Abstract
Murine gammaherpesvirus 68 (MHV-68) contains a ubiquitin (Ub)-specific cysteine protease (USP) domain embedded within the large tegument protein ORF64, as do all other herpesviruses. The biological role of this protease is still unclear, but for the alphaherpesvirus Marek's disease virus, its USP is involved in T-cell lymphoma formation. We here study the role of the MHV-68 USP, encoded by ORF64. By constructing a mutant virus with a single cysteine-to-alanine replacement in the active site of ORF64, we demonstrate that the USP activity of ORF64 is abolished. The mutant virus replicates less efficiently in vitro, and plaque size is reduced compared to that of a revertant virus. Electron microscopy of infected cells did not reveal any obvious differences in virion morphogenesis or differences in egress for the mutant and revertant viruses. Intraperitoneal infection of C57/BL6 mice demonstrates that the mutant virus is generally cleared by day 7, indicating a role for the USP in the persistence of MHV-68 infection or efficient replication. However, the USP activity in MHV-68 is unlikely to be involved in the establishment of latency or reactivation, since we observed no significant difference in viral DNA genome copy number in the spleen or in the number of cells that reactivate MHV-68 from latency. Our results for MHV-68 ORF64 are consistent with an enzymatic function of the tegument protein that is beneficial to the virus during acute infection, particularly in vivo.
The ubiquitin-proteasome system not only controls cytosolic proteolysis in eukaryotic cells but also regulates a number of diverse cellular processes, such as cell cycle progression, transcription regulation, and growth control. Unwanted proteins in the cell are tagged with multiple ubiquitin molecules and then targeted to the proteasome for degradation (reviewed in reference 13). Ubiquitination is reversible, and the removal of a ubiquitin-targeted protein substrate can be performed by ubiquitin-specific proteases (USPs) (23).
A number of viruses encode their own USPs, such as adenovirus (3) and severe acute respiratory syndrome coronavirus (4, 10, 20), as well as all members of the herpesvirus family (8, 14, 18, 19, 24, 40). Herpes simplex virus type 1 encodes a USP in the N-terminal portion of the large tegument protein UL36 (19), and this protease is the founding member of the herpesvirus USPs. All herpesviruses possess a large tegument protein with a USP activity embedded in the N-terminal portion. Although the extent of sequence similarity for the different family members is low in the USP domain, the residues implicated in the formation of the catalytic triad are strictly conserved (24). The structure of murine cytomegalovirus (MCMV) M48 USP in a complex with ubiquitin vinylmethylester (UbVME) demonstrates that the herpesvirus tegument USPs represent a distinct class of deubiquitinating enzymes (25). Furthermore, human CMV (HCMV) (40), murine gammaherpesvirus 68 (MHV-68) (14), Marek's disease virus (MDV) (18), and pseudorabies virus (PrV) (8) have all been shown to express the USP in the course of an active infection. Two additional deubiqutinating enzymes encoded by Epstein-Barr virus (EBV) have been identified by a bioinformatics approach (27), but whether these proteins are produced as enzymatically active species in the course of infection is not known. Herpesviruses rely on host cell machinery for replication. Given the key role of the ubiquitin-proteasome system in various cellular and immunological pathways, the ability of the virus to stabilize host proteins that are essential for—or remove those that interfere with—viral replication would be beneficial to the survival of the virus in the host, during both the acute phase and the latent phase of the infection.
MHV-68 is a natural rodent pathogen (5) and readily infects laboratory C57/BL6 mice. The genome of MHV-68 has been sequenced and shows a close genetic relationship to the human gammaherpesviruses Kaposi's sarcoma-associated herpesvirus (KSHV) and EBV (39). The functions of many MHV-68 gene products are similar to the corresponding gene products of human gammaherpesviruses, and therefore MHV-68 is widely used as a model for the pathogenesis of gammaherpesviruses. During in vivo infection, MHV-68 has an acute phase, following which the virus is cleared by 8 to 16 days postinfection (dpi) (9, 32, 41). After clearance, early latency is established, and by 6 weeks postinfection, a late latent phase is reached (37). The virus primarily infects and establishes latency in B cells in vivo (33, 41). However, differences between MHV-68 and the human gammaherpesviruses do exist, particularly in its ability to transform B cells.
We have demonstrated that the large tegument protein encoded by MHV-68 open reading frame 64 (ORF64) is indeed a functional deubiquitinating protease (14), based on an activity-based profiling approach that takes advantage of a ubiquitin-based active site-directed probe, UbVME, to target USPs (6, 7). Furthermore, we show that the recombinantly expressed amino-terminal segment of ORF64 displays deubiquitinating activity in vitro. We here investigate the biological role of the MHV-68-encoded USP by generating a mutant virus lacking USP activity. This is the first examination of the possible role of this activity in vivo for a gammaherpesvirus. We show that a single point mutation in the ORF64 active-site cysteine abolishes USP activity and decreases virus replication in vitro. We further show that the viral titers at early time points after infection are significantly lower in mice infected with the mutant virus, suggesting either that the infection is cleared faster in these mice or that the virus replicates less efficiently.
MATERIALS AND METHODS
Cells and virus stocks.
3T12 cells were cultured in Dulbecco's modified Eagle Medium (DMEM) supplemented with 10% fetal calf serum, penicillin (100 U/ml), streptomycin (100 μg/ml), and 2 mM l-glutamine at 37°C. Virus stocks of MHV-68 were prepared in 3T12 cells, and plaque assays on virus stocks were performed as previously described (9).
Construction of C→A and (C→A)REV viruses.
Two-step red-mediated recombination was performed as previously described (38). The primers used included 5′-CAGGCAGATTGTAAATTTGGAGAAAATGCAGGGGCTCAGGCCCTTAGCAACTGTATCATATAAGGATGACGACGATAAGTAGGG-3′ and 5′-GAAGTAGGAAGACATCAGGTATATGATACAGTTGCTAAGGGCCTGAGCCCCTGCATTTTCTCAACCAATTAACCAATTCTGATTAG-3′ for the C→A ORF64 MHV-68 mutant (C→A) and 5′-CAGGCAGATTGTAAATTTGGAGAAAATGCAGGGGCTCAGTGTCTTAGCAACTGTATCATATAAGGATGACGACGATAAGTAGGG-3′ and 5′-GAAGTAGGAAGACATCAGGTATATGATACAGTTGCTAAGACACTGAGCCCCTGCATTTTCTCAACCAATTAACCAATTCTGATTAG-3′ for revertant C→A ORF64 MHV-68 [(C→A)REV].
