Abstract
To test a previously coined “charge balance hypothesis” of human hepatitis B virus (HBV) capsid stability, we established an in vitro disassembly and reassembly system using bacterially expressed HBV capsids. Capsid disassembly can be induced by micrococcal nuclease digestion of encapsidated RNA. HBV core protein (HBc) mutants containing various amounts of arginine were constructed by serial truncations at the C terminus. Capsids containing smaller amounts of arginine (HBc 149, 154, and 157) remained intact after micrococcal nuclease digestion by native gel electrophoresis. Capsids containing larger amounts of arginine (HBc 159, 164, 169, and 171) exhibited reduced and more diffuse banding intensity and slightly upshifted mobility (HBc 159 and 164). Capsids containing the largest amounts of arginine (HBc 173, 175, and 183), as well as HBc 167, exhibited no detectable banding signal, indicating loss of capsid integrity or stability. Interestingly, capsid reassembly can be induced by polyanions, including oligonucleotides, poly-glutamic acid, and nonbiological polymer (polyacrylic acid). In contrast, polycations (polylysine and polyethylenimine) and low-molecular-weight anions (inositol triphosphate) induced no capsid reassembly. Results obtained by gel assay were confirmed by electron microscopy. Reassembled capsids comigrated with undigested parental capsids on agarose gels and cosedimented with undigested capsids by sucrose gradient ultracentrifugation. Taken together, the results indicate that HBV capsid assembly and integrity depend on polyanions, which probably can help minimize intersubunit charge repulsion caused mainly by arginine-rich domain III or IV in close contact. The exact structure of polyanions is not important for in vitro capsid reassembly. A large amount of independent experimental evidence for this newly coined “electrostatic interaction hypothesis” is discussed.
Chronic infection with hepatitis B virus (HBV) leads to the development of cirrhosis and hepatocellular carcinoma (6, 31, 36). HBV core protein (HBc) consists of the assembly domain (HBc amino acids 1 to 149) at the N terminus and the arginine-rich domain (ARD) at the C terminus (HBc amino acids 150 to 183) (33, 34). Escherichia coli-expressed HBc can spontaneously self-assemble into 28-nm capsid particles with a spherical appearance indistinguishable from that of human liver-derived capsid particles (7). Such capsid particles have been shown to package RNAs transcribed in E. coli (5, 8, 11, 28, 37). The four-helix bundle structure of HBV capsid particles is based on cryo-electron microscopy and X-ray crystallography using C-terminally truncated HBc (34, 38). At present, there is no known structure at the C terminus of HBc capsids (34, 41). HBc amino acids 150 to 183 contains four stretches of clustering arginine residues (ARD-I, -II, -III, and -IV) (Fig. 1). When the C-terminal domain of hepadnaviral core protein was serially truncated, a viral replication defect was observed (4, 19, 22, 27, 39). To date, it remains to be elucidated why the C terminus is so important for diverse biological activities, including RNA encapsidation and DNA replication. To address this issue, we proposed previously a so-called “charge balance hypothesis” (22), which highlights the importance of adequate electrostatic interactions between positive charge (basic residues) from HBc and negative charge from encapsidated RNA or DNA.
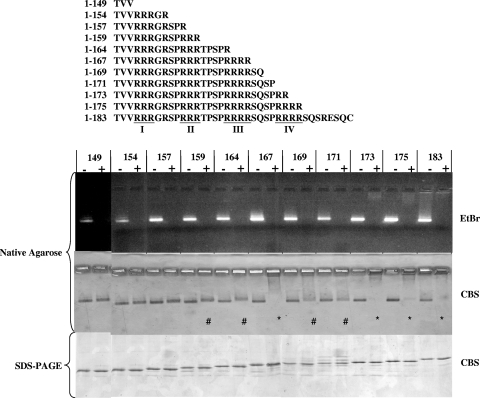
FIG. 1.
Effects of micrococcal nuclease treatment on HBV nucleocapsids with serially truncated C termini of core proteins. A series of HBV core expression vectors with different lengths and arginine contents were constructed in pET-Blue-1. The truncations are illustrated in the top panel, and the four ARDs (ARD-I, -II, -III, and -IV) are underlined. All constructs self-assembled into capsids when expressed in E. coli. The capsids run as a distinct band on 1% native agarose gels and can be stained with EtBr (upper panel) and Coomassie blue (middle panel). Untreated controls without micrococcal nuclease were incubated with the same buffer and conditions as for micrococcal nuclease-digested samples. In the upper panel, note the loss of the EtBr signal when the encapsidated RNAs were digested by micrococcal nuclease. In the middle panel, we observed three different groups of mutants with three different CBS patterns. Group 1 mutants, including HBc mutants 149, 154, and 157, exhibited no significant change in CBS banding pattern before or after micrococcal nuclease digestion. In group 2 capsids 159, 164, 169, and 171 (#), the CBS banding pattern became more diffuse, less intense, and slightly (yet reproducibly) upshifted. * indicates the near-complete loss of CBS banding in group 3 capsids 167, 173, 175, and 183. To confirm that this loss of CBS probably results from a loss of structural integrity of the capsid particle, rather than from a loss of the core protein per se, aliquots of samples were also run on denaturing SDS-PAGE (bottom panel).
