Abstract
Venezuelan equine encephalitis virus (VEEV) is an important human and veterinary pathogen causing sporadic epizootic outbreaks of potentially fatal encephalitis. The type I interferon (IFN) system plays a central role in controlling VEEV and other alphavirus infections, and IFN evasion is likely an important determinant of whether these viruses disseminate and cause disease within their hosts. Alphaviruses are thought to limit the induction of type I IFNs and IFN-stimulated genes by shutting off host cell macromolecular synthesis, which in the case of VEEV is partially mediated by the viral capsid protein. However, more specific strategies by which alphaviruses inhibit type I IFN signaling have not been characterized. Analyses of cells infected with VEEV and VEEV replicon particles (VRP) demonstrate that viral infection rapidly disrupts tyrosine phosphorylation and nuclear translocation of the transcription factor STAT1 in response to both IFN-β and IFN-γ. This effect was independent of host shutoff and expression of viral capsid, suggesting that VEEV uses novel mechanisms to interfere with type I and type II IFN signaling. Furthermore, at times when STAT1 activation was efficiently inhibited, VRP infection did not limit tyrosine phosphorylation of Jak1, Tyk2, or STAT2 after IFN-β treatment but did inhibit Jak1 and Jak2 activation in response to IFN-γ, suggesting that VEEV interferes with STAT1 activation by the type I and II receptor complexes through distinct mechanisms. Identification of the viral requirements for this novel STAT1 inhibition will further our understanding of alphavirus molecular pathogenesis and may provide insights into effective alphavirus-based vaccine design.
Venezuelan equine encephalitis virus (VEEV) is a mosquito-borne alphavirus in the family Togaviridae that is responsible for sporadic epidemics of encephalitis in equines and humans. Most cases of human and equine disease have been associated with epizootic VEEV strains (subtypes IAB and IC) that undergo efficient amplification within horses, but recent studies indicate that endemic transmission of equine avirulent strains (subtype ID) is responsible for many unreported cases in humans that live near habitats where enzootic transmission occurs (2, 46, 57). When infected via the mosquito vector, patients may present with malaise, fever, and headache (57). While fatalities are rare (<1%), patients that recover from encephalitis may suffer from permanent neurological sequelae (30).
The type I interferons (IFNs) α and β represent a crucial innate defense system against most viral pathogens, including alphaviruses. These cytokines act in autocrine and paracrine pathways to induce the expression of numerous IFN-stimulated genes (ISGs), such as 2′,5′-OAS, PKR, and Mx family members that are important for the control of viral infection (reviewed in reference 20). The signaling events that follow IFN stimulation have been well described (reviewed in references 31 and 42). In brief, when the type I IFNs bind the IFN-α/β receptor subunits IFNAR1 and IFNAR2, these subunits dimerize at the cell surface allowing the apposition of two protein tyrosine kinases (PTKs), Janus activated kinase 1 (Jak1) and tyrosine kinase 2 (Tyk2), that are associated with the receptor's cytoplasmic tails. Juxtaposed Jak1 and Tyk2 are then activated through auto- and/or transphosphorylation (12, 24, 38), and they in turn phosphorylate tyrosine residues present on the receptor tails, which serve as docking sites for the recruitment of various signal transducers and activators of transcription (STAT) factors. Jak1 and Tyk2 subsequently phosphorylate STAT1 and STAT2, which form heterodimers, and in association with IFN regulatory factor 9, the trimeric complex localizes to the nucleus where it binds promoters containing IFN-stimulated response elements to drive expression of ISGs. This sequence of events is mirrored when type II IFN (IFN-γ) binds its cell surface receptor subunits (IFN-γ receptor 1 [IFNGR1] and IFNGR2). Jak1 and Jak2 are activated at the IFNGR cytoplasmic tails, which in turn activate STAT1 by tyrosine phosphorylation. Unlike the response to type I IFN, IFN-γ stimulation results in the homodimerization of STAT1 molecules that translocate to the nucleus to bind ISG promoters containing IFN-γ activated sites.
Because the expression of ISGs is critical to limiting viral replication, viruses use numerous strategies to antagonize the IFN response. Control of alphavirus infection relies on an intact type I IFN system since various attenuated strains of VEEV, Sindbis virus (SINV), and Semliki Forest virus (SFV) become fully virulent in mice with disrupted IFN-α/β receptors (16, 28, 59). Despite its crucial role in protection, treatment of mice with type I IFN or poly(I:C), an IFN inducer, failed to protect animals from a subsequent challenge with virulent VEEV, suggesting the virus is partially resistant to these cytokines (28, 29), although administration of the more stable pegylated-IFN-α was successful (33). Previous studies indicate that sensitivity of different VEEV and Eastern equine encephalitis virus (EEEV) strains to IFN-α/β correlates with virulence potential (1, 4, 29, 54). Certain virulent epizootic strains of VEEV have been shown to be less sensitive to the effects of type I IFN than their less virulent enzootic progenitors (44, 54), suggesting that IFN sensitivity is a potential marker for epizootic potential. However, more recent studies challenge this association (7) and implicate mutations within the E2 glycoprotein that allow equine-avirulent enzootic strains (subtype ID) to emerge as subtype IC strains that achieve efficient equine amplification (6, 7, 26, 55). In general, although the role for IFN resistance in VEEV pathogenesis and/or emergence remains unclear, all alphaviruses demonstrate some sensitivity to type I IFNs and therefore must use mechanisms to limit either the induction or the cellular responses to these cytokines.
The major mechanism by which alphaviruses are believed to evade the antiviral effects of IFNs is through a global shutoff of host gene expression (19). Viruses containing mutations in the SINV nonstructural protein 2 (nsP2) carboxy terminus possess limited ability to shutoff RNA polymerase II-dependent cellular transcription and induce higher levels of IFN-α/β relative to wild-type viruses, suggesting that generalized inhibition of host macromolecular synthesis contributes to viral blockade of the type I IFN response (19, 22, 25). Expression of nsP2 from the Old World alphaviruses SINV and SFV is linked to host cell cytotoxicity; however, studies with VEEV and EEEV suggest that the New World alphaviruses mediate shutoff by viral capsid-dependent mechanisms (5, 21, 23, 41), although host shutoff also occurs in cells infected with VEEV replicon particles (VRP) that do not express the viral capsid protein (S. A. Montgomery, unpublished data). An amino-terminal portion of VEEV capsid is required for transcriptional shutoff and for the generation of host cell cytopathic effect through a mechanism proposed to involve the disruption of the nuclear import of cellular factors (8, 21). A similar region within EEEV capsid inhibits RNA polymerase II transcription and the antiviral effects of IFNs (3, 5, 8).
Although it is likely that host transcriptional and translational shutoff dampen the cellular antiviral response, these nonspecific mechanisms are not solely responsible for the inhibition of the IFN response by alphaviruses. A mutation within the nuclear localization signal of SFV nsP2 leads to significantly higher levels of type I IFN induction with no apparent effect on host cell transcriptional or translational shutoff (10). Since this mutant is significantly attenuated in vivo (16), alphaviruses likely use shutoff-independent mechanisms to antagonize the antiviral IFN response, and these mechanisms are important determinants of virulence.
In the present study, we tested whether infection with VEEV or VRP specifically inhibits IFN signaling through the Jak/STAT pathway. Recent findings demonstrate that nsP2 of VEEV specifically interacts with the nuclear importin molecule karyopherin α-1 (KPNA1) (36). Since KPNA1 is known to bind and shuttle STAT1 to the nuclear pore complex to direct its nuclear import (50), we investigated whether VEEV inhibits STAT1-dependent IFN signaling. Our results indicate that cells infected with VEEV or VRP, which lack the genes encoding the viral structural proteins, fail to respond to type I and II IFN, as demonstrated by decreased STAT1 tyrosine phosphorylation and nuclear localization. Since activation of the Jak/STAT pathway does not require de novo cellular gene transcription and translation, these findings provide additional evidence that alphaviruses are able to antagonize the IFN response by mechanisms independent of host transcriptional and translational shutoff.
MATERIALS AND METHODS
Cell culture and reagents.
