Abstract
We previously demonstrated that two closely spaced polyproline motifs, with the consensus sequence Pro-X-X-Pro-X-Lys/Arg, located between residues 343 to 356 of NS5A, mediated interactions with cellular SH3 domains. The N-terminal motif (termed PP2.1) is only conserved in genotype 1 isolates, whereas the C-terminal motif (PP2.2) is conserved throughout all hepatitis C virus (HCV) isolates, although this motif was shown to be dispensable for replication of the genotype 1b subgenomic replicon. In order to investigate the potential role of these motifs in the viral life cycle, we have undertaken a detailed mutagenic analysis of these proline residues in the context of both genotype 1b (FK5.1) or 2a subgenomic replicons and the genotype 2a infectious clone, JFH-1. We show that the PP2.2 motif is dispensable for RNA replication of all subgenomic replicons and, furthermore, is not required for virus production in JFH-1. In contrast, the PP2.1 motif is only required for genotype 1b RNA replication. Mutation of proline 346 within PP2.1 to alanine dramatically attenuated genotype 1b replicon replication in three distinct genetic backgrounds, but the corresponding proline 342 was not required for replication of the JFH-1 subgenomic replicon. However, the P342A mutation resulted in both a delay to virus release and a modest (up to 10-fold) reduction in virus production. These data point to critical roles for these proline residues at multiple stages in the HCV life cycle; however, they also caution against extrapolation of data from culture-adapted replicons to infectious virus.
Hepatitis C virus (HCV) is an enveloped RNA virus which is estimated to infect some 123 million individuals (24). In the majority of cases the virus establishes a chronic infection that can ultimately result in liver fibrosis, cirrhosis, or hepatocellular carcinoma. Thus, there is great interest in elucidating the mechanisms of viral replication, with a view to developing new chemotherapeutic agents. Since 1999, use of the subgenomic replicon system has led to significant progress in the understanding of the mechanism of viral RNA replication. It has been demonstrated that the five nonstructural proteins—NS3, NS4A, NS4B, NS5A, and NS5B—are necessary and sufficient to replicate an RNA molecule containing the 5′ and 3′ untranslated regions (UTRs) of the viral genome. However, apart from the RNA-dependent RNA polymerase (NS5B), the precise details of the roles of each of the nonstructural proteins in the process of RNA replication remain undefined. One problem associated with the subgenomic replicon system is the observation that the replicon RNA undergoes culture adaptation in which, as a result of the error-prone nature of the polymerase, mutations that confer enhanced replicative capacity are selected for in culture. Importantly, it has been shown using the chimpanzee model that, once engineered back into an infectious clone of the virus, such mutations may be attenuating in vivo (5). Recently, the HCV field has been revolutionized by the development of a cell culture infectious system based on a genotype 2a clone derived from a patient with fulminant hepatitis: the JFH-1 clone (30). JFH-1 is also unique in that subgenomic replicons derived from this clone are able to replicate efficiently without culture adaptation. This observation, as well as the fact that full-length genomes of JFH-1 are able to coordinate the coupling of RNA replication to packaging and release of infectious virus particles in Huh7 cells, points to fundamental differences between the RNA replication machinery of JFH-1 and that of the genotype 1b culture-adapted replicons.
The majority of mutations conferring culture adaptation map to the region coding for the NS5A protein. NS5A is a zinc-binding phosphoprotein that, as well as playing a critical role in RNA replication, also interacts with a plethora of cellular proteins (18). The protein has been demonstrated to consist of three domains separated by low-complexity sequences (LCS) (29). Of particular interest is the observation that within LCS2 (between domains II and III) (Fig. 1a), NS5A contains two closely spaced polyproline motifs that are able to bind to the SH3 domains of Src-family tyrosine kinases (16), as well as other SH3 domain containing proteins (e.g., Grb2 and amphiphysin II/Bin1) (23, 25, 33). These motifs, which we have termed PP2.1 and PP2.2 (Fig. 1a), conform to the consensus SH3 binding motif Pro-X-X-Pro-X-Arg/Lys, where X is any amino acid (21). Interestingly, although the C-terminal motif (PP2.2) is absolutely conserved in all HCV genotypes, we and others have shown that it is dispensable for HCV RNA replication because alanine substitution of three prolines in this motif (shown to abolish SH3 domain interactions [16]) within a culture-adapted subgenomic replicon had either no effect (19) or resulted in only a modest reduction in replicative capacity (23, 33). The N-terminal motif (PP2.1), however, is only conserved in genotype 1 isolates, although within this motif a proline at residue 346 in genotype 1b (342 in genotype 2a) is absolutely conserved throughout all genotypes, which suggests it has an important role in virus replication.
FIG. 1.
Polyproline motifs in NS5A. (a) Schematic of the structure of NS5A showing the endoplasmic reticulum-membrane associating amphipathic helix (gray box) (4), the position of the coordinated zinc ion, and the three domains with interlinking LCS (black boxes) (29). The lower part of this figure shows the amino acid sequence of the region from residues 343 to 356. These correspond to polyprotein residues 2315 to 2328 in the genotype 1b infectious clone J4 (31). Note that in JFH-1 the corresponding residues in the polyprotein are 2311 to 2325 (residues 339 to 352 within the NS5A sequence). The prolines and basic residues of the SH3 binding motifs (Pro-X-X-Pro-X-Arg/Lys) are in boldface. The accession numbers for the six sequences are as follows: 1a infectious clone H77 (AF009606), 1b infectious clone J4 (AF054247), FK5.1 culture-adapted subgenomic replicon (AJ242654) (13), Con1 isolate (AJ238799), 2a infectious clone JFH-1 (AB047639), and 3a isolate (D17763). (b) Schematic of mutants constructed in the present study. The wild-type FK5.1 sequence is on the top line, residues mutated to alanine indicated by A in the subsequent lines, hyphens indicate unchanged residues.
To shed more light on the role of these proline residues in the viral life cycle we have undertaken a mutagenic analysis both in the context of genotype 1b or 2a subgenomic replicons and the cell culture infectious JFH-1 clone. Our results point to key roles of these prolines in multiple stages of virus replication but highlight a surprising discrepancy between the requirements in the two systems.
