Abstract
The genome of measles virus (MV) is encapsidated by the nucleocapsid (N) protein and associates with RNA-dependent RNA polymerase to form the ribonucleoprotein complex. The matrix (M) protein is believed to play an important role in MV assembly by linking the ribonucleoprotein complex with envelope glycoproteins. Analyses using a yeast two-hybrid system and coimmunoprecipitation in mammalian cells revealed that the M protein interacts with the N protein and that two leucine residues at the carboxyl terminus of the N protein (L523 and L524) are critical for the interaction. In MV minigenome reporter gene assays, the M protein inhibited viral RNA synthesis only when it was able to interact with the N protein. The N protein colocalized with the M protein at the plasma membrane when the proteins were coexpressed in plasmid-transfected or MV-infected cells. In contrast, the N protein formed small dots in the perinuclear area when it was expressed without the M protein, or it was incapable of interacting with the M protein. Furthermore, a recombinant MV possessing a mutant N protein incapable of interacting with the M protein grew much less efficiently than the parental virus. Since the M protein has an intrinsic ability to associate with the plasma membrane, it may retain the ribonucleoprotein complex at the plasma membrane by binding to the N protein, thereby stopping viral RNA synthesis and promoting viral particle production. Consequently, our results indicate that the M protein regulates MV RNA synthesis and assembly via its interaction with the N protein.
Measles is an acute contagious disease characterized by high fever and a maculopapular rash (15). Measles virus (MV), the causative agent, is an enveloped virus classified as a member of the genus Morbillivirus in the family Paramyxoviridae. The virus has a nonsegmented negative-sense RNA genome, which contains six genes encoding single structural proteins, designated the nucleocapsid (N), phospho- (P), matrix (M), fusion (F), hemagglutinin (H), and large (L) proteins. The P gene encodes additional gene products, termed the V and C proteins, via an RNA editing process and an alternative translational initiation in a different reading frame, respectively (4, 9). The genome is encapsidated by the N protein and forms a nucleocapsid that exhibits helical symmetry. The amino-terminal region of the N protein (NCORE; amino acids [aa] 1 to 400) constitutes the core region of the helical nucleocapsid while the remaining carboxyl-terminal region (NTAIL; aa 401 to 525) is intrinsically disordered and located outside of the helical nucleocapsid core (36). A viral RNA-dependent RNA polymerase composed of the L and P proteins associates with the nucleocapsid, thereby forming the ribonucleoprotein (RNP) complex. The L protein possesses enzymatic activities that are required for nucleotide polymerization and the capping and polyadenylation of viral mRNAs while the P protein acts as an essential cofactor for the RNA-dependent RNA polymerase functions (15). The P protein directly interacts with NTAIL (2, 16) while the L protein indirectly associates with the nucleocapsid via its interaction with the P protein (10, 22). The RNP complex, but not the naked RNA genome, acts as a template for both transcription and replication. NTAIL has also been shown to interact with multiple host proteins, including the heat shock protein Hsp72, translation initiation factor eIF3-p40, interferon regulatory factor 3 and FcγRII (33, 48, 58, 62). The structural flexibility of the disordered NTAIL is probably responsible for its ability to interact with multiple partners.
The M protein plays a key role in virus assembly. Several lines of evidence indicate that the M protein associates with the inner surface of the plasma membrane (21, 46) as well as the cytoplasmic tails of the H and F glycoproteins (7, 8, 50, 52). The M protein has also been shown to interact with the RNP complex (20, 51) although its binding partner remains unknown. The M protein modulates viral RNA synthesis (51), and a small interfering RNA against M mRNA was reported to increase MV transcription levels in the small interfering RNA-treated cells (43).
In the present study, we analyzed the interactions among MV proteins using a yeast two-hybrid system and coimmunoprecipitation in mammalian cells. In addition to the known interactions, we detected a hitherto unreported interaction between the N and M proteins. Two leucine residues located at the carboxyl terminus of the N protein were found to be critical for this interaction. Further analyses using minigenome assays, plasmid transfection, and recombinant viruses indicated that the M protein regulates viral RNA synthesis and assembly via its interaction with the N protein.
MATERIALS AND METHODS
Cells and viruses.
Vero, CV-1, and HeLa cells constitutively expressing human signaling lymphocyte activation molecule (hSLAM) (Vero/hSLAM [41], CV-1/hSLAM [55], and HeLa/hSLAM [55] cells, respectively) were maintained in Dulbecco's modified Eagle's medium (DMEM; ICN Biomedicals, Aurora, OH) supplemented with 7.5% fetal bovine serum (FBS) and 500 μg/ml Geneticin (G418; Nacalai Tesque, Tokyo, Japan). 293T cells were maintained in DMEM supplemented with 7.5% FBS. B95a cells (31) and VV5-4 cells (a derivative of CHO cells) (1) were maintained in RPMI 1640 medium (ICN Biomedicals) supplemented with 7.5% FBS. Recombinant MVs based on the virulent IC-B strain were generated from cloned cDNAs as described previously (53, 57).
Reagents and antibodies.
A fusion-blocking peptide, Z-D-Phe-Phe-Gly (45), was purchased from the Peptide Institute (Osaka, Japan). A rabbit polyclonal antibody raised against the MV N protein was purchased from Novus Biologicals (Littleton, CO). Rabbit polyclonal and mouse monoclonal (clone E388) antibodies raised against the MV M protein were kindly provided by T. Kohama and T. A. Sato, respectively. The serum of a patient with subacute sclerosing panencephalitis (SSPE) (61) was kindly provided by M. B. A. Oldstone.
Construction of plasmids.
p(+)MV323-EGFP and p(+)MV323-Luci were derived from p(+)MV323, which encodes the full-length antigenomic cDNA of the virulent IC-B strain of MV (57). They encode an additional transcriptional unit of enhanced green fluorescence protein (EGFP) and Renilla luciferase, respectively (17, 56). p(+)MV-NΔ3-EGFP and p(+)MV-NΔ3-Luci were generated by deleting nine nucleotides encoding 3 aa at the carboxyl terminus of the N protein from p(+)MV323-EGFP and p(+)MV323-Luci, respectively. Three nucleotides (TAG) were inserted into the deletion site to maintain the genome length in multiples of six nucleotides. The recombinant MVs generated from p(+)MV323-EGFP, p(+)MV323-Luci, p(+)MV-NΔ3-EGFP, and p(+)MV-NΔ3-Luci were designated IC323-EGFP, IC323-Luci, IC-NΔ3-EGFP, and IC-NΔ3-Luci, respectively.
DNA fragments encoding mutant N proteins with carboxyl-terminal truncations of 3 and 15 aa (NΔ3 and NΔ15, respectively) were cloned into the eukaryotic cell expression vector pCA7, a derivative of pCAGGS (40), thereby generating pCA7-IC-NΔ3 and pCA7-IC-NΔ15, respectively. The expression plasmids pCA7-IC-C and pCA7-IC-M encoding the MV C and M proteins, respectively, were reported previously (39, 52). In yeast two-hybrid assays, a bait vector, pDBLeu (Invitrogen Life Technologies, Carlsbad, CA), and a prey vector, pPC86 (Invitrogen Life Technologies), were used. DNA fragments encoding the entire reading frames of the N, P, V, C, and M proteins of the MV IC-B strain were cloned into the pDBLeu bait vector. The cytoplasmic domains of the F protein (aa 518 to 550 at the carboxyl terminus [F518-550]) and H protein (aa 1 to 34 at the amino-terminus [H1-34]) of the IC-B strain were also cloned into pDBLeu. The L proteins of morbilliviruses have three conserved domains (D1, D2, and D3), which are linked by two variable hinges (H1 and H2) (13). DNA fragments encoding 1,707 aa of the amino terminus of the L protein (residues 1 to 1707 [L1-1707]; the region containing D1 and D2) and 476 aa at the carboxyl terminus of the L protein (residues 1708 to 2183 [L1708-2183]; the region containing D3) were cloned into the pDBLeu bait vector. DNA fragments encoding the N, P, V, C, and M proteins were also cloned into the pPC86 prey vector. DNA fragments encoding mutant N proteins with truncations of 3, 6, 9, 12, and 15 aa (NΔ3, NΔ6, NΔ9, NΔ12, and NΔ15, respectively) at the carboxyl-terminal end were cloned into both the pDBLeu bait and pPC86 prey vectors.
