Abstract
Human respiratory syncytial virus (RSV) contains a heavily glycosylated 90-kDa attachment glycoprotein (G). Infection of HEp-2 and Vero cells in culture depends largely on virion G protein binding to cell surface glycosaminoglycans (GAGs). This GAG-dependent phenotype has been described for RSV grown in HEp-2 cells, but we have found that it is greatly reduced by a single passage in Vero cells. Virions produced from Vero cells primarily display a 55-kDa G glycoprotein. This smaller G protein represents a post-Golgi compartment form that is lacking its C terminus, indicating that the C terminus is required for GAG dependency. Vero cell-grown virus infected primary well-differentiated human airway epithelial (HAE) cell cultures 600-fold less efficiently than did HEp-2 cell-grown virus, indicating that the C terminus of the G protein is also required for virus attachment to this model of the in vivo target cells. This reduced infectivity for HAE cell cultures is not likely to be due to the loss of GAG attachment since heparan sulfate, the primary GAG used by RSV for attachment to HEp-2 cells, is not detectable at the apical surface of HAE cell cultures where RSV enters. Growing RSV stocks in Vero cells could dramatically reduce the initial infection of the respiratory tract in animal models or in volunteers receiving attenuated virus vaccines, thereby reducing the efficiency of infection or the efficacy of the vaccine.
Human respiratory syncytial virus (RSV) is a negative-sense, single-stranded RNA virus in the family Paramyxoviridae, subfamily Pneumovirinae. RSV causes mild respiratory disease in all age groups, but the disease can be severe or fatal in infants and the elderly (4, 9, 11). Initial attempts to produce a killed vaccine were not successful, resulting instead in enhanced disease upon infection (26, 41). Efforts to produce a live attenuated vaccine are ongoing (6, 7, 51).
RSV produces three glycoproteins which are important for infection. The largest glycoprotein (G) is involved in attachment to the host cell (35), the fusion (F) glycoprotein mediates virion membrane fusion with the target cell membrane (2), and the small hydrophobic (SH) glycoprotein may attenuate apoptosis (15). The F protein is the only glycoprotein that is absolutely required for infection of cultured immortalized cells (27, 45) and syncytium formation, the most obvious cytopathic effect of RSV in immortalized cell culture. Although the G protein is not absolutely required for infection, it enhances infection and syncytium formation (45). The G protein attaches to cultured, immortalized cell lines (35) primarily via glycosaminoglycans (GAGs) on the cell surface (13, 22, 23, 30). GAGs are repeating disaccharide units of hexuronic acid and hexosamine that form unbranched polysaccharide chains and are found on the surface of most mammalian cells. The GAG type that appears most important for RSV infection of HEp-2 cells is heparan sulfate (HS) (23, 30).
The G protein is a type II integral membrane protein with its N terminus on the cytoplasmic side of the membrane and its C terminus as the extracellular ectodomain (49). An unglycosylated region in the center of the protein contains four cysteines held together by disulfide bonds in a cysteine noose (19, 24, 33), followed, to the C-terminal side, by a predicted heparin-binding domain (HBD) (12, 13). The 32-kDa G protein, while in the endoplasmic reticulum (ER), is modified by the addition of multiple N-linked carbohydrate chains, depending on the strain. These N-linked additions would increase the molecular mass of G to 45 to 60 kDa. Previous reports have found G protein forms of this size in cells and in virions at low levels (5, 20, 21, 50). All of these reports suggest that these smaller forms of the G protein are partially glycosylated processing intermediates.
Maturation of the N-linked carbohydrates of the G protein occurs in the Golgi compartment, where a large number of O-linked carbohydrate chains are added, resulting in an 84- to 92-kDa mature protein (14, 32, 35, 49). This size variation of the G protein is probably due, in part, to the difficulty in sizing heavily glycosylated molecules and variations in molecular mass markers.
The G protein shares no homology with the glycoproteins of paramyxoviruses outside the Pneumovirinae subfamily. The high serine and threonine content and the high O-linked glycosylation levels are similar to those found in mucins. The amount of O-linked glycosylation is partially dependent on the cell type used to produce the virus (18).
In the present study, we examined virus produced in HEp-2 and Vero cells, which are both commonly used to grow RSV in the laboratory, for dependence on GAGs by the ability to infect cells expressing GAG or deficient in GAG expression. We also examined the ability of the viruses to infect primary, well-differentiated human airway epithelial (HAE) cell cultures. In both systems, infectivity was greatly dependent upon the cell line used to grow the virus. Biochemical characterization of purified virus grown in these two cell lines revealed a smaller form of the RSV G protein in virions from Vero cells. Using C terminus-specific antibodies and a six-His tag at the C terminus of the G protein, we determined that the smaller G protein form was lacking its C terminus. These results highlight the importance of the C-terminal portion of the G protein and suggest that the cell line used to produce a virus can alter its infectivity.
MATERIALS AND METHODS
Viruses and cells.
The recombinant green fluorescent protein (GFP)-expressing RSV used in these experiments were rgRSV-SGF (strain A2) and mutants of this virus lacking one or more of the glycoprotein genes designated by the glycoproteins that they express, i.e., rgRSV-F, rgRSV-GF, and rgRSV-SF (45). The HEp-2 (American Type Culture Collection, Manassas, VA), MRC-5, A549, primary monkey kidney (PMK), and BSC-1 cell lines were grown in Opti-MEM I containing 2% fetal bovine serum (FBS) (Atlanta Biologicals, Norcross, GA), and Vero and World Health Organization (WHO)-approved Vero cells were grown in RPMI 1640 medium containing 5% FBS. The Chinese hamster ovary (CHO) K1 cell line and its mutant A745, which is severely deficient in the production of all GAGs due to a defective xylosyl transferase (10), were grown in F-12 containing 10% FBS. All media were purchased from Invitrogen (Carlsbad, CA). Cells were incubated at 37°C in 5% CO2. All virus stocks and cells tested negative for mycoplasma by PCR (Intronbio, Seongnam-Si, Korea).
GAG dependency index.
The GAG dependency index was derived by dividing the titer of the same virus sample determined on CHO K1 cells by its titer on CHO A745 cells. A volume of 40 μl of RSV whose titer had not been determined was added to 160 μl of medium as the first fivefold dilution. Additional serial fivefold dilutions of RSV were used to inoculate both CHO K1 and CHO A745 cells in 96-well plates that were ∼70% confluent. At 24 h, the infected cells were fixed in 3% paraformaldehyde for 20 min, permeabilized with 0.1% Triton X-100, stained with fluorescein isothiocyanate-labeled anti-RSV polyclonal antibodies (Virostat, Portland, ME), and counted.
Metabolic labeling of RSV.