The green fluorescent protein-containing bacterial artificial chromosome (BAC) cassette was removed by passaging the virus through Cre-expressing Vero cells. The removal of the BAC cassette was confirmed by fluorescence microscopy. The MHV-68 BAC was a kind gift from Ulrich Koszinoswski.
Antibodies.
The rabbit anti-gp150 antibody was generated by immunizing rabbits (Covance, Custom Immunology Services, Denver, PA) with two different keyhole limpet hemocyanin-conjugated peptides from gp150 (amino acid sequences AALGAAETVEGITSR and SNNNPTTIMRPPVAQNGES). The polyclonal rabbit antiserum to ORF64 was raised against the N-terminal 235 amino acids of ORF64 expressed in Escherichia coli Rosetta cells (14).
Metabolic labeling, probe labeling, immunoprecipitation, and detection.
Cells were infected with MHV-68 at a multiplicity of infection (MOI) of 5; after 24 h, cells were either metabolically labeled or harvested by simple removal from the plate. For labeling experiments, cells were incubated in methionine- and cysteine-free DMEM and then metabolically labeled by incubation with 110 μCi of [35S]methionine/cysteine (NEN-Dupont, Boston, MA) per milliliter at 37°C for 6 h. Labeled cells were lysed in NP-40 lysis buffer (50 mM Tris [pH 7.4], 0.5% NP-40, 5 mM MgCl2, 150 mM NaCl, 1 mM dithiothreitol, 1 mM phenylmethylsulfonylfluoride) for 20 min at 4°C. Following cell lysis, cell debris was removed by centrifugation at 14,000 rpm for 20 min. For nonradioactive experiments, cells were washed one time in phosphate-buffered saline (PBS) and then lysed in NP-40 lysis buffer. The protein concentration of the supernatants was determined by bicinchoninic acid assay. For active-site labeling experiments employing UbVME, 330 μg of protein (50,000 cpm) was used and diluted with 1 volume of 50 mM Tris-HCl (pH 8) and 100 mM NaCl and reacted with hemagglutinin (HA)-UbVME (6, 7) for 1 h at room temperature. Immunoprecipitation was performed by incubation of cell lysates with either anti-gp150 or anti-ORF64 for 2 h at 4°C, followed by addition of protein A beads for 2 h. The pelleted protein A beads were washed three times with buffer (0.5% NP-40 in 50 mM Tris [pH 7.4], 150 mM NaCl, 5 mM EDTA). The pellet was resuspended in sodium dodecyl sulfate (SDS) sample buffer, boiled, and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), followed by fluorography. For immunoblotting, proteins were transferred to polyvinylidene difluoride membrane, blocked in 5% (wt/vol) milk, and immunoblotted with a monoclonal rat anti-HA-horseradish peroxidase antibody (3F10; Roche, Mannheim, Germany) for 45 min. Following three washes in PBS-Tween 20 (0.1%), blots were developed by using the Western Lighting chemiluminescence reagent (PerkinElmer, Boston, MA).
Electron microscopy.
3T12 cells were infected with MHV-68 at an MOI of 5 for 16 or 24 h at 37°C. The cells were fixed in 2.5% gluteraldehyde-3% paraformaldehyde with 5% sucrose in 0.1 M sodium cacodylate buffer (pH 7.4), pelleted, and postfixed in 1% OsO4 in Veronal-acetate buffer. The cell pellet was stained en bloc overnight with 0.5% uranyl acetate in Veronal-acetate buffer (pH 6.0) and then dehydrated and embedded in Spurrs resin. Sections were cut on a Reichert Ultracut E microtome with a Diatome diamond knife at a thickness setting of 50 nm and stained with uranyl acetate and lead citrate. The sections were examined using a FEI Tecnai Spirit electron microscope at 80 kV and photographed with an AMT charge-coupled device camera.
Experimental animals and infections.
C57BL/6J mice were purchased from Jackson Laboratory and housed in accordance with institutional guidelines. Mice aged 6 to 10 weeks were infected intraperitoneally (i.p.) with 1 × 106 PFU C→A ORF64 MHV-68 or (C→A)REV ORF64 virus. All animal experiments were conducted in accordance with protocols approved by the MIT Committee on Animal Care.
Plaque assays.
The amount of lytic virus in the spleen was determined using a standard plaque assay on mouse 3T12 cells (9). Spleens obtained at 3 or 7 days postinfection were homogenized by sonication, and incubated for 1 h on 3T12 cells at 37°C, washed once in PBS, and overlaid with carboxymethyl cellulose-containing complete DMEM (Sigma-Aldrich). After 6 days of incubation, monolayers were fixed with methanol and stained with Giemsa stain.
Virus ex vivo reactivation assay.
Spleens were harvested at 16 and 42 dpi and dispersed into single-cell suspensions. Twofold serial dilutions of splenocytes (from 100,000 to 781 cells) were seeded on top of 5,000 mouse embryonic fibroblasts in 96-well plates as described previously (42). After 3 weeks, cytopathic effect was assessed microscopically. To measure the presence of preformed virus particles, cells were lysed prior to plating by one freeze-thaw cycle at −80°C.
Measurement of viral load by quantitative real-time PCR.
Viral load in the spleens of infected mice was quantified by real-time PCR using the ABI 7900 real-time PCR system (Applied Biosystems, Foster City, CA). DNA was extracted from the spleens using the DNeasy blood and tissue kit (Qiagen, Hilden, Germany). The TaqMan universal PCR master mix and universal cycling conditions (Applied Biosystems) were used for amplification of a 70-bp region of the MHV-68 gB gene, using the primers and probe described in reference 43. A standard curve was generated using known amounts of a plasmid containing the gB gene [pcDNA3.1(+)-gB]. The murine ribosomal protein L8 (rpl8) was amplified in parallel, using a primer and probe set described in reference 36, and used to normalize for input DNA between samples. A standard curve for rpl8 was constructed by serial dilution of a plasmid containing rpl8 (clone 5684024; Open Biosystems, Huntsville, AL). The data were analyzed using the SDS 2.2.3 program and are presented as viral genome copy number relative to the copy number of rpl8 after subtraction of background as determined from uninfected samples.
Cell preparation and surface marker staining.
