Abstract
The neuraminidase inhibitors zanamivir and oseltamivir are marketed for the treatment and prophylaxis of influenza and have been stockpiled by many countries for use in a pandemic. Although recent surveillance has identified a striking increase in the frequency of oseltamivir-resistant seasonal influenza A (H1N1) viruses in Europe, the United States, Oceania, and South Africa, to date there have been no reports of significant zanamivir resistance among influenza A (H1N1) viruses or any other human influenza viruses. We investigated the frequency of oseltamivir and zanamivir resistance in circulating seasonal influenza A (H1N1) viruses in Australasia and Southeast Asia. Analysis of 391 influenza A (H1N1) viruses isolated between 2006 and early 2008 from Australasia and Southeast Asia revealed nine viruses (2.3%) that demonstrated markedly reduced zanamivir susceptibility and contained a previously undescribed Gln136Lys (Q136K) neuraminidase mutation. The mutation had no effect on oseltamivir susceptibility but caused approximately a 300-fold and a 70-fold reduction in zanamivir and peramivir susceptibility, respectively. The role of the Q136K mutation in conferring zanamivir resistance was confirmed using reverse genetics. Interestingly, the mutation was not detected in the primary clinical specimens from which these mutant isolates were grown, suggesting that the resistant viruses either occurred in very low proportions in the primary clinical specimens or arose during MDCK cell culture passage. Compared to susceptible influenza A (H1N1) viruses, the Q136K mutant strains displayed greater viral fitness than the wild-type virus in MDCK cells but equivalent infectivity and transmissibility in a ferret model.
Two classes of antiviral drugs are currently available for the treatment and prophylaxis of influenza, the adamantanes and the neuraminidase (NA) inhibitors (NAIs). The adamantanes were the first agents to be recognized to have anti-influenza virus activities as early as 1964 (2) although the rapid emergence of drug-resistant influenza virus strains has limited their clinical effectiveness (12). The NAIs, zanamivir (Relenza) and oseltamivir (Tamiflu), were the first drugs to be specifically designed as anti-influenza virus agents and have been available on the market in many countries since 1999. During oseltamivir clinical trials, 1 to 4% of treated adults (6) and 5 to 6% of treated children were found to shed resistant viruses (30) although more recent studies have reported resistance in 16 to 18% of viruses from oseltamivir-treated children (20, 29). In contrast to the frequency of resistance seen following oseltamivir treatment, only one occurrence of significant zanamivir resistance has been observed following zanamivir treatment. The zanamivir-resistant strain, an influenza B virus with an R152K NA mutation, was isolated from an immunocompromised patient undergoing prolonged zanamivir treatment (7).
In addition to the analysis of influenza viruses isolated from patients undergoing either oseltamivir or zanamivir treatment, surveillance studies that analyze the NAI susceptibility of circulating viruses, predominantly from patients not undergoing NAI treatment, have also been conducted. Studies that have tested viruses isolated prior to the release of the NAIs (1996 to 1999) (23) and after the initiation of clinical use of these drugs (2000 to 2006) (16, 24) have found either no resistance or a very low frequency of resistance. In contrast, analysis of circulating seasonal influenza viruses from Europe during the 2007 to 2008 season revealed that 14% (59/437) of influenza A (H1N1) viruses had significantly decreased susceptibility to oseltamivir (21). Since this initial report, oseltamivir-resistant influenza A (H1N1) strains have spread throughout Europe (11) and have been detected at high frequencies in other countries including the United States (4), Japan (28), South Africa (1) and Oceania and Southeast Asia (17). These influenza A (H1N1) viruses have a mutation of histidine to tyrosine at residue 274 of the NA (N2 NA numbering; residue 275 by N1 NA numbering), which confers a high level of resistance to oseltamivir (10) but has no effect on susceptibility to zanamivir or to the adamantanes.
Prior to May 2008, when the oseltamivir-resistant variants became the dominant influenza A (H1N1) strain in Oceania and Southeast Asia (17), NAI sensitivity monitoring conducted at the WHO Collaborating Centre for Reference and Research on Influenza, Melbourne, identified a number of influenza A (H1N1) viruses with reduced zanamivir susceptibility. These viruses contained a previously undescribed mutation at residue 136 of the NA. Here, we report on the detection of these mutant viruses from geographically distinct locations, the in vitro and in vivo fitness of the strains, and the finding that the mutant viruses appear to have been preferentially propagated during viral culture in Madin-Darby canine kidney (MDCK) cells.
MATERIALS AND METHODS
Influenza virus isolates.
A total of 391 influenza A (H1N1) viruses collected between January 2006 and February 2008 from Australasia and Southeast Asia (Australia, 224; Macau, 13; Malaysia, 17; New Zealand, 27; Philippines, 66; Singapore, 22; Taiwan, 5; and Thailand, 17) were selected from strains collected through the WHO Global Influenza Surveillance Network. Viruses were chosen to achieve a geographical and temporal spread, with over 50 viruses being isolated from each of four different states within Australia. In addition, influenza A (H3N2) viruses (n = 475) and influenza B viruses (n = 275) collected between January 2006 and February 2008 were assayed.
Virus culture.
All viruses were isolated or repassaged (if they had been received as isolates) in MDCK cells (American Type Culture Collection CCL-34) maintained in maintenance medium (DMEM/Ham's F12 Coon's medium [Sigma-Aldrich] containing 3% sodium bicarbonate with the addition of 2 mM glutamine, 1% nonessential amino acids, 0.05% NaHCO3, 0.02 M HEPES, 4% penicillin and streptomycin, 2 μg/ml amphotericin B, and 4 μg/ml trypsin [CSL Biotherapies]). For serial passaging experiments, isolates were diluted at 1:100 and then reinoculated into MDCK cells.
NA inhibitors.
Oseltamivir carboxylate, the active form of the ethyl ester prodrug oseltamivir phosphate, was provided by Hoffmann-La Roche Ltd., Switzerland, and zanamivir was provided by GSK, Australia. Peramivir, an NAI currently undergoing clinical development in parenteral formulations, was provided by BioCryst, Birmingham, AL, and was used to test strains with increased zanamivir or oseltamivir 50% NA-inhibitory concentrations (IC50s).