The plasmids used for minigenome assays of MV (32), Sendai virus (SeV) (28), and parainfluenza virus type 5 (PIV5) (18, 34) were kindly provided by K. Komase, A. Kato, and B. He, respectively. Expression plasmids pCA7-SeV-C and pCA7-PIV5-V for the SeV C and PIV5 V proteins, respectively, were generated by inserting DNA fragments encoding these proteins into the pCA7 vector. The cDNAs encoding these proteins were provided by A. Kato and B. He, respectively.
Plaque assay.
Monolayers of Vero/hSLAM cells in 12-well cluster plates were infected with serially diluted virus samples. After 1 h of incubation at 37°C, the virus samples were removed, and the cells were overlaid with DMEM containing 7.5% FBS and 1% methylcellulose. At 5 days postinfection (p.i.), the cells were washed with phosphate-buffered saline (PBS). After cells were stained with 0.01% neutral red, the numbers of PFU were counted. Monolayers of CV-1/hSLAM cells in 12-well cluster plates were cultured with 100 μl of virus samples containing 50 PFU of MV for 1 h at 37°C, cultured with DMEM containing 7.5% FBS and 1% methylcellulose, and stained with 0.01% neutral red at 5 days p.i., as described above. After high-resolution digital images of the plaques were obtained, the sizes of the plaques were measured.
Minigenome assay.
Reporter gene expression from viral minigenomes encoding the luciferase gene was analyzed in VV5-4 cells (1). Monolayers of VV5-4 cells cultured in 24-well cluster plates were infected with vTF7-3 (a recombinant vaccinia virus expressing the T7 RNA polymerase) (14), at a multiplicity of infection (MOI) of 0.5 and then transfected with minigenome plasmids (p18MGFLuc01, pHvLuciRT4, and pMG-Rluc for MV, SeV and PIV5, respectively) together with appropriate support plasmids, which were described previously (18, 28, 32, 34, 39). At 2 or 3 days posttransfection, the enzymatic activities were measured by a luciferase assay system (Promega, Madison, WI) or Renilla luciferase assay system (Promega) and a luminometer (Mithras LB 940; Berthold Technologies, Bad Wildbad, Germany).
Yeast two-hybrid assay.
MaV203 yeast cells (Invitrogen Life Technologies) transformed with two plasmids comprising a bait plasmid (pDBLeu) and a prey plasmid (pPC86) were grown on synthetic complete (SC) medium plates lacking leucine and tryptophan (SC/−Leu/−Trp). After 2 days, yeast cells forming colonies on the plates were further selected on four different kinds of plates according to the manufacturer's instructions: SC/−Leu/−Trp plates lacking uracil (SC/−Leu/−Trp/−Ura), SC/−Leu/−Trp plates supplemented with 0.2% 5-fluoroorotic acid (SC/−Leu/−Trp/5FOA), SC/−Leu/−Trp plates lacking histidine and containing 10 mM of 3-aminotriazole (SC/−Leu/−Trp/−His/3AT), and YPAD medium (a rich medium for routine growth of yeast) plates supplemented with 5-bromo-5-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (YPAD/X-Gal). Retention of both the bait and prey vectors by yeast cells was confirmed by their growth on SC/−Leu/−Trp plates. Interactions between proteins fused to the GAL4 activation domain (AD) and the GAL4 DNA binding domain (BD) encoded in the prey and bait vectors, respectively, resulted in activation of transcription of the HIS3, URA3, and lacZ genes. When transcription of these genes was activated strongly, yeast cells produced blue colonies on the YPAD/X-Gal plates and formed colonies on the SC/−Leu/−Trp/−Ura and SC/−Leu/−Trp/−His/3AT plates but not on the SC/−Leu/−Trp/5FOA plates (yeast cells were killed by the toxicity of 5-fluorouracil converted from 5FOA). If the transcription of these genes was only weakly activated, yeast cells formed colonies on the SC/−Leu/−Trp/−His/3AT and SC/−Leu/−Trp/5FOA plates. However, they produced white colonies on YPAD/X-Gal plates and failed to form colonies on SC/−Leu/−Trp/−Ura plates.
Immunoprecipitation and Western blot analyses.
Subconfluent monolayers of 293T cells in six-well cluster plates were transfected with pCA7-IC-N (2 μg) or pCA7-IC-NΔ3 (2 μg) together with pCA7-IC-M (2 μg) (pCA7-IC-N has been referred to as pCAG-T7-IC-N in previous papers [38, 39]). At 48 h posttransfection, the cells were washed with PBS and lysed in 1 ml of coimmunoprecipitation buffer (10 mM HEPES, pH 7.4, 50 mM sodium pyrophosphate, 50 mM NaF, 50 mM NaCl, 5 mM EDTA, 5 mM EGTA, 100 μM sodium vanadate, 1% Triton X-100) (42) containing protease inhibitors (Sigma-Aldrich, St. Louis, MO). The lysates were centrifuged at 20,630 × g for 90 min at 4°C. A small amount (30 μl) of each supernatant was mixed with sodium dodecyl sulfate (SDS) loading buffer (50 mM Tris, pH 6.8, 100 mM dithiothreitol [DTT], 2% SDS, 0.1% bromophenol blue, 10% glycerol) and boiled for 5 min. The rest of the supernatant was incubated for 90 min at 4°C with a 1:1 mixture of protein A-Sepharose and protein G-Sepharose (GE Healthcare AB, Uppsala, Sweden), which had been pretreated with an anti-MV M protein monoclonal antibody (E388) for 90 min at 4°C. Complexes with the Sepharose were obtained by centrifugation and sequentially washed with buffer 1 (100 mM Tris, pH 7.6, 500 mM LiCl, 0.1% Triton X-100, 1 mM DTT) and buffer 2 (20 mM HEPES, pH 7.2, 2 mM EGTA, 10 mM MgCl2, 0.1% Triton X-100, 1 mM DTT) (42). The polypeptides in the precipitated complexes were fractionated by SDS-polyacrylamide gel electrophoresis (PAGE) using 10% polyacrylamide gels and electroblotted onto polyvinylidene difluoride membranes (Hybond-P; Amersham Biosciences, Piscataway, NJ). The membranes were incubated with the SSPE patient serum and an anti-M antibody, followed by incubation with horseradish peroxidase-conjugated anti-human immunoglobulin G (IgG) or anti-rabbit IgG (Invitrogen Life Technologies) for detection of the MV N and M proteins, respectively. The ECL plus reagent (Amersham Biosciences) was used to elicit chemiluminescent signals, and the signals on the membranes were detected and visualized using a VersaDoc 3000 imager (Bio-Rad, Hercules, CA).
Indirect immunofluorescence assay.
HeLa/hSLAM cells were seeded on coverslips in six-well cluster plates and infected with IC323-Luci or IC-NΔ3-Luci in the presence of the fusion-blocking peptide. CV-1/hSLAM cells were seeded on coverslips in six-well cluster plates and transfected with pCA7-IC-N or pCA7-IC-NΔ3, together with pCA7-IC-PΔC or pCA7-IC-PΔC plus pCA7-IC-M. VV5-4 cells seeded on coverslips in six-well cluster plates were infected with vTF7-3 (14) and transfected with an MV minigenome plasmid (p18MGFLuc01) and support plasmids (pCA7-IC-N or pCA7-IC-NΔ3, pCA7-IC-PΔC, and pGEMCR-9301B-L) with or without pCA7-IC-M. At 24 h p.i. or 48 h posttransfection, the cells were fixed and permeabilized with PBS containing 2.5% formaldehyde and 0.5% Triton X-100. The cells were then washed with PBS and incubated with a rabbit polyclonal antibody against the MV N protein (Novus Biologicals) and a mouse monoclonal antibody against the MV M protein (E388), followed by incubation with Alexa Fluor 488-conjugated donkey anti-rabbit IgG(H+L) and Alexa Fluor 594-conjugated donkey anti-mouse IgG(H+L) antibodies (Molecular Probes, Eugene, OR). The stained cells were observed using a confocal microscope (Radiance 2100; Bio-Rad).