Vero-81, BHK-21, HEK-293, and HeLa cells were grown under 5% CO2 at 37°C. Vero-81 cells (designated “Vero,” ATCC no. CCL-81) were obtained from the American Type Culture Collection and maintained in Dulbecco modified Eagle medium/F-12 medium (Gibco) supplemented with 10% fetal bovine serum (FBS; HyClone), l-glutamine (0.29 mg/ml; Gibco), nonessential amino acids (Gibco), penicillin-streptomycin (Gibco), and sodium bicarbonate adjusted to contain a final of 3 g/liter (Gibco). HeLa and 293 cells were maintained in Dulbecco modified Eagle medium (Gibco) supplemented with 10% FBS, l-glutamine, and penicillin-streptomycin. BHK-21 cells were maintained in alpha-MEM (Gibco) containing either 10% donor calf serum (HyClone) or 10% FBS (Lonza), 10% tryptose phosphate broth (Sigma), and supplemented with l-glutamine and penicillin-streptomycin. Actinomycin D (ActD) and cycloheximide (CHx) were purchased from Sigma. Recombinant human IFN-β was obtained from Calbiochem and resuspended according to the manufacturer's instructions. The biological activity was determined by using a type I IFN bioassay previously described using A549 cells, and relative protection from EMCV-induced cytopathic effect was compared to the NIH human IFN-β reference standard (Gb23-902-531) (51). Recombinant human IFN-γ was used according to the concentration given by the manufacturer (R&D Systems).
VEEV and VRP production.
The generation of a full-length cDNA clone derived from the wild-type Trinidad donkey VEEV isolate (pV3000) has been described previously (13). A cell culture-adapted mutant encoding two attenuating amino acid changes in the E1 and E2 glycloprotein, pV3014, was used in these studies due to its greater specific infectivity relative to the parental pV3000 (9, 27). The mechanism of attenuation of V3014 may involve its efficient binding of heparan sulfate, which enhances cell culture infectivity but could increase viral clearance in vivo (9). In vitro-transcribed RNA from this pV3014 cDNA was used to electroporate BHK-21 cells and, after 24 h, the supernatants were harvested, divided into aliquots, and stored at −80°C. Virus titers were quantified by plaque assay on BHK-21 cells. A split-helper system described previously (45) was used to generate VRP that express green fluorescent protein (GFP) or no transgene (VRP-empty). Supernatants of electroporated BHK-21 cells containing VRP were harvested and concentrated through 20% sucrose (wt/vol) at 72,000 × g for 4 h. Replicon titers were quantified as infectious units (IU) per ml by an indirect immunofluorescence staining assay of infected BHK-21 cells using antiserum raised against the viral nonstructural proteins (for the VRP-empty titer) or by counting GFP-positive cells (for VRP-GFP titer).
Immunoblot analysis.
For most experiments, Vero-81 cells were lysed in radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 1% Igepal CA-630, 0.1% sodium dodecyl sulfate [SDS], and 0.5% deoxycholate, supplemented with Complete miniprotease inhibitors (Roche) and phosphatase inhibitors [Sigma-P2850]) on ice for >5 min and scraped. Lysates were clarified by centrifugation for 5 to 10 min at 4°C, and the total protein was quantified by using a Coomassie Plus protein assay (Thermo). Equal amounts of total protein from each sample were denatured in SDS sample buffer and resolved by 8% SDS-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membrane (Bio-Rad) in transfer buffer (48 mM Tris, 39 mM glycine, 10% methanol), blocked in 3 to 5% dry nonfat milk (or bovine serum albumin) in PBST (137 mM NaCl, 2.7 mM KCl, 0.88 mM KH2PO4, 6.4 mM Na2HPO4, 0.1% Tween 20), and then incubated with the indicated primary antibody overnight at 4°C. Membranes were exposed to horseradish peroxidase (HRP)-conjugated secondary antibody and then developed by using ECL-Plus (Amersham) and exposed to film. The following primary antibodies were purchased from the indicated manufacturers: STAT1 (total), phospho-STAT1 (Tyr701), STAT3 (total), phospho-STAT3 (Tyr705), Tyk2 (total), phospho-Tyk2 (Tyr1054/1055), Jak2 (total), and phospho-Jak2 (Tyr1007/1008) from Cell Signaling; STAT2 and phospho-STAT2 (Tyr689 of mouse STAT2) from Upstate; actin and GRP78 from Santa Cruz; Jak1 (total) from BD transduction laboratories; and phospho-Jak1 (Tyr1022/1023) from Biosource. Goat anti-VEEV nsP2 was a gift from AlphaVax, Inc. Anti-mouse and anti-rabbit ECL HRP-conjugated immunoglobulin G (IgG) secondary antibodies were purchased from Amersham, and HRP-conjugated anti-goat IgG was obtained from Sigma. For detection of immunoprecipitated total Jak protein by Western, light-chain-specific HRP-conjugated anti-rabbit and anti-mouse IgG was used.
Immunoprecipitations.
For the detection of phosphorylated Jak tyrosine kinases, total Jak protein was immunoprecipitated and then probed by Western blotting with the appropriate phospho-specific antibody. Vero-81 cells were infected with VRP-GFP for 6 h and then stimulated with IFN for 20 min. Lysates were prepared as described above in phosphorylation lysis buffer (43) (0.5% Triton X-100, 150 mM NaCl, 10% glycerol, 1 mM EDTA, 50 mM HEPES [pH 7.4], 1.5 mM MgCl2, 200 μM sodium orthovanadate, 10 mM sodium pyrophosphate, 100 mM NaF, and Complete mini protease inhibitors). Lysates containing equal amounts of total protein (2.0 to 2.5 mg) were precleared with protein G-agarose (Sigma) and preimmune normal serum for 2 to 5 h at 4°C and then immunoprecipitated with protein G-agarose in the presence of the indicated antibody overnight at 4°C, washed four to five times with phosphorylation lysis buffer, and eluted in 2× SDS sample buffer for Western analysis. The anti-Jak kinase antibodies indicated above were used, except that Tyk2 (total) (BD Transduction Laboratories) was used for immunoprecipitation. Isotype-matched normal IgG was purchased from Santa Cruz (rabbit) or eBiosciences (mouse).
Subcellular fractionation.
Nuclear purification was performed as previously described (35), except the crude nuclear fraction was not banded, but rather was washed five consecutive times in 0.5 to 1.0 ml of homogenization medium prior to lysis in nuclear extraction buffer. Equal amounts of total protein from cytoplasmic and nuclear extracts were analyzed by Western blot as described above, and the purity of the nuclear extracts was determined by Western blotting to detect GRP78, which localizes to the endoplasmic reticulum and should not be found within nuclear fractions.
Immunofluorescence microscopy.
To assess subcellular localization of STAT1 by indirect immunofluorescence staining, Vero-81 cells were seeded on glass coverslips and infected with VRP-GFP for 6 h, treated with IFN-γ (200 U/ml for 40 min, rinsed in 1× PBS, fixed for 30 min in 4% paraformaldehyde, incubated for 15 min in PBS-glycine (100 mM [pH 7.2]), permeabilized with 0.2% Triton X-100, blocked 1 h with 10% normal goat serum in 1× immunofluorescence assay wash (3% bovine serum albumin, 0.05% Tween 20), stained for total STAT1 (Santa Cruz), and then stained with Alexa-Fluor 594-conjugated goat anti-rabbit IgG (Invitrogen) and DAPI (4′,6′-diamidino-2-phenylindole [Roche]; 10 μg/ml). Images were acquired with a Carl Zeiss LSM5 Pa confocal laser scanning microscope equipped with a 40×, 1.3 numerical aperture oil objective lens, using 505- to 530-nm band-pass (green fluorescence) or 560-nm long-pass (red fluorescence) filters. The pinhole diameter was set to 0.9 μm (equivalent to one area unit). Confocal images were averaged four times with imaging software (Zeiss) and formatted by using Adobe Photoshop.
Real-time PCR.