The first clue that such an electrostatic interaction could play an important role in RNA encapsidation is from the study of an engineered mutant, HBc 164. Despite its reduced arginine content relative to the wild-type (WT) full-length HBc 183 (Fig. 1), HBc mutant 164 can encapsidate, as efficiently as WT HBV, both 3.5-kb pregenomic RNA (pgRNA) and a spliced 2.2-kb subgenomic RNA (sgRNA) (19, 22). When WT HBV nucleocapsids (capsids) were treated with micrococcal nuclease, the encapsidated 3.5-kb pgRNA and its reverse-transcribing RNA template were resistant to micrococcal nuclease treatment. In contrast, when mutant 164 capsids were treated with micrococcal nuclease, the encapsidated 3.5-kb RNA was highly nuclease sensitive, while the encapsidated 2.2-kb sgRNA appeared to be nuclease resistant (19, 22).
To further elucidate the mechanism behind this phenomenon, we hypothesized that this result could be due to an abnormal or less stable capsid structure generated by a charge imbalance (insufficient positive charge or excessive negative charge) when arginine-deficient mutant 164 encapsidates the full-length 3.5-kb pgRNA. In contrast, a more normal or stable capsid structure can be generated when arginine-deficient mutant 164 encapsidates the 2.2-kb sgRNA (with a reduced negative charge content relative to the 3.5-kb pgRNA) (22). In addition to RNA encapsidation and capsid stability, electrostatic interaction could also play a role in HBV DNA synthesis and genome maturation. For example, when the truncated C terminus of HBc 164 was progressively restored, the core-associated viral DNA gradually increased in both size and signal intensity (22).
In this study, we designed an in vitro capsid disassembly/reassembly experiment which is complementary to the in vivo approach in tissue culture (22; P. K. Chua, F. M. Tang, J. Y. Huang, C. S. Suen, and C. Shih, submitted for publication). We investigated the relationship between the arginine content of HBV capsids and the encapsidated nucleic acid in maintaining capsid stability in vitro. In addition, we demonstrated that HBV capsids that are disassembled by depletion of encapsidated RNA can be efficiently reassembled in the presence of exogenous nucleic acid and non-nucleic acid polyanions but not in the presence of polycations or low-molecular-weight (low-MW) anions. Capsid disassembly induced by RNA digestion appears to be related to intersubunit charge repulsion between ARD-III or ARD-IV in close contact.
MATERIALS AND METHODS
Native agarose gel electrophoresis and EM.
Procedures for native agarose gel electrophoresis and electron microscopy (EM) were as described elsewhere (22, 28).
Production of HBV capsids in E. coli.
Expression, purification, and characterization of the E. coli-produced HBV capsids of ayw and adr subtype origins have been described previously (22, 28). WT full-length HBc (HBc 183) and C-terminally truncated mutant proteins (HBc 149, HBc 154, HBc 164, and HBc 167) were expressed from pET-Blue vector as detailed elsewhere (22, 28). Mutant HBc 157, HBc 159, HBc 169, HBc 171, and HBc 173 were constructed by PCR amplification using the full-length pET-HBc 183 DNA as a template, a common 5′ primer (5′-ATGGACATTGACCCGTATAA-3′), and their respective downstream primers containing complementary sequences to stop codons (underlined) at various positions: HBc 157, 5′-AGGGAGTTCTTCATCTAGGGGACCT-3′; HBc 159, 5′-GCGAGGCGAGGGTCATCTTCTTCTAGG-3′; HBc 169, 5′-CTGCGACGCGGCTATTGAGACCTTC-3′; HBc 171, 5′-GATCTTCTGCGTCACGGCGATTGAG-3′; HBc 173, 5′-GATTGAGATCATCAGCGACGCGGCG-3′; and HBc 175, 5′-TTCCCGAGATTGTCATCTTCTGCGAC-3′.
Micrococcal nuclease treatment of E. coli-derived HBV capsids.
Five micrograms of purified capsids (1 μl of a 5-mg/ml stock in Tris-buffered saline [0.1 M NaCl, 2 mM KCl, 25 mM Tris, pH 7.4]) was diluted into 20 μl of distilled water before incubation for 5 h to overnight at 37°C with excessive amount of micrococcal nuclease (Roche Applied Science Co. or New England BioLabs) in the presence of 8 mM CaCl2and 25 mM dithiothreitol. Micrococcal nuclease is an endonuclease that preferentially cleaves single-stranded substrates, but it will also cleave double-stranded DNA or RNA. The extent and effects of the micrococcal nuclease digestion were evaluated on 1% native agarose gels sequentially stained with ethidium bromide (EtBr) and Coomassie blue. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (15%) was used to confirm the retention of intact HBc monomers after nuclease treatment.
Reassembly of micrococcal nuclease-treated capsids.
Candidate chemicals were screened for their abilities to induce capsid reassembly from micrococcal nuclease-treated, disassembled WT HBV capsids (HBc 183). Non-nucleic acid compounds (1 μg/μl) were incubated with micrococcal nuclease-digested capsids for 1 h at 37°C with gentle agitation. Approximately 25 mM EDTA and 5 mM dithiothreitol were used for incubation at room temperature for at least 15 min before the addition of nucleic acid reassembly inducers. The final reaction mixture was incubated at 37°C for 5 h to overnight. Aliquots were assayed by native agarose gel electrophoresis, EM, or sucrose gradient centrifugation.