Reverse transcription-quantitative PCR.
Subconfluent monolayers of CV-1/hSLAM and Vero/hSLAM cells were infected with IC323-EGFP or IC-NΔ3-EGFP at an MOI of 0.001 or left uninfected in the presence of the fusion-blocking peptide. At 24 h p.i., total RNA was extracted from the cells using the TRIzol reagent (Invitrogen Life Technologies). The total RNA extracts were treated with RQ1 DNase (Promega) and then reverse transcribed into cDNAs using SuperScript III reverse transcriptase (Invitrogen Life Technologies) with an oligo(dT) primer. The amounts of cDNAs for the MV mRNAs were quantified using SYBR Premix Ex Taq II (TaKaRa Bio, Shiga, Japan) and a LightCycler instrument (Roche Diagnostics, Indianapolis, IN) as described previously (54). The levels of β-actin mRNA were quantified as an internal control, as reported previously (39).
Growth kinetics.
Monolayers of Vero/hSLAM or CV-1/hSLAM cells on six-well cluster plates were infected with IC323-EGFP or IC-NΔ3-EGFP at an MOI of 0.001 and cultured in 2 ml of medium. At various time intervals, the medium was harvested and centrifuged at 400 × g for 5 min at 4°C. The viral titer of the supernatant (cell-free titer) was determined by a plaque assay on Vero/hSLAM cells. After the medium was removed from the infected cells, 2 ml of fresh medium was added to each well. The infected cells were scraped into the medium, and the viral titer (cell-associated titer) was determined by a plaque assay.
RESULTS
The MV M protein interacts with the N protein.
We attempted to analyze the interactions among MV proteins comprehensively using a yeast two-hybrid system (Table 1). DNA fragments encoding the full-length N, P, V, C, and M proteins, cytoplasmic domains of the F and H proteins (F518-550 and H1-34, respectively), and the amino- and carboxyl-terminal regions of the L protein (L1-1707 and L1708-2183, respectively) were individually inserted in-frame downstream of the GAL4 DNA BD in the bait vector (pDBLeu) or the GAL4 AD in the prey vector (pPC86). The empty pDBLeu and pPC86 vectors were used as controls. MaV203 yeast cells grew on SC/−Leu/−Trp/−His/3AT plates when they were transfected with a bait vector encoding either the P or V protein (pDBLeu-P or -V) together with the empty pPC86 prey vector. A similar finding has been reported previously (11). Therefore, these bait vectors could not be used for further analyses. Experiments using pDBLeu-N as the bait indicated that, in addition to the known N-V (59) and N-N (2, 25) protein interactions, there was a weak, but significant, interaction between the N and M proteins (Table 1). Furthermore, when pDBLeu-N and pPC86-P were used, transcription of all three reporter genes, HIS3, lacZ, and URA3, was strongly activated (Table 1). This was consistent with the previously reported strong interaction between the N and P proteins (2, 11, 16). Similarly, data using pDBLeu-M and pPC86-N or -M confirmed the weak, but significant, N-M protein interaction and indicated the self-association of the M protein (47), which is similar to the case for the SeV M protein (19). Analysis using pPC86-P together with pDBLeu-L1-1707 or -L1708-2183 indicated that the P protein interacts with both the amino-terminal and carboxyl-terminal regions of the L protein. The interaction with the amino-terminal region was previously demonstrated by analyzing L protein mutants whose amino acid residues were progressively deleted from the carboxyl-terminal end (10, 22). On the other hand, the present study is the first to demonstrate that the P protein interacts with the carboxyl-terminal region (aa 1708 to 2183) of the L protein.
TABLE 1.
Interactions between MV proteins in yeast
| pDBLeu-encoded protein (DNA BD) | pPC86-encoded protein (AD) interactiona
|
|||||
|---|---|---|---|---|---|---|
| Empty | N | P | V | C | M | |
| Empty | − | − | − | − | − | − |
| N | − | + | +++ | + | − | + |
| P | + | ND | ND | ND | ND | ND |
| V | + | ND | ND | ND | ND | ND |
| C | − | − | − | − | − | − |
| M | − | + | − | − | − | + |
| F518-550 | − | − | − | − | − | − |
| H1-34 | − | − | − | − | − | − |
| L1-1707 | − | − | + | − | − | − |
| L1708-2183 | − | − | + | − | − | − |
+, weak interaction; +++, strong interaction; −, no interaction; ND, not done.
The carboxyl-terminal residues of the N protein are essential for the N-M protein interaction.
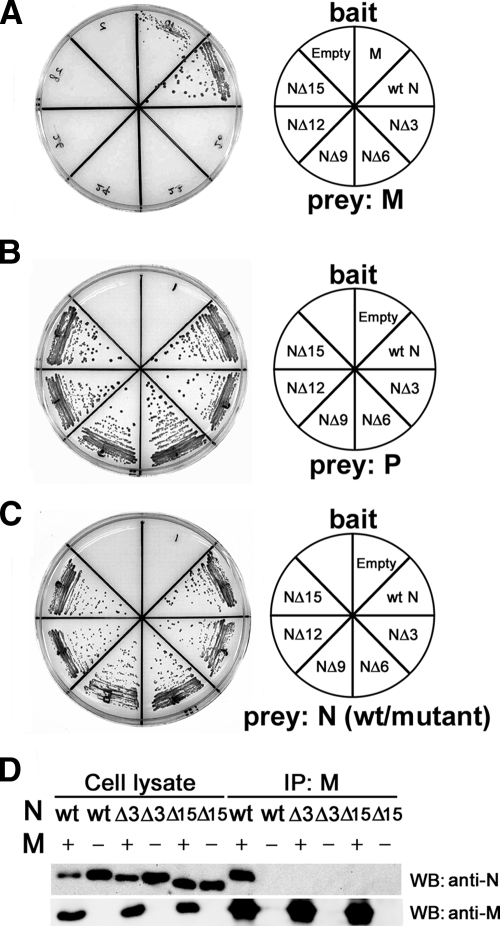
The N protein can be divided into two regions, NCORE and NTAIL (36). We predicted that the M protein may interact with NTAIL, like the P protein and multiple host factors. pDBLeu bait vectors expressing the mutant N proteins NΔ3, NΔ6, NΔ9, NΔ12, and NΔ15 with stepwise deletions from the carboxyl-terminal end were generated. The interactions of these mutant N proteins with the M protein were analyzed in the yeast two-hybrid system. All five mutants were unable to interact with the M protein (Fig. 1A). On the other hand, all of the mutants retained the abilities to interact with the P protein and to self-associate (Fig. 1B and C). Since even the NΔ3 protein was unable to interact with the M protein, the 3 aa residues at the carboxyl-terminal end of the N protein (two leucines and an aspartic acid, in the sequence order of LLD) were further examined. A single-amino acid deletion (NΔ1 [LL-]) did not affect the N-M protein interaction, whereas a 2-aa deletion (NΔ2 [L-]) abolished the interaction (Table 2). Substitution of an alanine for the individual leucine residues at positions 523 and 524 (N-ALD and N-LAD, respectively; substitutions are underlined) abolished the N-M protein interaction, while substitution of a serine or glutamic acid for the aspartic acid at position 525 (N-LLS and N-LLE, respectively) had little effect on the interaction (Table 2). These data indicate that the leucine residues at positions 523 and 524 in NTAIL are essential for the N-M protein interaction.
FIG. 1.