Total cellular RNA was harvested by using TRIzol and PurLink purification system (Invitrogen), and equivalent amounts of total RNA were reverse transcribed by using SuperScript III and random primers according to the manufacturer's instructions (Invitrogen). cDNA was then quantified by TaqMan real-time PCR using a 7300 real-time PCR system (Applied Biosystems). For each sample, 18S RNA and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) levels were quantified to calculate relative expression of the target genes by using the 2ΔΔCT method, and similar results were obtained with each reporter gene. The following primer-probe sets were used (Applied Biosystems): 18S (Hs_99999901_s1), GAPDH (Rh02621745_g1), guanylate binding protein 2 (Hs00894842_g1), and Trim 21(Hs00172616_m1).
Flow cytometry.
To determine the relative IFNGR1 surface expression, Vero-81 cells were seeded in six-well plates and infected with VRP or mock infected with diluent alone for 6 h. Cells were rinsed and then incubated on ice in enzyme-free cell dissociation buffer (Gibco) for 10 min. Mock- and VRP-infected cells were scraped and divided into aliquots into parallel groups for staining (2.5 × 106 to 5 × 106 cells/group). Each aliquot was then blocked with fluorescence-activated cell sorting staining buffer (1% donor calf serum, 1% normal rabbit serum [Sigma], and 0.1% NaN3 in 1× DPBS), stained according to the manufacturer's instructions with anti-human CD119 (IFNGR1) conjugated to phycoerythrin (eBiosciences). Parallel cell aliquots were left unstained. Cells were then washed and fixed in 1% formaldehyde. Samples were read with a CyAn cytometer and analyzed by using Summit software (Dako).
To verify the infectivity, VRP-infected cells in parallel wells containing glass coverslips were fixed in methanol and stained by indirect immunofluorescence assay using anti-VEEV-nonstructural protein mouse antiserum. In addition, separate wells were treated with IFN-γ, and cell lysates were harvested for analysis of STAT1 phosphorylation as described above.
Statistical analyses.
To determine whether VRP infection and/or IFN treatment resulted in a significant alteration of GBP-2 or Trim21 expression, we performed an analysis of variance by using R software (www.r-project.org), which demonstrated that IFN treatment and VRP infection both had significant effects on ISG expression. A post hoc comparison of the IFN-treated groups revealed that VRP infection resulted in a significant reduction (P < 0.05) in both GBP-2 and Trim21 expression after each treatment time.
RESULTS
VRP infection decreases ISG expression independently of the viral structural proteins.
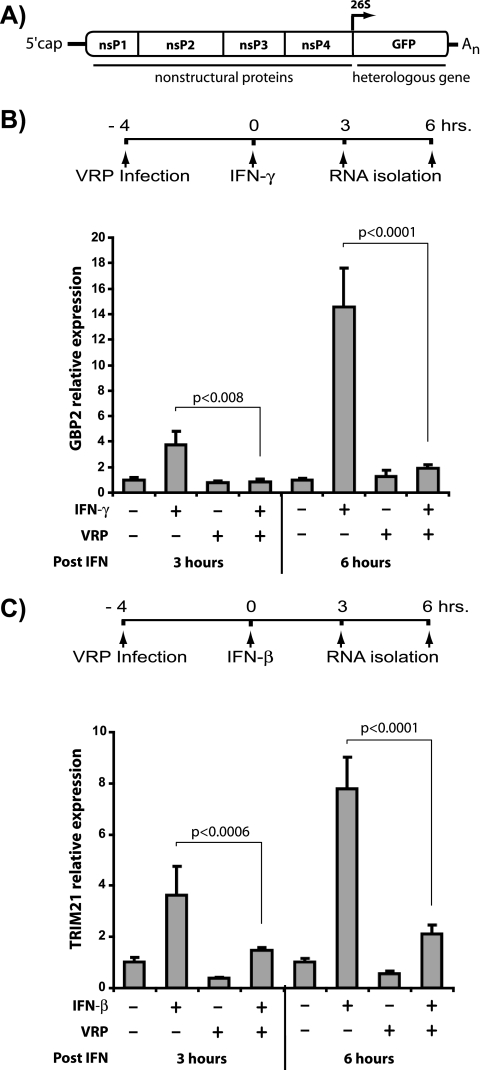
The reported interaction between nsP2 of VEEV and KPNA1 suggested that VEEV infection may disrupt IFN signaling by inhibiting STAT1 nuclear trafficking. To test this hypothesis, we first examined whether infection with VRP, in which the genes encoding the viral structural proteins have been replaced with a reporter gene (Fig. 1A), would interfere with STAT1-dependent, IFN-induced gene expression. Vero-81 cells, which cannot secrete endogenous type I IFN (15, 37), were either mock infected or infected with VRP-GFP and then treated with type I or type II IFN for either 3 or 6 h. Total RNA was harvested and cDNA was generated to measure the IFN-mediated induction of two ISGs: guanylate binding protein 2 (GBP-2), which is induced in a STAT1-dependent manner after type II IFN treatment (34, 47), and tripartite motif protein 21 (TRIM21), which can be induced by type I IFN treatment (14, 56). These studies demonstrated that VRP infection significantly decreased induction of both genes after either 3 or 6 h of IFN treatment assessed by real-time reverse transcription-PCR (Fig. 1B and C). Although the levels of housekeeping genes (18S RNA and GAPDH) were unaffected by VRP infection at these times, these studies cannot differentiate specific effects on IFN signaling from global transcriptional shutoff. Therefore, we next examined whether VRP or VEEV infection interferes with STAT1-mediated signal transduction.
FIG. 1.
VRP infection decreases ISG expression independently of the viral structural proteins. Vero-81 cells were infected with VRP, which express GFP in place of viral structural proteins from the subgenomic 26S promoter (genome organization shown in panel A), at an MOI of 10 IU/cell for 4 h and then stimulated with 1,000 U of IFN-γ (B) or IFN-β (C)/ml. Total RNA was isolated after 3 or 6 h of IFN treatment, and the relative guanylate binding protein 2 (GBP2) (B) or TRIM-21 (C) mRNA expression was determined by real-time PCR analysis of cDNA. Individual samples were normalized to GAPDH. The average of each group was calculated and is represented relative to mock-infected, untreated samples. Error bars indicate one standard deviation. To determine significance, an analysis of variance was performed. Post hoc comparisons of relevant groups at each time point are indicated by brackets, along with associated P values.
VRP and VEEV infection disrupts the nuclear localization of STAT1 in response to IFN.
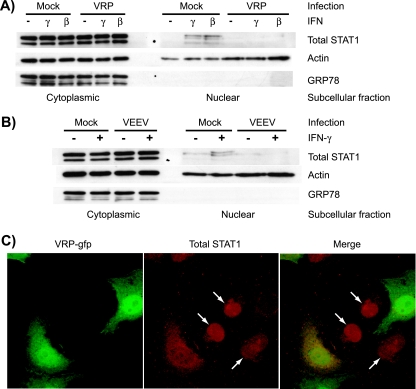
The translocation of STAT1 into the nucleus is an event critical to both type I and type II IFN signaling and requires the specific recognition of STAT1 by the importin molecule KPNA1 (50). Ebola virus VP24 interacts with this subfamily (NPI-1) of importins, an event that results in decreased STAT1 nuclear accumulation and IFN-induced gene expression (48, 49). Since nsP2 of VEEV directly interacts with KPNA1 (36), we tested whether STAT1 trafficking is affected by the expression of VEEV nonstructural proteins. Vero-81 cells were infected with VRP for 6 h and then treated for 20 min with 1,000 IU of IFN-β or IFN-γ/ml, after which subcellular fractions were harvested and subjected to Western blotting. As shown in Fig. 2A, total STAT1 was detected in the nuclear fractions in response to both IFN treatments in mock-infected, but not VRP-infected, lysates. The failure to detect STAT1 within nuclear fractions was not accompanied by any difference in total STAT1 levels within the cytoplasmic fractions, which suggests that the VRP-mediated block of nuclear import was not associated with a decrease in either the expression or stability of STAT1 protein. Similar results were found in cells infected with VRP for only 5 h prior to IFN stimulation (data not shown). To corroborate these findings in the context of VEEV infection, we repeated this assay in Vero-81 cells infected with a derivative of the wild-type Trinidad donkey VEEV strain (VEEV 3014) encoding the full-length genome. At 5 h postinfection, we were again unable to detect STAT1 in the nuclear fraction in response to 1,000 IU of IFN-γ/ml, a concentration that clearly yielded nuclear STAT1 in mock-infected cells (Fig. 2B), demonstrating that the VRP-mediated phenotype is relevant to cells infected with virus encoding the full-length VEEV genome. The same result was seen in cells infected with the wild-type V3000 virus (data not shown).