Chemical compounds.
The following chemicals were purchased from Sigma Co.: lower-MW dextran sulfate (MW, 10,000), inositol hexasulfate, inositol triphosphate, poly(I) (5′), and poly(C) (5′). Higher-MW dextran sulfate (MW, 500,000) was from Fisher Scientific Co. Heparan sulfate, fully de-O-sulfated heparin, and oversulfated heparin were purchased from Neoparin Inc. Synthetic polypeptides, i.e., poly-aspartic acid [poly(D)], poly-glutamic acid [poly(E)], polyasparagine [poly(N)], and polylysine [poly(K)], were obtained from Sigma Co. The nonbiological polymers polyacrylic acid (PAA), poly(sodium 4-styrenesulfonate (PSS), polyethylenimine, and poly(allylamine hydrochloride) (PAH) were from Aldrich Co. Nucleic acids screened in the capsid reassembly assay included a deoxynucleoside triphosphate (dNTP) mix, a random hexamer, and a control primer (15-mer) from a Novagen cDNA kit. The 20-, 25-, 50-, and 75-mer oligonucleotides were unrelated primers which happened to be available in the laboratory. Poly(G) and poly(T) were purchased from Tri-I Biotech, Inc., Taiwan, and characterized at 1 μg/μl in double-distilled water by matrix-assisted laser desorption ionization-time-of-flight mass spectrometry at Academia Sinica, Taiwan. The 2.7-kb plasmid DNA of pET-Blue 1 and the 4-kb plasmid DNA of pET Blue 1-149 were used as a source of double-stranded DNA. The 2.2-kb and 3.5-kb HBV RNAs were in vitro transcribed from pCH7-8T (35) and pGEM-4Z-HBV (40), respectively. Most chemical compounds were used at 0.5 to 1 μg/μl initially. In cases when no capsid reassembly was observed, tests were repeated with a wide range of concentrations of the tested chemicals (1 ng to 5 μg/μl).
Sucrose gradient ultracentrifugation.
Three micrograms each of undigested, micrococcal nuclease-digested, and reassembled full-length HBc 183 capsids (ayw or adr subtype) were adjusted to a 200-μl final volume with Tris-buffered saline and layered on top of a 4.6-ml 40 to 70% sucrose gradient. Ultracentrifugation was conducted in an MLS-50 rotor at 35,000 rpm for 18 h using a Beckman Optima MAX-XP ultracentrifuge. Approximately 0.25 ml per fraction was collected from the top, and 25 μl from each fraction was loaded on each lane of agarose gel, while 20 μl from each fraction was used for HBc/e enzyme-linked immunosorbent assay (ELISA) (International Immuno-Diagnostics, NV). The signal/cutoff data in Fig. 5 were processed as suggested by the vendor.
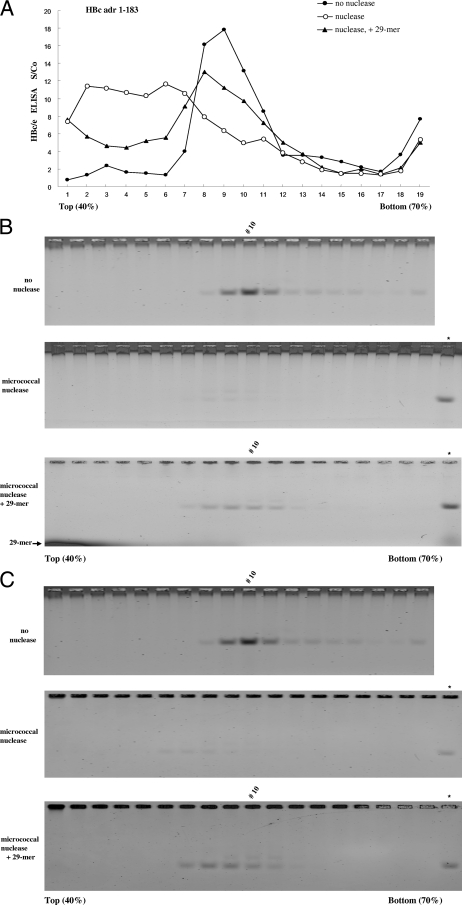
FIG. 5.
Sedimentation behaviors among undigested, micrococcal nuclease-digested, and reassembled HBc capsids (adr subtype) were compared using 40 to 70% sucrose density gradient centrifugation analysis. Aliquots of HBc capsids in different fractions collected from the gradient were subjected to ELISA for HBc/e (A), agarose gel electrophoresis and SYBR green II staining for nucleic acids (B), or SYPRO Ruby staining for proteins (C). (A) Reassembled HBc capsids cosedimented closely with undigested parental capsids around fractions 7 to 10. The profiles of digested capsids lost the peak around fractions 8 to 11 and shifted toward the top. Fractions 18 and 19 probably consist of aggregates of HBc particles near the bottom. Such aggregates were not observed in another independent experiment using freshly prepared HBc (ayw) (data not shown). S/Co, signal/cutoff ratio (see Materials and Methods). The symbol * in the last lanes of the middle and lower panels of panels B and C represents a positive control from HBc amino acid 1 to 183 capsids without micrococcal nuclease treatment. The darkened area at the bottom left corner of the lower panel of panel B reflects the 29-mer oligonucleotide. Very similar results were obtained using HBc capsids of ayw subtype (data not shown).
