cells were treated as described in panel A, labeled with anti-TLR3 antibody, counterstained with DAPI, and examined by confocal microscopy (Nikon TE2000 equipped with a confocal upgrade laser kit). Magnification, ×1,000. LPS, lipopolysaccharide.
TLR3 activation was further visualized by confocal microscopy (Fig. 4B). TLR3 expression was diffuse in the cytoplasm of the untreated hMSCs but was more focused along the edge of the BV-transduced cells, which was likewise observed in the poly(I:C)-treated hMSCs (Fig. 4B) (31). The results shown in Fig. 4, in conjunction with the microarray and PCR array data, concretely attested to TLR3 activation by BV transduction.
In immune cells, TLR3 activation induces TRIF expression and results in the nuclear translocation of phosphorylated IRF-3 and NF-κB (16). Western blot analyses of hMSCs (Fig. 5A) demonstrated that both BV transduction and poly(I:C) treatment stimulated a gradual increase in TRIF expression for 4 h and accumulation of phosphorylated IRF-3 and NF-κB in the nucleus. The nuclear trafficking of IRF-3 and NF-κB was further confirmed by confocal microscopy (Fig. 5B), which illustrated the absence of IRF-3 and NF-κB in the nuclei of untreated hMSCs and the presence of IRF-3 and NF-κB in the nuclei after BV transduction and poly(I:C) treatment.
FIG. 5.
BV transduction activated the TLR3 signaling pathway. (A) Activation of TRIF, IRF-3, and NF-κB. (B) Nuclear translocation of IRF-3 and NF-κB. The nuclear and cytoplasmic proteins were separately extracted from the transduced hMSCs (MOI of 100) and analyzed by Western blotting using primary antibodies specific for human TRIF, phosphorylated IRF-3 (p-IRF-3), phosphorylated NF-κB (p-NF-κB), or β-actin. Alternatively, cells were immunostained with antibodies specific for human IRF-3 or phosphorylated NF-κB for confocal microscopy. For comparison, the cells were left untreated or were treated with poly(I:C) for 24 h for the same analyses. Magnification, ×1,000.
TLR3 knockdown diminished BV-induced cytokine secretion and promoted migration.
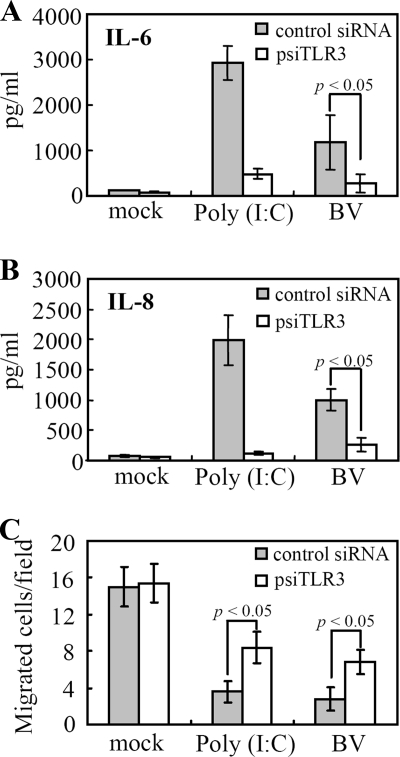
To correlate TLR3 activation and cytokine secretion, cells were nucleofected with a plasmid expressing the control siRNA or psiTLR3. After 48 h of culture, the cells were mock transduced, transduced, or treated with poly(I:C). As depicted in Fig. 6A and B, psiTLR3 treatment of hMSCs considerably abrogated poly(I:C)-induced IL-6 and IL-8 secretion, confirming the TLR3 knockdown by psiTLR3. Accordingly, TLR3 silencing by psiTLR3 treatment significantly (P < 0.05) attenuated BV-induced IL-6 and IL-8 secretion.
FIG. 6.