MATERIALS AND METHODS
Cell culture.
Huh7 and Huh7.5 cells were cultured in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, 100 IU of penicillin/ml, 100 μg of streptomycin/ml, 1% nonessential amino acids, and 20 mM HEPES buffer. For colony-forming assays, electroporated Huh7 cells were selected in the presence of G418 (1 mg/ml), and colonies were stained and counted after 2 weeks as described previously (19).
DNA constructs and manipulations.
Mutagenesis of NS5A within FK5.1 was achieved by using the GeneEditor kit (Promega) on an NsiI-NsiI subclone (nucleotides 3682 to 7122) of pFK5.1 (13), cloned into LITMUS28i (NEB). The NsiI-NsiI fragment was then reintroduced into both neomycin phosphotransferase and luciferase containing FK5.1 culture-adapted replicons. The mutated NsiI-NsiI fragments were also inserted into corresponding NsiI sites in pFK341repPIluc/5.1 (7). This construct contains the entire HCV 5′UTR (to nucleotide 341), followed by the poliovirus internal ribosome entry site, which then drives translation of the luciferase gene. To produce P346A mutants in the E1202G/T12080I/R2884G (ETR) and R2884G replicons, overlap PCR mutagenesis was used to generate a mutated NheI-BclI fragment with pFKrepPI-Luc-E1202G/T1280I/R2884G (13) as a template. The PCR fragments generated were ligated into pCR-Blunt and then excised using XhoI and BclI before being inserted into corresponding sites in either pFK-repPI-luc R2884G or pFK-repPI-luc ETR replicons. The presence of mutations was confirmed via diagnostic digest (EagI site) and sequencing. The JFH-1 mutations were generated by Kunkel mutagenesis on an NsiI-HindIII subclone of pJFH-1 (30) in LITMUS28i. Mutagenic primer sequences are available on request. RNA transcripts were generated from linearized plasmids and transfected into Huh7 or Huh7.5 cells as previously described (22).
Transient expression of HCV proteins.
Transient expression of the FK5.1 SGR was performed by infecting Huh7 cells with a recombinant vaccinia virus expressing T7 RNA polymerase, vTF7-3 (8), followed by transfection of the replicon plasmid. Briefly, Huh7 cells at 80% confluence in 6-cm dishes were infected with vTF7-3 at 10 PFU/cell in a final volume of 200 μl of phosphate-buffered saline (PBS) at 37°C for 50 min. Cells were washed twice with OptiMEM (Gibco) and transfected with 10 μg of replicon DNA using Lipofectin (Invitrogen). At 22 h postinfection the cells were washed with PBS and lysed in Glasgow lysis buffer (GLB) (9) prior to analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
Luciferase assay.
A total of 4 × 106 Huh7 or Huh7.5 cells were used per electroporation, and the cells were washed twice in diethyl pyrocarbonate-treated PBS and resuspended at 107 cells per ml. Then, 2 μg of replicon RNA was mixed with the cells, followed by electroporation at 950 μF and 270 V in a 0.4-cm cuvette. Cells were resuspended in complete Dulbecco modified Eagle medium, and 5 × 105 cells were seeded into six-well plates. Cells were harvested at 4, 24, 48, and 72 h posttransfection (hpt) in 200 μl of 1× passive lysis buffer (PLB; Promega). The luciferase activity of the cell lysates was determined as relative light units using LarI reagent (Promega) on a BMG plate reader.
Virus assays.
Cells were washed twice in diethyl pyrocarbonate-treated PBS and resuspended at 2 × 107 cells per ml. Then, 8 × 106 cells were used per electroporation, and 10 μg of full-length JFH-1 RNA was electroporated into cells as described above for luciferase assays. Cells were either seeded into T-75 flasks at 8 × 106 cells per flask or at 2.6 × 106 cells per T-25 flask. Cells and supernatants were harvested at 24, 48, and 72 hpt. Half of the cells were lysed in GLB, while the remaining cells were resuspended in 50 μl of PBS and lysed by repetitive freeze-thaw action (five times). The lysates were clarified by centrifugation at 2,800 × g in a microfuge for 5 min, and 50 μl of supernatant was then used to infect naive cells. Medium removed from virus-containing cells at 72 h was clarified by centrifugation at 200 × g for 5 min before being used to infect naive cells. At 48 to 72 h postinfection (with either supernatants or cell lysates) the cells were fixed with 4% paraformaldehyde for 20 min and permeabilized with 0.2% Triton X-100 for 7 min. The cells were then stained with sheep anti-NS5A at a 1:4,000 dilution for 1 h and then with the corresponding secondary fluorescent antibody. Foci were counted to determine virus titers. Cells were also stained with rabbit anti-Core at a 1:500 dilution as described above.
Western blotting.
Cells were harvested in GLB for full-length virus experiments and in PLB for luciferase assays. The protein concentration was determined by BCA assay (Pierce), and 10 μg of total protein was separated by SDS-PAGE, followed by semidry transfer onto polyvinylidene difluoride membranes. Membranes were blocked in 10% (wt/vol) dried skimmed milk powder in Tris-buffered saline with 0.1% (vol/vol) Tween 20 (TBS-T). Membranes were probed with either sheep anti-NS3 (1) or anti-NS5A (17), rabbit anti-core (a gift from J. McLauchlan), or anti-GAPDH (AbCam) in 5% (wt/vol) dried skimmed milk in TBS-T. The antibodies were detected with the relevant secondary horseradish peroxidase-conjugated antibody and in-house enhanced chemiluminescence reagent.
Quantitative reverse transcription-PCR (RT-PCR).