Single-cell suspensions were prepared from spleens by mechanical disruption in ice-cold RPMI 1640 medium (Invitrogen Life Technologies, Carlsbad, CA) containing 10% fetal calf serum. Erythrocytes were lysed with Sigma red blood cell lysis buffer (Sigma, St. Louis, MO). Dead cells were stained with ethidium monoazide (Invitrogen), prior to surface staining of splenocytes with saturating amounts of individual major histocompatibility complex (MHC) tetramers (prepared as described in reference 15) corresponding to the 22 known H-2b-restricted epitopes from MHV-68 (15) and antibodies against CD8α (BD Pharmingen, San Diego, CA). Cells were washed with PBS, fixed with 0.5% formaldehyde in PBS, and analyzed on an LSRII flow cytometer (BD Pharmingen), and the data were analyzed with FlowJo software (Tree Star).
RESULTS
Construction of MHV-68 ORF64 C→A and revertant viruses.
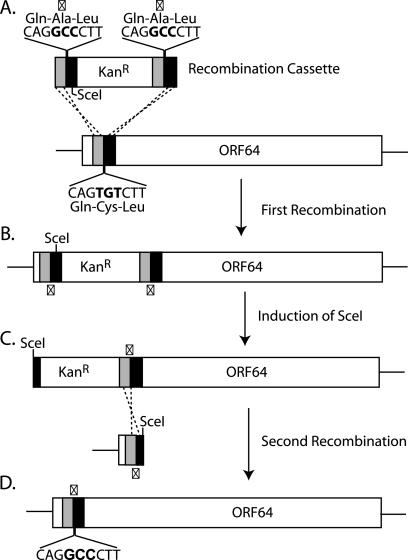
To explore the role of the deubiquitinating (USP) activity of ORF64 in vivo, we used recombination-based mutagenesis to alter the active-site cysteine (C33A) in ORF64 to alanine (C→A) (Fig. 1), starting with an infectious MHV-68 BAC clone as the substrate (2). We also generated a revertant virus [(C→A)REV], by restoring the wild-type (WT) amino acid sequence in MHV-68 ORF64 C→A virus. This allowed us to exclude effects of any potential second-site mutations. Correct mutagenesis was confirmed in all cases by restriction digestion and sequencing of the area surrounding the mutations (data not shown).
FIG. 1.
Construction of C→A MHV-68 and (C→A)REV MHV-68. Shown is a schematic representation of MHV-68 ORF64 C33A recombination. (A) A recombination cassette consisting of a kanamycin resistance gene flanked by homology arms to ORF64, which includes a TGT-to-GCC (C33A) mutation, was generated by PCR. (B) Recombination into the MHV-68 BAC ORF64 was followed by induction of SceI restriction enzyme (C) and a second recombination step to remove the kanamycin cassette (D), leaving behind the C33A mutation. The recombination process was repeated with a WT recombination cassette to generate a revertant BAC (not shown). X, point of mutation.
In vitro function of C→A.
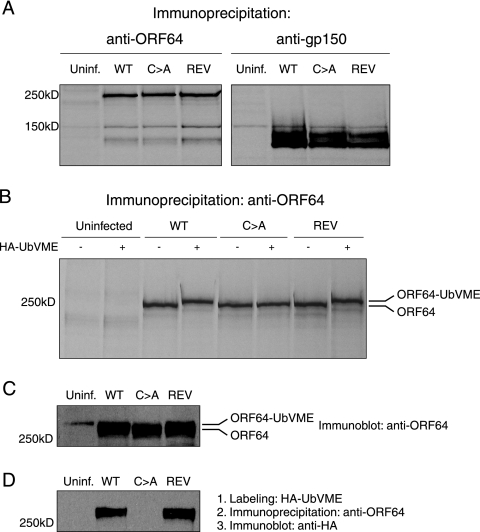
We first confirmed that ORF64 is still expressed in the C→A virus. We performed immunoprecipitations from radioactive [35S]methionine/cysteine-labeled cells that were uninfected or infected with WT, C→A, or (C→A)REV virus at 24 h p.i. with antibodies against ORF64 and gp150. For all three viruses, we detected similar levels of ORF64 and gp150. Furthermore, there was no obvious difference in the apparent molecular weights of ORF64 detected for these viruses (Fig. 2A and B). To confirm the lack of USP activity in the C→A virus, we took advantage of the activity-based HA-UbVME probe (7). Cell lysates were incubated with HA-UbVME and then immunoprecipitated with anti-ORF64. The resulting extracts were resolved by SDS-PAGE and immunoblotted for HA-UbVME-modified proteins using anti-HA antibodies. The input lysates for the immunoprecipitation were blotted with anti-ORF64. Active enzyme reacts with HA-UbVME, which results in an ∼7-kDa increase in apparent molecular mass, a mobility shift that is detectable even for proteins in the size range of full-length ORF64. We observed a size difference for the WT and (C→A)REV viruses as compared to C→A virus in samples exposed to HA-UbVME, corresponding to the addition of the HA-UbVME probe to the protein (Fig. 2C and D). We conclude that the C→A virus has indeed lost USP activity, as assessed by active-site labeling.
FIG. 2.
The C→A MHV-68 produces a full-length ORF64 protein and lacks deubiquitinating activity. (A) Lysates from uninfected (Uninf.) or WT, C→A, or (C→A)REV virus-infected radioactive [35S]methionine/cysteine-labeled 3T12 cells were immunoprecipitated with antibodies against ORF64 and gp150 followed by SDS-PAGE (8%). (B) Lysates from uninfected or WT, C→A, or (C→A)REV virus-infected radioactive [35S]methionine/cysteine-labeled 3T12 cells were treated with HA-UbVME, followed by immunoprecipitation with antibodies against ORF64, followed by SDS-PAGE (8%). (C) Lysates from uninfected or WT-, C→A-, or (C→A)REV-infected 3T12 cells were treated with HA-UbVME followed by immunoprecipitation with antibodies against ORF64 followed by SDS-PAGE (8%) and immunoblotting with anti-ORF64 antibody. (D) Lysates from uninfected or WT, C→A, or (C→A)REV virus-infected 3T12 cells were treated with HA-UbVME, followed by immunoprecipitation with antibodies against ORF64, followed by SDS-PAGE (8%) and immunoblotting with an anti-HA antibody.