NA inhibition assay.
The extent of NA inhibition was measured by the production of the fluorescent product 4-methylumbelliferone from the substrate 2-(4-methylumbelliferyl)-a-d-N-acetylneuraminic acid (Sigma) as a measure of NA activity (25) following a previously published protocol (16). IC50s were calculated using the logistic curve fit program Robosage kindly provided by GlaxoSmithKline, United Kingdom. Influenza A (H1N1) viruses with an IC50 greater than three standard deviations above the mean of NAI-susceptible circulating influenza A (H1N1) viruses were considered significant and were then retested in duplicate in three separate assays. Sensitive and resistant control strains were included in each assay and demonstrated variability in IC50 of 15% over 10 assays.
RT-PCR, sequencing, and clonal analysis.
Viral RNA was extracted from MDCK cell culture supernatants using either a QIAmp Viral RNA Mini Kit (Qiagen) or a MagnaPure extraction system (Roche). The NA and HA1 genes were amplified by reverse transcription-PCR (RT-PCR) and sequenced using standard techniques (15). The RT-PCR product was cloned using the Zero Blunt TOPO PCR Cloning Kit for sequencing (Invitrogen) and One Shot TOP10 competent cells (Invitrogen) according to the manufacturer's protocol. Colonies were screened by real-time PCR on an Applied Biosystems 7500 Real-Time PCR system using TaqMan Universal PCR Master Mix (Applied Biosystems). Each colony was assayed with both the Q136 (wild-type)- and the K136 (mutant)-specific primer/probe combinations. The Q136 and K136 RT-PCR assays shared the same forward primer (5′-TTGTCATAAGAGAACCTTTCATATCATGTT-3′) and probe (5′-TCACTTGGAATGCAGAAC-3′), but the reverse primer differed to provide the specificity for detection of either the Q136 (wild-type) genotype (5′-TGTTTGTCATTTAATAGAGCACCTTG-3′) or the K136 (mutant) genotype (5′-ATGTTTGTCATTTAATAGAGCACCTTT-3′).
Pyrosequencing.
RT-PCR of the NA genes was conducted as described previously (15), except that one of the PCR primers was biotinylated. Biotinylated PCR products (40 μl) were immobilized on streptavidin-coated Sepharose beads (Streptavidin Sepharose HP; Amersham Biosciences) in binding buffer (40 μl) by shaking at 1,000 rpm for 5 min at room temperature, according to the Pyromark ID System guidelines (Biotage, Sweden). The resultant single-stranded biotinylated DNAs were then transferred using a Pyromark Vacuum Prep Workstation (Biotage, Sweden) into 40 μl of annealing buffer containing pyrosequencing primers and annealed at 80°C for 2 min. The mixture was cooled to room temperature and then subjected to pyrosequencing reactions using Pyro Gold Reagents on a Pyromark ID System (Biotage AB, Sweden) following the protocol provided by the manufacturer. Relative proportions of wild-type and mutant genes were determined by the Pyromark ID, version 1.0, software following allele quantitation analysis.
Generation of recombinant viruses.
Recombinant viruses were generated using an eight-plasmid reverse genetics system with pHW2000 plasmids that contained the cDNA of the influenza viral gene segments (14). The NA gene of selected wild-type viruses was amplified by RT-PCR as previously described (14) and incorporated into the pHW2000 plasmid. The NA plasmid was then combined with other plasmids containing the seven remaining gene segments from A/Puerto Rico/8/1934 (H1N1) influenza virus (kindly supplied by Robert Webster from St. Jude Children's Research Hospital, Memphis, TN). All eight plasmids were transfected into a coculture of 293T and MDCK cells as previously described (14).
Replication kinetics.
MDCK cells were infected with viruses at a multiplicity of infection (MOI) of 0.01 50% tissue culture infectious doses (TCID50)/cell and incubated at 35°C for 1 h. The cells were then washed and overlaid with maintenance medium before being incubated at 35°C. A total of 300 μl of supernatant from each virus was collected at 12, 24, 36, 48, 60, and 72 h postinfection and stored at −70°C prior to the determination of viral infectivity titers.
Viral infectivity assay.
MDCK cells were seeded into 96-well plates (Cellstar; Greiner Bio-one) (1.5 × 104 cells per well) and grown to confluence overnight at 37°C in a 5% CO2 incubator. Monolayers were washed twice with Ca2+/Mg2+-free phosphate-buffered saline before inoculation with triplicate 10-fold dilutions of virus. After incubation for 4 days at 35°C in 5% CO2, each well was scored for virus growth by cytopathic effect, and the TCID50 was determined by the Reed-Meunch method (26).
Ferret studies.
Female ferrets aged approximately 6 months were equipped with microchips (LifeChip, Destron Fearing) and tested to ensure that they had no detectable preexisting antibodies to influenza A (H1N1), A (H3N2), or B viruses by a hemagglutination inhibition assay. The microchip has a unique electronic code number and also measures temperature. Both parameters were monitored by using a hand-held scanner (PocketReader; Destron Fearing). Infectivity titers of viruses were determined in MDCK cells, and four separate inocula (105, 104, 103, 102 TCID50s per 500 μl) of each virus were used to intranasally infect (500 μl) two ferrets (termed donors), which were then housed together in an individual HEPA-filtered enclosure. Twenty-four hours after infection, the cages were cleaned and disinfected, and a third uninfected ferret (termed the recipient) was put into each cage to determine virus transmissibility. Following light sedation with 2 mg/kg xylazine (Troy Laboratories, NSW, Australia) administered intramuscularly, nasal washes were taken from ferrets using an Optiva (Medex) 20-gauge intravenous catheter and 1 ml of phosphate-buffered saline containing 1% (wt/vol) bovine serum albumin, 100 μg/ml streptomycin, and 100 U/ml penicillin. Nasal washes were stored immediately at −70°C. Temperature and observations of wellness were obtained three times a day, and weights were measured daily. The ferrets were sacrificed on day 11. Ferret studies were approved by the CSL Limited/Pfizer Animal Ethics Committee.