For analysis of extracellular viral RNA, electroporated cells were split 1:2 at 48 hpt and grown for a further 72 h before the supernatants were harvested and filtered through a 0.45-μm-pore-size filter. A 4-ml portion of supernatant was pelleted through a 20% sucrose cushion at 150,000 × g for 4 h. The supernatant was removed, and 100 μl of RNase-free water was added to the pellet, which was then lysed in 300 μl of TRIzol, followed by incubation for 5 min. Next, 110 μl of chloroform was added, and the sample was mixed and incubated for 15 min at 20°C. Lysates were separated by centrifugation at 15,700 × g. The aqueous phase was removed, and the RNA was precipitated with 500 μl of isopropanol for 10 min at 20°C, pelleted, and washed with 75% ethanol. Pellets were air dried and resuspended in 20 μl of RNase-free water. For analysis of intracellular RNA, electroporated cells were trypsinized, washed in PBS, and pelleted. Cells were lysed in 750 μl of TRIzol and incubated for 5 min, 200 μl of chloroform was added, and the protocol described above was followed. cDNA was generated by using TaqMan reverse transcription reagents with random primers according to the manufacturer's recommendations. To create a standard, 1 μg in vitro-transcribed RNA was reverse transcribed. Serially diluted cDNA standards were analyzed in triplicate using TaqMan Fast Universal PCR master mix and no ampErase UNG (ABI). A FAM-MGB probe and outside primers in the 5′UTR were utilized, and real-time reactions were amplified under fast universal conditions on a 7500 Fast Real-Time PCR machine. The data were analyzed by using 7500 Fast System software (SDS v1.3.1) (ABI).
RESULTS
We previously generated an NS5A mutant termed PA2.2, in which three prolines in the PP2.2 motif of NS5A (residues 350, 353, and 354) were substituted by alanine. This NS5A mutant was unable to bind to a range of cellular SH3 domains, including the Src-family tyrosine kinases Fyn, Hck, and Lck and the adaptor protein Grb2 (16), and was unable to block signaling via the Ras-Erk mitogen-activated protein kinase pathway (17). However, when introduced into the genotype 1b culture-adapted replicon FK5.1, this mutation had no significant effect on colony formation (19). We were intrigued by this, given the conservation of sequences in and around this motif (Fig. 1a), so we decided to extend this analysis by generating a further set of mutants. First, the completely conserved arginine 356, also critical for SH3 binding of the PP2.2 motif (19), was mutated to alanine, either alone or in the context of the PA2.2 mutant (PA2.2/R356A). Second, to test whether proline 351 played any role, all four proline residues in the PP2.2 motif were mutated to alanine (ExtPA2.2) (Fig. 1b).
We also analyzed the role of the PP2.1 motif. In contrast to PP2.2, this motif is only present in genotype 1 isolates of HCV; in virtually all isolates of other HCV genotypes (apart from two 5a isolates) the first proline of the motif (residue 343) is substituted by alanine (14) (Fig. 1a). The lack of this residue is predicted to result in loss of the ability to bind SH3 domains. However, prolines 345 and 346 are completely conserved, suggesting a functional role. To test the role of the PP2.1 motif, prolines 343, 345, and 346 were mutated to alanine (termed PA2.1) in the context of either the wild-type genotype 1b replicon or the PA2.2 mutant.
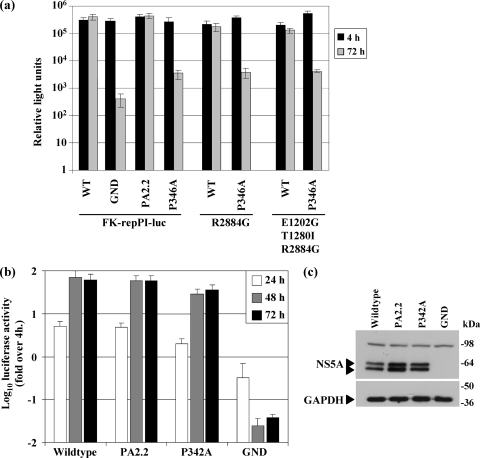
The ability of the mutant replicons to support RNA replication was first assessed by a colony formation assay. Consistent with our previous observation that the PA2.2 mutation had no effect on genotype 1b replicon function (19), other mutations within the PP2.2 motif (R356A and R356A/PA2.2) also had no significant effect on colony formation (Fig. 2a), although the ExtPA2.2 mutant exhibited a modest but reproducible 30% reduction in colony numbers. However, a mutation within the PP2.1 motif (PA2.1) reduced the ability of the replicon to establish G418-resistant colonies to control levels seen with the polymerase-defective GND replicon. These data were confirmed by the use of a transient, luciferase-based replicon in interferon-cured Huh7.5 cells (3); again, mutations in the PP2.2 motif had no effect on the levels of luciferase expressed from the replicons at 72 hpt. In contrast, the PA2.1 mutants showed an ∼1,000-fold reduction in luciferase at 72 h compared to the 4-h time point, and the same reduction was seen for the GND replicon (Fig. 2b).
FIG. 2.
Requirements of the PP2.1 and PP2.2 motifs for RNA replication of the FK5.1 culture-adapted replicon. (a) Colony formation assay. T7 transcripts (2 μg) were electroporated into Huh7 cells as previously described (19), 103 transfected cells were seeded into six-well plates and selected with G418 (1 mg/ml) from 24 hpt. Colony numbers were counted 14 days later and are illustrated graphically relative to the FK5.1 replicon. *PA2.2 data were taken from (19). (b) Luciferase replicon. Huh7.5 cells were electroporated with replicon RNA and harvested into PLB at 4 and 72 hpt. Luciferase activity was measured in a luminometer as previously described (17). The data from three independent experiments are shown.