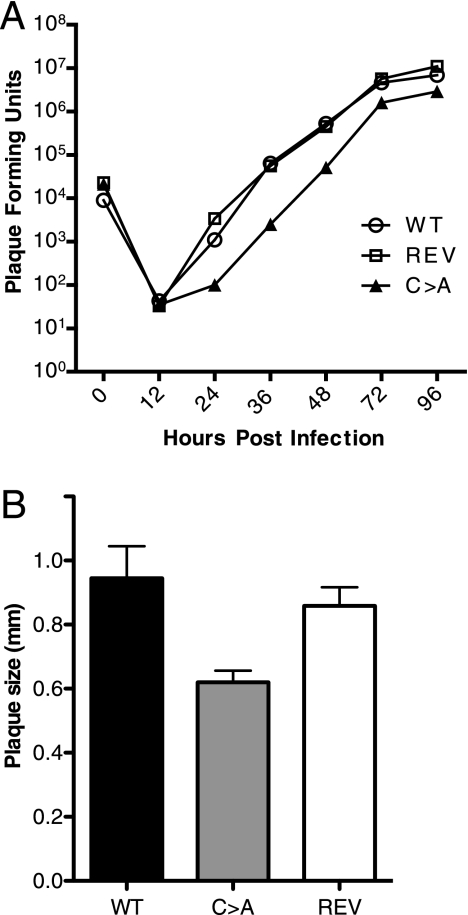
While we could propagate C→A virus in 3T12 cells, viral replication was delayed. To quantify this delay in replication, we performed a multistep growth curve for the WT, C→A, and (C→A)REV viruses, which indeed revealed a slight (∼10-fold) decrease in replication for the mutant virus (Fig. 3A). Furthermore, plaque size (diameter) was reduced by ∼20% in the C→A virus as compared to (C→A)REV and WT viruses (Fig. 3B). We conclude that USP activity is not essential for in vitro growth, yet a slight decrease in virus replication is seen in the C→A virus, especially at a low MOI. To examine virus replication in more detail, MHV-68-infected 3T12 cells were analyzed by electron microscopy at 16 and 24 h postinfection. The general appearance, distribution, and relative numbers of particles and particle types were similar for the WT, C→A, and (C→A)REV viruses; the only difference observed was that virus replication was slightly faster in the WT and (C→A)REV-infected cells, as also seen in the growth curves. We observed normal virion morphogenesis, including secondary envelopment and virion egress from the cells infected with the WT and (C→A)REV viruses, as well as from cells infected with C→A virus (Fig. 4). As expected, the (C→A)REV virus behaved like WT virus; therefore, we only used C→A and (C→A)REV MHV-68 for comparisons in all subsequent in vivo experiments.
FIG. 3.
Decreased growth ability and plaque size of C→A MHV-68. (A) Multistep growth kinetics. Replication of the WT, C→A, and (C→A)REV (REV) viruses was assessed after infection of 3T12 cells. Viral titers were determined by plaque assays, using the supernatant and lysed cells. (B) Determination of plaque size. Plaques of the WT, C→A, and (C→A)REV viruses on 3T12 cells were measured. The data are presented as mean plaque size in mm, and error bars represent standard deviations for three independent plaque assays where at least 10 plaques were measured per assay. Using a two-tailed t test, the P value for WT versus (C→A)REV virus is 0.2635 (not significant), that for WT versus C→A virus is 0.0061, and that for (C→A)REV versus C→A virus is 0.0038 (both significant, where P < 0.05 is significant).
FIG. 4.
Electron micrographs of 3T12 cells infected with WT, C→A, and (C→A)REV viruses. 3T12 cells were infected with MHV-68 and processed for electron microscopy as described in Materials and Methods. Depicted are the nuclear (N), cytoplasmic (C), and extracellular (EC) regions of cells infected with the WT (A), C→A (B), and (C→A)REV (C) viruses for 24 h. Panels D to F show magnification of enveloped viral particles between the nuclear membranes following primary envelopment during the egress process: WT in panel D, C→A virus in panel E, and (C→A)REV virus in panel F. For scaling, 500-nm bars are given.
Lack of USP activity leads to faster clearance of acute infection in the spleen.
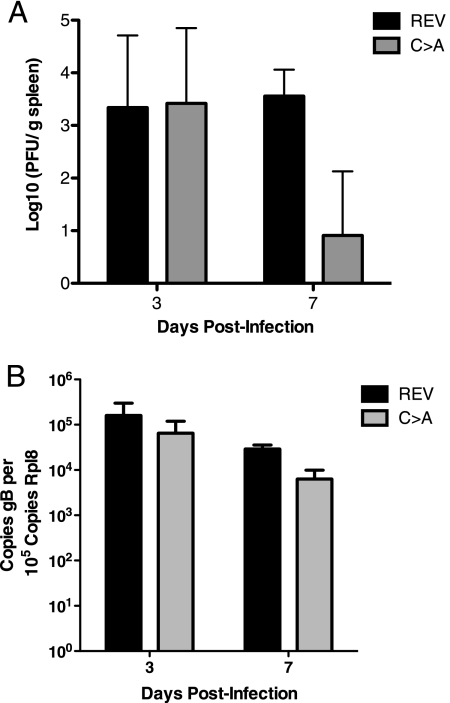
In MDV-infected chickens, USP activity is required for tumorigenesis in vivo (18). The lack of USP activity also delays onset of clinical signs of neuropathology in a small animal model using PrV (8). To date, studies that explore the effects of a deficiency in USP activity have been performed for two alphaherpesviruses, but there are no published in vivo data for beta- and gammaherpesviruses. We were therefore interested in assessing the role of USP activity during MHV-68 infection. MHV-68 infection in vivo is characterized by an acute phase, which is generally cleared by 10 to 15 days postinfection, and a latent phase (9, 32, 37, 41). We examined the effects of USP activity during acute infection and in the context of the establishment of latency. C57BL/6 mice were infected i.p. with C→A or (C→A)REV virus and sacrificed at 3 and 7 dpi, spleens were collected, and lysates were prepared to be used for plaque assays. Early after infection (3 days postinfection), both the C→A and (C→A)REV virus-infected groups had similar infectious viral loads, showing that input virus as well as early replication were similar for the two different viruses (Fig. 5A). However, after 7 days of infection, virus levels in spleens were significantly reduced (∼100-fold) for C→A virus compared to (C→A)REV virus (Fig. 5A), suggesting either increased clearance of MHV-68 or decreased replication of the virus in the spleen. Small sections of spleen were also collected for quantitative real-time PCR analysis of MHV-68 genome copy number, and—consistent with the infectious center assays—we found viremia to be reduced by ∼10 fold. These results, too, are consistent with either an increased ability for mice infected with virus lacking ORF64 USP activity to clear an acute MHV-68 infection or with reduced replication of the mutant virus (Fig. 5B).
FIG. 5.