Virion proteins were metabolically labeled with Tran35S-label (>70% 35S-labeled l-methionine and ∼15% 35S-labeled l-cysteine; MPBiomedical, Solon, OH) by adding 20 μCi/ml to Opti-MEM I with 2% FBS at 4 h postinoculation (p.i.). At 72 h p.i., virus was collected by scraping cells from the tissue culture dish with a rubber policeman, pipetting, and vortexing. Cells were removed by low-speed centrifugation (400 × g for 5 min). The supernatant was centrifuged at 20,000 × g for 90 min in 50-ml high-speed conical tubes (Sorvall) to pellet the virus in a Sorvall 600TC rotor. The pellet was resuspended in Hanks balanced salt solution with Ca2+, Mg2+, and 25 mM HEPES by repeated pipetting. This material was loaded onto a linear 25 to 55% sucrose gradient in SW28 tubes and centrifuged for 18 h at 4°C in an SW28 rotor at 125,000 × g. The gradient was fractionated from the bottom with a capillary tube connected to a peristaltic pump and a fraction collector. Samples of each fraction were analyzed with a Tri-Carb 2100TR scintillation counter (Packard). Fractions containing peak radioactivity were pooled, diluted, loaded onto a second linear 25 to 55% sucrose gradient, and purified a second time under the same conditions as the first spin. Fractions from the second gradient were similarly analyzed. The fractions were also tested for infectivity, and peak infectivity was found to coincide with peak radioactivity. The fractions containing peak radioactivity were pooled for analysis.
Western blotting.
Similar amounts of purified virions (15,000 cpm, metabolically labeled in parallel) were solubilized with lysis buffer that contained protease inhibitors (Pierce) and 2 mg/ml N-ethylmaleimide, reduced in Laemmli sample buffer with 5% β-mercaptoethanol, and boiled for 3 to 5 min, and proteins were separated by electrophoresis on a 10% polyacrylamide gel containing sodium dodecyl sulfate (SDS-PAGE) (31). The proteins were transferred at 200 mA for 1 h to an Immobilon (Millipore) membrane, blocked overnight with 5% nonfat milk-0.1% Tween 20, probed with primary antibody, washed twice with blocking solution, and probed with a horseradish peroxidase (HRP)-labeled goat anti-mouse antibody (Kirkegaard & Perry Laboratories). Lumi-Light Western blotting substrate (Roche) was added, and the membranes were exposed to film for 5 s or less to generate the images in the figures and separately scanned with a PhosphorImager (Typhoon; GE Healthcare). Quantification was done by averaging the density of bands from three different membranes. Primary antibodies used to stain the membranes were monoclonal antibody (MAb) L9 against the conserved central region of the RSV G protein; MAb D14 against the RSV nucleocapsid (N) protein (48); MAb 130-2G, which recognizes the C terminus of the G protein (1, 43); and MAb 5His (Qiagen, Valencia, CA), which recognizes the six-His tag.
Pulse-chase.
A 70% confluent six-well plate of Vero and HEp-2 cells was inoculated at a multiplicity of infection (MOI) of 3 with tipping for 1 h and then rinsed with PBS before fresh medium was added. At 24 h p.i., wells were washed three times with PBS before being pulsed with 20 μCi/ml Tran35S-label in Met/Cys-free Dulbecco modified Eagle medium for 15 min at 37°C, after which the medium was removed, the cells were rinsed with PBS, complete Dulbecco modified Eagle medium-10% FBS supplemented with 10 μM unlabeled methionine was added, and the mixture was incubated at 37°C. At various times after this pulse, the cells were solubilized with lysis buffer that contained protease inhibitors (Pierce) and 2 mg/ml N-ethylmaleimide. As a control that lacks Golgi compartment modifications, monensin (10 μM) was added 1 h before the pulse and maintained throughout the pulse and chase. Lysates were cleared of insoluble debris by centrifugation at 14,000 × g for 10 min. The cleared lysate was immunoprecipitated (IP) with MAb L9 and displayed by 10% SDS-PAGE. The gel was soaked overnight in a 20% methanol-3% glycerol solution to prevent cracking, followed by EN3HANCE (Perkin-Elmer) for 30 min, before drying and exposure to film.
Endoglycosidase H (Endo H) treatment.
Virions or IP samples were prepared and digested with 500 U of Endo H at 37°C for 2 h according to the manufacturer's instructions (New England BioLabs, Ipswich, MA).
Cell surface biotinylation.
A 60% confluent plate of Vero cells in RPMI medium and HEp-2 cells in Opti-MEM I was infected at an MOI of 0.1. At 72 h p.i., cell monolayers were rinsed three times with PBS to remove medium and FBS, incubated with 0.5 mg/ml Sulfo-NHS-Biotin (Pierce) at 4°C for 30 min, rinsed three times with PBS containing 1% FBS, and covered with lysis buffer that contained protease inhibitors (Pierce) and 2 mg/ml N-ethylmaleimide. The lysates were cleared of insoluble debris by centrifugation at 14,000 × g for 10 min, IP with MAb L9, separated by 10% SDS-PAGE, and transferred to an Immobilon membrane. The membrane was blocked with 2% FBS in PBS, probed with HRP-conjugated streptavidin (Kirkegaard & Perry Laboratories), treated with Lumi-Light Western blotting substrate (Roche), and exposed to film.
His-tagged recombinant G protein.
Six histidines were added to the C terminus of the G protein immediately before the terminal glutamine. Mutagenesis was performed by inverse PCR (3) with primers CATCACCATTAGTTACTTAAAAACATATTATCACAAAAGGCCTTGACCAACCG and AGGTGGGTTGTGTGGTGCGGTCGTAGTGGTA. The six-His sequence was inserted into SD-2, a plasmid containing the G protein sequence from the A2 strain flanked by the RSV gene start and gene stop transcription signals. Once modified, the G gene was inserted into RW30, a full-length, RSV cDNA carrying the gene for GFP in the first position (A. Kwilas and M. E. Peeples, unpublished data). Virus was recovered from this plasmid by the standard recovery protocol for rgRSV (45). The His-tagged G protein was detected by Western blotting and probing with MAb 5His.
Infection of HAE cell cultures.
Primary, well-differentiated HAE cell cultures grown on Transwell filters (Costar) at the air-liquid interface (16) were rinsed with PBS three times over a 30-min period to remove apical secretions and supplied with fresh basolateral medium prior to inoculation. The virus inoculum was diluted to 6.2 × 106 PFU in100 μl of Hanks balanced salt solution (∼MOI, 20) and applied to the apical surface of the HAE cell cultures for 2 h of incubation at 37°C, after which the inoculum was removed by aspiration and cultures were incubated at 37°C. At indicated times postinoculation, images were obtained with a Leica DMIRB inverted fluorescence microscope equipped with a cooled-color charge-coupled device digital camera (Retiga 1300; QImaging, Burnaby, British Columbia, Canada). The proportion of the epithelium positive for GFP was determined by pixilating a black-and-white image, inverting the image, and calculating the percentage of black pixels by computer for five images per culture and averaging the results.
To collect virus for the GAG dependency assay and Western blotting, 300 μl of serum-free medium was added to the apical surface of the HAE cell cultures. After 30 min of incubation at 37°C, the medium with released virus was collected and snap-frozen on dry ice. Because the sample volume in these cultures is too small to process in sucrose gradients, we performed low-speed centrifugation at 400 × g for 5 min to remove cells and debris. One-fifth of the virus sample was then centrifuged at 15,000 × g for 10 min at 4°C to pellet the virus for analysis.