HBc capsid fractions collected from sucrose gradients were analyzed by 1% native agarose gel electrophoresis in Tris-borate-EDTA buffer for 1.5 h at 100 V. Encapsidated nucleic acids were stained with SYBR green II (Invitrogen Co.) at a 1:5,000 dilution for 2 h and visualized using an Amersham Biosciences Typhoon 9410 imager. After destaining with 10% methanol and 7% glacial acetic acid for 2 h, the same gel was restained for 12 h with SYPRO Ruby (Sigma Co.) for protein signal.
Construction of arginine-to-alanine HBc ARD mutants.
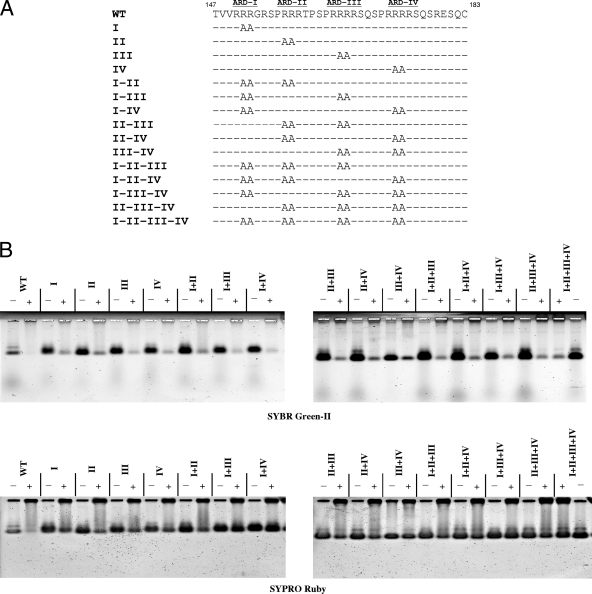
A total of 15 HBc ARD mutants were constructed by PCR amplification (QuikChange XL site-directed mutagenesis kit; Stratagene Co.), using the full-length WT (subtype adr) plasmid pET-HBc183 as a template. Arginine-to-alanine substitutions were introduced into HBc amino acids 151 and 152 of mutant ARD-I, 157 and 158 of mutant ARD-II, 165 and 166 of mutant ARD-III, and 173 and 174 of mutant ARD-IV. Other mutants, containing combinations of these four ARD mutations, are shown in Fig. 6. The sequences of mutagenesis primers, in the 5′-to-3′ direction and with alanine codons at mutation sites underlined, were as follows: ARD-I, CT ACT GTT GTT AGA GCC GCC GGC AGG TCC CC; ARD-II, GGC AGG TCC CCT GCC GCC AGA ACT CCC TCG CCT CGC; ARD-III, CT CCC TCG CCT CGC GCC GCC AGG TCT CAA TCG CCG CG; and ARD-IV, GG TCT CAA TCG CCG CGT GCC GCC AGA TCT CAA TCT CGG GAA TCT C. All mutants were confirmed by DNA sequencing.
FIG. 6.
Capsid stability was analyzed by micrococcal nuclease digestion of WT HBV capsids and 15 different mutant capsids containing various arginine-to-alanine (R-to-A) mutations at ARD of HBc (A). Unlike mutations at ARD-I and ARD-II, all R-to-A mutations involving ARD-III or ARD-IV significantly enhanced the capsid stability after nuclease digestion. Approximately 90% of HBc capsids were completely digested by micrococcal nuclease and measured by SYBR green-II staining (B). The residual 10% of incompletely digested capsids in most samples, visualized by SYBR green II staining, constituted some background noise level for the SYPRO Ruby staining assay in the bottom panel of panel B. To accurately interpret the data from SYPRO Ruby staining in a more quantitative manner, such background noise levels of incompletely digested samples have to be subtracted from SYPRO Ruby signals in all digested samples.
RESULTS
To assess the effect of adequate electrostatic interaction on capsid stability (integrity) and assembly, we used E. coli-expressed HBV capsids as an experimental model system. Previously, we discovered that when certain E. coli-expressed arginine-deficient mutant capsids were treated with micrococcal nuclease and DNase I, their packaged nucleic acids can be digested, resulting in alterations of capsid mobility on native agarose gels (22). To further elucidate the electrostatic interactions between the arginine residues (positive charge) and the encapsidated RNA (negative charge) of HBc capsids, here we used a more efficient micrococcal nuclease RNA digestion condition (lower-salt buffer, lower substrate concentration, no DNase I and Mg2+, and a larger amount of micrococcal nuclease) (see Materials and Methods). This new digestion condition resulted in almost complete removal of encapsidated RNAs from WT and several mutant HBc capsids. If adequate electrostatic interaction is indeed important for capsid stability, we should anticipate that the loss of encapsidated RNA by micrococcal nuclease digestion could lead to inadequate electrostatic interaction and thus to the loss of capsid stability.