Lack of USP activity leads to faster clearance of acute infection in the spleen. (A) Viral titers in spleens from C→A and (C→A)REV (REV) MHV-68-infected mice were determined by plaque assays at 3 and 7 dpi. The data are presented as 10 log PFU/g tissue, and the graph shows one representative experiment, which was repeated three times. Each time point represents data from at least eight animals, and error bars represent standard deviations. (B) Real-time PCR was performed on spleens from C→A and (C→A)REV MHV-68-infected mice at 3 and 7 dpi. The data are presented as viral genome copy number relative to the copy number of rpl8. Each time point represents samples from at least five animals, and error bars represent standard deviations.
The establishment of latency and reactivation of MHV-68 in the spleen are not dependent on ORF64 USP activity.
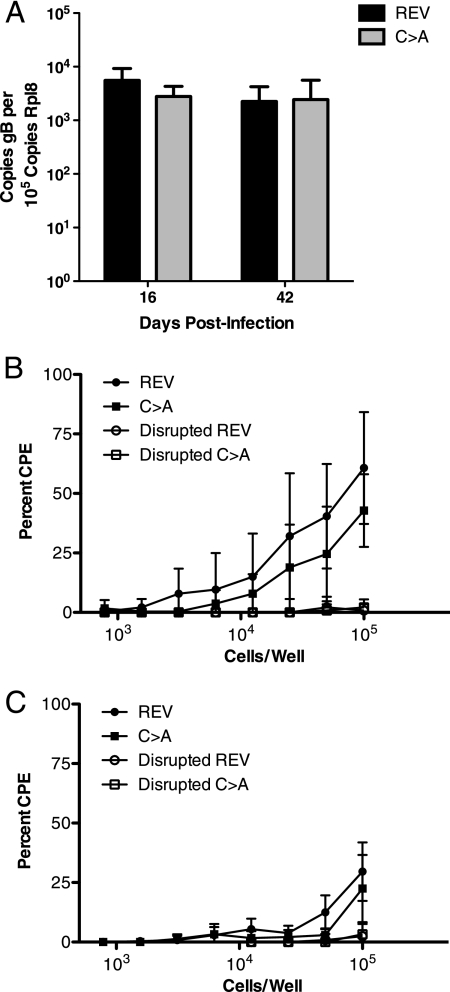
The more rapid viral clearance or reduced replication of virus lacking ORF64 USP activity upon acute infection might lead to lower latent virus loads in the spleen. The number of MHV-68 genomes was lower for the C→A mutant than for the (C→A)REV virus at 7 dpi, but the numbers were similar at 16 and 42 dpi (Fig. 6A). Viral persistence, replication, and latency at later stages of MHV-68 infection are therefore independent of ORF64 USP activity. Mice were further examined at 16 and 42 days following i.p. infection, and the frequencies of MHV-68-reactivating splenocytes did not differ significantly between C→A and (C→A)REV virus-infected mice (6B and C). Thus, ORF64 USP activity does not appear to play a role in the control of MHV-68 reactivation or in setting the latent viral load.
FIG. 6.
The establishment of latency and reactivation of MHV-68 in the spleen are not dependent on ORF64 USP activity. (A) Real-time PCR was performed on spleens from C→A and (C→A)REV (REV) MHV-68-infected mice at 16 and 42 dpi. The data are presented as viral genome copy number relative to the copy number of rpl8. Each time point represents samples from at least five animals. (B and C) An ex vivo limiting dilution reactivation assay was used to detect reactivation from latency in cells from the spleen at 16 dpi (B) and 42 dpi (C) from C→A and (C→A)REV virus-infected mice. The presence of preformed infectious virus, as measured by using disrupted cells, is shown as open symbols. CPE, cytopathic effect. The data represent the mean value from 10 mice per experimental group ± standard deviation.
The pattern of recognition of CD8+ T-cell epitopes does not differ between mice infected with C→A and (C→A)REV MHV-68.
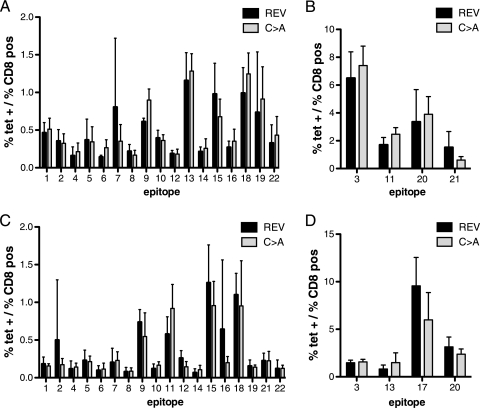
Since our data at 7 dpi could be explained either by faster clearance of the virus or by decreased replication of MHV-68 in the spleens at 7 dpi, we examined the pattern of display of MHV-68-specific CD8 T-cell epitopes in the C→A mutant and (C→A)REV viruses. We recently identified 19 new CD8 epitopes for MHV-68 for the H-2b haplotype (15) and used MHC class I tetramers to stain for all 22 currently known MHV-68 epitopes. We did not detect any differences in the CD8 epitope response patterns at either 7 or 16 dpi with C→A and (C→A)REV viruses (Fig. 7). This suggest that the clearance of the C→A mutant during acute infection is unlikely to be accounted for by increased expansion of any of the known MHV-68-specific CD8 T-cell populations.
FIG. 7.
The epitope patterns do not differ between mice infected with C→A MHV-68 and those infected with (C→A)REV (REV) MHV-68. Shown is cell surface staining of splenocytes from mice infected with C→A and (C→A)REV MHV-68 with the respective H-2Kb and H-2Db tetramers (tet) 7 and 16 days after infection. (A) The panel shows the mean percentage (0 to 2%) of epitope-positive CD8 T cells at 7 dpi for (C→A)REV and C→A virus-infected mice. (B) The panel shows the mean percentage (0 to 10%) of epitope-positive CD8 T cells at 7 dpi for (C→A)REV and C→A virus-infected mice. (C) The panel shows the mean percentage (0 to 2%) of epitope-positive CD8 T cells at 16 dpi for (C→A)REV and C→A virus-infected mice. (D) The panel shows the mean percentage (0 to 15%) of epitope-positive CD8 T cells at 16 dpi for (C→A)REV and C→A virus-infected mice. The data represent the mean value of three to six mice per experimental group ± standard deviation. The numbers in the graphs correspond to the following epitopes (peptide sequences): 1, RSYIYYAL; 2, AGYIYYQL; 3, KSLTYYKL; 4, TGFRYSYM; 5, SCLDYSHL; 6, AILKFKSL; 7, LGSVYYKL; 8, AIYSFRNA; 9, AVVQFIRV; 10, VNLVFPSV; 11, FVYLFHFM; 12, SVYGFTGV; 13, TNYKFSLV; 14, CNRIYARL; 15, ITYFFEMI; 16, LSPPMAHL; 17, TSINFVKI; 18, KNYIFEEKL; 19, SAITNHAAF; 20, AGPHNDMEI; 21, SAIENYETF; and 22, SAPMKTVTI.