RESULTS
A total of 391 influenza A (H1N1) virus isolates obtained from January 2006 to February 2008 were tested in a fluorescence-based NA inhibition assay to determine sensitivity to the NAIs. Of these influenza A (H1N1) viruses, nine strains (2.3%) had increased zanamivir IC50s compared to the mean IC50 of NAI-susceptible influenza A (H1N1) strains collected between 2006 and 2008 (n = 367) (Table 1), and two other viruses (0.5%) had significantly increased oseltamivir IC50s (due to a H274Y NA mutation) (data not shown). No influenza A (H3N2) viruses (n = 450) or influenza B viruses (n = 275) had significantly increased oseltamivir or zanamivir IC50s. Sequence analysis of the NA gene from the viruses with reduced zanamivir susceptibility revealed an amino acid mutation of glutamine (Q) to lysine (K) at the highly conserved residue 136 of the NA gene (N2 NA numbering) (Table 1) (sequences available in the GenBank database; accession numbers are listed in Table 1). The viral isolates A/Brisbane/297/2006, A/Brisbane/308/2007, A/Philippines/604/2006, A/Philippines/1279/2006, and A/Philippines/905/2006, which had zanamivir IC50s between 6.0 and 47.4 nM, demonstrated evidence of a mixed population of viruses with either Q or K at residue 136, while sequence chromatograms for the isolates with higher zanamivir IC50s (ranging from 97.5 to 238.8 nM) revealed a homogeneous population of viruses with the K136 mutation (data not shown). The isolates with reduced zanamivir susceptibility were recovered from individuals who had not been undergoing either zanamivir or oseltamivir treatment, and there was no epidemiological information to suggest any link between the cases.
TABLE 1.
Characteristics of viruses containing a Q136K mutation
| Virus(es) | Passage history | Sample date | NAI IC50 (nM)a
|
Adamantane sensitivityb | GenBank accession no.
|
|||
|---|---|---|---|---|---|---|---|---|
| Zanamivir | Oseltamivir carboxylate | Peramivir | HA sequence | NA sequence | ||||
| All sensitive influenza A (H1N1) virusesc | 0.4 ± 0.2 | 0.5 ± 0.3d | 0.2 ± 0.1 | |||||
| A/Philippines/905/2006 | MDCK3 | 30 May 2006 | 24.2 ± 3.0 | 0.2 ± 0.1 | 5.8 ± 1.0 | Sensitive | CY030867 | CY030868 |
| A/Philippines/1279/2006 | MDCK3 | 26 June 2006 | 6.0 ± 0.5 | 0.6 ± 0.1 | 1.3 ± 0.2 | Sensitive | CY030863 | CY030864 |
| A/Philippines/604/2006 | MDCK3 | 27 June 2006 | 47.4 ± 6.7 | 0.5 ± 0.1 | 10.0 ± 0.7 | Sensitive | CY030865 | CY030866 |
| A/Brisbane/297/2006 | MDCK3 | 26 November 2006 | 46.6 ± 3.8 | 0.3 ± 0.1 | 9.6 ± 0.8 | Sensitive | CY030869 | EU124179 |
| A/Christchurch/62/2007 | MDCK3 | 30 August 2007 | 161.8 ± 24.7 | 0.3 ± 0.1 | 25.3 ± 4.1 | Sensitive | CY030876 | CY030877 |
| A/Brisbane/308/2007 | MDCK3 | 2 September 2007 | 45.2 ± 10.4 | 0.3 ± 0.1 | 7.2 ± 1.9 | Sensitive | CY030874 | CY030875 |
| A/Brisbane/334/2007 | MDCK4 | 11 September 2007 | 238.8 ± 28.9 | 0.2 ± 0.1 | 35.2 ± 6.0 | Sensitive | CY030870 | CY030871 |
| A/Thailand/39/2008 | MDCK3 | 2 January 2008 | 97.5 ± 5.8 | 0.3 ± 0.1 | 12.9 ± 0.8 | Sensitive | CY030872 | CY030873 |
| A/Macau/229/2008 | MDCK3 | 21 February 2008 | 85.0 ± 7.8 | 0.4 ± 0.1 | 16.9 ± 2.1 | Sensitive | FJ899924 | FJ899925 |
Mean IC50 value ± 1 standard deviation calculated using values derived from three separate assays performed in duplicate.
Adamantane sensitivity is based on sequence analysis of the M2 gene.
Mean IC50s obtained with wild-type susceptible influenza A (H1N1) viruses (n = 370) collected between January 2006 and March 2008.
Mean value does not include the two oseltamivir-resistant viruses.
To confirm the effect of the Q136K NA mutation on NAI sensitivity in a homogenous population, the A/Philippines/1279/2006 isolate was plaque purified, and its NA gene was cloned for reverse genetics experiments. The zanamivir IC50s for the plaque-purified and reverse genetics-derived Q136K mutant viruses were 98.1 and 124.2 nM, respectively, equivalent to a 248- and 327-fold reduction in zanamivir susceptibility compared to the wild-type strain (Table 2). The plaque-purified and recombinant Q136K mutant viruses also demonstrated 64- and 75-fold reductions in peramivir susceptibility, but they remained susceptible to oseltamivir (Table 2), thus confirming the role of the Q136K mutation.
TABLE 2.
Impact of Q136K NA mutation on the NAI susceptibility of plaque-purified and reverse genetics-derived viruses
| Virus name (type)a | NA mutation | IC50 (nM)b
|
||
|---|---|---|---|---|
| Zanamivir | Oseltamivir carboxylate | Peramivir | ||
| PP-1 (Q136 wild type) | None | 0.3 ± 0.1 | 0.6 ± 0.2 | 0.3 ± 0.1 |
| PP-2 (K136 mutant) | Q136K | 98.1 ± 12.8 | 0.4 ± 0.1 | 22.5 ± 6.5 |
| RG-1 (Q136 wild type) | None | 0.5 ± 0.1 | 0.8 ± 0.1 | 0.3 ± 0.1 |
| RG-2 (K136 mutant) | Q136K | 124.2 ± 24.9 | 0.2 ± 0.1 | 19.3 ± 3.3 |
PP-1 and PP-2 are twice-plaque-purified viruses derived from the A/Philippines/1279/2006 isolate. Viruses derived by reverse genetics (RG) contained seven segments from A/Puerto Rico/34/1934 and the NA segment from the A/Philippines/1279/2006 isolate.