As shown in Fig. 1, upper panel, we constructed a series of C-terminally truncated HBc mutants. These mutants contain various amounts of arginine residues. The digestion of encapsidated RNAs was monitored by EtBr staining of the native agarose gel containing both digested capsid samples and undigested (mock-digested) controls. Overall, there is a general trend of stronger EtBr staining for those undigested mutants with higher arginine contents (and thus larger amounts of encapsidated RNA). To monitor capsid integrity, the same gel was later reexamined with Coomassie blue staining (CBS). We observed three different groups of mutants displaying three different CBS patterns. Group 1 mutants, including HBc mutants 149, 154, and 157, exhibited no apparent change in CBS banding pattern before versus after micrococcal nuclease digestion. Group 2 mutants, including HBc 159, 164, 169, and 171, exhibited a more diffuse and reduced CBS banding intensity after micrococcal nuclease digestion. In mutants 159 and 164, the diffuse bandings are also slightly upshifted, suggesting an altered charge/mass ratio of less-structured capsids after micrococcal nuclease digestion. Group 3 mutants, including HBc 167, 173, 175, and WT full-length 183, almost completely lost CBS banding signals after micrococcal nuclease digestion, suggesting a complete loss of capsid integrity.
The loss of CBS signal in the group 3 mutants after micrococcal nuclease treatment could be caused either by the loss of HBc protein per se or the loss of capsid integrity. To distinguish between these two possibilities, we performed SDS-PAGE. As shown in the lower panel of Fig. 1, the micrococcal nuclease-treated core protein monomer on denaturing SDS-PAGE comigrated with the untreated control, indicating that the capsid monomer remained intact. Therefore, the loss of CBS signal in group 3 mutants appears to be due to the loss of specific capsid architecture rather than to protein degradation caused by protease contamination. To confirm this conclusion, we compared nuclease-treated and untreated samples under EM. Consistent with the gel assay results in Fig. 1, group 1 mutants, including HBc 149 and 154 (Fig. 2) and 157 (data not shown), did not reveal any significant changes in the numbers and morphology of capsid particles with or without micrococcal nuclease (S7) treatment. In contrast, very few particles can be found under EM in micrococcal nuclease-treated group 3 mutants, including HBc 167 and 183 (Fig. 2) and 173 and 175 (data not shown). The morphological change of HBc 164 after micrococcal nuclease treatment is very subtle, if any. There might be more “thick-wall” particles in untreated than treated HBc 164 (Fig. 2), suggesting the conversion after nuclease treatment from “thick-wall” capsids (less interior space by negative staining of 2% uranyl acetate) to “thin-wall” capsids (more interior space) (28).
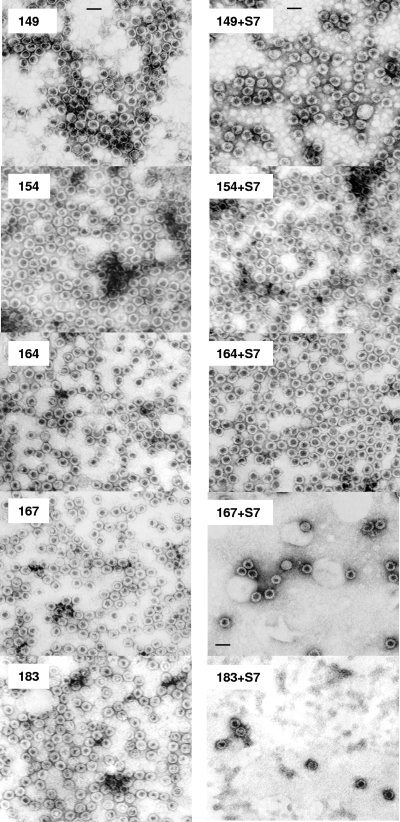
FIG. 2.
EMs of HBV capsids with or without micrococcal nuclease (S7) treatment. Sample 183 is the WT HBV capsids with full-length core protein. Mutants 149, 154, 164, and 167 are serially truncated at the carboxyl terminus (Fig. 1, top panel). These capsids were negatively stained and micrographs produced using a Philips EM 201 EM. Note the significant loss (10-fold) of capsid numbers in the micrococcal nuclease-treated capsids 167 and 183. In contrast, mutant capsids 149, 154, and 164 did not display a similar phenotype after micrococcal nuclease treatment. All pictures have the same magnification (scale bar, 40 nm).