FIG. 2.
VRP and VEEV infection disrupts the nuclear localization of STAT1 in response to IFN. Vero-81 cells were infected for 6 h with VRP at an MOI of 5 IU/cell (A and C) or for 5 h with VEEV 3014 at 10 PFU/cell (B) and then stimulated with IFN-γ or IFN-β for 20 to 40 min (200 to 1,000 U/ml). Subcellular extracts were prepared and analyzed by Western blotting to determine total STAT1 distribution (A and B). GRP 78, a protein found within the endoplasmic reticulum, verifies the purity of the nuclear fractions. In an indirect immunofluorescence staining assay (C), cells were fixed after IFN treatment and stained for total STAT1 protein. Infected cells expressing GFP and STAT1 subcellular distribution (red) were detected by confocal microscopy and demonstrate the nuclear redistribution of STAT1 in uninfected cells but not in VRP-infected cells.
To confirm a defect in STAT1 nuclear accumulation, we used indirect immunofluorescence staining for STAT1 and confocal microscopy to assess the subcellular localization of STAT1 in VRP-infected Vero-81 cells. At a low multiplicity of infection (MOI; 3 IU/cell), the major population of cells showed both diffuse STAT1 staining, as well as GFP fluorescence, indicating that these cells were infected with GFP-expressing VRP (Fig. 2C). In contrast, cells with distinct nuclear STAT1 staining, indicating they were IFN responsive, were GFP negative (arrows in Fig. 2C).
Taken together, these experiments indicate that infection with VRP and VEEV reduced the nuclear accumulation of a transcription factor central to the IFN response. Previous reports indicate that an N-terminal region within the capsid protein of the New World alphaviruses VEEV and EEEV is important for the shutoff of host gene expression (3, 5, 21, 23) and that this activity may involve the disruption of nuclear import of host factors (8, 21). It is possible that the viral capsid protein contributes to the disruption of STAT1 nuclear import in cells infected with viruses encoding the full-length VEEV genome (Fig. 2B), but this mechanism cannot explain the results found in VRP-infected cells that do not express the capsid protein. Finally, diminished STAT1 nuclear accumulation was not associated with a decrease in total STAT1 levels. Thus, if shutoff of macromolecular synthesis is required for the disruption of STAT1 trafficking, it must affect targets other than STAT1, such as components of the IFN receptor complexes required for STAT1 activation.
Tyrosine phosphorylation of STAT1 in reponse to IFN-β and IFN-γ is reduced by VRP and VEEV infection.
The nuclear translocation of transcriptionally active STAT1 dimers requires prior activation by phosphorylation at tyrosine 701 (52, 53, 58). Since the reduced nuclear trafficking of STAT1 in VRP- and VEEV-infected cells could be due to defective STAT1 activation, we next determined whether STAT1 is tyrosine phosphorylated normally in infected cells. Surprisingly, the accumulation of STAT1 phosphorylated at tyrosine 701 (p-STAT1) was appreciably reduced in VRP-infected Vero-81 cells stimulated with various doses of IFN-β and IFN-γ (Fig. 3A). Very similar results were achieved in two IFN-competent human cell lines, HeLa and HEK-293 (data not shown). This inhibition was not accompanied by any detectable difference in total STAT1 levels (compare lanes 9 through 12 of Fig. 3A). At large IFN doses, a doublet band is seen in total STAT1 blots from mock-infected cells (lanes 5, 7, 13, and 15 of Fig. 3A). This doublet corresponds to phosphorylated and nonphosphorylated STAT1 and does not indicate an increase in total STAT1 levels. The equivalent total STAT1 levels between mock- and VRP-infected cells again indicated that the VRP-mediated blockade involved neither a failure to express de novo STAT1 nor its specific degradation. Interestingly, phosphorylation of STAT2 at tyrosine 690, which also occurs at the cytoplasmic tails of the IFN-α/β receptor complex, was unaffected at this time postinfection (6 h) (Fig. 3B), although its activation was reduced at later times (Fig. 4). Although the activation of STAT1 and STAT2 is thought to be crucial during IFN signaling, a third factor, STAT3, can also be activated by the same receptor complexes to initiate separate signaling events (42). As seen with STAT1, VRP-infected cells had less tyrosine-phosphorylated STAT3, but total STAT3 levels equivalent to those of mock-infected cells (Fig. 3C).
FIG. 3.
Tyrosine phosphorylation of STAT1 and STAT3 in response to IFN-β and IFN-γ is reduced by VRP infection. Vero-81 cells were infected with VRP (MOI = 5 IU/cell) for 6 h and treated for 20 min with various concentrations of IFN-γ and IFN-β. Whole-cell extracts were harvested and 20 μg of total protein from each lysate was resolved by SDS-polyacrylamide gel electrophoresis and analyzed in three separate blots to assess activation of STAT1 (A), STAT2 (B), and STAT3 (C) at the indicated phosphotyrosine residues. Blots were then stripped and reprobed using the respective total STAT antibody. All actin loading control blots were similar to that shown in panel C. *, The anti-mouse pTyr689-STAT2 antibody used (Upstate) cross-reacts with the corresponding human pTyr690-STAT2.
FIG. 4.
Activation of STAT1, but not STAT2, is limited by VEEV and VRP at early times postinfection. Vero-81 cells were infected for the indicated times with either VRP (MOI = 5 IU/cell) or VEEV 3014 (MOI = 20 PFU/cell) and then stimulated with IFN-β (500 U/ml) for 20 min. Samples were analyzed as described in Fig. 3. To compare the kinetics and amount of nonstructural protein accumulation, a separate Western immunoblot indicates fully processed VEEV nonstructural protein 2 (VEEV nsP2).
To determine the kinetics by which VRP inhibits the activation of STATs and whether infection with virus encoding the full-length VEEV genome also limits STAT activation, we treated VRP- or VEEV-infected Vero-81 cells with IFN-β at various times postinfection and found that STAT1 activation was reduced as early as 4 h postinfection (Fig. 4). Infection with VEEV also severely reduced STAT1 activation, but at later times relative to VRP infection. These kinetics correlate with greater and more rapid accumulation of viral nonstructural proteins in cells infected with VRP relative to levels seen in VEEV-infected cells (Fig. 4 and data not shown), although it is clear that large amounts of fully processed VEEV nsP2 accumulate by 4 h postinfection prior to any detectable effect on STAT1 activation in VEEV-infected cells. While the VEEV nsP2-KPNA1 interaction may play a role in the disruption of STAT1 nuclear import (Fig. 2), these results indicate that defects in upstream signaling events likely contribute to the reduced levels of nuclear STAT1 and the decreased accumulation of STAT1-driven gene transcripts in infected cells.
Viral replication, but not de novo host gene expression, is required for the VRP-mediated blockade of STAT1 activation.
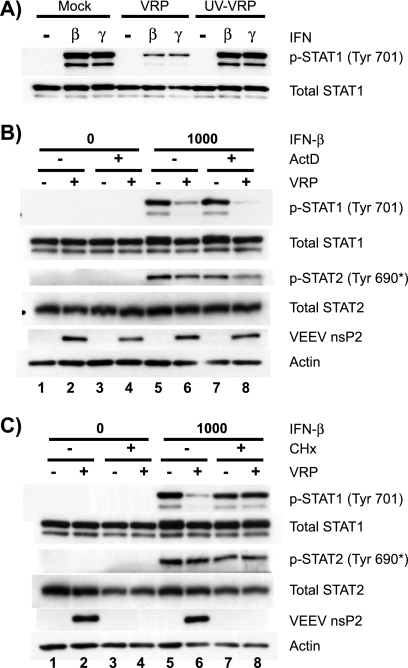
At early times (3 h) postinfection, we failed to detect a defect in STAT1 activation within VRP-infected cells (data not shown), but the inhibition was nearly maximal at later times (4 to 5 h postinfection) (Fig. 4). These kinetics suggested that viral replication is required for the blockade to occur, which was confirmed by demonstrating that cells infected with UV-inactivated VRP (Fig. 5A) were able to activate STAT1 to the same degree as mock-infected cells in response to IFN.