RESULTS
Quantification of GAG dependency.
We and others have previously reported that RSV infection of HEp-2 and other immortalized cell lines is largely dependent upon the presence of GAGs on the cell surface (12, 22, 23, 30, 36, 46). This conclusion was based in part on the much greater sensitivity to RSV infection of CHO K1 cells than a mutant derivative, CHO A745 (10). The CHO A745 cell line is defective in xylosyl transferase, the enzyme that initiates GAG synthesis by linking xylose to a serine or threonine in the proper context. The result of this defect is a severe deficiency in total GAG expression (10). To quantify GAG usage by RSV, we have titrated the same virus sample on these two cell lines. The ratio of the CHO K1 titer to the CHO A745 titer is the GAG dependency index. A GAG dependency index of 1 means no preference for GAG-expressing cells, whereas values of >1 indicate an increasing use of GAG for infection.
We used the GAG dependency index to compare the GAG usage of recombinant GFP-expressing strain A2 RSV expressing all three of the viral glycoproteins, rgRSV-SGF, and mutants of this virus that are missing the G or SH glycoprotein gene or both. rgRSV-SGF and rgRSV-GF grown in HEp-2 cells displayed 17-fold and 14-fold GAG dependency, respectively (Fig. 1A). The two viruses lacking the G protein, rgRSV-F and rgRSV-SF, both displayed low GAG dependency, fourfold and twofold, respectively. These results confirm previous findings that the G protein is the major GAG-binding protein while the F protein plays a minor role in GAG binding.
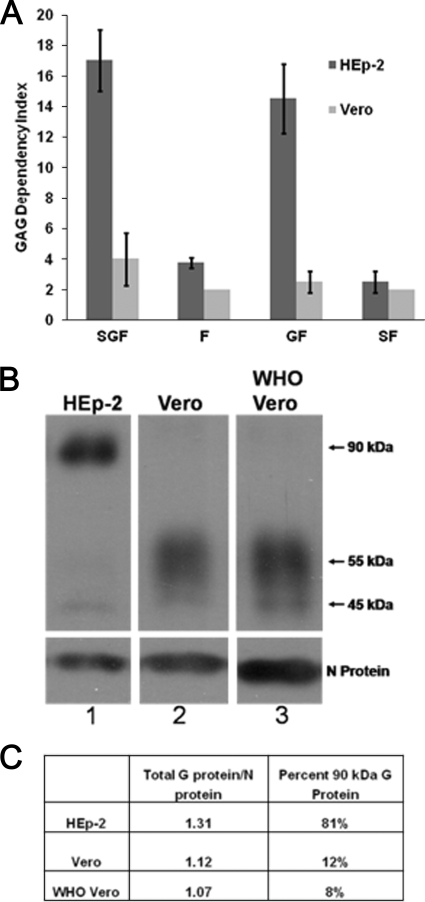
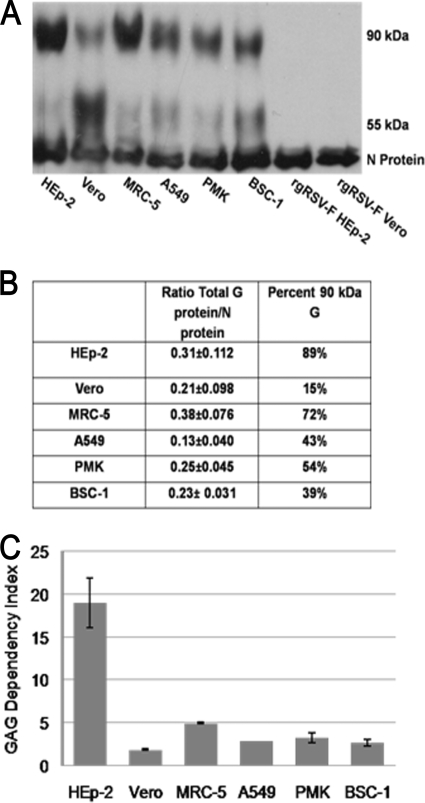
FIG. 1.
Importance of cell surface GAGs for infection and size of the G protein in virions produced in HEp-2 and Vero cells. (A) GAG dependency of recombinant RSVs from strain A2 expressing different combinations of viral glycoproteins that were grown in HEp-2 or Vero cells. Virus titers were determined on CHO K1 and CHO A745 cells and compared as follows: CHO K1/CHO A745 = GAG dependency index. (B) Western blot assay of sucrose-purified virions (15,000 cpm) probed with MAbs L9 (G protein) and D14 (N protein). All lanes are from the same gel, from which irrelevant lanes were removed. (C) The percentage of total virion G protein in the 90-kDa form was divided by the amount of virion N protein to normalize the amount of G protein in the virions produced by HEp-2 and Vero cells.
We also determined the GAG dependency index of virus grown in Vero cells. Surprisingly, rgRSV-SGF and rgRSV-GF grown in Vero cells showed 4-fold and 2-fold GAG dependency, in contrast to the 17-fold and 14-fold GAG dependency of the same viruses grown in HEp-2 cells (Fig. 1A). The GAG dependency of the Vero cell-grown viruses expressing the G protein was similar to that of viruses lacking the G protein, suggesting that the G protein in Vero cell-grown RSV has severely reduced function, at least with regard to the GAG dependency phenotype.
The G protein in virions.
To compare the amount of G protein in similar amounts of virions derived from HEp-2 and Vero cells, we added Tran35S-label to the culture medium during virus production, collected the supernatants after 72 h, and purified these metabolically labeled virions twice by sucrose density gradient. Similar amounts of virions based on the amount of incorporated radioactivity were separated by SDS-PAGE for Western blot analysis. The blot was probed with L9, a MAb against the G protein. MAb D14, against the N protein, was included as an internal virion control for loading variation. As expected, the mature, 90-kDa form of the G protein was readily detected in virions grown in HEp-2 cells. Minor amounts of a 55-kDa and a 45-kDa form were also detected (Fig. 1B, lane 1). The Vero cell-grown virions contained primarily the 55-kDa form of the G protein, with a minor amount of the 90-kDa and 45-kDa forms (Fig. 1B, lane 2). To determine whether the Vero cell line that we used was unique in this regard, we examined virions produced in the WHO-approved Vero cells used for vaccine production. Virions produced by these Vero cells also contained predominantly the 55-kDa G protein (Fig. 1B, lane 3).
If the virus from one cell line had more G protein than the other, it might have an advantage in GAG binding. To examine this possibility, we quantified the level of G protein in virions from each cell line, normalized to the N protein (Fig. 1C). There was minimal difference in the amounts of G protein in virions from HEp-2 and Vero cells. However, the predominant form of the G protein was very different. The 90-kDa form represented 81% of the G protein in HEp-2 cell-grown virions but only 12% of the G protein in Vero cell-grown virions, most of the remainder in each case being the 55-kDa form. The low abundance of the 90-kDa form of G protein in virions from Vero cells correlates with the decreased GAG dependency of this virus.