TLR3 silencing diminished BV-induced cytokine secretion and promoted migration. (A) IL-6 expression. (B) IL-8 expression. (C) Cell migration. hMSCs were nucleofected with a control siRNA plasmid or with psiTLR3, which expresses the TLR3 siRNA. After 48 h of culture, the cells were mock transduced, transduced with BV, or treated with poly(I:C). Twenty-four hours later, the medium was collected for ELISAs, and the trypsinized hMSCs (5 × 104) were loaded onto the transwell inserts. The migrated cells were labeled with DAPI-containing mounting medium and counted under a fluorescence microscope. Data are expressed as the average number of migrated cells/field.
Additionally, we examined the effect of TLR3 knockdown on BV-induced migration by the transwell migration assay. The results shown in Fig. 6C indicate that the migration of cells treated with the control siRNA was remarkably impeded by both poly(I:C) treatment and BV transduction, but psiTLR3 treatment significantly (P < 0.05) ameliorated the migration of poly(I:C)-treated and BV-transduced hMSCs.
DISCUSSION
The present study primarily aimed to explore the hMSC response to BV transduction and to decipher the molecular pathway. We determined that most hMSC surface markers remained undisturbed after BV transduction (Fig. 1), suggesting that hMSC characteristics are retained. This response contrasted markedly with the evident BV-induced upregulation of surface molecules (e.g., HLA-II) in dendritic cells (29) but was in line with the negligible perturbation of hMSC marker expression (e.g., CD34 and CD105) after poly(I:C) treatment (18). BV transduction only slightly upregulated HLA-I, which is desirable since HLA-I is responsible for presenting endogenously synthesized proteins to CD8+ T cells. BV transduction also marginally downregulated CD73, but the physiological significance of this is unknown.
We identified 816 known genes that were significantly perturbed by BV transduction. Among all TLR genes, TLR3 expression showed the most pronounced upregulation. Concurrent with the TLR3 pathway (see introduction), BV transduction upregulated not only TLR3 but its downstream genes such as TRIF, TRAF6, NFKB1A (encoding IκB), IL-6, IL-8, IL12A, CCL2, CCL5, and CXCL2 (Table 1; see also Tables S1 and S2 in the supplemental material). At the protein level, BV elicited transient IL-6 and IL-8 production in a dose-dependent manner (Fig. 2 and 3), which concurred with the activation of TLR3 (Fig. 4) and its signaling molecules like TRIF, IRF-3, and NF-κB (Fig. 5). Critically, silencing TLR3 expression considerably abolished BV-induced cytokine secretion and augmented hMSC migration (Fig. 6). These data collectively confirmed the activation of the TLR3 signaling pathway by BV.
However, BV transduction provoked no secretion of IL-1β, IFN-γ, IL-12, and TNF-α (Fig. 2A). These proteins were highly expressed by BV-transduced dendritic cells (1) but were not robustly secreted by the poly(I:C)-treated hMSCs (18, 31). Nor did we detect IFN-β secretion from 0.25 to 24 h after BV transduction or poly(I:C) treatment (see Table S9 in the supplemental material). IFN-β is the signature IFN induced after TLR3 activation in murine cells (2, 15), but its expression was not reported in studies that treated hMSCs with poly(I:C) (18, 31). In contrast, Opitz et al. recently showed that poly(I:C) treatment of hMSCs induced detectable IFN-β secretion and a subsequent signaling loop (24). One key difference was the poly(I:C) dose (50 μg/ml) these investigators used, which was markedly higher than amounts used in this (10 μg/ml) and other studies. As such, it appears that in hMSCs TLR3 ligation could elicit IFN-β secretion but at a fairly low magnitude. This suggests that in hMSCs certain pathways downstream of IRF-3 might be lacking or blocked unless potently stimulated. In this study, the virus dose (MOI of 100) used is sufficient to transduce 80 to 90% of hMSCs (12) and induce ectopic bone formation in vivo when hMSCs express an osteogenic factor (4). Given that these IFNs and cytokines are pivotal in establishing the antiviral state and immune responses, the undetectable induction of these proteins at an MOI of 100 is instrumental for the safe use of BV-transduced hMSCs for tissue regeneration.