The carboxyl-terminal end of the N protein is essential for the N-M protein interaction. (A to C) Yeast two-hybrid assay. MaV203 yeast cells were transfected with a bait vector expressing the GAL4 DNA BD fused with the M, wt N, NΔ3, NΔ6, NΔ9, NΔ12, or NΔ15 protein or the GAL4 DNA BD alone (Empty) (the bait used is shown in the explanatory figure on the right) together with a prey vector expressing the GAL4 AD fused with the M protein (A), P protein (B), or wt N protein or corresponding N protein deletion mutant (C). Self-association of the wt N protein and its deletion mutants was examined. Reporter gene (HIS3) expression, which results in the growth of MaV203 cells on SC/−Leu/−Trp/−His/3AT plates, was analyzed. (D) Coimmunoprecipitation assay. 293T cells were transfected with a plasmid expressing the wt N, NΔ3, or NΔ15 protein with (+) or without (−) a plasmid expressing the M protein. At 48 h posttransfection, small amounts of cell lysates obtained from the transfected cells (Cell lysate) were subjected to SDS-PAGE and Western blotting (WB) for detection of the N and M proteins using a human polyclonal antibody against the N protein and a rabbit polyclonal antibody against the M protein. The remaining cell lysates were incubated with protein A/G-conjugated Sepharose beads, which had been pretreated with a mouse monoclonal antibody against the M protein (E388). The immune complexes were obtained by centrifugation. The precipitated immune complexes were subjected to SDS-PAGE and Western blotting for detection of the N and M proteins. IP, immunoprecipitation.
TABLE 2.
Effect of the C-terminal mutations of the N protein on interactions in yeast
| pDBLeu-encoded protein (DNA BD)a | pPC86-encoded protein (AD) interactionb
|
||
|---|---|---|---|
| Empty | M | P | |
| Empty | − | − | − |
| wt N [LLD] | − | + | +++ |
| NΔ1 [LL-] | − | + | +++ |
| NΔ2 [L-] | − | − | +++ |
| NΔ3 [—] | − | − | +++ |
| N-ALD | − | − | +++ |
| N-LAD | − | − | +++ |
| N-LLS | − | + | +++ |
| N-LLE | − | + | +++ |
Substituted nucleotides are underlined. The carboxyl-terminal deletions of the N mutant proteins are indicated by dashes.
+, weak interaction; +++, strong interaction; −, no interaction.
The N-M protein interaction was also assessed in mammalian cells. 293T cells were individually transfected with pCA7 vectors encoding the wild-type (wt) N, NΔ3, and NΔ15 proteins with or without a pCA7 plasmid encoding the M protein. When the whole-cell lysates were subjected to SDS-PAGE and Western blotting, all the proteins were detected at high levels (Fig. 1D). In another experiment, the cell lysates were incubated with protein A/G conjugated to Sepharose beads, which had been pretreated with a monoclonal antibody against the M protein, and immune complexes were retrieved by centrifugation. When the immune complexes were subjected to SDS-PAGE and Western blotting, the wt N protein, but not the NΔ3 and NΔ15 proteins, was detected in complexes that contained the M protein (Fig. 1D). These data indicate that the N protein also interacts with the M protein in mammalian cells and that its carboxyl-terminal residues are essential for the interaction.
The MV M protein inhibits RNA synthesis in the MV minigenome system via its interaction with the N protein.
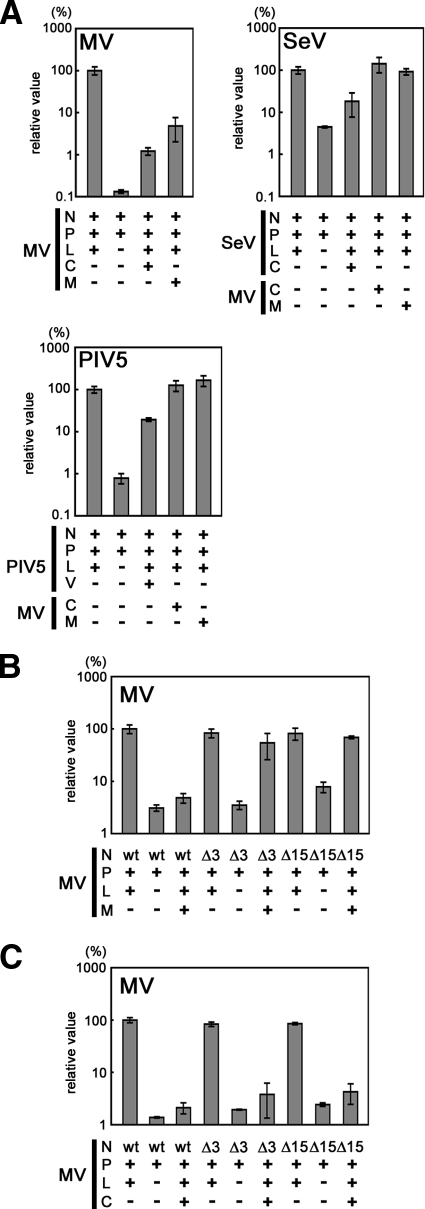
Next, we examined the activity of the M protein in viral RNA synthesis. Consistent with previous reports that the M protein inhibits MV RNA synthesis (51), our results confirmed that coexpression of the M protein reduced the level of the reporter (luciferase) gene expression from an MV minigenome by ∼95% (Fig. 2A). The C protein also inhibited minigenome RNA synthesis, which is consistent with previous reports (3, 39, 44). In contrast, the MV M protein hardly affected reporter gene expression levels in minigenome systems of other paramyxoviruses (SeV and PIV5) while the SeV C protein and PIV5 V protein inhibited reporter gene expression from their own minigenomes (Fig. 2A), as reported previously (26, 27, 34). These data indicate that the ability of the MV M protein to inhibit minigenome expression is specific for MV RNA.
FIG. 2.
The interaction with the carboxyl-terminal end of the N protein is required for the M protein to inhibit MV minigenome gene expression. (A) The MV M and C proteins specifically inhibit MV minigenome gene expression. VV5-4 cells were infected with vTF7-3 at an MOI of 0.5 and then transfected with a minigenome plasmid (p18MGFLuc01 [32], pHvLuciRT4 [28], and pMG-Rluc [34] for MV, SeV, and PIV5, respectively) and three support plasmids expressing the N, P, and L proteins of each virus. +, expression of the protein; −, no expression of the protein. The L protein expression plasmid was omitted from the transfection mixtures for negative control cells (L−). The MV M and C protein expression plasmids (pCA7-IC-M and pCA7-IC-C, respectively) were included in the transfection mixtures for some cells (MV M+ and MV C+, respectively). At 72 h posttransfection, luciferase activity was measured. The luciferase activity in control cells transfected only with a minigenome plasmid and three support plasmids (N, P, and L) for each virus was set to 100%. Data represent the means ± standard deviation of triplicate samples. (B) Effects of carboxyl-terminal deletion of the N protein on the activity of the M protein to inhibit MV minigenome gene expression. VV5-4 cells infected with vTF7-3 were transfected with the MV minigenome plasmid (p18MGFLuc01) and three support plasmids (pCA7-IC-N, pCA7-IC-PΔC, and pGEMCR-9301B-L). As replacements for pCA7-IC-N (wt), pCA7-IC-NΔ3 (Δ3) and pCA7-IC-NΔ15 (Δ15) were also used as support plasmids. pGEMCR-9301B-L was omitted from the transfection mixture for negative control cells (L−). pCA7-IC-M was added to the transfection mixture for some cells (M+). At 48 h posttransfection, luciferase activity was measured. The luciferase activity in cells transfected with p18MGFLuc01, pCA7-IC-N, pCA7-IC-PΔC, and pGEMCR-9301B-L (N wt, P+, L+, and M−) was set to 100%. Data represent the means ± standard deviations of triplicate samples. (C) Effects of carboxyl-terminal deletion of the N protein on the activity of the C protein to inhibit MV minigenome gene expression. Experiments were performed as described for panel B, except that pCA7-IC-C was used instead of pCA7-IC-M.
We hypothesized that the N-M protein interaction may be involved in this specific inhibitory activity of the M protein. The NΔ3 and NΔ15 proteins induced reporter gene expression from the MV minigenome as efficiently as the wt N protein when they were expressed together with the P and L proteins (Fig. 2B). However, coexpression of the M protein no longer inhibited reporter gene expression from the RNP complexes formed with the NΔ3 and NΔ15 proteins (Fig. 2B). These results indicate that the M protein regulates MV RNA synthesis by interacting with the carboxyl-terminal residues of the N protein. In contrast, the MV C protein still efficiently inhibited reporter gene expression from the RNP complexes formed with the NΔ3 and NΔ15 proteins (Fig. 2C), indicating that the M and C proteins regulate viral RNA synthesis by different means.