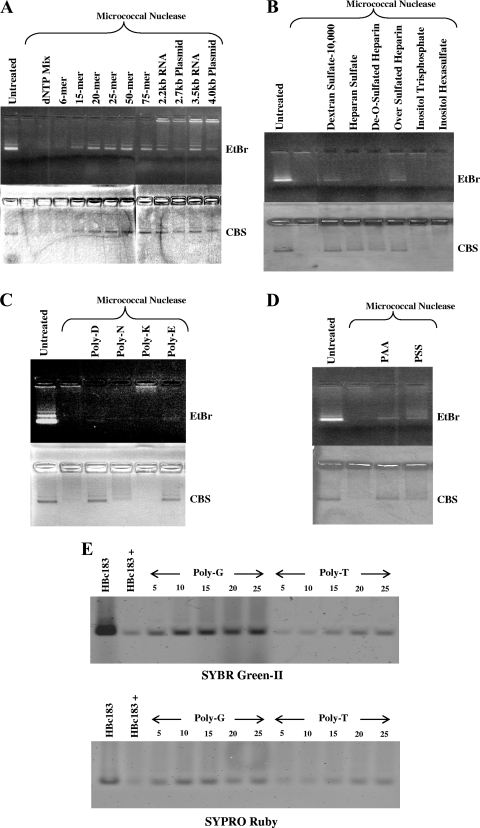
A logical question to follow from the results in Fig. 1 and 2 is whether the disassembled capsids can be reassembled by providing fresh exogenous RNA or DNA. Previously, it has been possible to catalyze in vitro capsid assembly from truncated HBc amino acids 1 to 149 or 13 to 183 using nonphysiological high-salt conditions (16, 37). To date, high-salt treatment (0.15 to 0.25 M NaCl, pH 7.5) remains the only successful method for inducing HBc capsid assembly (34). Here, we established a new capsid reassembly system using the micrococcal nuclease-treated full-length HBc amino acid 1 to 183 capsids, a low salt concentration (5 mM NaCl), and exogenous polyanion inducers. Because the enzymatic activity of micrococcal nuclease depends on the presence of calcium ion, one can inactivate micrococcal nuclease by chelating the calcium ion via the addition of EDTA. As shown in Fig. 3A, upon the addition of RNA (2.2 kb and 3.5 kb) or plasmid double-stranded DNA (2.7 kbp and 4.0 kbp), discrete banding by CBS staining reappeared on the native agarose gel. In addition, 15-mer, 20-mer, 25-mer, 50-mer, and 75-mer oligonucleotides as well as poly(C) and poly(I) (data not shown) also induced capsid reassembly. In contrast, a 6-mer oligonucleotide and dNTP mix induced no reassembly, even at high concentrations (Fig. 3A).
FIG. 3.
Reassembly of micrococcal nuclease-treated full-length HBc amino acid 1 to 183 capsids was assayed by native agarose gel electrophoresis. Capsids were treated with micrococcal nuclease to remove the nucleic acids packaged during spontaneous assembly in E. coli. Following the digestion, the disassembled capsids were incubated with a variety of compounds and allowed to reassemble. (A) The micrococcal nuclease-treated capsids were allowed to incubate with nucleic acids of different sizes and compositions. The 2.2-kb and 3.5-kb RNAs were made by in vitro transcription. (B) Typical results with several negatively charged polysaccharides, glycosaminoglycans, and inositol derivatives. The dextran sulfate shown here has an average MW of 10,000 Da. It should be noted that de-O-sulfated heparin still contains N-sulfated groups. (C) Synthetic polypeptides. (D) Two nonbiological polymers, PAA and PSS, were tested. EtBr stainings at loading wells of panels A to D probably are random aggregates of HBc proteins and particles. (E) Further testing of the effects of length versus sequence per se of homopolymer oligonucleotide inducers for capsid reassembly. SYBR green II staining for encapsidated nucleic acids and SYPRO Ruby staining for capsid proteins are more sensitive assays than EtBr staining or CBS. Using the same 8 μM concentration of all homopolymer samples, we detected weak, yet reproducible, reassembly of a 5-mer poly(G) and a 15-mer poly(T). HBc183 in the first lane is WT HBc capsids without treatment by micrococcal nuclease. HBc183+ represents samples of HBc 183 after nuclease treatment. Note that reassembly efficiencies of different inducers can be compared only when the assembly reaction for each inducer is performed at the respective optimal concentration.
Encouraged by the results in Fig. 3A, we asked if negative charge from non-nucleic acid can also facilitate capsid reassembly. Interestingly, sulfated polysaccharides (heparan sulfate, oversulfated heparin, and dextran sulfate, with an average MW of near 10,000), can induce capsid reassembly (Fig. 3B), while dextran sulfate, with an average MW near 500,000, cannot induce reassembly (data not shown). This result suggests that an inducer with too high an MW, despite its negative charge, could not serve as a good inducer for reassembly. On the other hand, inducers with too low an MW, such as inositol triphosphate and inositol hexasulfate, induced no reassembly either, despite their negative charges (Fig. 3B). We also observed weak reassembly by de-O-sulfated heparin. We speculate that this result could be related to its remaining negatively charged N-sulfate groups.
In addition to polynucleotides and polysaccharides, we also tested the potential effect of synthetic polypeptides on capsid assembly. As shown in Fig. 3C, both negatively charged poly(D) and poly(E) can catalyze reassembly, but not positively charged poly(K), with an average MW similar to those of poly(D) and poly(E). Furthermore, we observed very weak signal or background noise by more neutrally charged poly(N), with an average MW similar to those of poly(D) and poly(E). Finally, we tested nonbiological polyanions, such as PAA and PSS. As shown in Fig. 3D, PAA induced better reassembly than PSS. In contrast, we observed no capsid reassembly by the polycations polyethylenimine and PAH over a very wide range of concentrations (1 pg to 5 μg/μl) (data not shown). The negative results of PAH cannot be attributed to a problem with the MW, since PAH and PSS have similar MWs.
In Fig. 3A, we observed efficient reassembly using a 15-mer oligonucleotide and staining with EtBr and CBS. To test further the effects of length versus sequence per se on capsid reassembly, we used homopolymer poly(G) or poly(T) with variable lengths as inducers for reassembly, as well as more sensitive methods of SYBR green II staining for encapsidated nucleic acids and SYPRO Ruby staining for capsid proteins. As shown in Fig. 3E, we detected reassembly of a 5-mer poly(G) and a 15-mer poly(T). Because the molar concentrations of poly(T) and poly(G) used here were always the same (8 μM final), the difference between poly(G) and poly(T) in SYBR green II staining could reflect a difference in their respective affinities for SYBR green II, due to the different intrinsic physical properties of these two homopolymers. Furthermore, the SYPRO Ruby staining intensities of capsid samples reassembled by poly(G) are always stronger than those of capsids reassembled by poly(T) (Fig. 3E, lower panel), indicating a real difference between poly(G) and poly(T) in inducing capsid reassembly.