DISCUSSION
The highly conserved catalytic triad in the large tegument protein of all herpesviruses suggests an important function for the USP encoded by it. We and others have demonstrated deubiquitinating activity in the large tegument protein for a number of herpesviruses (14, 18, 19, 40). Recombinant viruses that lack this deubiquitinating activity have also been engineered (8, 18, 40). To date, further investigation of the in vivo function for the USPs in the large tegument proteins has been performed only with two alphaherpesviruses, owing to the availability of suitable animal models: MDV (18) and PrV (8). When USP activity in the MDV large tegument protein was abolished, replication of MDV was diminished and the ability of the virus to form T-cell lymphomas was strikingly reduced in infected chickens (18), thus implying a role for USP activity in the establishment and/or maintenance of T-cell transformation. In the case of PrV, USP activity was shown to be important for virus replication and secondary envelopment late in infection (8). Mice infected with the PrV USP mutant demonstrated delayed neuroinvasion kinetics (8). Furthermore, in vitro, short hairpin RNA to EBV BPLF1 diminished viral genome copy number (44).
Here we explored the role of the herpesviral USP for a gammaherpesvirus, MHV-68 (14). To do so, we generated a mutant virus that lacks the USP activity in ORF64, by altering the active-site cysteine at position 33 in ORF64 to alanine. We also generated the corresponding revertant virus. The C→A mutation in ORF64 abolished all deubiquitinating activity, but as expected, full-length protein was still produced. For neither of the alphaherpesviruses MDV (18) and PrV (8) nor the betaherpesvirus HCMV (40) is the absence of USP activity lethal, and lack of USP activity only slightly diminishes viral replication in cell culture. For MHV-68, we likewise saw decreased viral replication (by ∼10-fold) in cell culture, as well as reduced plaque size for the C→A virus. Electron microscopy failed to show obvious defects in virion morphogenesis after infection, as seen also for HCMV and the corresponding viral active-site cysteine mutant (40). In contrast, for the PrV active-site cysteine mutant, accumulation of cytoplasmic nonenveloped nucleocapsids was observed (8). This PrV mutant also consisted of a single point mutation in which the full-length protein was expressed; however, the cysteine was mutated to serine rather than alanine (8).
Replacement of the active-site cysteine in the MDV deubiquitinating protein DUB led to a slight reduction in replication and plaque size in vitro, while early replication in chickens in vivo was unaffected (18). An impairment of viral neuroinvasion was observed in mice infected with PrV lacking the USP active-site cysteine, a delay that parallels the replication defect in cell culture (8). The fact that MHV-68 is a natural pathogen of wild rodents (5) allowed us to use the mouse to study this herpesvirus infection in its natural host. In i.p.-infected mice, we observed similar levels of virus in the spleen at 3 dpi for C→A virus and its revertant, indicating that virus input was similar for the two viruses. At 7 dpi, the amount of virus was significantly reduced for the C→A mutant, as compared to the revertant (100×). Viral DNA copy numbers in the spleen were also lower for the mutant virus (10×), suggesting faster clearance or decreased replication efficiency. Given that we see a decrease in replication efficiency of the virus in vitro, it is likely that this difference is also mirrored in vivo at 7 dpi. The fact that we see a 100× decrease in infectivity of the C→A virus compared to its revertant and only a 10× decrease in viral genomes suggests that this is due to the production of increased numbers of noninfectious particles, which would be detected in Fig. 5B but not 5A. However, we cannot rule out that the C→A virus is also cleared more efficiently than the revertant by the immune system.
MHV-68 is usually cleared by 8 to 16 dpi during in vivo infection (9, 32, 41). Since we observed similar virus levels at day 3, and the genome copy numbers were higher at day 3 than day 7 for the mutant virus, we hypothesized that the difference in the viral titers observed at day 7 might be attributable to more rapid clearance of the mutant virus during acute infection. Several host mechanisms contribute to clearing of MHV-68, one of which is the CD8+ T-cell response (11, 41). We have described two distinct patterns of MHV-68-specific CD8 T-cell involvement in C57BL/6 mice: an early phase that peaks relatively early after infection, around 6 dpi, and a slightly later phase that peaks at 10 dpi (15). Could differences in the CD8+ T-cell response be responsible for the difference in virus levels at 7 dpi? To address this question, we examined the frequencies of CD8+ T cells specific for MHV-68 (15, 21, 28). Recognition of the 22 currently known CD8+ T-cell epitopes for MHV-68 did not show any differences in CD8+ T-cell reactivity for the mutant virus compared to the revertant.
Although virus levels were lower in mice infected with C→A MHV-68 at 7 dpi, this difference was no longer apparent at days 16 and 42, nor did the frequencies of reactivating splenocytes differ significantly between C→A and (C→A)REV virus-infected mice. The peak of acute-phase replication depends on viral dose: an i.p. dose of 10E6 PFU peaks at 7 dpi in the spleen (37). However, establishment of latency does not depend on the dose of infection (37). Latent infection is established early on during the acute response, and viral replication is not absolutely required to establish latency. When the lytic cycle is inhibited early after infection, as is the case for a replication-deficient virus, latency is still established (12). This observation may well explain why the viral genome copy numbers are the same during latency for both C→A virus and C→A revertant virus, even if the kinetics of replication for the C→A MHV-68 are slower and the acute-phase titers are lower than for the revertant.