Among the cytokines investigated, we detected only IL-6 and IL-8. This is conceivable as they are constitutively expressed by hMSCs (19) and are potently stimulated after poly(I:C) treatment (Fig. 6) (18, 31). IL-6 dictates the regulation of both inflammatory responses and hematopoiesis (26), and its overproduction relates to the pathology of autoimmune diseases and tissue remodeling. Conversely, IL-8 is present in the inflammatory milieu during tissue repair (34). The robust secretion of IL-6 and IL-8 thus suggests that BV transduction might impact hMSC differentiation and potentiate the inflammatory response after transplantation. To date, the consequences of TLR3 activation and IL-6/8 expression on hMSCs remain elusive. It was reported that TLR3 activation in hMSCs promotes migration (31) and hampers immunosuppressive properties but interferes with neither the phenotype nor the differentiation potential (18). However, it was also shown that TLR3 activation enhances the immunosuppressive properties of hMSCs (24). The discrepancy likely stems from the differences in experimental procedures, poly(I:C) dose, and duration of ligand treatment (24). For example, hMSCs have been incubated with poly(I:C) for 5 days (18) or 24 h (24) prior to evaluation of the immunosuppressive properties. In our hands, BV transduction of hMSCs did not impair long-term proliferation (11), differentiation (12), and immunosuppressive properties (5). The disparity in the immunosuppressive properties could arise from the differences in the protocols because the cells were exposed to BV for only 4 h, after which the virus was withdrawn. Also the BV-induced cytokine response was transient, precipitously dropping after 24 hpt and vanishing at 96 hpt (Fig. 2E and F). As a result, unlike results of previous studies that continuously stimulated hMSCs with poly(I:C), BV transduction only transiently activated the TLR3-mediated responses, which accounts for the intangible adverse effects. Our data, however, suggest that hMSCs be transplanted after cytokine responses wane in order to circumvent the disturbance of hMSC functions and elicitation of immune responses in vivo.
Our findings also raised an intriguing question: how did BV, a DNA virus, trigger the TLR3 pathway that is generally regarded as a sensor of dsRNA? Since BV genes (e.g., immediate-early gene ie1) can be expressed at low levels in mammalian cells (20), the most likely explanation is that some BV genes were transcribed in hMSCs and that the RNA intermediates were recognized by TLR3. However, the underlying mechanism(s) awaits further investigation. Also intriguing is that BV DNA activated the TLR9 pathway in mouse immune cells (1), yet only TLR3 activation was detected in hMSCs. Because hMSCs express high levels of TLR3 and TLR4 but low levels of TLR1, TLR2, TLR5, and TLR6 and negligible levels of TLR7 to TLR10 (18), the undetectable activation of TLR7 to TLR9 may be explained by the lack of viral DNA-sensing (TLR9) and single-stranded RNA-sensing (TLR7) receptors.
In summary, hMSCs can be genetically engineered with various viral vectors (8) and serve as a promising cell and gene therapy vehicle, yet little is known about how hMSCs respond to viral vector transduction. This study, for the first time, systematically explored the cellular responses of hMSCs to virus transduction at the molecular level. We revealed that BV transduction of hMSCs barely perturbed surface marker expression even while altering the expression of genes implicated in several pathways. We also provided the first evidence that a DNA viral vector can activate the TLR3 pathway in hMSCs, leading to a cytokine expression profile distinct from that in immune cells. Although TLR3 has been implicated in controlling the infection of two DNA viruses (vaccinia virus [10] and mouse cytomegalovirus [30]), there was no direct evidence confirming the induction of the TLR3 pathway by a DNA virus until the recent discovery that Kaposi's sarcoma-associated herpesvirus triggers the TLR3 pathway in human monocytes (35). Since DNA vectors including adenovirus, herpes simplex virus, and adeno-associated virus have been employed for genetically modifying hMSCs, our findings underscore the importance of evaluating whether these vectors also provoke the TLR3 signaling cascade and downstream immune responses. Our data also indicate that BV transduction elicits only mild and transient responses, thereby easing the safety concerns of using BV for hMSC engineering.