The observation that the PP2.1 motif appeared to play a critical role in RNA replication was unexpected since we had previously demonstrated that this motif was dispensable for binding to cellular SH3 domains. It was only able to bind to the SH3 domain of the Src-family tyrosine kinase, Lyn (16), whereas the PP2.2 motif bound to Lyn, Fyn, Lck, Hck, the adaptor protein Grb2 (25), and amphiphysin II (33). In addition to this, the first proline in this motif (residue 343) is only conserved in genotype 1 isolates of HCV; the lack of this proline would disrupt the SH3-binding consensus, suggesting that if the prolines within PP2.1 did play a role in the viral replication cycle, it was unrelated to the ability to bind SH3 domains. Therefore, in order to further investigate the requirements for RNA replication of the three prolines in this motif (residues 343, 345, and 346), we generated three mutant genotype 1b replicons in which each of these prolines was individually substituted by alanine (P343A, P345A, and P346A) and a mutant in which both prolines 343 and 346 were alanine substituted (P343/6A). As expected, given the lack of conservation of the first proline residue, the P343A mutation had no significant effect on the ability of the replicon to establish G418-resistant colonies (Fig. 2a). P345A had an intermediate effect, reducing colony formation by ca. 70%. In contrast, the P346A mutation, either singly or in combination with P343A, completely abrogated colony formation. Again, these data were confirmed by the use of a transient, luciferase-based replicon in Huh7.5 cells (Fig. 2b), since P343A had no effect, P345A had an intermediate effect (reducing luciferase expression ∼10-fold), and P346A exhibited levels of luciferase expression comparable to the GND control replicon.
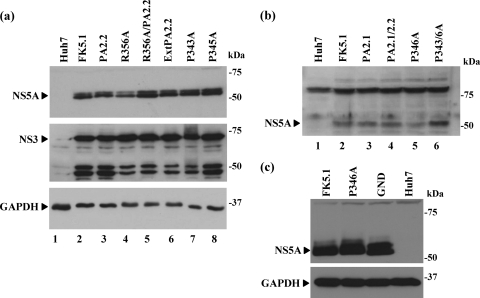
We established polyclonal cell populations from the five mutant replicon transfections described above that formed colonies: R356A, R356A/PA2.2, ExtPA2.2, P343A, and P345A. Sequence analysis of RT-PCR products derived from these cells revealed that the mutations were maintained after selection (data not shown), and there is thus no selective pressure for reversion to the wild type. As shown in Fig. 3a, all of these lines expressed similar levels of both NS3 and NS5A, indicating that these mutations had no effect on either polyprotein translation or proteolytic processing. Of note, there appeared to be no effect on the levels of NS5A hyperphosphorylation, as judged by the presence of the p58 form. Although we were not able to establish stable polyclonal populations of the mutants within the PP2.1 motif that abrogated colony formation (PA2.1, PA2.1/2.2, P346A, and P343/6A), we considered that it was important to verify that these replicons were competent for both polyprotein translation and proteolytic processing. To confirm this, we transfected the in vitro-transcribed replicon RNAs into Huh7 cells and lysed the cells at 4 h. Western blot analysis of these cell lysates revealed that similar low levels of NS5A could be detected in all replicons apart from P346A which exhibited a marked reduction in the abundance of NS5A protein (Fig. 3b). To confirm that this reduction in P346A abundance was not due to instability of the mutated NS5A protein, we took advantage of the presence of a T7 promoter driving transcription of the replicon constructs. Huh7 cells were infected with a recombinant vaccinia virus expressing T7 RNA polymerase (vTF7-3) (8) and then transfected with replicon plasmids. Cells were lysed at 22 h postinfection and analyzed by Western blotting for the presence of NS5A. As shown in Fig. 3c, both wild-type and P346A forms of FK5.1 NS5A were equally abundant, indicating that the functional defect in the P346A replicons was not due to defects in translation, polyprotein processing, or protein stability and are most likely therefore explained by the inability of the mutant forms of NS5A to function in the process of RNA replication. As a control, we also show that NS5A from the GND replicon was expressed as a stable protein (lane 3). It is noteworthy that this control is a Con1 replicon, not the culture-adapted FK5.1, and therefore exhibits both a slightly higher mobility in SDS-PAGE and a higher level of p58 hyperphosphorylated NS5A.
FIG. 3.
Expression of HCV nonstructural proteins by PP2.1/2.2 motif mutant replicons. (a) Analysis of stable polyclonal cell populations. Polyclonal populations of cells transfected with the indicated replicons were maintained in the presence of G418 (500 μg/ml). Cell lysates were analyzed by Western blotting with polyclonal antisera to NS5A (17) or NS3 (1) or a monoclonal antibody to GAPDH (glyceraldehyde-3-phosphate dehydrogenase), followed by the appropriate horseradish peroxidase-conjugated secondary antibodies (Sigma) and enhanced chemiluminescence. The appropriate bands for NS5A, NS3, and GAPDH are indicated (▸), the lower-molecular-mass bands in the NS3 blot represent proteolytic degradation products. Lane 1 is a lysate from control, untransfected Huh7 cells. (b) Analysis of input RNA translation of replication-incompetent replicons. For replicons that did not establish G418 resistant cell clones, Huh7 cells (106) were transfected with 2 μg of RNA and harvested at 4 hpt. Lysates (20 μg of protein) were analyzed by Western blotting with a polyclonal antisera to NS5A. The NS5A-specific band is indicated (▸); the higher-molecular-mass band is a cellular protein that cross-reacts with the NS5A antiserum and is always detected upon extended exposure of Western blots. (c) Huh7 cells were infected with vTF7-3 (8) (10 PFU/cell) and transfected with the indicated replicon plasmid DNAs. At 22 h postinfection lysates (10 μg of protein) were analyzed by Western blotting for NS5A or GAPDH.
The FK5.1 culture-adapted replicon contains seven coding mutations in comparison to the parental Con1 replicon sequence: E1202G and T1280I in NS3; L1757I in NS4B; and N2109D, S2197P, P2327S, and K2350E in NS5A (13). It was conceivable that the inability of the P346A mutant to replicate in the context of the FK5.1 replicon was due to incompatibility with the existing set of mutations. This has previously been shown to be the case for certain combinations of adaptive mutations; for example, single substitutions S2204R or S2197P (both in NS5A) are adaptive but in combination yield a replicon with dramatically reduced replication efficiency (15). We therefore cloned the P346A mutation into two further luciferase-based culture-adapted replicons that contained fewer mutations and, importantly, did not contain mutations within the NS5A coding region. These were a replicon termed ETR that contained three adaptive mutations (E1202G and T1280I in NS3 and R2884G in NS5B), and a replicon containing the single R2884G adaptive mutation in NS5B (15). Since these replicons contained a chimeric HCV/poliovirus 5′UTR (7), we also recreated the PA2.2 and P346A mutations in the context of the FK5.1 replicon containing the chimeric HCV/poliovirus 5′UTR (FK-repPI) (7) to confirm that the phenotype of these mutations was preserved in the context of the hybrid 5′UTR.