The N-M protein interaction causes the redistribution of the N protein within the cell.
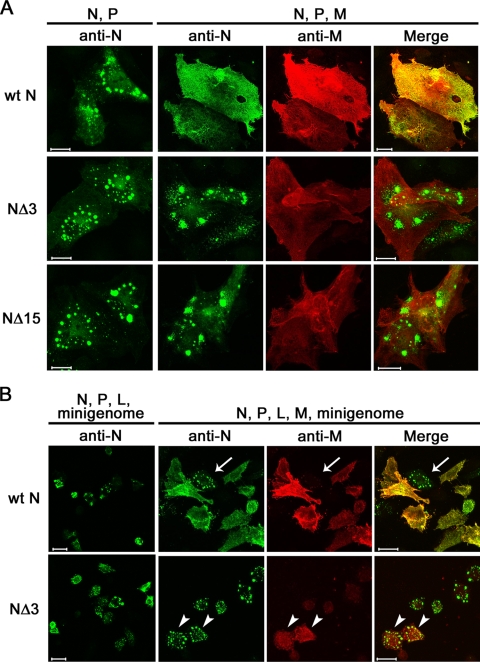
Many lines of evidence have indicated that paramyxovirus M proteins play major roles in virus assembly (49). The MV M protein, but not the N protein, has an intrinsic ability to associate with the lipid membrane or lipid raft (37, 46, 60). It has been proposed that the M protein recruits the nucleocapsid to the plasma membrane (47) or lipid raft for assembly (60). To assess their intracellular distributions, the wt N, NΔ3, and NΔ15 proteins were expressed in CV1/hSLAM cells, together with the P protein. The P protein is required for cytoplasmic retention of the N protein (23). The wt N, NΔ3, and NΔ15 proteins were all mainly localized in the cytoplasm, forming small dots in the perinuclear area (Fig. 3A). When the M protein was coexpressed, the wt N protein was colocalized with the M protein and became redistributed. In contrast, the intracellular distributions of the NΔ3 and NΔ15 proteins were hardly affected by coexpression of the M protein (Fig. 3A).
FIG. 3.
Intracellular distributions of the N and M proteins analyzed by indirect immunofluorescence and confocal microscopy. (A) CV-1/hSLAM cells were transfected with pCA7-IC-N (wt N), pCA7-IC-NΔ3 (NΔ3), or pCA7-IC-NΔ15 (NΔ15) together with pCA7-IC-PΔC (N, P). Cells were also transfected with pCA7-IC-N (wt N), pCA7-IC-NΔ3 (NΔ3), or pCA7-IC-NΔ15 (NΔ15) together with pCA7-IC-PΔC and pCA7-IC-M (N, P, M). At 48 h posttransfection, the intracellular distributions of the N and M proteins were analyzed by indirect immunofluorescence and confocal microscopy. The primary antibodies used were a rabbit polyclonal antibody against the N protein and a mouse monoclonal antibody against the M protein. The secondary antibodies were Alexa Fluor 488-conjugated anti-rabbit and Alexa Fluor 594-conjugated anti-mouse antibodies, respectively. Bar, 20 μm. (B) VV5-4 cells infected with vTF7-3 were transfected with the MV minigenome plasmid (p18MGFLuc01) together with three support plasmids expressing the N, P, and L proteins (N, P, L, minigenome). Either pCA7-IC-N (wt N) or pCA7-IC-NΔ3 (NΔ3) was used as the N protein expression plasmid. Cells were also transfected with pCA7-IC-M together with the minigenome plasmid and three support plasmids (N, P, L, M, minigenome). At 48 h posttransfection, the intracellular distributions of the N and M proteins were analyzed by indirect immunofluorescence and confocal microscopy as described for panel A. Arrows and arrowheads indicate cells expressing M protein at low and high levels, respectively. Bar, 20 μm.
Similar experiments were performed in VV5-4 cells in which the MV minigenome was replicated and transcribed by the N, P, and L proteins, as shown in Fig. 2B. Both the wt N and NΔ3 proteins were mostly localized in the perinuclear area, forming small dots (Fig. 3B). When the M protein was coexpressed, the wt N protein became redistributed (Fig. 3B). Notably, in cells expressing the M protein at low levels, the wt N protein remained as small dots in the perinuclear area (Fig. 3B). In contrast, the intracellular distribution of the NΔ3 protein remained unaltered even in cells highly expressing the M protein (Fig. 3B). Therefore, the M protein neither inhibits viral RNA synthesis nor causes redistribution of the N protein when it is unable to interact with the N protein (e.g., NΔ3).
The N-M protein interaction is required for efficient production of infectious MV particles.
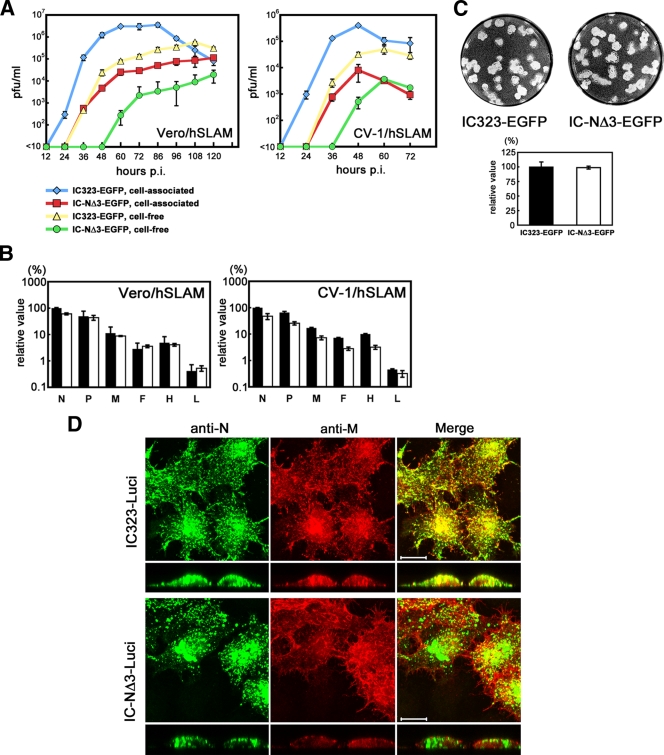
The relevance of the N-M protein interaction was analyzed in the context of viral particles using reverse genetics. A recombinant MV possessing the NΔ3 protein (IC-NΔ3-EGFP) replicated much less efficiently than the parental virus possessing the wt N protein (IC323-EGFP) in Vero/hSLAM and CV1/hSLAM cells (Fig. 4A), indicating that the N-M protein interaction is indeed important for efficient virus production. The MV transcript levels examined in the presence of the fusion-blocking peptide were comparable between IC323-EGFP- and IC-NΔ3-EGFP-infected Vero/hSLAM cells, whereas the transcript levels in IC-NΔ3-EGFP-infected CV-1/hSLAM cells tended to be severalfold lower than those in IC323-EGFP-infected cells (Fig. 4B). The difference in the transcript levels between the two viruses did not appear to account for the large difference in viral growth. Since only very low titers of IC-NΔ3-EGFP were obtained, we could not reliably compare the viral protein production levels between IC323-EGFP- and IC-NΔ3-EGFP-infected cells using Western blot analysis. Instead, we compared the sizes of the plaques produced by the two viruses. In both CV-1/hSLAM and Vero/hSLAM cells, IC-NΔ3-EGFP produced large plaques similar to those of IC323-EGFP (Fig. 4C and data not shown). These results suggest that the multiplication of IC-NΔ3-EGFP is adversely affected at a step later than viral protein production, presumably virus assembly, by the impaired interaction between the N and M proteins.
FIG. 4.