FIG. 5.
Viral replication, but not de novo host gene expression, is required for the VRP-mediated STAT1 blockade. (A) Vero-81 cells were either mock infected or infected with untreated VRP or VRP that had been exposed to UV light for 1 min (UV-VRP). At 6 h postinfection, cells were stimulated with 1,000 U of IFN/ml and analyzed as in Fig. 3. (B and C) Vero-81 cells were pretreated for 1 h in the absence or presence of the inhibitor ActD or CHx prior to infection with VRP (MOI = 10 IU/cell) or diluent (mock). Virus was allowed to bind to cells in the indicated treatment groups in the presence of each inhibitor for 1 h, after which the inoculum was replaced with medium containing the inhibitor. At 6 h postinfection (7 h of inhibitor treatment), cells were stimulated with IFN-β (1,000 U/ml) for 20 min, and STAT phosphorylation was assessed as described in Fig. 3.
Since STAT1 and STAT2 require the same upstream factors for activation, it is unlikely that nonspecific mechanisms such as generalized host macromolecular synthesis shutoff would prevent activation of STAT1 but not STAT2 at a given time (4 to 6 h postinfection). Specific antagonism may involve a direct interaction between a viral protein and a component of the Jak/STAT apparatus, or it may indirectly involve negative regulation of Jak/STAT signaling by a host factor that is induced or potentiated by VRP infection. Such negative regulators include cellular protein tyrosine phosphatases and suppressors of cytokine signaling proteins. To determine whether de novo host transcription is required for VRP to mediate the blockade of STAT1 activation, we treated Vero-81 cells with ActD (1.0 μg/ml), which specifically inhibits host, but not viral, transcription. As expected, mock-infected cells were able to respond to IFN in the presence of ActD since de novo gene expression is not required for the activation of latent STAT factors (Fig. 5B, lane 7). The inhibition of STAT1 activation by VRP was not affected in ActD-treated cells (Fig. 5B, lanes 6 and 8), indicating that the mechanism does not require de novo expression of a host gene that may be induced upon infection.
To inhibit host and viral translation, we treated Vero-81 cells with CHx (0.5 μg/ml) and found that STAT1 was activated similarly in the presence or absence of the inhibitor in mock-infected cells (Fig. 5C, lanes 5 and 7). Furthermore, when cells were infected with VRP in the presence of CHx, the VRP-mediated inhibition of STAT1 phosphorylation was lost (Fig. 5C, lanes 7 and 8), again indicating that productive replication and translation of viral proteins is required for the inhibition of STAT1 activation. Moreover, proteins such as viral capsid, which are introduced into the cell in limited quantities by the incoming replicon particles, are not sufficient to mediate the Jak/STAT signaling blockade in the absence of productive replication. Taken together, these data indicate that viral replication results in decreased STAT1 activation, an effect that cannot be achieved in cells treated with inhibitors of host macromolecular synthesis.
VRP infection inhibits the activation of Janus kinases associated with the type II IFN receptor but not with the type I IFN receptor.
Stimulation of either the type I or type II IFN receptor results in the phosphorylation of STAT1 at Tyr701. The decrease in STAT1 phosphorylation in VRP-infected cells may reflect a defect at the level of STAT1 itself, such as an interaction with a viral protein that prevents STAT1 from being phosphorylated, or the virus-induced activation of a host cell protein tyrosine phosphatase that inactivates STAT1. Alternatively, the activation of the type I and type II IFN receptor complexes, which is essential for IFN-induced tyrosine phosphorylation of STAT1, may be disrupted in VRP-infected cells. After type I IFN stimulation, Jak1 and Tyk2 become tyrosine phosphorylated at the IFNAR cytoplasmic tails. Similarly, after type II IFN stimulation, Jak1 and Jak2 become phosphorylated at the IFNGR subunits. Therefore, inhibition of Jak1 could lead to signaling defects in response to both IFN classes and result in decreased STAT1 (and STAT3) phosphorylation.
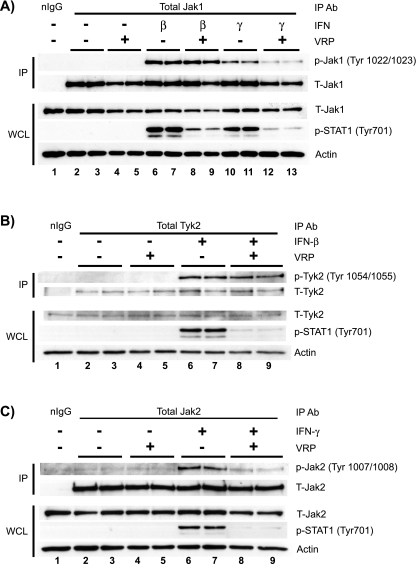
We assessed the activation of Jak kinases in VRP-infected Vero-81 cells by immunoprecipitation of Jak1, Tyk2, and Jak2, followed by Western blot analysis using phospho-specific antibodies that specifically recognize their activated epitopes. As shown in Fig. 6A, Jak1 became phosphorylated at tyrosines 1022 and 1023 in response to both IFN treatments. Interestingly, VRP infection had no effect on this activation after treatment with IFN-β (Fig. 6A, IP lanes 8 and 9), but there was a notable decrease in tyrosine-phosphorylated Jak1 after treatment with IFN-γ (Fig. 6A, IP lanes 12 and 13). Although we have obtained very consistent results indicating that VRP infection disrupts Jak1 activation after treatment with IFN-γ, but not IFN-β, we did not detect any consistent difference in total Jak1 protein levels between mock and VRP-infected cells (Fig. 6A, WCL lanes 2 to 12). To further examine whether VRP differentially modulates signaling at the type I versus type II IFN receptors, we assessed the activation of Tyk2 and Jak2 at each receptor complex, respectively. Again, activation of the type I IFN receptor components occurred normally in VRP-infected cells as indicated by phospho-Tyk2 levels equivalent to those in mock-infected cells. These events appear to be disrupted at the type II IFN receptor, which showed decreased Jak2 tyrosine phosphorylation (Fig. 6B and C) but normal levels of total Jak2.
FIG. 6.
VRP infection inhibits the activation of Janus kinases associated with the type II, but not the type I, IFN receptor. Vero-81 cells were mock infected or infected with VRP for 6 h and then stimulated for 20 min with IFN. Each infection and treatment was performed in duplicate. Whole-cell lysates (WCL) were prepared, and equivalent amounts of total protein were subjected to immunoprecipitation with antibodies recognizing total Jak1 (A), Tyk2 (B), and Jak2 (C) or with isotype matched normal IgG (nIgG). To assess Jak protein activation, the immunoprecipitates were then analyzed by Western blotting using the indicated phospho-specific Jak antibodies (IP panels). These membranes were then stripped and reprobed with the corresponding antibody recognizing total Jak protein. A portion of the input whole-cell lysates (50 μg of total protein) from each sample was also analyzed by Western blotting to detect effects that VRP has on total cellular Jak levels, as well as on STAT1 phosphorylation (WCL panels).
Surface expression of IFNGR1 subunits is moderately reduced in VRP-infected cells.
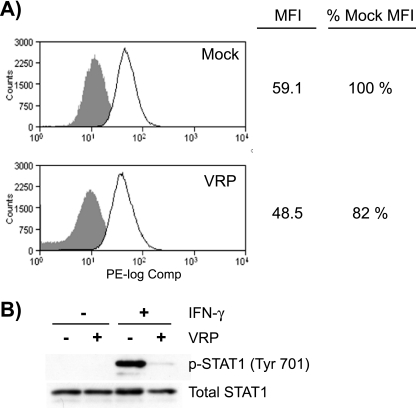
Given differential effects of VRP infection on type I and type II IFN receptor complexes (Fig. 6), we hypothesized that VRP infection reduces the cell surface expression of the IFNGR. Accordingly, we measured the relative surface expression of the IFNGR1 subunit between mock- and VRP-infected Vero-81 cells by flow cytometry. Surface IFNGR1 expression was slightly, but consistently, lower in VRP-infected cells (82 to 90% of that in mock-infected cells) in three independent experiments (Fig. 7A). Despite this minimal decrease in IFNGR1 surface expression, a large decrease in STAT1 phosphorylation was detected in parallel VRP-infected cells stimulated with IFN-γ (Fig. 7B). Although it was consistent, the minimal decrease in IFNGR1 surface expression is unlikely to account for the dramatic defect of downstream STAT1, Jak1, and Jak2 phosphorylation seen in VRP-infected cells in response to IFN-γ, suggesting that additional mechanisms are involved.