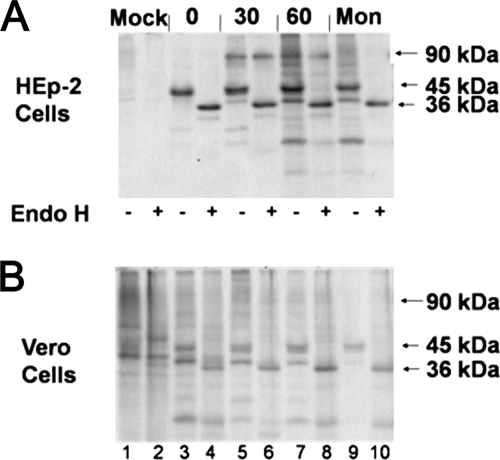
Production and maturation of the G protein.
G protein forms ranging from 45 kDa to 60 kDa have previously been described in infected cells and virions and suggested to be G protein processing intermediates (5, 14, 20, 21, 50). To determine whether the G protein intermediates correspond to the 55-kDa form found in Vero cell-grown RSV, we performed a pulse-chase experiment. At 24 h p.i., infected cells were pulsed with Tran35S-label for 15 min, chased with excess unlabeled methionine for the times indicated, and lysed and the G protein was IP with MAb L9. In HEp-2 cells, a 45-kDa processing intermediate was present in the pulse (Fig. 2A, lane 3), while the 90-kDa fully O-glycosylated, mature 90-kDa form of the G protein first appeared in the 30-min chase (Fig. 2A, lane 5). The 45-kDa G protein intermediate appeared similarly in Vero cells (Fig. 2B, lane 3), but the 90-kDa form was less prominent.
FIG. 2.
Pulse-chase of G protein in RSV-infected cells. (A) HEp-2 and (B) Vero cells were pulsed with Tran35S-label for 15 min. Cells were lysed and then IP with MAb L9 at the times indicated (in minutes). After IP, the samples were treated with 500 U of Endo H for 2 h at 37°C (+) or left untreated (−). After SDS-PAGE separation, the gel was fixed and dried. The gel was exposed to film at −80°C for 1 week before film development. Mon, monensin.
Endo H digestion of these pulse-chase samples (Fig. 2A and B, lanes 4, 6, 8, and 10) revealed that the 45-kDa G protein was Endo H sensitive while the 90-kDa form was not. Endo H sensitivity indicates the presence of immature N-linked sugars, while resistance indicates that the N-linked sugars have been matured in the Golgi compartment. This result confirms that the 45-kDa G protein intermediate contains immature N-linked carbohydrate chains, as expected for glycoproteins in the ER, prior to maturation in the Golgi compartment. The Endo H resistance of the 90-kDa form indicates that it has migrated through the Golgi compartment. The size increase in the G protein, from 45 kDa to 90 kDa, is consistent with the addition of many O-linked carbohydrates, a process that also takes place in the Golgi compartment.
To confirm that the increase in size requires transport to the Golgi compartment, infected HEp-2 cells were treated with monensin, a drug that blocks the transport of glycoproteins from the ER to the Golgi compartment. Monensin treatment prevented the appearance of the 90-kDa G protein (Fig. 2A and B, lanes 9), and the 45-kDa form remained Endo H sensitive (Fig. 2A and B, lanes 10), as expected. The 90-kDa species of the G protein in these cell lysates is similar to the size of the predominant form of the G protein in virions produced by HEp-2 cells. The 45-kDa species of the G protein in these cell lysates appears to be somewhat smaller than the 55-kDa form that predominates in virions produced by Vero cells. The lack of a 55-kDa species following a 1-h pulse indicates that it is generated by a process that occurs more than 1 h postsynthesis.
Maturation state of the G protein in virions.
Purified virions from both HEp-2 and Vero cells were treated with Endo H to determine whether their 45- to 55-kDa forms were processing intermediates. The minor 45-kDa species from both HEp-2 and Vero cell-grown viruses was sensitive to Endo H digestion, increasing in migration to 36 kDa (Fig. 3B, lanes 2 and 4), the size of unglycosylated G protein. This minor 45-kDa G protein species is therefore an intermediate that has not been processed in the Golgi compartment. In contrast, the 90-kDa and 55-kDa species were resistant to Endo H, indicating the presence of mature N-linked sugars on these forms, the result of processing in the Golgi compartment. Since the 55-kDa form has passed through the Golgi compartment, this form could be the result of inefficient O-linked glycosylation or of cleavage of the 90-kDa G protein with the loss of its C terminus.
FIG. 3.
Glycan maturity and presence of the C terminus in virion G protein. (A) Schematic of the RSV G protein, an N-terminally anchored type II glycoprotein. The soluble form of the G protein (sG) starts at Met-48 and is released from the cell surface by cleavage. A central conserved domain with four conserved cysteines (CCCC) is followed by a predicted HBD. The G protein is modified by N-linked glycans (N) and many O-linked glycans (potential sites are indicated by stalks with filled circles). The binding areas for MAbs L9, 130-2G, and 232-1F are designated by arrows. The six-His tag at the COOH terminus is also shown. TM, transmembrane domain. (B) Western blot assay of viruses treated with Endo H and probed with MAb L9. Samples containing each virus (15,000 cpm) were treated with 500 U of Endo H for 2 h at 37°C. (C) HEp-2 and Vero cell-grown viruses were probed with MAb L9. The same blot was stripped, blocked, and reprobed with MAb 130-2G. (D) Western blot assay of rgRSV-G-6His purified virions (15,000 cpm) from HEp-2 and Vero cells probed with MAb L9 or 5His.
Analysis of the C terminus of the G protein.
To determine whether the C terminus of the G protein is present in the 55-kDa form, we analyzed HEp-2 and Vero cell-purified virions by Western blot assay. The blot was probed with MAb L9, which binds within the central, conserved cysteine region of the G protein (Fig. 3A) (47). MAb L9 detected the 90-kDa, 55-kDa, and 45-kDa forms, indicating that they are all present in these virions (Fig. 3C, lanes 1 and 2). The blot was stripped and reprobed with MAb 130-2G, which recognizes the C terminus of the G protein (Fig. 3A) (43). This MAb stained the 90-kDa and 45-kDa forms of the G protein but not the 55-kDa form (Fig. 3C, lanes 3 and 4), indicating that the C terminus is missing. A second C terminus-specific MAb, 232-1F (43), had the same reactivity pattern (data not shown).
To confirm the loss of the C terminus of the G protein in the 55-kDa form, we constructed and rescued a virus with a six-His tag at the C terminus of the G protein, rgRSV-G-6His (Fig. 3A). This virus was grown in HEp-2 and Vero cells, and purified virions were subjected to Western blot analysis with MAbs L9 and 5His. MAb L9 reacted with both the 55-kDa and 90-kDa forms (Fig. 3D, lanes 1 and 2) from both virus preparations, whereas MAb 5His only reacted with the 90-kDa form (Fig. 3D, lanes 3 and 4). These results confirm that the 55-kDa G protein found in virions from both cell lines is missing its C terminus.