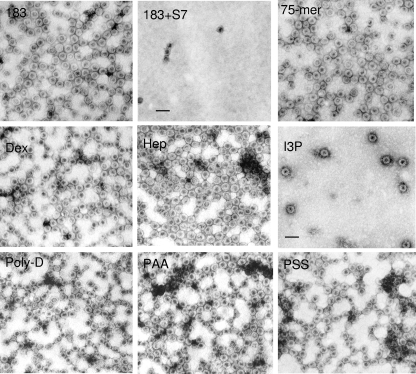
Some of the results in Fig. 3A to D obtained by gel assay were also confirmed by EM (Fig. 4). To further compare physical properties between reassembled and undigested capsids, we performed sucrose density gradient centrifugation analysis (Fig. 5). Indeed, reassembled capsids induced by a 29-mer oligonucleotide cosedimented closely with undigested parental capsids (Fig. 5A). In general, micrococcal nuclease digestion shifted the profiles to the left (top), while reassembly shifted the profile back to the original pattern of undigested capsids. A similar result was observed when poly(E) was used as an inducer (data not shown). The results in Fig. 5A obtained by ELISA were confirmed by native agarose gel electrophoresis (Fig. 5B and C). We noted that the peak of reassembled capsids could sometimes deviate from that of undigested capsids by one fraction to the top or bottom (Fig. 5A, B, and C). It remains unclear if this is due simply to experimental variations or to subtle differences in capsid conformations caused by their respective inducers [RNA versus 29-mer versus poly(E)]. Taking the results together, reassembled capsids appeared to have a size, shape, and density similar to those of their undigested counterpart. The results in Fig. 5 lend further support for the electrostatic interaction hypothesis.
FIG. 4.
EMs of micrococcal nuclease (S7)-treated WT full-length amino acid 1 to 183 capsids reassembled upon induction with various compounds (see Materials and Methods). Dex, dextran sulfate (10 kDa); Hep, heparan sulfate; I3P, inositol triphosphate. See Fig. 3 legend for other denotations. All pictures have the same magnification (scale bar, 50 nm).
DISCUSSION
Charge repulsion and electrostatic interaction hypothesis of capsid stability.
By studying a series of C-terminally truncated HBc capsids, we noted differences in capsid stability following micrococcal nuclease treatment (Fig. 1). Our previously coined “charge balance hypothesis” assumes that the positive charge of ARD can bind to the negatively charged phosphodiester links of encapsidated RNA. Depletion of the encapsidated RNA by micrococcal nuclease could result in positive charge repulsion, presumably caused by ARD in close contact in the context of full-length icosahedral particles. Such a repulsion force could outweigh the intersubunit hydrophobic interactions between HBc residues 113 to 143 (20) or residues 128 to 139 (34, 38), which contribute to capsid stability. Therefore, when the encapsidated RNA is digested, group I mutants containing fewer arginines are more stable due to reduced positive charge repulsion. Conversely, group 3 mutants containing more arginines are less stable due to increased positive charge repulsion after micrococcal nuclease digestion (Fig. 1).
This charge repulsion hypothesis can be tested further by capsid disassembly assay. When arginine to alanine (R-to-A) substitutions are introduced into ARD-III or -IV in the full-length HBc 183 context (Fig. 6A), capsid stability after micrococcal nuclease digestion is increased significantly, as measured by SYPRO Ruby protein staining (Fig. 6B, lower panel). Presumably, the charge repulsion force caused by ARD of full-length HBc in close contact must be reduced in these R-to-A mutants. In contrast, R-to-A mutations at ARD-I or -II, have little effect on capsid stability after nuclease digestion (Fig. 6B, lower panel). These results suggest that charge repulsion is largely due to ARD-III and ARD-IV.
An RNA scaffold interpretation.
In addition to minimizing charge repulsion, polyanions could serve as a scaffold for capsid assembly. Like a “beads-on-a-string model,” a scaffold hypothesis envisions linking together HBc subunits (beads) via their electrostatic interactions with a common polyanion molecule (string). Once HBc subunits are brought into close proximity by their bindings to a common polyanion molecule, they can polymerize into particles more easily. Similarly, some kinds of polyanions could absorb more water solvent and thus increase the relative concentration of HBc to above the threshold level required for capsid assembly (23). While the scaffold effect, if any, might contribute to the efficiency of capsid reassembly, it cannot by itself explain the results of capsid disassembly in Fig. 1 and 2. Our electrostatic interaction hypothesis and the RNA scaffold interpretation are not mutually exclusive. It is possible that pgRNA can function as a scaffold to facilitate capsid formation, in addition to providing adequate electrostatic interaction for the maintenance of capsid stability.
The low-MW anions (dNTP and inositol triphosphate), even at very high concentrations, are not good inducers for reassembly (Fig. 3 and data not shown). However, a 10-mer of poly(G) can be an efficient inducer (Fig. 3E). Most likely, very-low-MW anions do not have sufficient length or charge to recruit a sufficient number of HBc molecules. The reason that high-MW dextran sulfate (average MW, 500,000) cannot induce assembly is less clear (data not shown). It is possible that a bulky size or excessive negative charge cannot be tolerated by stable capsids.