FIG. 7.
Surface expression of IFNGR1 subunits is moderately reduced in VRP-infected cells. Vero-81 cells were infected with mock or VRP (MOI = 5) for 6 h, which resulted in the infection of 98.8% of cells as determined by an indirect immunofluorescence staining assay that detects the viral nonstructural proteins (data not shown). (A) Mock- or VRP-infected cells were pooled, counted, and then separated into replicate groups for staining. Cells were either stained with anti-CD119 (IFNGR1) directly conjugated to phycoerythrin (solid line) or left unstained (shaded histogram). Cells in each group were then washed, fixed, and analyzed by flow cytometry. Mean fluorescence intensities (MFI) of mock- and VRP-infected samples were compared, and the decrease in IFNGR1 surface expression was comparable between three independent experiments (10 to 18% decrease). (B) Mock- and VRP-infected cells in parallel cultures were stimulated with 1,000 U of IFN-γ/ml for 20 min and harvested and analyzed as described in Fig. 3 to measure the inhibition of STAT1 phosphorylation.
DISCUSSION
Alphaviruses are proposed to inhibit the type I IFN system through global shutoff of host transcription and translation, which would prevent the induction of all type I IFN classes and ISGs. The New World alphaviruses EEEV and VEEV utilize their capsid proteins to shutoff host transcription, which requires an N-terminal region within this protein (3, 5, 8, 21, 23). Although the mechanisms by which the viral capsid inhibits transcription are unclear, this activity is associated with defects in type I IFN and ISG induction. In the present study, we found that VEEV also disrupts the cellular response to type I and type II IFN by specific mechanisms that do not require expression of the viral capsid gene and most likely act distinctly from, but in concert with, generalized host shutoff to downregulate the host antiviral response.
We report that cells infected with VRP fail to activate the transcription factor STAT1 normally, a blockade that correlates with failed STAT1 nuclear localization and decreased STAT1-dependent gene transcription. This blockade did not require viral capsid protein since (i) no structural genes are expressed from the replicon genome and (ii) components of incoming particles were not sufficient for STAT1 inhibition in either CHx-treated cells or cells infected with UV-inactivated VRP (Fig. 5). Thus, de novo viral gene expression is required for the inhibition of STAT1 activation, which likely involves the expression of the viral nonstructural proteins or requires productive viral RNA synthesis. We cannot rule out the possibility that the small amount of capsid introduced into the cell by incoming virions and viral replication are both required for STAT1 inhibition; however, preliminary results suggest that electroporation of VEEV replicon RNA alone is sufficient to disrupt STAT1 nuclear localization (data not shown).
Although the shutoff of host macromolecular synthesis plays an important role in alphavirus pathogenesis (3, 19), several lines of evidence suggest that this function is most likely not required for the VRP-mediated Jak/STAT signaling inhibition. First, the decrease in STAT1 phosphorylation was not associated with any effect on total STAT1 levels, which indicates the blockade was not due to decreased synthesis (or increased degradation) of this factor (Fig. 3A, lanes 9 to 12). Second, although the IFNAR complex fails to phosphorylate STAT1 after 5 to 6 h of VRP infection (Fig. 3 and 4), the defect is specific for STAT1 since this complex activates Jak1, Tyk2, and STAT2 normally, indicating that the IFNAR complex is fully functional at these times (Fig. 4 and 6). Finally, since it is possible an unidentified host factor with a short half-life is required for STAT1 activation by type I IFN, we pharmacologically induced host shutoff with CHx or ActD but failed to recapitulate the magnitude of STAT1 inhibition achieved by VRP (Fig. 5). Thus, even if VRP induces host shutoff by these early times postinfection, the VRP must use an additional strategy to block STAT1 phosphorylation since STAT1 activation can occur in the absence of de novo cellular transcription and translation (17, 18, 40).
Although our findings indicate that STAT1 activation is inhibited in the absence of de novo host gene expression or viral structural protein synthesis, the particular mechanisms and viral factors required for this inhibition remain to be identified. Accumulation of tyrosine-phosphorylated STATs is determined by the rates of phosphorylation and dephosphorylation, and VEEV infection could affect either or both processes. One possibility is that a particular viral nonstructural protein binds STAT1 (and STAT3) or the receptor complexes preventing its phosphorylation by Jak kinases. Alternatively, VRP may activate latent host proteins that regulate these processes, such as host protein tyrosine phosphatases. If this latter explanation is correct and there is rapid STAT dephosphorylation after IFN stimulation, the data presented in Fig. 5 demonstrate that such a host factor is not transcriptionally induced upon VRP infection and therefore must be present in a latent form at sufficient levels to counteract the very rapid STAT phosphorylation.
In the case of the IFNAR, we found that the receptor complex remains functional even at times when STAT1 inhibition is nearly maximal (Fig. 6). One possible explanation is that the VEEV-mediated mechanism(s), such as the direct binding of a viral nonstructural protein, is acting at the level of specific STAT factors, preventing their association with the IFNAR and/or their activation by Jak1/Tyk2. The relative resistance of STAT2 to the VEEV-mediated blockade, in this case, could be explained by a reduced affinity of this nonstructural protein for STAT2. However, it is clear that STAT2 is affected at later times (after 6 h of infection, Fig. 4). STAT2 associates with the IFNAR2 cytoplasmic tail prior to its engagement with the phosphorylated IFNAR1 subunit upon receptor ligation (32, 39). The orientation and duration of this preassociation may allow STAT2 to become phosphorylated more efficiently than STAT1 and thus be less sensitive to the VEEV-mediated mechanism. However, a potential role for STAT2-IFNAR2 preassociation in this phenotype remains to be determined.
In contrast to the events at the IFNAR complex, we detected defects in the activation of IFNGR components in VRP-infected cells, namely, reduced Jak1 and Jak2 activation (Fig. 6). While IFNGR surface expression was modestly reduced (Fig. 7), it is unlikely that reduced receptor surface expression alone fully explains the large decrease in Jak1, Jak2, and STAT1 activation. It is possible that VEE prevents the activation of STAT1 by both receptor complexes through common mechanisms but, in the case of the IFNGR, additional mechanisms contribute to the defect in Jak1 and Jak2 activation. Taken together, our data suggest that signaling steps downstream of receptor surface expression, such as the kinase activities of Jak proteins and the phosphorylation of the critical tyrosine residues present on the receptor cytoplasmic tails, are disrupted by VEEV infection, and further studies are under way to define the mechanism(s) behind this process. In addition, it will be important to further assess whether VEEV infection interferes with the STAT1 signaling cascade at steps downstream of STAT1 tyrosine phosphorylation, since the previously demonstrated nsP2-KPNA1 interaction may further contribute to the inhibition of STAT1-dependent signaling.
These studies indicate that, in addition to its global interference with host macromolecular synthesis, VEEV is able to antagonize the type I and type II IFN response by specific mechanisms. It remains to be determined whether the inhibition of Jak/STAT signaling is specific for VEEV, or whether other alphaviruses use similar strategies. Recent work by Griffin et al. indicates that the noncytolytic clearance of SINV from infected neuronal cells requires functional IFN-γ and Jak1 signaling (11), which would suggest that this activity is not shared by all alphaviruses or that it may be cell type dependent. It is important to note that VEEV is sensitive to the effects of type I IFN. For example, IFNAR-deficient mice succumb to infection with VEEV much earlier than wild-type mice. The disruption of IFN signaling identified in these studies, therefore, may simply dampen the magnitude of the IFN response or may be relevant to particular cell types important for initiating the innate immune response in the animal. Identification of the viral determinants that mediate the inhibition of Jak/STAT signaling in VEEV will provide a more complete understanding of the determinants for IFN sensitivity, viral virulence, and pathogenesis and may ultimately enable better design of alphavirus-based vaccines.