Ubiquitination and deubiquitination regulate essential cellular processes. Viral interference with cellular physiology could create a host environment more favorable for virus replication and dissemination. MHV-68 interferes with both ubiquitination and deubiquitination. Both KSHV and MHV-68 encode a ubiquitin ligase, K3, that downregulates class I MHC expression (29, 30), and MHV-68 possesses a deubiquitinating enzyme, ORF64 (14), the substrates and function of which remain unknown. Tumor viruses may modulate the ubiquitin-proteasome machinery for different purposes (as reviewed in reference 26). The EBV EBNA1 protein interacts with host-derived USP7 (also called “HAUSP”) (17) and is believed to interfere with normal regulation of the mdm2-p53 pathway (reviewed in reference 16). The MDV USP contributes to oncogenic transformation of infected chickens (18). Gammaherpesviruses can also cause cancers: EBV γ-1 is associated with multiple human malignancies, including nasopharyngeal carcinoma and B-cell lymphomas (22), and KSHV γ-2 has been associated with Kaposi's sarcoma and B-cell lymphomas (1). MHV-68 is commonly used as a model for the pathogenesis of gammaherpesviruses as MHV-68 infects and establishes latency in B cells (33, 41). Chronically infected inbred mice develop a lymphoproliferative disease, similar to that seen in EBV-infected patients (31). However, unlike EBV infection, MHV-68 genome-positive cells are detected to a variable degree in the tumor cells, suggesting that direct transformation is not responsible for the induction of malignancies in these models (31, 35). It is possible that the ORF64 USP is involved in lymphomagenesis in MHV-68-infected mice. This could be explored in lymphoma-prone models (31, 34, 35), although it remains unclear how this attribute of the virus could contribute to its evolutionary fitness.
The conservation of the tegument-encoded USP activity in every member of the herpesvirus family does suggest an important function for this activity. In the two alphaherpesvirus animal models that have thus far been investigated, there was a clear in vivo effect on the pathogenesis of the virus for the respective tegument protein USP mutants. Our in vivo model for gammaherpesvirus did not use disease or pathogenesis as a readout but rather examined acute and latent virus levels, and we did indeed find a difference in acute viral titers. It is possible that lymphomagenesis as a readout in the animal model for the USP function of MHV-68 (31, 35) might reveal effects similar to the ones seen in animal models for MDV (18) and PrV (8). Other factors, such as the route of infection, could also affect clinical outcome. We performed our in vivo infections i.p., a route commonly used for experimental infection with MHV-68. Although intranasal infections are also used, at least the establishment and maintenance of latency do not appear to depend on the route of infection (37).
In summary, we show that a single mutation of the active-site cysteine in the deubiquitinating enzyme ORF64 in MHV-68 does not affect ORF64 protein expression but does eliminate its deubiquitinating activity. This is consistent with the structural model, based on the analysis of the single herpesvirus USP domain for which a structure has been solved (25), the MCMV M48 product.
A more complete picture of the biological role of the herpesvirus ubiquitin-specific proteases will require identification of their natural substrates. The first such candidate for the EBV large tegument protein USP was recently identified: the N-terminal region of EBV DUB was shown to interact with the small subunit of the EBV ribonucleotide reductase (RR) (44). Furthermore, the large subunit of EBV RR was shown to be ubiquitinated and its activity was decreased when the enzymatically active N-terminal region of the EBV DUB was expressed, suggesting that the EBV DUB deubiquitinates the RR (44). The MHV-68 mutant described here may help us identify the natural substrate(s) for the ORF64 DUB.
Acknowledgments
This study was supported by grants from the NIH (to H.L.P.). S.G.-R. is supported by a postdoctoral grant from the Wenner-Gren Foundations (Sweden).
We thank the Osterrieder lab for the recombination reagents and Neil Margulis for assistance in making the point mutant C→A virus.
Footnotes
Published ahead of print on 12 August 2009.
REFERENCES
- 1.Ablashi, D. V., L. G. Chatlynne, J. E. Whitman, Jr., and E. Cesarman. 2002. Spectrum of Kaposi's sarcoma-associated herpesvirus, or human herpesvirus 8, diseases. Clin. Microbiol. Rev. 15:439-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balakirev, M. Y., M. Jaquinod, A. L. Haas, and J. Chroboczek. 2002. Deubiquitinating function of adenovirus proteinase. J. Virol. 76:6323-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barretto, N., D. Jukneliene, K. Ratia, Z. Chen, A. D. Mesecar, and S. C. Baker. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 79:15189-15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blasdell, K., C. McCracken, A. Morris, A. A. Nash, M. Begon, M. Bennett, and J. P. Stewart. 2003. The wood mouse is a natural host for Murid herpesvirus 4. J. Gen. Virol. 84:111-113. [DOI] [PubMed] [Google Scholar]
- 6.Borodovsky, A., B. M. Kessler, R. Casagrande, H. S. Overkleeft, K. D. Wilkinson, and H. L. Ploegh. 2001. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 20:5187-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borodovsky, A., H. Ovaa, N. Kolli, T. Gan-Erdene, K. D. Wilkinson, H. L. Ploegh, and B. M. Kessler. 2002. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 9:1149-1159. [DOI] [PubMed] [Google Scholar]
- 8.Böttcher, S., C. Maresch, H. Granzow, B. G. Klupp, J. P. Teifke, and T. C. Mettenleiter. 2008. Mutagenesis of the active-site cysteine in the ubiquitin-specific protease contained in large tegument protein pUL36 of pseudorabies virus impairs viral replication in vitro and neuroinvasion in vivo. J. Virol. 82:6009-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardin, R. D., J. W. Brooks, S. R. Sarawar, and P. C. Doherty. 1996. Progressive loss of CD8+ T cell-mediated control of a gamma-herpesvirus in the absence of CD4+ T cells. J. Exp. Med. 184:863-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Z., Y. Wang, K. Ratia, A. D. Mesecar, K. D. Wilkinson, and S. C. Baker. 2007. Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J. Virol. 81:6007-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehtisham, S., N. P. Sunil-Chandra, and A. A. Nash. 1993. Pathogenesis of murine gammaherpesvirus infection in mice deficient in CD4 and CD8 T cells. J. Virol. 67:5247-5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flano, E., Q. Jia, J. Moore, D. L. Woodland, R. Sun, and M. A. Blackman. 2005. Early establishment of gamma-herpesvirus latency: implications for immune control. J. Immunol. 174:4972-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glickman, M. H., and A. Ciechanover. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373-428. [DOI] [PubMed] [Google Scholar]