Mean IC50 value ± 1 standard deviation calculated using values derived from three separate assays performed in duplicate.
As some of the isolates were mixed populations of Q136 wild-type and K136 mutant viruses, it was important to determine the presence of the mutation in the clinical specimens from which the isolates were grown. RT-PCR products for the NA gene were successfully amplified from only three of the primary original specimens that were available for analysis (viruses A/Christchurch/62/2007, A/Macau/229/2008, and A/Brisbane/308/2007), while the other specimens were RT-PCR negative, presumably due to nucleic acid degradation. Conventional sequencing analysis of the viral population in the primary clinical specimens revealed only the presence of the Q136 wild-type genotype. To achieve greater sensitivity in detecting minor viral proportions, the PCR products from these original specimens were cloned, and the colonies were screened for the presence or absence of the Q136K mutation. Surprisingly, even though over 100 colonies were screened for each specimen, only Q136 wild-type NA clones were detected (Table 3). This suggested either that the Q136K mutant virus was not present in the original specimen or that it was present at a frequency of less than 1%. In comparison with the cloning results from the original specimens, when the MDCK-grown isolate of A/Christchurch/62/2007 (MDCK passage 4 [MDCK4]) was cloned, 34 out of 43 colonies were found to contain the Q136K mutation (Table 3), confirming the ability of these methods to identify mutant clones.
TABLE 3.
Molecular clonal analysis of viruses to determine the presence or absence of the Q136K NA mutation
| Virus | Passage history | No. of clones screened | No. of Q136 clones detected | No. of K136 clones detected |
|---|---|---|---|---|
| A/Christchurch/62/2007 | Original specimen | 135 | 135 | 0 |
| A/Macau/229/2008 | Original specimen | 125 | 125 | 0 |
| A/Brisbane/308/2007 | Original specimen | 176 | 176 | 0 |
| A/Christchurch/62/2007 | MDCK4 | 43 | 9 | 34 |
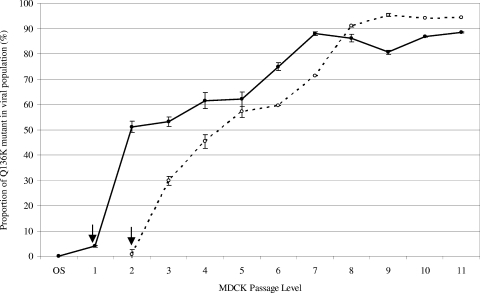
To determine how the proportion of the Q136K mutant may have changed upon MDCK culture, the A/Christchurch/62/2007 and A/Macau/229/2008 strains were serially passaged, and the viral populations were analyzed (Fig. 1). Based on pyrosequencing quantitation data, the proportion of the Q136K mutant virus in the A/Christchurch/62/2007 viral population increased from undetectable (<1%) in the original specimen and 3.9% in MDCK1 to 51.1% by MDCK2. The proportion of mutant virus continued to increase until passage 7, after which it remained between 80% and 90% until the final passage (Fig. 1). A similar trend was observed with the A/Macau/229/2008 virus, where the proportion of mutant started at a very low level at early passage (MDCK2, 0.6%), increased dramatically in the next few passages (MDCK3, 29.8%; MDCK4, 45.3%) and then plateaued between 90% and 95% in the isolates from passage 8 to the passage 11 (Fig. 1). In contrast, pyrosequencing analysis of eight NAI-sensitive influenza A (H1N1) isolates that had been passaged five to six times in MDCK cells revealed either undetectable levels of the Q136K mutant (0.0% for six strains) or very low levels (0.2% and 0.5%) that were undetectable in repeat assays.
FIG. 1.
Proportion of Q136K mutant in viral isolates following serial passage in MDCK cells. Means and standard deviations of the proportion of Q136K mutant virus in populations were determined following pyrosequencing analysis of amplified PCR product from three separate RT-PCR assays performed on separate occasions. Viruses A/Christchurch/62/2007 (•) from the original specimen (OS) to MDCK11 and A/Macau/229/2008 (○) from MDCK2 to MDCK11 were assayed (no result was obtained for the A/Macau/229/2008 original specimen, and the A/Macau/229/2008 MDCK1 isolate was not available for testing). Arrows indicate MDCK isolates that were obtained in the Christchurch or Macau laboratories. All subsequent isolates were obtained in the WHO Collaborating Centre for Reference and Research on Influenza, Melbourne.
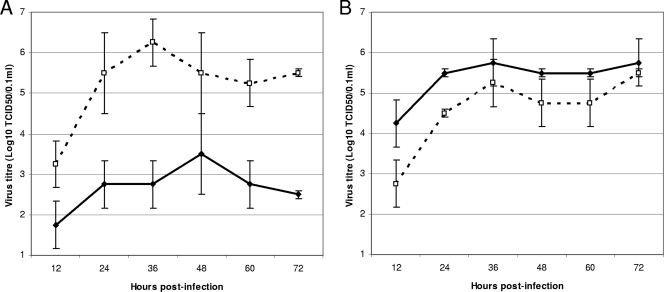
The growth rates of the plaque-purified A/Philippines/1279/2006 viruses and the recombinant strains containing the Q136K mutation (Table 2) were tested in MDCK cells (Fig. 2). The plaque-purified PP-2 (K136 mutant) virus with reduced zanamivir susceptibility grew to a significantly higher titer and at a faster rate in MDCK cells than the PP-1 (Q136 wild-type) zanamivir-susceptible virus (Fig. 2A). The superior growth characteristics of the mutant strain compared to the wild-type virus in MDCK cells provides support for the serial passaging data of the A/Christchurch/62/2007 and A/Macau/229/2008 viruses, where the mutant virus outgrew the wild-type within the mixed viral population. In contrast, the infectivity and transmissibility of the same two strains in a ferret model were similar. Both strains, even at the lowest inoculum (102 TCID50), infected all inoculated ferrets and was transmitted readily to naïve ferrets (Table 4). Titers of the two viruses in nasal washes of ferrets were also not significantly different; the mean titer (± standard deviation) of nasal washes from days 4 and 6 was 2.1 ± 1.0 log10 TCID50/0.1 ml for the PP-1 (Q136 wild-type) zanamivir-susceptible strain compared with 2.5 ± 1.1 log10 TCID50/0.1 ml for the PP-2 (K136 mutant) strain. Throughout the experiment both viruses appeared stable, with no hemagglutinin or NA sequence differences detected between the viruses used to infect the donor ferrets and those collected following nasal washing. Assessments of wellness and measurements of weight loss and temperature changes were also not significantly different between ferrets infected with the zanamivir-susceptible and -resistant strains (data not shown).