Acknowledgments
We thank Martha Collier and Bianca Trollinger for expert technical assistance with VRP production and cell culture. We also thank Nancy Davis, Matthew Frieman, Reed Shabman, Catherine Cruz, and Clayton Beard for helpful scientific discussions and editorial comments on the manuscript.
This study was supported by NIH grant R01 AI067641 to M.T.H. J.D.S. was supported by NIH grant T32 GM008719.
Footnotes
Published ahead of print on 5 August 2009.
REFERENCES
- 1.Aguilar, P. V., A. P. Adams, E. Wang, W. Kang, A. S. Carrara, M. Anishchenko, I. Frolov, and S. C. Weaver. 2008. Structural and nonstructural protein genome regions of eastern equine encephalitis virus are determinants of interferon sensitivity and murine virulence. J. Virol. 82:4920-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar, P. V., I. P. Greene, L. L. Coffey, G. Medina, A. C. Moncayo, M. Anishchenko, G. V. Ludwig, M. J. Turell, M. L. O'Guinn, J. Lee, R. B. Tesh, D. M. Watts, K. L. Russell, C. Hice, S. Yanoviak, A. C. Morrison, T. A. Klein, D. J. Dohm, H. Guzman, A. P. Travassos da Rosa, C. Guevara, T. Kochel, J. Olson, C. Cabezas, and S. C. Weaver. 2004. Endemic Venezuelan equine encephalitis in northern Peru. Emerg. Infect. Dis. 10:880-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilar, P. V., L. W. Leung, E. Wang, S. C. Weaver, and C. F. Basler. 2008. A five-amino-acid deletion of the eastern equine encephalitis virus capsid protein attenuates replication in mammalian systems but not in mosquito cells. J. Virol. 82:6972-6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aguilar, P. V., S. Paessler, A. S. Carrara, S. Baron, J. Poast, E. Wang, A. C. Moncayo, M. Anishchenko, D. Watts, R. B. Tesh, and S. C. Weaver. 2005. Variation in interferon sensitivity and induction among strains of eastern equine encephalitis virus. J. Virol. 79:11300-11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aguilar, P. V., S. C. Weaver, and C. F. Basler. 2007. Capsid protein of eastern equine encephalitis virus inhibits host cell gene expression. J. Virol. 81:3866-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anishchenko, M., R. A. Bowen, S. Paessler, L. Austgen, I. P. Greene, and S. C. Weaver. 2006. Venezuelan encephalitis emergence mediated by a phylogenetically predicted viral mutation. Proc. Natl. Acad. Sci. USA 103:4994-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anishchenko, M., S. Paessler, I. P. Greene, P. V. Aguilar, A. S. Carrara, and S. C. Weaver. 2004. Generation and characterization of closely related epizootic and enzootic infectious cDNA clones for studying interferon sensitivity and emergence mechanisms of Venezuelan equine encephalitis virus. J. Virol. 78:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atasheva, S., N. Garmashova, I. Frolov, and E. Frolova. 2008. Venezuelan equine encephalitis virus capsid protein inhibits nuclear import in mammalian but not in mosquito cells. J. Virol. 82:4028-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernard, K. A., W. B. Klimstra, and R. E. Johnston. 2000. Mutations in the E2 glycoprotein of Venezuelan equine encephalitis virus confer heparan sulfate interaction, low morbidity, and rapid clearance from blood of mice. Virology 276:93-103. [DOI] [PubMed] [Google Scholar]
- 10.Breakwell, L., P. Dosenovic, G. B. Karlsson Hedestam, M. D'Amato, P. Liljestrom, J. Fazakerley, and G. M. McInerney. 2007. Semliki Forest virus nonstructural protein 2 is involved in suppression of the type I interferon response. J. Virol. 81:8677-8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burdeinick-Kerr, R., D. Govindarajan, and D. E. Griffin. 2009. Noncytolytic clearance of Sindbis virus infection from neurons by gamma interferon is dependent on Jak/STAT signaling. J. Virol. 83:3429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colamonici, O. R., H. Uyttendaele, P. Domanski, H. Yan, and J. J. Krolewski. 1994. p135tyk2, an interferon-alpha-activated tyrosine kinase, is physically associated with an interferon-alpha receptor. J. Biol. Chem. 269:3518-3522. [PubMed] [Google Scholar]
- 13.Davis, N. L., L. V. Willis, J. F. Smith, and R. E. Johnston. 1989. In vitro synthesis of infectious Venezuelan equine encephalitis virus RNA from a cDNA clone: analysis of a viable deletion mutant. Virology 171:189-204. [DOI] [PubMed] [Google Scholar]
- 14.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diaz, M. O., S. Ziemin, M. M. Le Beau, P. Pitha, S. D. Smith, R. R. Chilcote, and J. D. Rowley. 1988. Homozygous deletion of the alpha- and beta 1-interferon genes in human leukemia and derived cell lines. Proc. Natl. Acad. Sci. USA 85:5259-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fazakerley, J. K., A. Boyd, M. L. Mikkola, and L. Kaariainen. 2002. A single amino acid change in the nuclear localization sequence of the nsP2 protein affects the neurovirulence of Semliki Forest virus. J. Virol. 76:392-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman, R. L., S. P. Manly, M. McMahon, I. M. Kerr, and G. R. Stark. 1984. Transcriptional and posttranscriptional regulation of interferon-induced gene expression in human cells. Cell 38:745-755. [DOI] [PubMed] [Google Scholar]
- 18.Friedman, R. L., and G. R. Stark. 1985. alpha-Interferon-induced transcription of HLA and metallothionein genes containing homologous upstream sequences. Nature 314:637-639. [DOI] [PubMed] [Google Scholar]
- 19.Frolova, E. I., R. Z. Fayzulin, S. H. Cook, D. E. Griffin, C. M. Rice, and I. Frolov. 2002. Roles of nonstructural protein nsP2 and Alpha/Beta interferons in determining the outcome of Sindbis virus infection. J. Virol. 76:11254-11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Sastre, A., and C. A. Biron. 2006. Type 1 interferons and the virus-host relationship: a lesson in detente. Science 312:879-882. [DOI] [PubMed] [Google Scholar]
- 21.Garmashova, N., S. Atasheva, W. Kang, S. C. Weaver, E. Frolova, and I. Frolov. 2007. Analysis of Venezuelan equine encephalitis virus capsid protein function in the inhibition of cellular transcription. J. Virol. 81:13552-13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garmashova, N., R. Gorchakov, E. Frolova, and I. Frolov. 2006. Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription. J. Virol. 80:5686-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garmashova, N., R. Gorchakov, E. Volkova, S. Paessler, E. Frolova, and I. Frolov. 2007. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J. Virol. 81:2472-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gauzzi, M. C., L. Velazquez, R. McKendry, K. E. Mogensen, M. Fellous, and S. Pellegrini. 1996. Interferon-alpha-dependent activation of Tyk2 requires phosphorylation of positive regulatory tyrosines by another kinase. J. Biol. Chem. 271:20494-20500. [DOI] [PubMed] [Google Scholar]
- 25.Gorchakov, R., E. Frolova, and I. Frolov. 2005. Inhibition of transcription and translation in Sindbis virus-infected cells. J. Virol. 79:9397-9409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greene, I. P., S. Paessler, L. Austgen, M. Anishchenko, A. C. Brault, R. A. Bowen, and S. C. Weaver. 2005. Envelope glycoprotein mutations mediate equine amplification and virulence of epizootic Venezuelan equine encephalitis virus. J. Virol. 79:9128-9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grieder, F. B., N. L. Davis, J. F. Aronson, P. C. Charles, D. C. Sellon, K. Suzuki, and R. E. Johnston. 1995. Specific restrictions in the progression of Venezuelan equine encephalitis virus-induced disease resulting from single amino acid changes in the glycoproteins. Virology 206:994-1006. [DOI] [PubMed] [Google Scholar]
- 28.Grieder, F. B., and S. N. Vogel. 1999. Role of interferon and interferon regulatory factors in early protection against Venezuelan equine encephalitis virus infection. Virology 257:106-118. [DOI] [PubMed] [Google Scholar]