As shown in Fig. 4a, consistent with the phenotype of the P346A mutant in FK5.1 (Fig. 2), this mutant also exhibited a defect in RNA replication in both the ETR and R2884G backgrounds, as well as the FK-repPI replicon. Replication of these RNAs was reduced 100-fold compared to the corresponding wild-type replicons. Although significantly impaired, it is interesting that the P346A defect was not as pronounced in the context of the repPI constructs compared to the 1,000-fold reductions seen for FK5.1 (Fig. 2b), suggesting that the higher replicative capacity of the repPI replicons might partially compensate for the P346A defect. In contrast the PA2.2 mutant retained wild-type replication in the repPI context. All of the constructs were competent for translation and polyprotein processing, as judged by NS5A expression at 4 hpt (data not shown). Accordingly, we conclude that the inability of P346A to replicate was not due to incompatibility with adaptive mutations in the FK5.1 replicon.
FIG. 4.
Requirements of proline 346/342 in the context of alternative genotype 1b or 2a subgenomic replicons. (a) Genotype 1b replicons. Huh7.5 cells were electroporated with the indicated replicon RNAs and harvested into PLB at 4 and 72 hpt. The luciferase activity was measured in a luminometer as previously described (17). (b) Genotype 2a (JFH-1) replicon. Huh7 cells were electroporated with the indicated replicon RNAs and harvested at 4, 24, 48, and 72 hpt. Luciferase activity was measured as described above. The data from three independent experiments are shown. Values are shown as the fold increase luciferase activity over the 4-hpt time point. (c) The 72-hpt samples from panel b were analyzed by Western blotting with antibodies to NS5A (17) or GAPDH.
During the course of the present study a cell culture infectious clone of HCV, JFH-1, became available, so we used this system to investigate the role of the polyproline motifs in the context of the complete viral life cycle. In common with all genotype 2a isolates, JFH-1 does not contain an intact PP2.1 motif (Fig. 1a), since the first proline (P343) is substituted by alanine. However, the proline shown to be critical for RNA replication of the culture-adapted subgenomic replicon, P346, is present in JFH-1, as is the intact PP2.2 motif. Due to a 4-amino-acid deletion (residues 277 to 280) within JFH-1 NS5A relative to the Con1 (1b) sequence, the residue corresponding to P346 in JFH-1 is P342. We therefore created two mutant forms of JFH-1 NS5A: P342A and PA2.2 (P347A, P350A, and P351A). The RNA replication capacity of these two mutants was first tested in the context of a luciferase-based JFH-1 subgenomic replicon (SGR-luc-JFH-1) (26). Somewhat surprisingly, the replication of both mutants was indistinguishable from that of the wild type (Fig. 4b). Furthermore, analysis of NS5A levels by Western blotting at 72 hpt also showed no significant difference between the mutants and the wild type (Fig. 4c). Thus, in the context of a genotype 2a subgenomic replicon P342 is not required for RNA replication.
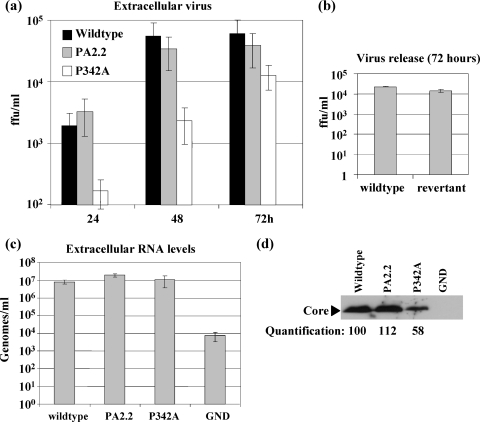
A number of recent studies have demonstrated a role for NS5A in virus assembly (2, 20, 27). We therefore analyzed the effects of the P342A and PA2.2 mutations on the production of infectious virus. As shown in Fig. 5a, the PA2.2 mutant was indistinguishable from the wild type both in terms of absolute amounts of infectious virus produced and the kinetics of virus release. However, the P342A mutant showed a 10-fold reduction in virus production at 24 and 48 hpt. Although the difference between the wild type and the P342A mutant was less marked at 72 hpt (∼5-fold reduction), it was still evident at 120 hpt (data not shown). This observation is consistent with a defect in release of infectious virus rather than a delay. To confirm that the apparent modest rebound in P342A titer at 72 hpt was not due to reversion or acquisition of a compensatory mutation, we amplified the NS5A coding sequence by RT-PCR from cell lysates prepared at 72 hpt. Sequence analysis of five clones revealed that not only was the P342A mutation maintained in all cases but there were also no additional mutations within NS5A. As further confirmation that the phenotype of the P342A mutant was not due to the acquisition of an additional mutation during the cloning process, we generated a revertant virus in which the wild-type NS5A coding sequence was reengineered back into the P342A mutant. As shown in Fig. 5b, virus production by the revertant was restored to wild-type levels.
FIG. 5.
Requirement for proline 342 in virus release. (a) Huh7 cells were electroporated with the indicated full-length viral RNAs and virus release into the culture supernatant measured by a focus-forming assay at the indicated time points. (b) The release of virus into culture supernatant was measured at 72 hpt for the wild-type and P342 revertant viruses. (c) Genomic RNA levels in virus preparations purified by sedimentation from culture supernatants through a sucrose cushion were measured at 120 hpt by quantitative RT-PCR. The data from six independent experiments are shown. (d) Representative anti-Core Western blot from equal amounts of virus preparations purified by sedimentation from culture supernatants through a sucrose cushion. The relative levels of Core were determined by densitometric analysis of Western blots using ImageJ software.