Analyses of recombinant MVs possessing carboxyl-terminal deletion of the N protein. (A) Growth kinetics. Vero/hSLAM and CV-1/hSLAM cells were infected with recombinant MVs (IC323-EGFP and IC-NΔ3-EGFP) at an MOI of 0.001. The infectious titers in culture medium (cell free) and cells (cell associated) were determined at various time points. (B) Quantification of viral mRNAs. Vero/hSLAM and CV-1/hSLAM cells were infected with IC323-EGFP (black bar) or IC-NΔ3-EGFP (white bar) at an MOI of 0.001 in the presence of a fusion-blocking peptide. At 24 h p.i., the levels of the N, P, M, F, H, and L mRNAs of MV in the infected cells were analyzed by reverse transcription-quantitative PCR. Data were normalized by the levels of β-actin mRNA and represent the means ± standard deviations of triplicate samples. The N mRNA level in IC323-EGFP-infected cells was set to 100%. (C) Plaque assays. Monolayers of CV-1/hSLAM cells on 12-well cluster plates were infected with 50 PFU of IC323-EGFP or IC-NΔ3-EGFP and overlaid with DMEM containing 7.5% FBS and 1% methylcellulose. At 5 days p.i., the cells were stained with 0.01% neutral red, and the sizes of the plaques were measured. The mean diameters ± standard deviations are shown in the bar graph. (D) HeLa/hSLAM cells were infected with IC323-Luci (upper panels) or IC-NΔ3-Luci (lower panels). At 24 h p.i., the intracellular distributions of the N and M proteins were analyzed by indirect immunofluorescence and confocal microscopy as described in the legend of Fig. 3A. Longitudinal sections are also shown at the bottom. Bar, 10 μm.
The intracellular distributions of the N and M proteins were examined in cells infected with recombinant MVs possessing the wt N or NΔ3 protein (Fig. 4D). Since a green fluorescent Alexa Fluor 488-conjugated secondary antibody was used to detect the N protein, IC323-Luci and IC-NΔ3-Luci, rather than IC323-EGFP and IC-NΔ3-EGFP, respectively, were used for this experiment. In IC323-Luci-infected cells, the wt N protein was mostly colocalized with the M protein at the cell surface or as small dots in the perinuclear area (as observed in transfected cells expressing low levels of the M protein). Although the NΔ3 protein was also localized in small dots in infected cells, its distribution was totally unrelated to that of the M protein.
DISCUSSION
In the present study, we have demonstrated that the M protein regulates MV RNA synthesis and assembly via its direct interaction with the N protein. The N protein is biologically divided into two regions. NCORE has a rigid structure and contains all of the regions necessary for self-assembly and binding to viral genomic and antigenomic RNA (2, 25, 35). On the other hand, the intrinsically disordered NTAIL protrudes from the viral nucleocapsid (25, 29, 36) and interacts with many viral and cellular proteins (2, 16, 33, 62). The present study indicates that NTAIL also binds to the M protein. NTAIL contains an α-helical molecular recognition element (aa 488 to 499), which interacts with three α-helical strands of the carboxyl-terminal module (XD; aa 459 to 507) of the P protein (24, 30). Surface plasmon resonance analyses suggested that the BOX3 region (aa 517 to 525) of the N protein also interacts with the P protein (6). However, another study using nuclear magnetic resonance reported that the BOX3 region does not directly contribute to the interaction with the P protein (5). Our present study supports the findings of the latter study since deletion of up to 15 aa residues from the carboxyl-terminal end of the N protein did not affect its interaction with the P protein, as examined by the yeast two-hybrid system. Then, the question arises as to whether the carboxyl-terminal region of the N protein participates in viral RNA synthesis. We performed minigenome assays and found that the levels of viral RNA synthesis did not change when a mutant N protein lacking the carboxyl-terminal 15 aa residues was used for the assay. This finding is consistent with a previous report that truncation of the carboxyl-terminal 24 aa residues of the MV Edmonston N protein does not diminish transcription and replication of the minigenome (62).
Using the minigenome assay, we further found that the ability of the M protein to regulate viral RNA synthesis was mediated by its interaction with the N protein. The C protein of MV inhibits viral RNA synthesis (3, 39, 44). However, in our yeast two-hybrid system, we found that the C protein did not interact with any of the MV proteins including the N protein. Furthermore, the C protein still exerted its inhibitory activity when the mutant N proteins that were unable to interact with the M protein were used for the minigenome assay. Therefore, the C protein is likely to regulate viral RNA synthesis in a different manner from the M protein. The host protein Hsp72 has been shown to stimulate MV minigenome reporter gene expression by interacting with the carboxyl-terminal BOX3 region of the N protein (62). It is possible that the M protein exerts its inhibitory activity by modulating the interaction between the N protein and Hsp72.
Although MV assembly has been studied for many years, its precise mechanisms, including the viral and host proteins involved, are largely unknown. In the Paramyxoviridae, the M protein underlies the viral envelope and forms electron-dense layers in electron micrographs (15). The M protein has been shown to interact with the viral RNP complex (20, 51) as well as the cytoplasmic domains of viral envelope glycoproteins (7, 8, 50, 52). Although the recombinant MV lacking the M protein (MV-ΔM) was successfully rescued, virus titers were reduced ∼250-fold compared with the parental virus (7), as observed for IC-NΔ3-EGFP (possessing a mutant N protein that was incapable of interacting with the M protein) in the present study (2-log reduction). Furthermore, the inability of defective M proteins to associate with the viral nucleocapsid is thought to be responsible for the low level of infectious viral particle production in patients with SSPE (20). Consequently, the M protein and its interaction with the RNP complex are believed to play important roles in MV assembly. The present study has clearly demonstrated that the N protein is the RNP complex component that interacts with the M protein. A similar finding has been obtained for another paramyxovirus, human PIV1 (hPIV1). When cells were infected with SeV and transfected with the nucleoprotein (NP) cDNA of hPIV1, nucleocapsids composed of a mixture of NP molecules as well as those composed of NPs solely from SeV or solely from hPIV1 were detected in the cytoplasm (12). However, most of the NPs in the nucleocapsids of the progeny SeV virions were found to be from SeV. Coexpression of the hPIV1 M protein greatly increased the incorporation of the nucleocapsids containing hPIV1 NP into SeV virions. Analysis using SeV-hPIV1 chimera NP cDNAs indicated that the hPIV1 M protein caused specific incorporation of the nucleocapsid containing hPIV1 NP into SeV virions by interacting with the carboxyl-terminal domain of hPIV1 NP (aa position 420 to 466) (12).
Interestingly, previous studies have shown that MV-ΔM and viruses bearing the H and F proteins with shortened cytoplasmic tails are more efficient in inducing cell-cell fusion than the parental MV (7, 8). In contrast, IC-NΔ3-EGFP and IC323-EGFP produced similar sizes of plaques. The observations suggest that the interaction between the M and envelope proteins, but not that between the M and N proteins, critically affects MV-induced membrane fusion.
The present study has also shed new light on the mechanism by which the M protein promotes MV particle production. The N and M proteins were expressed in cultured cells by transfection with expression plasmids or by infection with recombinant viruses, and their intracellular distributions were investigated by indirect immunofluorescence staining and confocal microscopy. Our results revealed that in the presence of the P protein, the N protein formed small dots in the perinuclear area when it was expressed without the M protein or when it was incapable of interacting with the M protein. However, when the N protein was expressed in the presence of the P protein and in a form that was able to interact with the M protein, the N protein colocalized with the M protein in the cytoplasm as well as at the plasma membrane. We further found that IC-NΔ3-EGFP produced similar or slightly reduced levels of viral RNA and comparable sizes of plaques (and therefore probably comparable levels of viral proteins) compared with the parental IC323-EGFP although its production of infectious viral particles was markedly impaired. Taken together, these observations may suggest that the M protein retains the viral RNP complex at the plasma membrane by interacting with the N protein and promotes the production of infectious viral particles.
Unlike the recombinant MV lacking the C protein (39), IC-NΔ3-EGFP did not produce an increased level of viral RNA in infected cells. This observation may be explained if we presume that, in addition to the interaction between the N and M proteins, retention of the RNP complex by the M protein at the plasma membrane is required to stop viral RNA synthesis. If this hypothesis is correct, the M protein inhibits RNA synthesis only from the RNP complexes that are to become incorporated into progeny virions at the plasma membrane and does not affect viral RNA synthesis from the RNP complexes in the cytoplasm. In contrast, the nonstructural C protein may inhibit overall viral RNA synthesis in the cells. In MV-infected cells, the level of the M protein may be regulated (unlike the case in transfected cells) such that viral RNA synthesis continues at a sufficient level in the perinuclear area of the cytoplasm.