Cell and cell surface forms of the G protein.
As a first step in determining the location of the 55-kDa form within cells, we compared infected total cell lysate with cell surface proteins identified by biotinylation. The samples were electrophoresed and probed with MAb L9 in order to visualize the forms and relative proportions of the G protein in each sample. Infected HEp-2 cell lysates and cell surface and purified virions (Fig. 4) all displayed the 90-kDa form as the major form, with minor amounts of the 55-kDa and 45-kDa forms. Cell lysates from infected Vero cells (Fig. 4) also displayed more of the 90-kDa form than of the 55-kDa form. In striking contrast, most of the G protein on the cell surface was the 55-kDa form, with a minor amount of the 90-kDa form, similar to the virions. The difference between the Vero cell lysate and cell surface suggests that the G protein is processed to the 55-kDa form at the cell surface or in the Golgi compartment just before arriving at the cell surface. Therefore, virions do not selectively incorporate the 55-kDa form instead of the 90-kDa form as they are budding from infected Vero cells. Instead, they incorporate the cell surface G protein, which is predominantly the 55-kDa form.
FIG. 4.
Comparison of G proteins from total cell lysates, the cell surface, and purified virions. Lanes 1, 4, 5, and 8 were stained with MAb L9. Lanes 2, 3, 6, and 7 were processed separately to detect the biotinylated cell surface proteins that had been IP with MAb L9 and developed with streptavidin-HRP and a chemiluminescent substrate.
Phenotype of virions from other cell lines.
To determine whether Vero cells are unique in the production of virions with major amounts of a cleaved G protein, we analyzed rgRSV-G-6His virions produced in other cell types used to grow RSV. The MRC-5 human diploid fibroblast cell line is also approved for vaccine production. PMK cells are often used in clinical virology laboratories to isolate virus from patients for identification, and similar cells were used to grow the original formalin-inactivated RSV vaccine (26). BSC-1 cells were used in early studies to identify RSV proteins, but the 90-kDa protein was difficult to detect (52). Virions produced from these cell lines incorporated less 90-kDa protein and more 55-kDa protein than did the HEp-2-grown virus (Fig. 5A and B), and all of these virions displayed low GAG dependency (Fig. 5C).
FIG. 5.
Forms of the G protein in virions produced by other cell lines. (A) Western blot assay of purified rgRSV-G-6His virions probed with MAbs L9 and D14. (B) Ratio of the total G protein versus N protein and the percent 90-kDa G protein. Ratios were determined from triplicate blots. (C) GAG dependency of rgRSV-G-6His grown in different cells. Virus titers on CHO K1/CHO A745 = GAG dependency index.
HEp-2 and Vero cell-grown rgRSV-G-6His had a GAG dependency similar to that of rgRSV-SGF (Fig. 1A), indicating that addition of the His tag does not affect GAG binding. The ratios of total G protein to N protein were also generally similar (Fig. 5B). The 90-kDa G protein of virus grown in MRC-5 cells represents 72% of the total G protein, compared to 89% for HEp-2 cells, but its GAG dependency was reduced by more than 3.5-fold (from 18-fold to 5-fold). These results suggest that a threshold amount of 90-kDa G protein is needed to determine GAG dependency. It is possible that the G protein is oligomeric and that it does not function if one or more of the monomers is the 55-kDa form.
Phenotype of virions produced in HAE cell cultures.
As shown above, the cleavage and function of the virion G protein depend on the cell line producing the virion. We have previously used primary well-differentiated HAE cell cultures as a model to study RSV pathogenesis (54). The apical surface of HAE cell cultures includes ciliated cells and mucus-producing goblet cells. HAE cell cultures were inoculated with rgRSV-SGF grown in HEp-2 cells, and virus was harvested from the apical surface at 3 days p.i. The HAE cell-grown virus was highly GAG dependent (Fig. 6A), similar to HEp-2 cell-grown virus rather than to Vero cell-grown virus. To assess the size of the G protein in HAE cell culture-grown virus, we examined virions released from the apical surface of HAE cell cultures. Because of the small sample size, these virions could not be sucrose purified; however, virions were partially purified by removing cell debris by low-speed centrifugation and by pelleting virions at a higher speed. The virus pellet was reduced prior to SDS-PAGE. Western blotting identified a predominant, novel form of the G protein migrating at approximately 180 kDa (Fig. 6B, lanes 2 and 3) in addition to a small amount of the 90-kDa form and a minor amount of a smaller G protein form or breakdown product. The inoculum virus (Fig. 6B, lane 1) was grown in HEp-2 cells and displays primarily the 90-kDa G protein.
FIG. 6.
(A) GAG dependency indexes of RSVs produced in HEp-2, Vero, and HAE cells. Virus titers were determined on CHO K1 and CHO A745 cells and compared as follows: CHO K1/CHO A745 = GAG dependency index. (B) Western blot assay of crude virus released from HAE cell cultures, probed with MAb L9. HEp-2 virus initial inoculum bound to HAE cell culture at 0 days p.i. (lane 1), HAE cell virus culture no. 1 (lane 2), HAE cell virus culture no. 2 (lane 3). (C) Western blot assay of rgRSV-G-6His virions from HEp-2 cells and HAE cell cultures probed with MAb L9 or 5His.
We also examined rgRSV-G-6His virus from HAE cell cultures to determine if this 180-kDa form contains full-length G protein. MAb L9 (Fig. 6C, lanes 1 and 2) and MAb 5His (Fig. 6C, lanes 3 and 4) both reacted with the 180-kDa form from HAE cells, as well as the 90-kDa HEp-2 cell-grown virus control. This result clearly demonstrates that the C terminus of the G protein is present in the 180-kDa form and therefore that the G protein is full length. This conclusion is consistent with the results above (Fig. 6A) demonstrating that the HAE cell culture-grown virus retains high GAG dependency. We hypothesize that this “supersized” G protein is either a dimer of the 90-kDa G protein or the 90-kDa form with additional or more extensive O-linked carbohydrate chains. In the absence of reducing agent or boiling, the protein also migrates at 180 kDa (data not shown). These results imply that the protein is not an aggregated form of the G protein.
Infection of HAE cell cultures with Vero cell-grown virus.
We have demonstrated that RSV grown in Vero cells does not use GAGs efficiently for infection of immortalized cells and that the cause of this reduced infection is loss of the C terminus of the G protein. The loss of GAG binding should not affect infection of HAE cell cultures because these cells do not express HS on their apical surface (53) and RSV infects only via the apical surface (54). Nevertheless, loss of the G protein C terminus may affect its function, attachment to respiratory epithelial cells. We tested this possibility by inoculating HAE cell cultures with rgRSV-SGF produced in HEp-2 or Vero cells. Equivalent numbers of infective units of each virus were used to inoculate the HAE cell cultures. The virus titers used to calculate the infectivity of the stock virus was determined on GAG-deficient CHO A745 cells. In this way, the titer advantage that GAG binding provides HEp-2 cell virus when titrated on a GAG expressing cell line was eliminated.