To investigate whether the 10-fold reduction in virus infectivity seen in the P342A mutant was reflected in a concomitant reduction in virus RNA release, we purified extracellular virus by sedimentation through a sucrose cushion and measured the levels of genomic RNA by quantitative RT-PCR. Surprisingly, as shown in Fig. 5c, the levels of released viral RNA were similar for the wild-type and both mutant viruses. Interestingly, Western blot analysis of the purified virus preparation revealed that there was a twofold reduction in the amount of Core protein in the case of the P342A virus compared to either the wild type or PA2.2 (Fig. 5d). The most appropriate interpretation of these results is that the amount of virus released by the P342A mutant is similar to that of the wild type, but the infectivity of this virus is reduced. However, in the absence of a methodology for accurate quantitation of HCV particles and measurement of particle/infectivity ratios, we cannot absolutely rule out that the P342A mutant releases fewer virus particles than does the wild type.
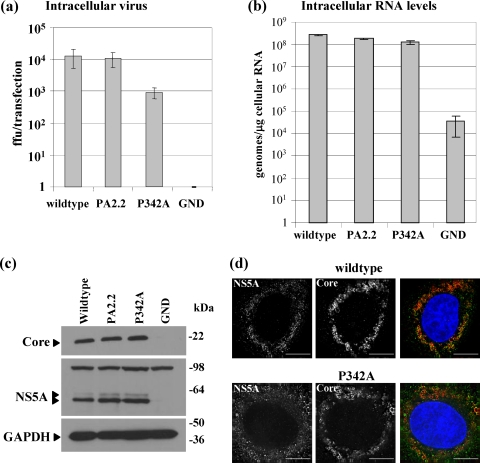
To determine whether the reduction in extracellular infectious virus seen in the P342A mutant was the result of a defect in virus assembly or release from the cell, we examined the levels of intracellular virus at 72 hpt. This analysis (Fig. 6a) revealed that the titer of intracellular infectious virus was 10-fold lower in the P342A mutant than in the wild type or PA2.2 mutant, confirming that the defect in the virus was at the level of virus assembly rather than at release. Again, this was restored to wild-type levels in the revertant (data not shown). As was the case for extracellular virus release, the defect in assembly of P342A was not reflected by a reduction in the amount of genomic RNA present within the cells. As shown in Fig. 6b, both mutants showed levels of intracellular genomic RNA very similar to that of the wild type. Furthermore, when we examined the intracellular accumulation of viral proteins by Western blotting, we observed that, for both mutants, the levels of Core and NS5A were comparable to those of the wild type. The intracellular distribution of Core and NS5A was also not affected, as shown by the immunofluorescence images in Fig. 6d. We conclude that the P342A mutation within NS5A has a modest, but significant, effect on the assembly of infectious HCV particles within Huh7 cells.
FIG. 6.
Requirement for proline 342 in virus assembly. (a) Huh7 cells were electroporated with the indicated full-length viral RNAs and intracellular virus levels measured following cell disruption by repetitive freeze-thaw at 72 hpt. (b) Intracellular genomic RNA levels were measured at 120 hpt by quantitative RT-PCR. The data from three independent experiments are shown. (c) Representative Western blots for Core, NS5A, and GAPDH at 72 hpt. (d) Immunofluorescence analysis of NS5A (green) and Core (red) in Huh7 cells 72 hpt. Scale bars, 10 μm.
DISCUSSION
In this study we present a detailed analysis of the role of a cluster of proline residues situated between domains 2 and 3 of NS5A. Our data clearly show that a completely conserved motif with the consensus sequence Pro-X-X-Pro-X-Arg/Lys (termed PP2.2) is in fact dispensable for all aspects of the virus life cycle in a cell culture-based system. Given that this motif is absolutely required for the interactions between NS5A and the Src-family tyrosine kinases Lck, Hck, and Fyn (16), as well as the adaptor proteins Grb2 (25) and amphiphysin II/Bin1 (23, 33), we conclude that the interactions between NS5A and these cellular proteins are not required for HCV replication. We cannot rule out that these cellular proteins play roles in virus replication that are not dependent on interactions with the PP2.2 motif of NS5A. However, the conservation of this motif in all HCV isolates strongly suggests a functional role, and it may be that interactions between PP2.2 and cellular SH3 domains are involved in pathogenesis or persistence of the virus in the host—aspects of the virus life cycle that are not addressable using an in vitro replication system. In this regard it is interesting that a genotype 1b full-length clone containing the PA2.2 mutation failed to establish infection in a chimpanzee (23), although it is difficult to conclude that this supports a role for the PP2.2 motif in viral replication from a negative result in a single animal.
Alanine substitution of proline 343 had no effect on the replication of the FK5.1 replicon (Fig. 2a). This result is in contrast to the observation of Tellinghuisen et al. (28), whose data demonstrated a complete abrogation of replication for this mutant in the context of either a GIT (E1202G and T1280I in NS3, together with K1846T in NS4B) or S2204I replicon. Although the discrepancy in these results is difficult to reconcile, it should be noted that when the GIT replicon P343A mutant was expressed by a vaccinia virus-T7 system no NS5A expression was seen (28), suggestive of a major defect in NS5A translation or stability. In contrast, NS5A in cells stably harboring the FK5.1 P343A mutant could readily be detected by Western blotting (Fig. 3a). Thus, the lack of replication of the GIT or S2201I P343A mutants is likely due to a defect in NS5A expression rather than an inherent inability to function in RNA replication. Our data are thus consistent with the hypothesis that this proline is not required for NS5A function. This conclusion is supported by the fact that this proline residue is only present in genotype 1 HCV isolates (and the two documented genotype 5a isolates) (14); in all other isolates it is replaced by an alanine and is thus clearly not universally required for NS5A protein stability or function.