FIG. 2.
Virus growth curves of plaque-purified viruses and recombinant viruses in MDCK cells. (A) Growth curves of A/Philippines/1279/2006 plaque-purified viruses PP-1 (Q136 wild-type) (•) and PP-2 (K136 mutant) (○) in MDCK cells over 72 h. (B) Growth curves of recombinant viruses RG-1 (Q136 wild-type) (•) and RG-2 (K136 mutant) (○) in MDCK cells over 72 h.
TABLE 4.
Ferret infectivity and transmission of plaque-purified and recombinant viruses
| Virusa | Dose (TCID50) | Ferretb | Infectivity titer on the indicated day (log10 TCID50/0.1ml)c
|
Serologyd | |||||
|---|---|---|---|---|---|---|---|---|---|
| 2 | 4 | 6 | 8 | 10 | 11 | ||||
| PP-1 Q136 wild type | 105 | D1 | 2.5 | 1.0 | — | — | — | — | 160 |
| D2 | 2.5 | 3.0 | 3.0 | — | — | — | 160 | ||
| R | — | 2.5 | 0.5 | 0.5 | — | — | 160 | ||
| 104 | D1 | 2.5 | 2.5 | 2.5 | — | — | — | 160 | |
| D2 | 3.0 | 0.5 | — | — | — | — | 160 | ||
| R | 0.5 | 2.5 | 2.0 | — | — | — | 160 | ||
| 105 | D1 | 4.0 | 2.5 | 2.5 | — | — | — | 160 | |
| D2 | 3.5 | 3.2 | 2.0 | — | — | — | 160 | ||
| R | — | 3.0 | 2.5 | 0.5 | — | — | 160 | ||
| 102 | D1 | 5.5 | 2.0 | 2.0 | — | — | — | 160 | |
| D2 | 2.5 | 3.0 | 3.5 | 2.5 | — | — | 160 | ||
| R | 0.5 | 3.0 | 2.0 | 3.0 | — | — | 160 | ||
| PP-2 K136 mutant | 105 | D1 | 3.5 | 4.5 | 3.5 | — | — | — | 160 |
| D2 | 3.0 | 2.0 | 1.0 | — | — | — | 320 | ||
| R | — | 2.0 | 2.5 | 1.0 | 0.5 | — | 160 | ||
| 104 | D1 | 2.5 | 3.0 | 2.5 | — | — | — | 160 | |
| D2 | 1.5 | 1.0 | 2.0 | — | — | — | 160 | ||
| R | — | 3.0 | 2.0 | 2.0 | — | — | 160 | ||
| 105 | D1 | 4.0 | 2.0 | 4.0 | — | — | — | 160 | |
| D2 | 4.5 | 2.0 | 4.5 | — | — | — | 160 | ||
| R | — | 1.5 | 3.0 | 3.0 | — | — | 160 | ||
| 102 | D1 | 2.0 | 3.0 | 3.0 | 2.5 | — | — | 160 | |
| D2 | 4.0 | 2.5 | 2.5 | 0.5 | — | —- | 160 | ||
| R | — | — | 2.5 | 3.0 | 1.0 | — | 160 | ||
| RG-1 Q136 wild type | 105 | D1 | 3.5 | 1.5 | 2.0 | — | — | — | 160 |
| D2 | 2.5 | 1.5 | — | — | — | — | 160 | ||
| R | — | 1.0 | — | — | — | — | 20 | ||
| 104 | D1 | 2.0 | — | — | — | — | — | 80 | |
| D2 | 5.0 | — | — | — | — | — | 160 | ||
| R | — | — | — | — | — | — | <10 | ||
| 105 | D1 | 2.5 | 1.0 | 1.0 | — | — | — | 160 | |
| D2 | 4.5 | 1.5 | 1.0 | — | — | — | 160 | ||
| R | — | 1.5 | — | — | — | — | 160 | ||
| 102 | D1 | — | 3.5 | 1.0 | — | — | — | 160 | |
| D2 | — | 2.0 | — | — | — | — | 160 | ||
| R | — | — | — | — | — | — | <10 | ||
| RG-2 K136 mutant | 105 | D1 | — | — | — | — | — | — | 160 |
| D2 | 3.0 | — | — | — | — | — | 160 | ||
| R | — | — | — | — | — | — | <10 | ||
| 104 | D1 | 1.5 | — | — | — | — | — | 160 | |
| D2 | 1.5 | — | — | — | — | — | 160 | ||
| R | — | — | — | — | — | — | <10 | ||
| 105 | D1 | 1.0 | — | — | — | — | — | 160 | |
| D2 | 1.0 | 1.5 | — | — | — | — | 160 | ||
| R | — | — | — | — | — | — | <10 | ||
| 102 | D1 | — | — | — | — | — | — | 10 | |
| D2 | — | — | — | — | — | — | <10 | ||
| R | — | — | — | — | — | — | <10 | ||
Virus was isolated from nasal washes of ferrets on the indicated days after infection with either plaque-purified viruses PP-1 (Q136 wild type) or PP-2 (K136 mutant) or recombinant viruses RG-1 (Q136 wild type) or RG-2 (K136 mutant).
Ferrets are identified as donor (D; D1 is donor ferret number 1) or recipient (R). Recipient ferrets were placed with infected ferrets 24 h postinfection of donor ferrets.
Values indicate the infectivity titers of nasal washes; dashes indicate that influenza virus could not be isolated.
Hemagglutination titers of serum taken from ferrets on day 11 postinfection.