At 1 day p.i. with rgRSV-SGF grown in Vero cells, the level of GFP expression was 600- to 1,800-fold less than that of HAE cell cultures inoculated with virus from HEp-2 cells (Fig. 7). The finding that Vero cell-grown RSV is extremely inefficient at infecting HAE cell cultures compared to HEp-2 cell-grown RSV indicates that the complete 90-kDa G protein is important for infection of primary respiratory cells. In the same experiment, we inoculated HAE cell cultures with rgRSV-F, a virus completely lacking the G and SH genes, grown in HEp-2 cells. This virus also initiated infection of HAE cell cultures inefficiently, 9- to 13-fold less than rgRSV-SGF grown in HEp-2 cells.
FIG. 7.
Infection of primary HAE cell cultures inoculated with rgRSV-SGF grown in HEp-2 and Vero cells and rgRSV-F grown in HEp-2 cells (6.2 × 106 PFU in 100 μl [∼MOI, 0.05]). The percent GFP was calculated from five random pictures obtained from each HAE cell culture. Each time point represents three HAE cell cultures.
At 2 days p.i., all of the viruses had begun to spread, including Vero cell-grown rgRSV-SGF. The form of the G protein in the virions produced by HAE cells would be determined by the HAE cell cultures at this point, not by Vero cells. rgRSV-SGF spread more rapidly than rgRSV-F. This result is consistent with progeny virions from HAE cell cultures that contain the full-size (or supersized) G protein which enables subsequent infection and spread in HAE cell cultures.
DISCUSSION
In early biochemical studies, the large RSV glycoprotein, eventually named the G protein, was found in virions produced from infected HeLa cells (34, 40) but not in virions from BSC-1 cells (52). BSC-1 cells were derived from African green monkey kidneys, as are Vero cells. A study to resolve this dramatic difference between American- and Scottish-grown RSVs found that both cell lines produced the 85-kDa form, though the BSC-1 cells produced less (42). In the present report, we have identified an associated phenotype: RSV produced in HEp-2 cells (or HeLa cells [data not shown]) are highly dependent on GAGs for virus entry, while RSV grown in Vero, PMK, MRC-5, A549, or BSC-1 cells is much less dependent on GAGs for infection.
We have found that while virions from Vero cells have low levels of the 90-kDa G protein, they contain much more of a smaller, 55-kDa version of the G protein. Using our GAG dependency assay, we have determined that this Vero cell-grown RSV infects GAG-expressing CHO cells fourfold less efficiently than virus produced in HEp-2 cells and containing primarily the 90-kDa full-length G protein. It appears that the 90-kDa form of the G protein uses GAGs on the surface of immortalized cells to initiate infection much more efficiently than the 55-kDa form.
Estimating the cleavage position in the 90-kDa G protein that would generate a 55-kDa G protein is challenging. Identifying the site is helped by knowing that the cleavage site is conserved between the A2 and Long strains of RSV because virions from Long also contain primarily the 55-kDa G protein when produced in Vero cells (data not shown). The real difficulty in estimating the position is due to the large amount of carbohydrates attached to this protein. The G protein contains seven potential N-linked glycosylation sites, but the difference of 9 kDa between the 45-kDa intermediate and its size following Endo H treatment, 36 kDa, suggests that only three or four of these sites are used. Approximately half of the final 90-kDa molecular mass is due to O-linked glycosylation. We used the program NetNGlyc 1.0 to predict the three or four N-linked sites most likely to be used and the NetOGlyc 3.1 (25) program to predict the most likely O-glycosylation sites (ExPASy Proteomics Server, http://ca.expasy.org/tools/#ptm). Using these programs to predict molecular weight, we estimate that the cleavage site would be C terminal to the central disulfide-bonded region (13), within or just C terminal to the HBD (Fig. 4A). HBDs are defined by a high concentration of basic amino acids. Cleavage in or near the HBD might well destroy the ability of the G protein to bind GAGs. One or more of the many lysines in this area could be a target for a plasmin-like enzyme, or the lone arginine could be the substrate for a trypsin-like enzyme.
The original, formalin-inactivated RSV vaccine that led to increased pathology rather than protection upon challenge was produced in cells derived from the same source as Vero cells, African green monkey kidneys (17, 26, 29). Vero cells have been used to produce similar vaccines for many of the animal studies that have focused on the cause of the increased pathology. Along with the increased pathology, the vaccine induced nonfunctional antibodies. Deletions from the G protein C terminus result in a loss of recognition by human, rabbit, and murine antibodies (32, 38, 39), suggesting that the C terminus of the G protein contains its major antigenic determinants. The reduced amount of the C-terminal antigenic determinants and the presence of a possibly misfolded, truncated G protein may have also contributed to the decrease in neutralizing antibodies and the increase in nonfunctional antibodies in children who received the formalin-inactivated RSV vaccine (37).
While GAG interactions are important for infection of immortalized cells in culture, it is unlikely that they are critical for infection of airway cells in vivo. The well-differentiated HAE cell cultures that appear and function like normal airway epithelium do not express HS on their apical surface (53). We anticipate, therefore, that the ability of the RSV G protein to bind to GAGs will not be important for the initiation of RSV infection of HAE cell cultures or of airway cells in vivo. However, the loss of the C-terminal region of the G protein might well lead to a loss of attachment function. Indeed, we found that the infectivity of Vero cell-grown virus for HAE cell cultures is much lower (600- to 1,800-fold) than that of HEp-2 cell-grown virus (Fig. 7, 1 day p.i.).
Since HAE cell cultures closely model the human airway epithelium, attenuated vaccines grown in Vero cells would also be expected to infect humans inefficiently. In trials of attenuated virus vaccines, escalating doses are used to identify the lowest dose that induces an adequate antibody response. Vero cell-grown attenuated vaccine candidates do induce neutralizing antibodies in infants. However, these vaccine candidates have caused respiratory symptoms in a small number of infants and these symptoms have been deemed unacceptable (28).
We predict that if such vaccines were produced in a different cell line, less inoculum would be needed to induce the same antibody response. In addition to increased efficiency of vaccine production, a lower inoculum would also reduce exposure to nonreplicating virus antigens, cell culture-derived cytokines, chemokines, and other possible contaminants in the inoculum. Exposure to local high levels of these proteins could contribute to inflammation and symptoms in the minority of infants who have been vaccinated. After the initial infection of HAE cell cultures, the source of the virus would not play a role, as progeny virus from the infected HAE cells would contain primarily intact G protein (Fig. 6B). This virus spreads at a rate similar to infection initiated by HEp-2 cell-grown RSV (Fig. 7), after a 3-day lag. The same pattern would likely be true in vaccinees.
The G protein is important for infection in vivo, since RSV lacking its G gene replicates poorly in rodents and nonhuman primates (8). In model animal experiments, as in vaccine trials, Vero cell-grown virus could reduce the efficiency of the initial infection. In fact, Vero cell-grown virus has been shown to infect mice poorly compared to HeLa cell-grown virus (44), consistent with our data and predictions.