Mutagenesis of proline 346 in the context of three different genotype 1b subgenomic replicons (FK5.1, ETR, and R2884G) identified a key role for this residue in RNA replication, since all of these replicons were severely defective. It is noteworthy that the P346A defect was less apparent in the context of a replicon with a chimeric HCV 5′UTR/poliovirus internal ribosome entry site (repPI) than the wild-type HCV 5′UTR. This suggests that the higher replicative capacity of the repPI might partially compensate for the P346A defect. Recently, it has been published that, in the context of the GIT replicon (containing culture adaptive mutations E1202G and T1280I in NS3 and K1846T in NS4B), the P346A mutation showed only a modest (10-fold) reduction in colony formation (28). The only mutation in the GIT replicon that is not represented in ETR is K1846T. Given that NS4B has been shown to influence NS5A hyperphosphorylation (18), these data suggest that proline 346 may be involved in a direct protein-protein interaction between NS4B and NS5A. In the context of FK5.1, ETR, and R2884G this interaction may require a lysine at residue 1846 in NS4B. However, in the GIT replicon the K1846T mutation may allow this interaction to occur in the absence of proline 346.
The situation is further complicated by the observation that mutation of the corresponding residue in the genotype 2a JFH-1 subgenomic replicon, proline 342, had no effect on RNA replication (Fig. 4b). A simplistic explanation, consistent with the observation that the phenotype of genotype 1b P346A was less apparent in the repPI background, is that the higher efficiency of JFH-1 RNA replication compared to genotype 1b in some way abrogates the replication defect of the proline-to-alanine mutation. However, a more attractive explanation is that one or more of the amino acid differences between Con1 and JFH-1 results in a replication phenotype that is independent of proline 342. The molecular basis for this remains to be elucidated but, as discussed above, it might be that this residue is critical for the interaction of NS5A with one of the other nonstructural proteins of HCV (e.g., NS4B) or an as-yet-undefined cellular protein during the process of RNA replication. For the genotype 2a JFH-1 isolate, the ability to replicate more efficiently than genotype 1b may mean that it is not dependent on this interaction or that it is mediated via an alternative binding site within NS5A.
NS5A plays key roles in multiple aspects of the virus life cycle. As well as an involvement in genome replication (mainly mapping to domain I and part of domain II) (28), the protein has recently been shown to be important in the assembly of infectious virus particles (2, 20, 27). The latter role of NS5A maps to domain III, in particular, a number of conserved serines toward the C-terminal end of the protein (11). In addition to this, our data point to a role for proline 342 in virus assembly, since this mutation resulted in an ∼10-fold reduction in the assembly of infectious virus particles. The mechanistic details of the involvement of NS5A in the process of virus assembly remain obscure, although an interaction with Core has been proposed (20). Intriguingly the level of Core in preparations of extracellular P342A virus is approximately half that of wild-type virus, whereas the levels of genomic RNA are comparable, which is again consistent with a defect in virus assembly. Proline 342 might either be involved in a direct protein-protein interaction (e.g., with Core) or, since it is present within the LCS, acting as a linker between domains II and III, its inherent rigidity may serve to position these two domains with respect to each other to facilitate such interactions. Alternatively, this proline may be the subject of posttranslational modification, e.g., by cis-trans isomerization or hydroxylation. In this regard, HCV RNA replication is known to require the peptidyl-prolyl isomerase cyclophilin B (CyPB) and is inhibited by the CyPB inhibitor cyclosporine. Interestingly, mutations that conferred resistance to cyclosporine treatment mapped to both NS5A and NS5B (6), with NS5A mutations having a greater effect than NS5B. Furthermore, it has been shown that, unlike genotype 1b, JFH-1 RNA replication did not require CyPB (12), although this conclusion is controversial as another study reported that both genotypes required CyPA (but not CyPB) (32). Prolines in domain II of NS5A have recently been shown to be substrates for both CyPA and CyPB (10); however, it has not yet been determined whether the cluster of prolines in LCS2 between domains II and III are possible cyclophilin substrates, although they are predicted to be surface exposed. It is thus conceivable that the differential requirement for proline 346/342 in genotypes 1b and 2a reflects the targeting of this proline by CyPB. In other words, in genotype 2a it may be that this proline is not a substrate for CyPB such that JFH-1 is not dependent on CyPB for RNA replication. The involvement of P342 in virus assembly presumably reflects an alternative role for NS5A. In conclusion, we believe the data presented here suggest that, in addition to the requirement for domain III (2, 27, 20), there is also a role for residues within LCS2 between domains II and III in the process of virus assembly. This is the subject of ongoing studies in our laboratory.
Acknowledgments
This study was supported by grants to Mark Harris from the Wellcome Trust (0671250) and Medical Research Council (G0401577). Stephen Griffin is the recipient of a Medical Research Council New Investigator Award (G0700124). Mair Hughes is supported by a Cooperative Awards in Science and Engineering (CASE) Ph.D. studentship from the Biotechnology and Biological Sciences Research Council and Arrow Therapeutics.
We thank Ralf Bartenschlager and Volker Lohmann (University of Heidelberg) for the FK5.1 and repPI replicon constructs, Takaji Wakita (National Institute for Infectious Diseases, Tokyo, Japan) for pJFH-1, John McLauchlan (MRC Virology Unit, Glasgow, United Kingdom) for the SGR-luc-JFH-1 construct and Core antiserum, and Charles Rice (The Rockefeller University, New York, NY) for the Huh7.5 cells. We are grateful to Cheryl Walters and John Barr (University of Leeds) for providing the vTF7-3 virus stock and for help with the assay and Andrew Macdonald (University of Leeds) for advice and critical reading of the manuscript.
Footnotes
Published ahead of print on 5 August 2009.