Acknowledgments
We thank T. Kohama, T. A. Sato, M. B. A. Oldstone, K. Komase, A. Kato, B. He and W. Chang for reagents; K. Amako and A. Takade for invaluable discussions; and the staff of the Research Support Center, Faculty of Medicine, Kyushu University, for technical support.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology and the Ministry of Health, Labor and Welfare of Japan.
Footnotes
Published ahead of print on 5 August 2009.
REFERENCES
- 1.Bair, C. H., C. S. Chung, I. A. Vasilevskaya, and W. Chang. 1996. Isolation and characterization of a Chinese hamster ovary mutant cell line with altered sensitivity to vaccinia virus killing. J. Virol. 70:4655-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bankamp, B., S. M. Horikami, P. D. Thompson, M. Huber, M. Billeter, and S. A. Moyer. 1996. Domains of the measles virus N protein required for binding to P protein and self-assembly. Virology 216:272-277. [DOI] [PubMed] [Google Scholar]
- 3.Bankamp, B., J. Wilson, W. J. Bellini, and P. A. Rota. 2005. Identification of naturally occurring amino acid variations that affect the ability of the measles virus C protein to regulate genome replication and transcription. Virology 336:120-129. [DOI] [PubMed] [Google Scholar]
- 4.Bellini, W. J., G. Englund, S. Rozenblatt, H. Arnheiter, and C. D. Richardson. 1985. Measles virus P gene codes for two proteins. J. Virol. 53:908-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernard, C., S. Gely, J. M. Bourhis, X. Morelli, S. Longhi, and H. Darbon. 2009. Interaction between the C-terminal domains of N and P proteins of measles virus investigated by NMR. FEBS Lett. 583:1084-1089. [DOI] [PubMed] [Google Scholar]
- 6.Bourhis, J. M., V. Receveur-Brechot, M. Oglesbee, X. Zhang, M. Buccellato, H. Darbon, B. Canard, S. Finet, and S. Longhi. 2005. The intrinsically disordered C-terminal domain of the measles virus nucleoprotein interacts with the C-terminal domain of the phosphoprotein via two distinct sites and remains predominantly unfolded. Protein Sci. 14:1975-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cathomen, T., B. Mrkic, D. Spehner, R. Drillien, R. Naef, J. Pavlovic, A. Aguzzi, M. A. Billeter, and R. Cattaneo. 1998. A matrix-less measles virus is infectious and elicits extensive cell fusion: consequences for propagation in the brain. EMBO J. 17:3899-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cathomen, T., H. Y. Naim, and R. Cattaneo. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cattaneo, R., K. Kaelin, K. Baczko, and M. A. Billeter. 1989. Measles virus editing provides an additional cysteine-rich protein. Cell 56:759-764. [DOI] [PubMed] [Google Scholar]
- 10.Cevik, B., D. E. Holmes, E. Vrotsos, J. A. Feller, S. Smallwood, and S. A. Moyer. 2004. The phosphoprotein (P) and L binding sites reside in the N-terminus of the L subunit of the measles virus RNA polymerase. Virology 327:297-306. [DOI] [PubMed] [Google Scholar]
- 11.Chen, M., J. C. Cortay, and D. Gerlier. 2003. Measles virus protein interactions in yeast: new findings and caveats. Virus Res. 98:123-129. [DOI] [PubMed] [Google Scholar]
- 12.Coronel, E. C., T. Takimoto, K. G. Murti, N. Varich, and A. Portner. 2001. Nucleocapsid incorporation into parainfluenza virus is regulated by specific interaction with matrix protein. J. Virol. 75:1117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duprex, W. P., F. M. Collins, and B. K. Rima. 2002. Modulating the function of the measles virus RNA-dependent RNA polymerase by insertion of green fluorescent protein into the open reading frame. J. Virol. 76:7322-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffin, D. E. 2007. Measles virus, p. 1551-1585. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5 ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 16.Harty, R. N., and P. Palese. 1995. Measles virus phosphoprotein (P) requires the NH2- and COOH-terminal domains for interactions with the nucleoprotein (N) but only the COOH terminus for interactions with itself. J. Gen. Virol. 76:2863-2867. [DOI] [PubMed] [Google Scholar]
- 17.Hashimoto, K., N. Ono, H. Tatsuo, H. Minagawa, M. Takeda, K. Takeuchi, and Y. Yanagi. 2002. SLAM (CD150)-independent measles virus entry as revealed by recombinant virus expressing green fluorescent protein. J. Virol. 76:6743-6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He, B., R. G. Paterson, C. D. Ward, and R. A. Lamb. 1997. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology 237:249-260. [DOI] [PubMed] [Google Scholar]
- 19.Heggeness, M. H., P. R. Smith, and P. W. Choppin. 1982. In vitro assembly of the nonglycosylated membrane protein (M) of Sendai virus. Proc. Natl. Acad. Sci. USA 79:6232-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirano, A., M. Ayata, A. H. Wang, and T. C. Wong. 1993. Functional analysis of matrix proteins expressed from cloned genes of measles virus variants that cause subacute sclerosing panencephalitis reveals a common defect in nucleocapsid binding. J. Virol. 67:1848-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirano, A., A. H. Wang, A. F. Gombart, and T. C. Wong. 1992. The matrix proteins of neurovirulent subacute sclerosing panencephalitis virus and its acute measles virus progenitor are functionally different. Proc. Natl. Acad. Sci. USA 89:8745-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horikami, S. M., S. Smallwood, B. Bankamp, and S. A. Moyer. 1994. An amino-proximal domain of the L protein binds to the P protein in the measles virus RNA polymerase complex. Virology 205:540-545. [DOI] [PubMed] [Google Scholar]
- 23.Huber, M., R. Cattaneo, P. Spielhofer, C. Orvell, E. Norrby, M. Messerli, J. C. Perriard, and M. A. Billeter. 1991. Measles virus phosphoprotein retains the nucleocapsid protein in the cytoplasm. Virology 185:299-308. [DOI] [PubMed] [Google Scholar]
- 24.Johansson, K., J. M. Bourhis, V. Campanacci, C. Cambillau, B. Canard, and S. Longhi. 2003. Crystal structure of the measles virus phosphoprotein domain responsible for the induced folding of the C-terminal domain of the nucleoprotein. J. Biol. Chem. 278:44567-44573. [DOI] [PubMed] [Google Scholar]
- 25.Karlin, D., S. Longhi, and B. Canard. 2002. Substitution of two residues in the measles virus nucleoprotein results in an impaired self-association. Virology 302:420-432. [DOI] [PubMed] [Google Scholar]
- 26.Kato, A., Y. Ohnishi, M. Hishiyama, M. Kohase, S. Saito, M. Tashiro, and Y. Nagai. 2002. The amino-terminal half of Sendai virus C protein is not responsible for either counteracting the antiviral action of interferons or down-regulating viral RNA synthesis. J. Virol. 76:7114-7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato, A., Y. Ohnishi, M. Kohase, S. Saito, M. Tashiro, and Y. Nagai. 2001. Y2, the smallest of the Sendai virus C proteins, is fully capable of both counteracting the antiviral action of interferons and inhibiting viral RNA synthesis. J. Virol. 75:3802-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kato, A., Y. Sakai, T. Shioda, T. Kondo, M. Nakanishi, and Y. Nagai. 1996. Initiation of Sendai virus multiplication from transfected cDNA or RNA with negative or positive sense. Genes Cells 1:569-579. [DOI] [PubMed] [Google Scholar]
- 29.Kingston, R. L., W. A. Baase, and L. S. Gay. 2004. Characterization of nucleocapsid binding by the measles virus and mumps virus phosphoproteins. J. Virol. 78:8630-8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kingston, R. L., D. J. Hamel, L. S. Gay, F. W. Dahlquist, and B. W. Matthews. 2004. Structural basis for the attachment of a paramyxoviral polymerase to its template. Proc. Natl. Acad. Sci. USA 101:8301-8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobune, F., H. Sakata, and A. Sugiura. 1990. Marmoset lymphoblastoid cells as a sensitive host for isolation of measles virus. J. Virol. 64:700-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komase, K., T. Nakayama, M. Iijima, K. Miki, R. Kawanishi, and H. Uejima. 2006. The phosphoprotein of attenuated measles AIK-C vaccine strain contributes to its temperature-sensitive phenotype. Vaccine 24:826-834. [DOI] [PubMed] [Google Scholar]