A comparison of the viral fitness of the two recombinant viruses RG-1 (Q136 wild-type) and RG-2 (K136 mutant), which contained the NA from either the PP-1 (Q136 wild-type) or the PP-2 (K136 mutant) virus and the remaining seven genes from A/Puerto Rico/8/1934 virus, revealed different results from those obtained with the plaque-purified viruses. The RG-2 recombinant strain with reduced zanamivir susceptibility grew more poorly in MDCK cells (Fig. 2B) and was less infectious and transmissible in ferrets (Table 4) than the zanamivir-susceptible RG-1 virus. This suggested that one or more compensatory amino acid mutations, in addition to the Q136K NA mutation, may be responsible for the plaque-purified mutant's having greater viral fitness than the recombinant virus. Although no NA sequence differences apart from the Q136K were detected between the plaque-purified wild-type viruses and the mutant strains from the A/Philippines/1279/2006, A/Christchurch/62/2007, and A/Macau/229/2008 isolates, sequence analysis of the HA gene from the same strains did identify some amino acid differences (I131T in A/Philippines/1279/2006, H47P and N48H in A/Christchurch/62/2007, and K75E in A/Macau/229/2008). No sequence differences were observed between the matrix genes from the wild-type strains compared with the mutant strains.
DISCUSSION
Here, we report a novel NAI resistance mutation that was detected in 2.3% of influenza A (H1N1) virus isolates obtained between 2006 and early 2008. The Q136K mutation, which has not been identified or reported previously, confers reductions in zanamivir and peramivir susceptibility but has no effect on oseltamivir susceptibility. Surprisingly, the mutant virus was not detected in the available primary clinical specimens of the mutant Q136K isolates, which suggested either that the mutant viruses were present at a low frequency (<1%) in the clinical specimens and had a selective advantage in MDCK cell culture or that the Q136K mutation was not present in viruses in the clinical specimens and occurred spontaneously in progeny viruses during MDCK cell culture. However, if the latter is correct, then it is surprising that we do not observe the K136 mutant more often, particularly given the observed fitness advantage of the mutant compared to the wild-type during MDCK culture. Since 2001, when the majority of the WHO Collaborating Centres for Influenza started conducting NAI susceptibility testing, we estimate that over 20,000 influenza A (H1N1) viruses have been isolated in MDCK cells but, apart from those described in this study, no zanamivir-resistant Q136K mutants have previously been reported. In addition, examination of the 2,264 influenza A (H1N1) virus NA sequences currently available in the GenBank revealed that only 10 viruses had a K at residue 136, and nine of these are the strains reported in this study. Our observation that such mutants can preferentially expand in MDCK cells highlights the importance of testing the clinical specimens of isolates with reduced NAI susceptibility to determine the presence or absence of resistance mutations.
The K136 mutant virus demonstrated a similar level of fitness to the Q136 wild-type in the ferret model, indicating the potential for viruses with this resistance mutation to circulate in the human population. The recent spread of oseltamivir-resistant influenza A (H1N1) viruses with an H274Y NA mutation has shown that NAI-resistant viruses can spread rapidly and circulate widely in the global community (17) even though previous ferret model data showed that earlier H274Y mutant viruses had a reduced viral fitness (13, 19). Given that both the Q136K and H274Y mutations have occurred in N1 NAs from contemporary influenza A (H1N1) viruses with no apparent impact on viral fitness, there is concern that one or both of the mutations may occur in highly pathogenic avian influenza A (H5N1) viruses, thereby reducing the effectiveness of NAI treatment of infected patients. To date, influenza A (H5N1) viruses with an H274Y NA mutation have been isolated from three patients undergoing oseltamivir treatment (two of these patients died) (3, 22). Analysis of sequences from the GenBank found no influenza A (H5N1) viruses with a Q136K mutation, but nine strains did have a Q136H mutation, and one virus had a Q136R mutation. The drug sensitivity of these mutants has not yet been assessed. Both oseltamivir and zanamivir have been stockpiled by many countries worldwide for use in the event of an influenza pandemic. It is therefore essential that the NAI sensitivity of avian influenza A (H5N1) viruses is constantly monitored so that the potential effectiveness of available treatments is known.
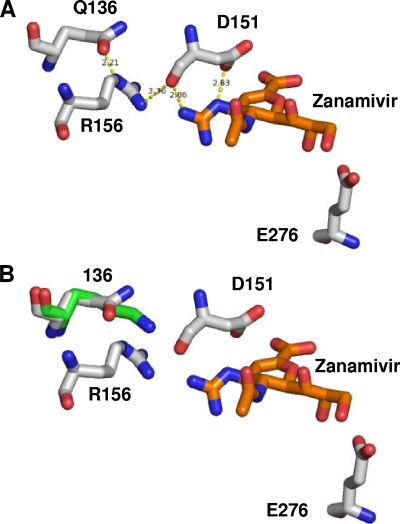
The crystal structure of N1 NA, isolated from influenza A (H5N1) viruses, complexed to zanamivir (27) reveals a possible molecular basis for the susceptibility of the Q136K mutation reported here. The side chain of Q136 is involved in a hydrogen bonding network, whereby it interacts with the side chain of R156, which in turn interacts with the backbone of D151. The latter residue forms two hydrogen bonds with the guanidino moiety of zanamivir (Fig. 3). R156 also interacts directly with zanamivir via van der Waals interactions. Mutation of residue 136 to a lysine residue would break down the hydrogen bonding network, possibly leading to increased mobility of R156 and D151, disturbing the aforementioned interactions with the drug. Similarly, peramivir would be affected by the mutation since it has the same guanidino moiety as zanamivir. In contrast, oseltamivir does not form any interactions with R156 or the D151 backbone (27) and hence would be predicted to be unaffected by the mutation. Since the side chain of D151 interacts with the amino moiety of oseltamivir (27), the data here suggest that this interaction is retained in the Q136K mutation. X-ray crystallography reveals that group 1 NAs, which include N1, N4, N5, and N8 subtypes, have a 150-loop (residues 147 to 152) conformation that opens up a cavity adjacent to the active site (termed the 150-cavity), which is not present in group 2 NAs (27). The size of the 150-cavity is determined by the interaction of the residue 136 and the 150-loop. In the study by Russell et al., it was proposed that it may be possible to design new NAIs to exploit this 150-cavity (27). In light of our work it would be of interest to determine how a Q136K mutation may affect the ability of such NA inhibitors to bind inside the 150-cavity.