In summary, efficient RSV infection requires the intact G protein, particularly infection of primary respiratory epithelial target cells. Most of the G protein in virions produced from Vero cells is truncated by cleavage. This truncated form of the G protein does not appear to be functional in either GAG attachment to immortalized cells or attachment to primary HAE cell cultures, since the initiation of infection is reduced in both cases. This loss of attachment function would most likely also result in poor infection initiation in vivo, negatively impacting on both animal experiments and attenuated-vaccine studies with volunteers.
Acknowledgments
We thank Barb Newton for excellent technical help, Peter Collins for the recombinant RSV system, Larry Anderson for the C-terminal antibodies for the G protein, Russell Durbin for helpful discussions, Cliff Beall for the WHO-approved Vero cells, and Jeff Esko for the A745 cells.
This work was supported by NIH grants AI047213 and HL051818.
Footnotes
Published ahead of print on 5 August 2009.
REFERENCES
- 1.Anderson, L. J., P. Bingham, and J. C. Hierholzer. 1988. Neutralization of respiratory syncytial virus by individual and mixtures of F and G protein monoclonal antibodies. J. Virol. 62:4232-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barretto, N., L. K. Hallak, and M. E. Peeples. 2003. Neuraminidase treatment of respiratory syncytial virus-infected cells or virions, but not target cells, enhances cell-cell fusion and infection. Virology 313:33-43. [DOI] [PubMed] [Google Scholar]
- 3.Byrappa, S., D. K. Gavin, and K. C. Gupta. 1995. A highly efficient procedure for site-specific mutagenesis of full-length plasmids using Vent DNA polymerase. Genome Res. 5:404-407. [DOI] [PubMed] [Google Scholar]
- 4.Chanock, R. M., and L. Finberg. 1957. Recovery from infants with respiratory illness of a virus related to chimpanzee coryza agent (CCA). II. Epidemiological aspects of infection in infants and young children. Am. J. Hyg. 66:291-300. [DOI] [PubMed] [Google Scholar]
- 5.Collins, P. L., and G. Mottet. 1992. Oligomerization and post-translational processing of glycoprotein G of human respiratory syncytial virus: altered O-glycosylation in the presence of brefeldin A. J. Gen. Virol. 73:849-863. [DOI] [PubMed] [Google Scholar]
- 6.Collins, P. L., and B. R. Murphy. 2005. New generation live vaccines against human respiratory syncytial virus designed by reverse genetics. Proc. Am. Thorac. Soc. 2:166-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins, P. L., and B. R. Murphy. 2002. Respiratory syncytial virus: reverse genetics and vaccine strategies. Virology 296:204-211. [DOI] [PubMed] [Google Scholar]
- 8.Crowe, J. E., Jr., P. T. Bui, C. Y. Firestone, M. Connors, W. R. Elkins, R. M. Chanock, and B. R. Murphy. 1996. Live subgroup B respiratory syncytial virus vaccines that are attenuated, genetically stable, and immunogenic in rodents and nonhuman primates. J. Infect. Dis. 173:829-839. [DOI] [PubMed] [Google Scholar]
- 9.Dowell, S. F., L. J. Anderson, H. E. Gary, Jr., D. D. Erdman, J. F. Plouffe, T. M. File, Jr., B. J. Marston, and R. F. Breiman. 1996. Respiratory syncytial virus is an important cause of community-acquired lower respiratory infection among hospitalized adults. J. Infect. Dis. 174:456-462. [DOI] [PubMed] [Google Scholar]
- 10.Esko, J. D., T. E. Stewart, and W. H. Taylor. 1985. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc. Natl. Acad. Sci. USA 82:3197-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falsey, A. R., and E. E. Walsh. 2000. Respiratory syncytial virus infection in adults. Clin. Microbiol. Rev. 13:371-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldman, S. A., S. Audet, and J. A. Beeler. 2000. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J. Virol. 74:6442-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feldman, S. A., R. M. Hendry, and J. A. Beeler. 1999. Identification of a linear heparin binding domain for human respiratory syncytial virus attachment glycoprotein G. J. Virol. 73:6610-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernie, B. F., G. Dapolito, P. J. Cote, Jr., and J. L. Gerin. 1985. Kinetics of synthesis of respiratory syncytial virus glycoproteins. J. Gen. Virol. 66(Pt. 9):1983-1990. [DOI] [PubMed] [Google Scholar]
- 15.Fuentes, S., K. C. Tran, P. Luthra, M. N. Teng, and B. He. 2007. Function of the respiratory syncytial virus small hydrophobic protein. J. Virol. 81:8361-8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fulcher, M. L., S. Gabriel, K. A. Burns, J. R. Yankaskas, and S. H. Randell. 2005. Well-differentiated human airway epithelial cell cultures. Methods Mol. Med. 107:183-206. [DOI] [PubMed] [Google Scholar]
- 17.Fulginiti, V. A., J. J. Eller, O. F. Sieber, J. W. Joyner, M. Minamitani, and G. Meiklejohn. 1969. Respiratory virus immunization. I. A field trial of two inactivated respiratory virus vaccines; an aqueous trivalent parainfluenza virus vaccine and an alum-precipitated respiratory syncytial virus vaccine. Am. J. Epidemiol. 89:435-448. [DOI] [PubMed] [Google Scholar]
- 18.García-Beato, R., I. Martinez, C. Franci, F. X. Real, B. Garcia-Barreno, and J. A. Melero. 1996. Host cell effect upon glycosylation and antigenicity of human respiratory syncytial virus G glycoprotein. Virology 221:301-309. [DOI] [PubMed] [Google Scholar]
- 19.Gorman, J. J., B. L. Ferguson, D. Speelman, and J. Mills. 1997. Determination of the disulfide bond arrangement of human respiratory syncytial virus attachment (G) protein by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Protein Sci. 6:1308-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruber, C., and S. Levine. 1985. Respiratory syncytial virus polypeptides. IV. The oligosaccharides of the glycoproteins. J. Gen. Virol. 66:417-432. [DOI] [PubMed] [Google Scholar]
- 21.Gruber, C., and S. Levine. 1985. Respiratory syncytial virus polypeptides. V. The kinetics of glycoprotein synthesis. J. Gen. Virol. 66:1241-1247. [DOI] [PubMed] [Google Scholar]
- 22.Hallak, L., D. Spillman, P. L. Collins, and M. E. Peeples. 2000. Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection. J. Virol. 74:10508-10513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hallak, L. K., P. L. Collins, W. Knudson, and M. E. Peeples. 2000. Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology 271:264-275. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, P. R., M. K. Spriggs, R. A. Olmsted, and P. L. Collins. 1987. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc. Natl. Acad. Sci. USA 84:5625-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Julenius, K., A. Molgaard, R. Gupta, and S. Brunak. 2005. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15:153-164. [DOI] [PubMed] [Google Scholar]