- 33.Laine, D., M. C. Trescol-Biemont, S. Longhi, G. Libeau, J. C. Marie, P. O. Vidalain, O. Azocar, A. Diallo, B. Canard, C. Rabourdin-Combe, and H. Valentin. 2003. Measles virus (MV) nucleoprotein binds to a novel cell surface receptor distinct from FcγRII via its C-terminal domain: role in MV-induced immunosuppression. J. Virol. 77:11332-11346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, Y., F. Horvath, J. A. Aligo, R. Wilson, and B. He. 2005. The role of simian virus 5 V protein on viral RNA synthesis. Virology 338:270-280. [DOI] [PubMed] [Google Scholar]
- 35.Liston, P., R. Batal, C. DiFlumeri, and D. J. Briedis. 1997. Protein interaction domains of the measles virus nucleocapsid protein (NP). Arch. Virol. 142:305-321. [DOI] [PubMed] [Google Scholar]
- 36.Longhi, S., V. Receveur-Brechot, D. Karlin, K. Johansson, H. Darbon, D. Bhella, R. Yeo, S. Finet, and B. Canard. 2003. The C-terminal domain of the measles virus nucleoprotein is intrinsically disordered and folds upon binding to the C-terminal moiety of the phosphoprotein. J. Biol. Chem. 278:18638-18648. [DOI] [PubMed] [Google Scholar]
- 37.Manie, S. N., S. de Breyne, S. Vincent, and D. Gerlier. 2000. Measles virus structural components are enriched into lipid raft microdomains: a potential cellular location for virus assembly. J. Virol. 74:305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakatsu, Y., M. Takeda, S. Ohno, R. Koga, and Y. Yanagi. 2006. Translational inhibition and increased interferon induction in cells infected with C protein-deficient measles virus. J. Virol. 80:11861-11867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakatsu, Y., M. Takeda, S. Ohno, Y. Shirogane, M. Iwasaki, and Y. Yanagi. 2008. Measles virus circumvents the host interferon response by different actions of the C and V proteins. J. Virol. 82:8296-8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 41.Ono, N., H. Tatsuo, Y. Hidaka, T. Aoki, H. Minagawa, and Y. Yanagi. 2001. Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J. Virol. 75:4399-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plemper, R. K., A. L. Hammond, D. Gerlier, A. K. Fielding, and R. Cattaneo. 2002. Strength of envelope protein interaction modulates cytopathicity of measles virus. J. Virol. 76:5051-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reuter, T., B. Weissbrich, S. Schneider-Schaulies, and J. Schneider-Schaulies. 2006. RNA interference with measles virus N, P, and L mRNAs efficiently prevents and with matrix protein mRNA enhances viral transcription. J. Virol. 80:5951-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reutter, G. L., C. Cortese-Grogan, J. Wilson, and S. A. Moyer. 2001. Mutations in the measles virus C protein that up regulate viral RNA synthesis. Virology 285:100-109. [DOI] [PubMed] [Google Scholar]
- 45.Richardson, C. D., A. Scheid, and P. W. Choppin. 1980. Specific inhibition of paramyxovirus and myxovirus replication by oligopeptides with amino acid sequences similar to those at the N-termini of the F1 or HA2 viral polypeptides. Virology 105:205-222. [DOI] [PubMed] [Google Scholar]
- 46.Riedl, P., M. Moll, H. D. Klenk, and A. Maisner. 2002. Measles virus matrix protein is not cotransported with the viral glycoproteins but requires virus infection for efficient surface targeting. Virus Res. 83:1-12. [DOI] [PubMed] [Google Scholar]
- 47.Runkler, N., C. Pohl, S. Schneider-Schaulies, H. D. Klenk, and A. Maisner. 2007. Measles virus nucleocapsid transport to the plasma membrane requires stable expression and surface accumulation of the viral matrix protein. Cell Microbiol. 9:1203-1214. [DOI] [PubMed] [Google Scholar]
- 48.Sato, H., M. Masuda, M. Kanai, K. Tsukiyama-Kohara, M. Yoneda, and C. Kai. 2007. Measles virus N protein inhibits host translation by binding to eIF3-p40. J. Virol. 81:11569-11576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmitt, A. P., and R. A. Lamb. 2004. Escaping from the cell: assembly and budding of negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283:145-196. [DOI] [PubMed] [Google Scholar]
- 50.Spielhofer, P., T. Bachi, T. Fehr, G. Christiansen, R. Cattaneo, K. Kaelin, M. A. Billeter, and H. Y. Naim. 1998. Chimeric measles viruses with a foreign envelope. J. Virol. 72:2150-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suryanarayana, K., K. Baczko, V. ter Meulen, and R. R. Wagner. 1994. Transcription inhibition and other properties of matrix proteins expressed by M genes cloned from measles viruses and diseased human brain tissue. J. Virol. 68:1532-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tahara, M., M. Takeda, and Y. Yanagi. 2007. Altered interaction of the matrix protein with the cytoplasmic tail of hemagglutinin modulates measles virus growth by affecting virus assembly and cell-cell fusion. J. Virol. 81:6827-6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takeda, M., S. Ohno, F. Seki, K. Hashimoto, N. Miyajima, K. Takeuchi, and Y. Yanagi. 2005. Efficient rescue of measles virus from cloned cDNA using SLAM-expressing Chinese hamster ovary cells. Virus Res. 108:161-165. [DOI] [PubMed] [Google Scholar]
- 54.Takeda, M., S. Ohno, F. Seki, Y. Nakatsu, M. Tahara, and Y. Yanagi. 2005. Long untranslated regions of the measles virus M and F genes control virus replication and cytopathogenicity. J. Virol. 79:14346-14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takeda, M., S. Ohno, M. Tahara, H. Takeuchi, Y. Shirogane, H. Ohmura, T. Nakamura, and Y. Yanagi. 2008. Measles viruses possessing the polymerase protein genes of the Edmonston vaccine strain exhibit attenuated gene expression and growth in cultured cells and SLAM knock-in mice. J. Virol. 82:11979-11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takeda, M., M. Tahara, T. Hashiguchi, T. A. Sato, F. Jinnouchi, S. Ueki, S. Ohno, and Y. Yanagi. 2007. A human lung carcinoma cell line supports efficient measles virus growth and syncytium formation via a SLAM- and CD46-independent mechanism. J. Virol. 81:12091-12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takeda, M., K. Takeuchi, N. Miyajima, F. Kobune, Y. Ami, N. Nagata, Y. Suzaki, Y. Nagai, and M. Tashiro. 2000. Recovery of pathogenic measles virus from cloned cDNA. J. Virol. 74:6643-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.tenOever, B. R., M. J. Servant, N. Grandvaux, R. Lin, and J. Hiscott. 2002. Recognition of the measles virus nucleocapsid as a mechanism of IRF-3 activation. J. Virol. 76:3659-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tober, C., M. Seufert, H. Schneider, M. A. Billeter, I. C. Johnston, S. Niewiesk, V. ter Meulen, and S. Schneider-Schaulies. 1998. Expression of measles virus V protein is associated with pathogenicity and control of viral RNA synthesis. J. Virol. 72:8124-8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vincent, S., D. Gerlier, and S. N. Manie. 2000. Measles virus assembly within membrane rafts. J. Virol. 74:9911-9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yanagi, Y., B. A. Cubitt, and M. B. A. Oldstone. 1992. Measles virus inhibits mitogen-induced T cell proliferation but does not directly perturb the T cell activation process inside the cell. Virology 187:280-289. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, X., C. Glendening, H. Linke, C. L. Parks, C. Brooks, S. A. Udem, and M. Oglesbee. 2002. Identification and characterization of a regulatory domain on the carboxyl terminus of the measles virus nucleocapsid protein. J. Virol. 76:8737-8746. [DOI] [PMC free article] [PubMed] [Google Scholar]