FIG. 3.
Modeling of resistance mutations based on the crystal structure of the H5N1-zanamivir complex. (A) Stick representation of wild-type H5N1 (white sticks) with zanamivir (orange sticks). (B) Mutation Q136K (green sticks) results in a loss of the hydrogen bond network. Figures were produced with the modeling software package PyMol (W. L. DeLano, DeLano Scientific, Paolo Alto, CA) using the sequence of the Protein Data Bank code 2HTQ protein (27) downloaded from the RCS Protein Data Bank.
Although it was not possible in this study to confirm that the Q136K mutation was present in the primary specimens from patients from whom the mutant isolates were generated, the results demonstrate that this mutation has the potential to occur in N1 viruses. The likelihood for the mutation to occur under zanamivir selective pressure is unknown, and there have been no studies reporting the effects of culturing influenza A (H1N1) viruses in the presence of increasing zanamivir concentrations, but it is notable that serial passage of influenza A (H5N1) viruses under zanamivir selective pressure did not generate any viruses with the Q136K mutation (18). No viruses with this change have been detected in samples from zanamivir-treated patients. To date there has been only one report of a resistant virus being isolated from a zanamivir-treated patient (7). The virus was an influenza B strain with an R152K NA mutation which had a similar zanamivir IC50 (100 nM) to the plaque-purified or recombinant Q136K mutants from this study (9). The zanamivir IC50s for the Q136K mutants were also similar to the previously reported influenza A (H3N2) virus zanamivir mutants, E119G (100 nM) and E119D (150 nM) (9), which were generated during passage of virus in the presence of zanamivir (8). Although zanamivir has not been widely used in many countries, sales in Japan, the country with the highest per capita use of NAIs, have increased considerably in the past 2 years as a result of the concerns surrounding possible psychoneurological effects reported in Japanese teenagers being treated with oseltamivir (5). In addition, zanamivir use is likely to increase if the oseltamivir resistance in influenza A (H1N1) viruses now seen globally persists or predominates. In light of these findings it remains important that testing of commonly circulating and potential pandemic strains is conducted globally to monitor sensitivity to not only oseltamivir but also zanamivir and that the Q136 NA residue be recognized as a key site that can affect NAI susceptibility.
Acknowledgments
We thank the Research Institute for Tropical Medicine (Manila, Philippines), Queensland Health Services (Brisbane, Australia), Canterbury Health Services (Christchurch, New Zealand), and the WHO National Influenza Centre, National Institute of Health (Bangkok, Thailand), for submitting the viruses investigated in this study. In addition we thank all of the other laboratories that have submitted samples used in this study to the WHO Collaborating Centre for Reference and Research on Influenza, Melbourne, Australia. We also thank Naomi Komadina, Yi-Mo Deng, and Pina Iannello for undertaking the sequence analysis of some isolates and Robert Shaw, Chris Durrant, and Helen Sjogren for the culture of isolates used in the surveillance study.
The WHO Collaborating Centre for Reference and Research on Influenza is supported by the Australian Government Department of Health and Ageing.
Footnotes
Published ahead of print on 29 July 2009.
REFERENCES
- 1.Besselaar, T. G., D. Naidoo, A. Buys, V. Gregory, J. McAnerney, J. M. Manamela, L. Blumberg, and B. D. Schoub. 2008. Widespread oseltamivir resistance in influenza A viruses (H1N1), South Africa. Emerg. Infect. Dis. 14:1809-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies, W. L., R. R. Grunert, R. F. Haff, J. W. McGahen, E. M. Neumayer, M. Paulshock, J. C. Watts, T. R. Wood, E. C. Hermann, and C. E. Hoffmann. 1964. Antiviral activity of 1-adamantanamine (Amantadine). Science 144:862-863. [DOI] [PubMed] [Google Scholar]
- 3.de Jong, M. D., T. T. Tran, H. K. Truong, M. H. Vo, G. J. Smith, V. C. Nguyen, V. C. Bach, T. Q. Phan, Q. H. Do, Y. Guan, J. S. Peiris, T. H. Tran, and J. Farrar. 2005. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N. Engl. J. Med. 353:2667-2672. [DOI] [PubMed] [Google Scholar]
- 4.Dharan, N. J., L. V. Gubareva, J. J. Meyer, M. Okomo-Adhiambo, R. C. McClinton, S. A. Marshall, G. K. St, S. Epperson, L. Brammer, A. I. Klimov, J. S. Bresee, and A. M. Fry. 2009. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. JAMA 301:1034-1041. [DOI] [PubMed] [Google Scholar]
- 5.Fuyuno, I. 2007. Tamiflu side effects come under scrutiny. Nature 446:358-359. [DOI] [PubMed] [Google Scholar]
- 6.Gubareva, L. V., L. Kaiser, M. N. Matrosovich, Y. Soo-Hoo, and F. G. Hayden. 2001. Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J. Infect. Dis. 183:523-531. [DOI] [PubMed] [Google Scholar]
- 7.Gubareva, L. V., M. N. Matrosovich, M. K. Brenner, R. C. Bethell, and R. G. Webster. 1998. Evidence for zanamivir resistance in an immunocompromised child infected with influenza B virus. J. Infect. Dis. 178:1257-1262. [DOI] [PubMed] [Google Scholar]
- 8.Gubareva, L. V., M. J. Robinson, R. C. Bethell, and R. G. Webster. 1997. Catalytic and framework mutations in the neuraminidase active site of influenza viruses that are resistant to 4-guanidino-Neu5Ac2en. J. Virol. 71:3385-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gubareva, L. V., R. G. Webster, and F. G. Hayden. 2001. Comparison of the activities of zanamivir, oseltamivir, and RWJ-270201 against clinical isolates of influenza virus and neuraminidase inhibitor-resistant variants. Antimicrob. Agents Chemother. 45:3403-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gubareva, L. V., R. G. Webster, and F. G. Hayden. 2002. Detection of influenza virus resistance to neuraminidase inhibitors by an enzyme inhibition assay. Antivir. Res. 53:47-61. [DOI] [PubMed] [Google Scholar]
- 11.Hauge, S. H., S. Dudman, K. Borgen, A. Lackenby, and O. Hungnes. 2009. Oseltamivir-resistant influenza viruses A (H1N1), Norway, 2007-08. Emerg. Infect. Dis. 15:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayden, F. G. 1996. Amantadine and rimantadine: clinical aspects, p. 78-92. In D. D. Richman (ed.), Antiviral drug resistance. John Wiley & Sons, Hoboken, NJ.