Supplementary Material
Acknowledgments
We acknowledge the support from the VTY Joint Research Program, Tsou's Foundation (VGHUST98-P5-17), National Tsing Hua University Booster Program (98N2901E1), National Science Council (NSC 97-2627-B-007-014), Ministry of Economic Affairs (98-EC-17-A-17-R7-0525), and National Health Research Institutes (NHRI-EX97-9412EI), Taiwan.
We declare that we have no competing financial interests.
G.-Y.C., H.-C.S., and Y.-C.H. designed the research; G.-Y.C., H.-C.S., C.-Y.C., S.-M.H., Y.-J.C., W.-H.L., C.-K.C., J.-L.H., and H.-J.S. performed the research; S.-M.H., H.-J.S. and Y.-J.C. contributed new reagents or analytic tools; G.-Y.C., H.-C.S., and H.-J.S. analyzed data; and G.-Y.C., H.-C.S., S.-M. H., and Y.-C.H. wrote the paper.
Footnotes
Published ahead of print on 5 August 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abe, T., H. Hemmi, H. Miyamoto, K. Moriishi, S. Tamura, H. Takaku, S. Akira, and Y. Matsuura. 2005. Involvement of the Toll-like receptor 9 signaling pathway in the induction of innate immunity by baculovirus. J. Virol. 79:2847-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124:783-801. [DOI] [PubMed] [Google Scholar]
- 3.Chen, H.-C., L.-Y. Sung, W.-H. Lo, C.-K. Chuang, Y.-H. Wang, J.-L. Lin, and Y.-C. Hu. 2008. Combination of baculovirus-mediated BMP-2 expression and rotating-shaft bioreactor culture synergistically enhances cartilage formation. Gene Ther. 15:309-317. [DOI] [PubMed] [Google Scholar]
- 4.Chuang, C.-K., L.-Y. Sung, S.-M. Hwang, W.-H. Lo, H.-C. Chen, and Y.-C. Hu. 2007. Baculovirus as a new gene delivery vector for stem cells engineering and bone tissue engineering. Gene Ther. 14:1417-1424. [DOI] [PubMed] [Google Scholar]
- 5.Chuang, C.-K., T.-H. Wong, S.-M. Hwang, Y.-H. Chang, Y.-H. Chen, Y.-C. Chiu, S.-F. Huang, and Y.-C. Hu. 2009. Baculovirus transduction of mesenchymal stem cells: in vitro responses and in vivo immune responses after cell transplantation. Mol. Ther. 17:889-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Condreay, J. P., S. M. Witherspoon, W. C. Clay, and T. A. Kost. 1999. Transient and stable gene expression in mammalian cells transduced with a recombinant baculovirus vector. Proc. Natl. Acad. Sci. USA 96:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Draghici, S., P. Khatri, A. L. Tarca, K. Amin, A. Done, C. Voichita, C. Georgescu, and R. Romero. 2007. A systems biology approach for pathway level analysis. Genome Res. 17:1537-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans, C. H., S. C. Ghivizzani, and P. D. Robbins. 2009. Orthopedic gene therapy in 2008. Mol. Ther. 17:231-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fong, S., M. Shoemaker, J. Cadaoas, A. Lo, W. Liao, M. Tagliaferri, I. Cohen, and E. Shtivelman. 2008. Molecular mechanisms underlying selective cytotoxic activity of BZL101, an extract of Scutellaria barbata, towards breast cancer cells. Cancer Biol. Ther. 7:577-586. [DOI] [PubMed] [Google Scholar]
- 10.Harte, M. T., I. R. Haga, G. Maloney, P. Gray, P. C. Reading, N. W. Bartlett, G. L. Smith, A. Bowie, and L. A. O'Neill. 2003. The poxvirus protein A52R targets Toll-like receptor signaling complexes to suppress host defense. J. Exp. Med. 197:343-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho, Y.-C., Y.-C. Chung, S.-M. Hwang, K.-C. Wang, and Y.-C. Hu. 2005. Transgene expression and differentiation of baculovirus-transduced human mesenchymal stem cells. J. Gene Med. 7:860-868. [DOI] [PubMed] [Google Scholar]