- 26.Kapikian, A. Z., R. H. Mitchell, R. M. Chanock, R. A. Shvedoff, and C. E. Stewart. 1969. An epidemiologic study of altered clinical reactivity to respiratory syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am. J. Epidemiol. 89:405-421. [DOI] [PubMed] [Google Scholar]
- 27.Karron, R. A., D. A. Buonagurio, A. F. Georgiu, S. S. Whitehead, J. E. Adamus, M. L. Clements-Mann, D. O. Harris, V. B. Randolph, S. A. Udem, B. R. Murphy, and M. S. Sidhu. 1997. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc. Natl. Acad. Sci. USA 94:13961-13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karron, R. A., P. F. Wright, R. B. Belshe, B. Thumar, R. Casey, F. Newman, F. P. Polack, V. B. Randolph, A. Deatly, J. Hackell, W. Gruber, B. R. Murphy, and P. L. Collins. 2005. Identification of a recombinant live attenuated respiratory syncytial virus vaccine candidate that is highly attenuated in infants. J. Infect. Dis. 191:1093-1104. [DOI] [PubMed] [Google Scholar]
- 29.Kim, H. W., J. G. Canchola, C. D. Brandt, G. Pyles, R. M. Chanock, K. Jensen, and R. H. Parrott. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 89:422-434. [DOI] [PubMed] [Google Scholar]
- 30.Krusat, T., and H. J. Streckert. 1997. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch. Virol. 142:1247-1254. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 32.Lambert, D. M. 1988. Role of oligosaccharides in the structure and function of respiratory syncytial virus glycoproteins. Virology 164:458-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langedijk, J. P., W. M. Schaaper, R. H. Meloen, and J. T. van Oirschot. 1996. Proposed three-dimensional model for the attachment protein G of respiratory syncytial virus. J. Gen. Virol. 77(Pt. 6):1249-1257. [DOI] [PubMed] [Google Scholar]
- 34.Levine, S. 1977. Polypeptides of respiratory syncytial virus. J. Virol. 21:427-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levine, S., R. Klaiber-Franco, and P. R. Paradiso. 1987. Demonstration that glycoprotein G is the attachment protein of respiratory syncytial virus. J. Gen. Virol. 68:2521-2524. [DOI] [PubMed] [Google Scholar]
- 36.Martínez, I., and J. A. Melero. 2000. Binding of human respiratory syncytial virus to cells: implication of sulfated cell surface proteoglycans. J. Gen. Virol. 81(Pt. 11):2715-2722. [DOI] [PubMed] [Google Scholar]
- 37.Murphy, B. R., G. A. Prince, E. E. Walsh, H. W. Kim, R. H. Parrott, V. G. Hemming, W. J. Rodriguez, and R. M. Chanock. 1986. Dissociation between serum neutralizing and glycoprotein antibody responses of infants and children who received inactivated respiratory syncytial virus vaccine. J. Clin. Microbiol. 24:197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palomo, C., P. A. Cane, and J. A. Melero. 2000. Evaluation of the antibody specificities of human convalescent-phase sera against the attachment (G) protein of human respiratory syncytial virus: influence of strain variation and carbohydrate side chains. J. Med. Virol. 60:468-474. [PubMed] [Google Scholar]
- 39.Palomo, C., B. Garcia-Barreno, C. Penas, and J. A. Melero. 1991. The G protein of human respiratory syncytial virus: significance of carbohydrate side-chains and the C-terminal end to its antigenicity. J. Gen. Virol. 72(Pt. 3):669-675. [DOI] [PubMed] [Google Scholar]
- 40.Peeples, M., and S. Levine. 1979. Respiratory syncytial virus polypeptides: their location in the virion. Virology 95:137-145. [DOI] [PubMed] [Google Scholar]
- 41.Prince, G. A., S. J. Curtis, K. C. Yim, and D. D. Porter. 2001. Vaccine-enhanced respiratory syncytial virus disease in cotton rats following immunization with Lot 100 or a newly prepared reference vaccine. J. Gen. Virol. 82:2881-2888. [DOI] [PubMed] [Google Scholar]
- 42.Pringle, C. R., P. V. Shirodaria, H. B. Gimenez, and S. Levine. 1981. Antigen and polypeptide synthesis by temperature-sensitive mutants of respiratory syncytial virus. J. Gen. Virol. 54:173-183. [DOI] [PubMed] [Google Scholar]
- 43.Sullender, W. 1995. Antigenic analysis of chimeric and truncated G proteins of respiratory syncytial virus. Virology 209:70-79. [DOI] [PubMed] [Google Scholar]
- 44.Taylor, G., E. J. Stott, M. Hughes, and A. P. Collins. 1984. Respiratory syncytial virus infection in mice. Infect. Immun. 43:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Techaarpornkul, S., N. Barretto, and M. E. Peeples. 2001. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 75:6825-6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Techaarpornkul, S., P. L. Collins, and M. E. Peeples. 2002. Respiratory syncytial virus with the fusion protein as its only viral glycoprotein is less dependent on cellular glycosaminoglycans for attachment than complete virus. Virology 294:296-304. [DOI] [PubMed] [Google Scholar]
- 47.Walsh, E. E., A. R. Falsey, and W. M. Sullender. 1998. Monoclonal antibody neutralization escape mutants of respiratory syncytial virus with unique alterations in the attachment (G) protein. J. Gen. Virol. 79(Pt. 3):479-487. [DOI] [PubMed] [Google Scholar]
- 48.Walsh, E. E., C. B. Hall, J. J. Schlesinger, M. W. Brandriss, S. Hildreth, and P. Paradiso. 1989. Comparison of antigenic sites of subtype-specific respiratory syncytial virus attachment proteins. J. Gen. Virol. 70(Pt. 11):2953-2961. [DOI] [PubMed] [Google Scholar]
- 49.Wertz, G. W., P. L. Collins, Y. Huang, C. Gruber, S. Levine, and L. A. Ball. 1985. Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc. Natl. Acad. Sci. USA 82:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wertz, G. W., M. Krieger, and L. A. Ball. 1989. Structure and cell surface maturation of the attachment glycoprotein of human respiratory syncytial virus in a cell line deficient in O glycosylation. J. Virol. 63:4767-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wright, P. F., R. A. Karron, R. B. Belshe, J. Thompson, J. E. Crowe, Jr., T. G. Boyce, L. L. Halburnt, G. W. Reed, S. S. Whitehead, E. L. Anderson, A. E. Wittek, R. Casey, M. Eichelberger, B. Thumar, V. B. Randolph, S. A. Udem, R. M. Chanock, and B. R. Murphy. 2000. Evaluation of a live, cold-passaged, temperature-sensitive, respiratory syncytial virus vaccine candidate in infancy. J. Infect. Dis. 182:1331-1342. [DOI] [PubMed] [Google Scholar]
- 52.Wunner, W. H., and C. R. Pringle. 1976. Respiratory syncytial virus proteins. Virology 73:228-243. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, L., A. Bukreyev, C. I. Thompson, B. Watson, M. E. Peeples, P. L. Collins, and R. J. Pickles. 2005. Infection of ciliated cells by human parainfluenza virus type 3 in an in vitro model of human airway epithelium. J. Virol. 79:1113-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, L., M. E. Peeples, R. C. Boucher, P. L. Collins, and R. J. Pickles. 2002. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J. Virol. 76:5654-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]