- 13.Herlocher, M. L., R. Truscon, S. Elias, H. L. Yen, N. A. Roberts, S. E. Ohmit, and A. S. Monto. 2004. Influenza viruses resistant to the antiviral drug oseltamivir: transmission studies in ferrets. J. Infect. Dis. 190:1627-1630. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann, E., S. Krauss, D. Perez, R. Webby, and R. Webster. 2002. Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine 20:3165. [DOI] [PubMed] [Google Scholar]
- 15.Hurt, A. C., and I. G. Barr. 2008. Influenza viruses with reduced sensitivity to the NA inhibitor drugs in untreated young children. Commun. Dis. Intell. 32:57-62. [DOI] [PubMed] [Google Scholar]
- 16.Hurt, A. C., I. G. Barr, G. Hartel, and A. W. Hampson. 2004. Susceptibility of human influenza viruses from Australasia and South East Asia to the neuraminidase inhibitors zanamivir and oseltamivir. Antivir. Res. 62:37-45. [DOI] [PubMed] [Google Scholar]
- 17.Hurt, A. C., J. Ernest, Y. Deng, P. Iannello, T. G. Besselaar, C. Birch, P. Buchy, M. Chittaganpitch, S. C. Chiu, D. E. Dwyer, A. Guigon, B. Harrower, I. P. Kei, T. Kok, C. Lin, K. McPhie, A. Mohd, R. Olveda, T. Panayotou, W. Rawlinson, L. Scott, D. Smith, H. D'Souza, N. Komadina, R. Shaw, A. Kelso, and I. G. Barr. 2009. Emergence and spread of oseltamivir-resistant A (H1N1) influenza viruses in Oceania, South East Asia and South Africa. Antivir. Res. 83:90-93. [DOI] [PubMed] [Google Scholar]
- 18.Hurt, A. C., J. K. Holien, and I. G. Barr. 2009. In vitro generation of neuraminidase inhibitor resistance in A (H5N1) influenza viruses. Antimicrob. Agents Chemother. doi: 10.1128/AAC.00334-09. [DOI] [PMC free article] [PubMed]
- 19.Ives, J. A. L., J. A. Carr, D. B. Mendel, C. Y. Tai, R. Lambkin, L. Kelly, J. S. Oxford, F. G. Hayden, and N. A. Roberts. 2002. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antivir. Res. 55:307-317. [DOI] [PubMed] [Google Scholar]
- 20.Kiso, M., K. Mitamura, Y. Sakai-Tagawa, K. Shiraishi, C. Kawakami, K. Kimura, F. G. Hayden, N. Sugaya, and Y. Kawaoka. 2004. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet 364:759-765. [DOI] [PubMed] [Google Scholar]
- 21.Lackenby, A., O. Hungnes, S. Dudman, A. Meijer, W. Pajet, A. J. Hay, and M. C. Zambon. 2008. Emergence of resistance to oseltamivir among influenza A (H1N1) viruses in Europe. Euro. Surveill. 13:5. [DOI] [PubMed] [Google Scholar]
- 22.Le, Q. M., M. Kiso, K. Someya, Y. T. Sakai, T. H. Nguyen, K. H. Nguyen, N. D. Pham, H. H. Ngyen, S. Yamada, Y. Muramoto, T. Horimoto, A. Takada, H. Goto, T. Suzuki, Y. Suzuki, and Y. Kawaoka. 2005. Avian flu: isolation of drug-resistant H5N1 virus. Nature 437:1108. [DOI] [PubMed] [Google Scholar]
- 23.McKimm-Breschkin, J., T. Trivedi, A. Hampson, A. Hay, A. Klimov, M. Tashiro, F. Hayden, and M. Zambon. 2003. Neuraminidase sequence analysis and susceptibilities of influenza virus clinical isolates to zanamivir and oseltamivir. Antimicrob. Agents Chemother. 47:2264-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monto, A. S., J. L. McKimm-Breschkin, C. Macken, A. W. Hampson, A. Hay, A. Klimov, M. Tashiro, R. G. Webster, M. Aymard, F. G. Hayden, and M. Zambon. 2006. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob. Agents Chemother. 50:2395-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potier, M., L. Mameli, M. Belisle, L. Dallaire, and S. B. Melancon. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-d-N-acetylneuraminate) substrate. Anal. Biochem. 94:287-296. [DOI] [PubMed] [Google Scholar]
- 26.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 27.Russell, R. J., L. F. Haire, D. J. Stevens, P. J. Collins, Y. P. Lin, G. M. Blackburn, A. J. Hay, S. J. Gamblin, and J. J. Skehel. 2006. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 443:45-49. [DOI] [PubMed] [Google Scholar]
- 28.Tamura, D., K. Mitamura, M. Yamazaki, M. Fujino, M. Nirasawa, K. Kimura, M. Kiso, H. Shimizu, C. Kawakami, S. Hiroi, K. Takahashi, M. Hatta, H. Minagawa, Y. Kimura, S. Kaneda, S. Sugita, T. Horimoto, N. Sugaya, and Y. Kawaoka. 2009. Oseltamivir-resistant influenza A viruses circulating in Japan. J. Clin. Microbiol. 47:1424-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward, P., I. Small, J. Smith, P. Suter, and R. Dutkowski. 2005. Oseltamivir (Tamiflu) and its potential for use in the event of an influenza pandemic. J. Antimicrob. Chemother. 55(Suppl. 1):i5-i21. [DOI] [PubMed] [Google Scholar]
- 30.Whitley, R. J., F. G. Hayden, K. S. Reisinger, N. Young, R. Dutkowski, D. Ipe, R. G. Mills, and P. Ward. 2001. Oral oseltamivir treatment of influenza in children. Pediatr. Infect. Dis. J. 20:127-133. [DOI] [PubMed] [Google Scholar]