- 12.Ho, Y.-C., H.-P. Lee, S.-M. Hwang, W.-H. Lo, H.-C. Chen, C.-K. Chung, and Y.-C. Hu. 2006. Baculovirus transduction of human mesenchymal stem cell-derived progenitor cells: variation of transgene expression with cellular differentiation states. Gene Ther. 13:1471-1479. [DOI] [PubMed] [Google Scholar]
- 13.Horwitz, E. M., D. J. Prockop, L. A. Fitzpatrick, W. W. K. Koo, P. L. Gordon, M. Neel, M. Sussman, P. Orchard, J. C. Marx, R. E. Pyeritz, and M. K. Brenner. 1999. Transplantability and therapeutic effects of bone marrow-derived mesenchymal cells in children with osteogenesis imperfecta. Nat. Med. 5:309-313. [DOI] [PubMed] [Google Scholar]
- 14.Hu, Y.-C. 2008. Baculoviral vectors for gene delivery: a review. Curr. Gene Ther. 8:54-65. [DOI] [PubMed] [Google Scholar]
- 15.Ishii, K. J., S. Koyama, A. Nakagawa, C. Coban, and S. Akira. 2008. Host innate immune receptors and beyond: making sense of microbial infections. Cell Host Microbe 3:352-363. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, Z., T. W. Mak, G. Sen, and X. Li. 2004. Toll-like receptor 3-mediated activation of NF-κB and IRF3 diverges at Toll-IL-1 receptor domain-containing adapter inducing IFN-β. Proc. Natl. Acad. Sci. USA 101:3533-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kost, T. A., J. P. Condreay, and D. L. Jarvis. 2005. Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat. Biotechnol. 23:567-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liotta, F., R. Angeli, L. Cosmi, L. Fili, C. Manuelli, F. Frosali, B. Mazzinghi, L. Maggi, A. Pasini, V. Lisi, V. Santarlasci, L. Consoloni, M. L. Angelotti, P. Romagnani, P. Parronchi, M. Krampera, E. Maggi, S. Romagnani, and F. Annunziato. 2008. Toll-like receptors 3 and 4 are expressed by human bone marrow-derived mesenchymal stem cells and can inhibit their T-cell modulatory activity by impairing Notch signaling. Stem Cells 26:279-289. [DOI] [PubMed] [Google Scholar]
- 19.Liu, C. H., and S. M. Hwang. 2005. Cytokine interactions in mesenchymal stem cells from cord blood. Cytokine 32:270-279. [DOI] [PubMed] [Google Scholar]
- 20.Liu, C. Y. Y., C. H. Wang, J. C. Wang, and Y. C. Chao. 2007. Stimulation of baculovirus transcriptome expression in mammalian cells by baculoviral transcriptional activators. J. Gen. Virol. 88:2176-2184. [DOI] [PubMed] [Google Scholar]
- 21.Lo, W.-H., S.-M. Hwang, C.-K. Chuang, C.-Y. Chen, and Y.-C. Hu. 2009. Development of a hybrid baculoviral vector for sustained transgene expression. Mol. Ther. 17:658-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mack, G. S. 2009. Osiris seals billion-dollar deal with Genzyme for cell therapy. Nat. Biotechnol. 27:106-107. [DOI] [PubMed] [Google Scholar]
- 23.O'Neill, L. A., and A. G. Bowie. 2007. The family of five: TIR-domain-containing adaptors in Toll-like receptor signaling. Nat. Rev. Immunol. 7:353-364. [DOI] [PubMed] [Google Scholar]
- 24.Opitz, C. A., U. M. Litzenburger, C. Lutz, T. V. Lanz, I. Tritschler, A. Koppel, E. Tolosa, M. Hoberg, J. Anderl, W. K. Aicher, M. Weller, W. Wick, and M. Platten. 2009. Toll-Like receptor engagement enhances the immunosuppressive properties of human bone marrow-derived mesenchymal stem cells by inducing indoleamine-2,3-dioxygenase-1 via interferon-β and protein kinase R. Stem Cells 27:909-919. [DOI] [PubMed] [Google Scholar]
- 25.O'Reilly, D., L. Miller, and V. Luckow. 1992. Baculovirus expression vectors: a laboratory manual. W. H. Freeman and Co., New York, NY.