- 29.Jahrling, P. B., E. Navarro, and W. F. Scherer. 1976. Interferon induction and sensitivity as correlates to virulence of Venezuelan encephalitis viruses for hamsters. Arch. Virol. 51:23-35. [DOI] [PubMed] [Google Scholar]
- 30.Leon, C. A. 1975. Sequelae of Venezuelan equine encephalitis in humans: a four year follow-up. Int. J. Epidemiol. 4:131-140. [DOI] [PubMed] [Google Scholar]
- 31.Levy, D. E., and J. E. Darnell, Jr. 2002. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell. Biol. 3:651-662. [DOI] [PubMed] [Google Scholar]
- 32.Li, X., S. Leung, I. M. Kerr, and G. R. Stark. 1997. Functional subdomains of STAT2 required for preassociation with the alpha interferon receptor and for signaling. Mol. Cell. Biol. 17:2048-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukaszewski, R. A., and T. J. Brooks. 2000. Pegylated alpha interferon is an effective treatment for virulent Venezuelan equine encephalitis virus and has profound effects on the host immune response to infection. J. Virol. 74:5006-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacMicking, J. D. 2004. IFN-inducible GTPases and immunity to intracellular pathogens. Trends Immunol. 25:601-609. [DOI] [PubMed] [Google Scholar]
- 35.Montgomery, S. A., P. Berglund, C. W. Beard, and R. E. Johnston. 2006. Ribosomal protein S6 associates with alphavirus nonstructural protein 2 and mediates expression from alphavirus messages. J. Virol. 80:7729-7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montgomery, S. A., and R. E. Johnston. 2007. Nuclear import and export of Venezuelan equine encephalitis virus nonstructural protein 2. J. Virol. 81:10268-10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mosca, J. D., and P. M. Pitha. 1986. Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis. Mol. Cell. Biol. 6:2279-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muller, M., J. Briscoe, C. Laxton, D. Guschin, A. Ziemiecki, O. Silvennoinen, A. G. Harpur, G. Barbieri, B. A. Witthuhn, C. Schindler, et al. 1993. The protein tyrosine kinase JAK1 complements defects in interferon-alpha/beta and -gamma signal transduction. Nature 366:129-135. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen, V. P., A. Z. Saleh, A. E. Arch., H. Yan, F. Piazza, J. Kim, and J. J. Krolewski. 2002. Stat2 binding to the interferon-alpha receptor 2 subunit is not required for interferon-alpha signaling. J. Biol. Chem. 277:9713-9721. [DOI] [PubMed] [Google Scholar]
- 40.Pearse, R. N., R. Feinman, K. Shuai, J. E. Darnell, Jr., and J. V. Ravetch. 1993. Interferon gamma-induced transcription of the high-affinity Fc receptor for IgG requires assembly of a complex that includes the 91-kDa subunit of transcription factor ISGF3. Proc. Natl. Acad. Sci. USA 90:4314-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petrakova, O., E. Volkova, R. Gorchakov, S. Paessler, R. M. Kinney, and I. Frolov. 2005. Noncytopathic replication of Venezuelan equine encephalitis virus and eastern equine encephalitis virus replicons in mammalian cells. J. Virol. 79:7597-7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Platanias, L. C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signaling. Nat. Rev. Immunol. 5:375-386. [DOI] [PubMed] [Google Scholar]
- 43.Platanias, L. C., S. Uddin, P. Domanski, and O. R. Colamonici. 1996. Differences in interferon alpha and beta signaling. Interferon beta selectively induces the interaction of the alpha and betaL subunits of the type I interferon receptor. J. Biol. Chem. 271:23630-23633. [DOI] [PubMed] [Google Scholar]
- 44.Powers, A. M., A. C. Brault, R. M. Kinney, and S. C. Weaver. 2000. The use of chimeric Venezuelan equine encephalitis viruses as an approach for the molecular identification of natural virulence determinants. J. Virol. 74:4258-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pushko, P., M. Parker, G. V. Ludwig, N. L. Davis, R. E. Johnston, and J. F. Smith. 1997. Replicon-helper systems from attenuated Venezuelan equine encephalitis virus: expression of heterologous genes in vitro and immunization against heterologous pathogens in vivo. Virology 239:389-401. [DOI] [PubMed] [Google Scholar]
- 46.Quiroz, E., P. V. Aguilar, J. Cisneros, R. B. Tesh, and S. C. Weaver. 2009. Venezuelan equine encephalitis in panama: fatal endemic disease and genetic diversity of etiologic viral strains. PLoS Negl. Trop. Dis. 3:e472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramsauer, K., M. Farlik, G. Zupkovitz, C. Seiser, A. Kroger, H. Hauser, and T. Decker. 2007. Distinct modes of action applied by transcription factors STAT1 and IRF1 to initiate transcription of the IFN-gamma-inducible gbp2 gene. Proc. Natl. Acad. Sci. USA 104:2849-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reid, S. P., L. W. Leung, A. L. Hartman, O. Martinez, M. L. Shaw, C. Carbonnelle, V. E. Volchkov, S. T. Nichol, and C. F. Basler. 2006. Ebola virus VP24 binds karyopherin alpha1 and blocks STAT1 nuclear accumulation. J. Virol. 80:5156-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reid, S. P., C. Valmas, O. Martinez, F. M. Sanchez, and C. F. Basler. 2007. Ebola virus VP24 proteins inhibit the interaction of NPI-1 subfamily karyopherin alpha proteins with activated STAT1. J. Virol. 81:13469-13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sekimoto, T., N. Imamoto, K. Nakajima, T. Hirano, and Y. Yoneda. 1997. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 16:7067-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shabman, R. S., T. E. Morrison, C. Moore, L. White, M. S. Suthar, L. Hueston, N. Rulli, B. Lidbury, J. P. Ting, S. Mahalingam, and M. T. Heise. 2007. Differential induction of type I interferon responses in myeloid dendritic cells by mosquito and mammalian-cell-derived alphaviruses. J. Virol. 81:237-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shuai, K., C. M. Horvath, L. H. Huang, S. A. Qureshi, D. Cowburn, and J. E. Darnell, Jr. 1994. Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell 76:821-828. [DOI] [PubMed] [Google Scholar]
- 53.Shuai, K., G. R. Stark, I. M. Kerr, and J. E. Darnell, Jr. 1993. A single phosphotyrosine residue of Stat91 required for gene activation by interferon-gamma. Science 261:1744-1746. [DOI] [PubMed] [Google Scholar]
- 54.Spotts, D. R., R. M. Reich, M. A. Kalkhan, R. M. Kinney, and J. T. Roehrig. 1998. Resistance to alpha/beta interferons correlates with the epizootic and virulence potential of Venezuelan equine encephalitis viruses and is determined by the 5′ noncoding region and glycoproteins. J. Virol. 72:10286-10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, E., R. Barrera, J. Boshell, C. Ferro, J. E. Freier, J. C. Navarro, R. Salas, C. Vasquez, and S. C. Weaver. 1999. Genetic and phenotypic changes accompanying the emergence of epizootic subtype IC Venezuelan equine encephalitis viruses from an enzootic subtype ID progenitor. J. Virol. 73:4266-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, J., and I. L. Campbell. 2005. Innate STAT1-dependent genomic response of neurons to the antiviral cytokine alpha interferon. J. Virol. 79:8295-8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weaver, S. C., C. Ferro, R. Barrera, J. Boshell, and J. C. Navarro. 2004. Venezuelan equine encephalitis. Annu. Rev. Entomol. 49:141-174. [DOI] [PubMed] [Google Scholar]
- 58.Wenta, N., H. Strauss, S. Meyer, and U. Vinkemeier. 2008. Tyrosine phosphorylation regulates the partitioning of STAT1 between different dimer conformations. Proc. Natl. Acad. Sci. USA 105:9238-9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White, L. J., J. G. Wang, N. L. Davis, and R. E. Johnston. 2001. Role of alpha/beta interferon in Venezuelan equine encephalitis virus pathogenesis: effect of an attenuating mutation in the 5′ untranslated region. J. Virol. 75:3706-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]