- 14.Gredmark, S., C. Schlieker, V. Quesada, E. Spooner, and H. L. Ploegh. 2007. A functional ubiquitin-specific protease embedded in the large tegument protein (ORF64) of murine gammaherpesvirus 68 is active during the course of infection. J. Virol. 81:10300-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gredmark-Russ, S., E. J. Cheung, M. K. Isaacson, H. L. Ploegh, and G. M. Grotenbreg. 2008. The CD8 T-cell response against murine gammaherpesvirus 68 is directed towards a broad repertoire of epitopes from both early and late antigens. J. Virol. 82:12205-12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holowaty, M. N., and L. Frappier. 2004. HAUSP/USP7 as an Epstein-Barr virus target. Biochem. Soc. Trans. 32:731-732. [DOI] [PubMed] [Google Scholar]
- 17.Holowaty, M. N., M. Zeghouf, H. Wu, J. Tellam, V. Athanasopoulos, J. Greenblatt, and L. Frappier. 2003. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J. Biol. Chem. 278:29987-29994. [DOI] [PubMed] [Google Scholar]
- 18.Jarosinski, K., L. Kattenhorn, B. Kaufer, H. Ploegh, and N. Osterrieder. 2007. A herpesvirus ubiquitin-specific protease is critical for efficient T cell lymphoma formation. Proc. Natl. Acad. Sci. USA 104:20025-20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kattenhorn, L. M., G. A. Korbel, B. M. Kessler, E. Spooner, and H. L. Ploegh. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19:547-557. [DOI] [PubMed] [Google Scholar]
- 20.Lindner, H. A., N. Fotouhi-Ardakani, V. Lytvyn, P. Lachance, T. Sulea, and R. Menard. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 79:15199-15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, L., E. J. Usherwood, M. A. Blackman, and D. L. Woodland. 1999. T-cell vaccination alters the course of murine herpesvirus 68 infection and the establishment of viral latency in mice. J. Virol. 73:9849-9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pattle, S. B., and P. J. Farrell. 2006. The role of Epstein-Barr virus in cancer. Expert Opin. Biol. Ther. 6:1193-1205. [DOI] [PubMed] [Google Scholar]
- 23.Reyes-Turcu, F. E., and K. D. Wilkinson. 2009. Polyubiquitin binding and disassembly by deubiquitinating enzymes. Chem. Rev. 109:1495-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlieker, C., G. A. Korbel, L. M. Kattenhorn, and H. L. Ploegh. 2005. A deubiquitinating activity is conserved in the large tegument protein of the Herpesviridae. J. Virol. 79:15582-15585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlieker, C., W. A. Weihofen, E. Frijns, L. M. Kattenhorn, R. Gaudet, and H. L. Ploegh. 2007. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol. Cell 25:677-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shackelford, J., and J. S. Pagano. 2004. Tumor viruses and cell signaling pathways: deubiquitination versus ubiquitination. Mol. Cell. Biol. 24:5089-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sompallae, R., S. Gastaldello, S. Hildebrand, N. Zinin, G. Hassink, K. Lindsten, J. Haas, B. Persson, and M. G. Masucci. 2008. Epstein-Barr virus encodes three bona fide ubiquitin-specific proteases. J. Virol. 82:10477-10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevenson, P. G., G. T. Belz, J. D. Altman, and P. C. Doherty. 1999. Changing patterns of dominance in the CD8+ T cell response during acute and persistent murine gamma-herpesvirus infection. Eur. J. Immunol. 29:1059-1067. [DOI] [PubMed] [Google Scholar]
- 29.Stevenson, P. G., S. Efstathiou, P. C. Doherty, and P. J. Lehner. 2000. Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc. Natl. Acad. Sci. USA 97:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevenson, P. G., J. S. May, X. G. Smith, S. Marques, H. Adler, U. H. Koszinowski, J. P. Simas, and S. Efstathiou. 2002. K3-mediated evasion of CD8(+) T cells aids amplification of a latent gamma-herpesvirus. Nat. Immunol. 3:733-740. [DOI] [PubMed] [Google Scholar]
- 31.Sunil-Chandra, N. P., J. Arno, J. Fazakerley, and A. A. Nash. 1994. Lymphoproliferative disease in mice infected with murine gammaherpesvirus 68. Am. J. Pathol. 145:818-826. [PMC free article] [PubMed] [Google Scholar]
- 32.Sunil-Chandra, N. P., S. Efstathiou, J. Arno, and A. A. Nash. 1992. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J. Gen. Virol. 73:2347-2356. [DOI] [PubMed] [Google Scholar]
- 33.Sunil-Chandra, N. P., S. Efstathiou, and A. A. Nash. 1992. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J. Gen. Virol. 73:3275-3279. [DOI] [PubMed] [Google Scholar]
- 34.Tarakanova, V. L., F. Kreisel, D. W. White, and H. W. Virgin IV. 2008. Murine gammaherpesvirus 68 genes both induce and suppress lymphoproliferative disease. J. Virol. 82:1034-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarakanova, V. L., F. Suarez, S. A. Tibbetts, M. A. Jacoby, K. E. Weck, J. L. Hess, S. H. Speck, and H. W. Virgin IV. 2005. Murine gammaherpesvirus 68 infection is associated with lymphoproliferative disease and lymphoma in BALB β2 microglobulin-deficient mice. J. Virol. 79:14668-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thakur, N. N., S. El-Gogo, B. Steer, K. Freimuller, A. Waha, and H. Adler. 2007. A gammaherpesviral internal repeat contributes to latency amplification. PLoS ONE 2:e733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tibbetts, S. A., J. Loh, V. van Berkel, J. S. McClellan, M. A. Jacoby, S. B. Kapadia, S. H. Speck, and H. W. Virgin IV. 2003. Establishment and maintenance of gammaherpesvirus latency are independent of infective dose and route of infection. J. Virol. 77:7696-7701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tischer, B. K., J. von Einem, B. Kaufer, and N. Osterrieder. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 40:191-197. [DOI] [PubMed] [Google Scholar]
- 39.Virgin, H. W., IV, P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, J., A. N. Loveland, L. M. Kattenhorn, H. L. Ploegh, and W. Gibson. 2006. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 80:6003-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weck, K. E., M. L. Barkon, L. I. Yoo, S. H. Speck, and H. W. Virgin IV. 1996. Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J. Virol. 70:6775-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weck, K. E., S. S. Kim, H. W. Virgin IV, and S. H. Speck. 1999. B cells regulate murine gammaherpesvirus 68 latency. J. Virol. 73:4651-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinberg, J. B., M. L. Lutzke, R. Alfinito, and R. Rochford. 2004. Mouse strain differences in the chemokine response to acute lung infection with a murine gammaherpesvirus. Viral Immunol. 17:69-77. [DOI] [PubMed] [Google Scholar]
- 44.Whitehurst, C. B., S. Ning, G. L. Bentz, F. Dufour, E. Gershburg, J. Shackelford, Y. Langelier, and J. S. Pagano. 2009. The Epstein-Barr virus (EBV) deubiquitinating enzyme BPLF1 reduces EBV ribonucleotide reductase activity. J. Virol. 83:4345-4353. [DOI] [PMC free article] [PubMed] [Google Scholar]