- 26.Pevsner-Fischer, M., V. Morad, M. Cohen-Sfady, L. Rousso-Noori, A. Zanin-Zhorov, S. Cohen, I. R. Cohen, and D. Zipori. 2007. Toll-like receptors and their ligands control mesenchymal stem cell functions. Blood 109:1422-1432. [DOI] [PubMed] [Google Scholar]
- 27.Phinney, D. G., and L. Isakova. 2005. Plasticity and therapeutic potential of mesenchymal stem cells in the nervous system. Curr. Pharm. Des. 11:1255-1265. [DOI] [PubMed] [Google Scholar]
- 28.Sarkis, C., C. Serguera, S. Petres, D. Buchet, J. L. Ridet, L. Edelman, and J. Mallet. 2000. Efficient transduction of neural cells in vitro and in vivo by a baculovirus-derived vector. Proc. Natl. Acad. Sci. USA 97:14638-14643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strauss, R., A. Huser, S. Ni, S. Tuve, N. Kiviat, P. S. Sow, C. Hofmann, and A. Lieber. 2007. Baculovirus-based vaccination vectors allow for efficient induction of immune responses against Plasmodium falciparum circumsporozoite protein. Mol. Ther. 15:193-202. [DOI] [PubMed] [Google Scholar]
- 30.Tabeta, K., P. Georgel, E. Janssen, X. Du, K. Hoebe, K. Crozat, S. Mudd, L. Shamel, S. Sovath, J. Goode, L. Alexopoulou, R. A. Flavell, and B. Beutler. 2004. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc. Natl. Acad. Sci. USA 101:3516-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomchuck, S. L., K. J. Zwezdaryk, S. B. Coffelt, R. S. Waterman, E. S. Danka, and A. B. Scandurro. 2008. Toll-like receptors on human mesenchymal stem cells drive their migration and immunomodulating responses. Stem Cells 26:99-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uccelli, A., L. Moretta, and V. Pistoia. 2008. Mesenchymal stem cells in health and disease. Nat. Rev. Immunol. 8:726-736. [DOI] [PubMed] [Google Scholar]
- 33.Vercammen, E., J. Staal, and R. Beyaert. 2008. Sensing of viral infection and activation of innate immunity by Toll-like receptor 3. Clin. Microbiol. Rev. 21:13-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, J. P., G. N. Bowen, C. Padden, A. Cerny, R. W. Finberg, P. E. Newburger, and E. A. Kurt-Jones. 2008. Toll-like receptor-mediated activation of neutrophils by influenza A virus. Blood 112:2028-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.West, J., and B. Damania. 2008. Upregulation of the TLR3 pathway by Kaposi's sarcoma-associated herpesvirus during primary infection. J. Virol. 82:5440-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.