REFERENCES
- 1.Aoubala, M., J. Holt, R. A. Clegg, D. J. Rowlands, and M. Harris. 2001. The inhibition of cAMP-dependent protein kinase by full-length hepatitis C virus NS3/4A complex is due to ATP hydrolysis. J. Gen. Virol. 82:1637-1646. [DOI] [PubMed] [Google Scholar]
- 2.Appel, N., M. Zayas, S. Miller, J. Krijnse-Locker, T. Schaller, P. Friebe, S. Kallis, U. Engel, and R. Bartenschlager. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. [DOI] [PubMed] [Google Scholar]
- 5.Bukh, J., T. Pietschmann, V. Lohmann, N. Krieger, K. Faulk, R. E. Engle, S. Govindarajan, M. Shapiro, M. St. Claire, and R. Bartenschlager. 2002. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc. Natl. Acad. Sci. USA 99:14416-14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandes, F., D. S. Poole, S. Hoover, R. Middleton, A. C. Andrei, J. Gerstner, and R. Striker. 2007. Sensitivity of hepatitis C virus to cyclosporine A depends on nonstructural proteins NS5A and NS5B. Hepatology 46:1026-1033. [DOI] [PubMed] [Google Scholar]
- 7.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris, M., and K. Coates. 1993. Identification of cellular proteins that bind to the human immunodeficiency virus type 1 nef gene product in vitro: a role for myristylation. J. Gen. Virol. 74:1581-1589. [DOI] [PubMed] [Google Scholar]
- 10.Hanoulle, X., A. Badillo, J. M. Wieruszeski, D. Verdegem, I. Landrieu, R. Bartenschlager, F. Penin, and G. Lippens. 2009. Hepatitis C virus NS5A protein is a substrate for the peptidyl-prolyl cis/trans isomerase activity of cyclophilins A and B. J. Biol. Chem. 284:13589-13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes, M., S. Griffin, and M. Harris. 2009. Domain III of NS5A contributes to both RNA replication and assembly of hepatitis C virus particles. J. Gen. Virol. 90:1329-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishii, N., K. Watashi, T. Hishiki, K. Goto, D. Inoue, M. Hijikata, T. Wakita, N. Kato, and K. Shimotohno. 2006. Diverse effects of cyclosporine on hepatitis C virus strain replication. J. Virol. 80:4510-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuiken, C., K. Yusim, L. Boykin, and R. Richardson. 2005. The Los Alamos hepatitis C sequence database. Bioinformatics 21:379-384. [DOI] [PubMed] [Google Scholar]
- 15.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macdonald, A., K. Crowder, A. Street, C. McCormick, and M. Harris. 2004. The hepatitis C virus NS5A protein binds to members of the Src family of tyrosine kinases and regulates kinase activity. J. Gen. Virol. 85:721-729. [DOI] [PubMed] [Google Scholar]
- 17.Macdonald, A., K. Crowder, A. Street, C. McCormick, K. Saksela, and M. Harris. 2003. The hepatitis C virus NS5A protein inhibits activating protein-1 (AP1) function by perturbing Ras-ERK pathway signalling. J. Biol. Chem. 278:17775-17784. [DOI] [PubMed] [Google Scholar]
- 18.Macdonald, A., and M. Harris. 2004. Hepatitis C virus NS5A: tales of a promiscuous protein. J. Gen. Virol. 85:2485-2502. [DOI] [PubMed] [Google Scholar]
- 19.Macdonald, A., S. Mazaleyrat, C. McCormick, A. Street, N. J. Burgoyne, R. M. Jackson, V. Cazeaux, H. Shelton, K. Saksela, and M. Harris. 2005. Further studies on hepatitis C virus NS5A-SH3 domain interactions: identification of residues critical for binding and implications for viral RNA replication and modulation of cell signalling. J. Gen. Virol. 86:1035-1044. [DOI] [PubMed] [Google Scholar]
- 20.Masaki, T., R. Suzuki, K. Murakami, H. Aizaki, K. Ishii, A. Murayama, T. Date, Y. Matsuura, T. Miyamura, T. Wakita, and T. Suzuki. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82:7964-7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayer, B. J. 2001. SH3 domains: complexity in moderation. J. Cell Sci. 114:1253-1263. [DOI] [PubMed] [Google Scholar]
- 22.McCormick, C. J., L. Challinor, A. Macdonald, D. J. Rowlands, and M. Harris. 2004. Introduction of replication-competent hepatitis C virus transcripts using a tetracycline-regulable baculovirus delivery system. J. Gen. Virol. 85:429-439. [DOI] [PubMed] [Google Scholar]
- 23.Nanda, S. K., D. Herion, and T. J. Liang. 2006. Src homology 3 domain of hepatitis C virus NS5A protein interacts with Bin1 and is important for apoptosis and infectivity. Gastroenterology 130:794-809. [DOI] [PubMed] [Google Scholar]
- 24.Shepard, C. W., L. Finelli, and M. J. Alter. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558-567. [DOI] [PubMed] [Google Scholar]
- 25.Tan, S. L., H. Nakao, Y. P. He, S. Vijaysri, P. Neddermann, B. L. Jacobs, B. J. Mayer, and M. G. Katze. 1999. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc. Natl. Acad. Sci. USA 96:5533-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Targett-Adams, P., and J. McLauchlan. 2005. Development and characterization of a transient-replication assay for the genotype 2a hepatitis C virus subgenomic replicon. J. Gen. Virol. 86:3075-3080. [DOI] [PubMed] [Google Scholar]
- 27.Tellinghuisen, T. L., K. L. Foss, and J. Treadaway. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tellinghuisen, T. L., K. L. Foss, J. C. Treadaway, and C. M. Rice. 2008. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J. Virol. 82:1073-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tellinghuisen, T. L., J. Marcotrigiano, A. E. Gorbalenya, and C. M. Rice. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J. Biol. Chem. 279:48576-48587. [DOI] [PubMed] [Google Scholar]
- 30.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yanagi, M., M. StClaire, M. Shapiro, S. U. Emerson, R. H. Purcell, and J. Bukh. 1998. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology 244:161-172. [DOI] [PubMed] [Google Scholar]
- 32.Yang, F., J. M. Robotham, H. B. Nelson, A. Irsigler, R. Kenworthy, and H. Tang. 2008. Cyclophilin A is an essential cofactor for hepatitis C virus infection and the principal mediator of cyclosporine resistance in vitro. J. Virol. 82:5269-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zech, B., A. Kurtenbach, N. Krieger, D. Strand, S. Blencke, M. Morbitzer, K. Salassidis, M. Cotten, J. Wissing, S. Obert, R. Bartenschlager, T. Herget, and H. Daub. 2003. Identification and characterization of amphiphysin II as a novel cellular interaction partner of the hepatitis C virus NS5A protein. J. Gen. Virol. 84:555-560. [DOI] [PubMed] [Google Scholar]