Experimental evidence in support of the electrostatic interaction hypothesis.
Because of the lack of a known structure of the HBc tail (34, 38, 41), it is not clear whether all of the 16 arginines at the C terminus (Fig. 1) are making equal or unequal contributions to the electrostatic interaction with encapsidated RNA or DNA during the life cycle of HBV. It is therefore less straightforward to calculate mathematically any putative stoichiometry between the positive and negative charges of HBc capsids. However, there are at least 11 independent indications in support of this notion. (i) Bacterially expressed capsids contain low-MW RNA in amounts approximately equivalent to the 3.5-kb pgRNA (25, 37). These results imply that a roughly constant amount of negative charges may be optimal for forming stable capsids, although the packaged RNA is not of the authentic 3.5-kb pgRNA in natural infection. In filamentous bacteriophage fd, a stoichiometry-like relationship between positively charged coat protein and the negatively charged genome has also been observed (14). We noted as well that it has been reported in the literature that 1% NP-40-treated retroviral core particles can also be disrupted by RNase treatment, and all particles appeared to contain a roughly constant amount of RNA (26). Conceptually, this stoichiometry-like phenomenon resembles our electrostatic interaction hypothesis for HBV. (ii) The more phosphorylation there is at the C terminus of HBc (i.e., more negative charge) in vitro, the less RNA encapsidation there is in vitro by E. coli-expressed capsids (16). This result suggests again that a constant amount of negative charge may be optimal for most efficient in vitro RNA encapsidation. (iii) Similarly, when all three major serine phosphorylation sites of HBc were replaced with aspartic acid, the amount of packaged RNA was greatly reduced (9). When all three serine phosphorylation sites were replaced with glutamic acids, this triple mutant preferentially packaged in vivo the 2.2-kb spliced sgRNAs (19). These results suggest that extensive serine phosphorylation can reduce the efficiency and size of encapsidated RNA during in vivo RNA packaging. Paradoxically, when all three major serine phosphorylation sites were replaced with alanine, there was only little detectable pgRNA encapsidation (9, 21). We entertain the interpretation that pgRNA by itself may not have sufficient negative charge for productive assembly of stable capsids. A minimal degree of phosphorylation of HBc protein could supplement HBc capsids with more negative charge and thus be required for optimal pgRNA encapsidation and capsid stability in mammalian cells. However, since E. coli is not known to contain phosphorylation activity, then how could bacterial capsids package a total amount of 3.5-kb RNA similar to that of mammalian capsids (25, 37)? Among the several possible scenarios, our favorite explanation is that mammalian capsids can package one quantum amount of RNA (i.e., one 3.5-kb pgRNA molecule per capsid particle), while E. coli can make up to the total amount of negative charge equivalent to 3.5-kb RNA by packaging multiple smaller core-specific and nonspecific RNA species. It is conceivable that the folded structure of the 3.5-kb pgRNA in mammalian capsids must be different from that of the lower-MW RNA packaged by E. coli capsids. For example, the 3.5-kb pgRNA in mammalian capsids must fold into a specific higher-order structure in order to be recognized by viral polymerase (2, 12, 15, 24). Therefore, the actual amount of total effective negative charge of pgRNA in direct contact with arginines in mammalian capsids must be less than 3.5 kb, since some of the RNA moiety will loop out without being in contact with arginines of HBc. The “deficit” in effective negative charge required for optimal capsid stability can be offset by a small degree of phosphorylation of HBc before or during RNA encapsidation and capsid assembly. In contrast, the core-specific small mRNAs packaged by E. coli capsids are without the encapsidation signal (ɛ). They must fold into a different secondary structure of their own, and multiple copies of the encapsidated RNA in E. coli capsids could add up to a total amount of effective negative charge required for capsid stability without the supplementary negative charge from HBc phosphorylation. In this scenario, a constant total amount of effective negative charge, contributed by pgRNA, HBc phosphorylation, or copackaged lower-MW RNA, is favored by HBc for efficient capsid assembly and stability. (iv) As mentioned in the introduction, arginine-deficient HBc mutant 164 preferentially packaged the shorter 2.2-kb spliced sgRNAs (19, 22). Similarly, HBc mutant Cd163, which contains only one arginine less than mutant 164, appeared to package only RNA species shorter than 1.7 kb in nuclease-treated, immunoprecipitated core particles (4). Finally, mosaic particles containing WT and various amounts of mutant cores with reduced positive charge content (mutants C144Arg and C144Lys) also selectively packaged a shorter pregenome for encapsidation (13). Taken together, the reduction of arginine content (i.e., positive charge) of HBc appears to correlate with the reduction of the size of encapsidated RNA (i.e., negative charge). It appears that the stubborn requirement of a constant amount of encapsidated RNA by WT capsids can be negotiated flexibly, once the arginine content is varied in mutant capsids. (v) When the missing arginine residues were systematically restored to the C terminus of mutant 164, core particle-associated DNA and RNase-resistant RNA gradually increased in both size and intensity (22). Therefore, the increase of the arginine content of HBc appears to correlate with the increase of capsid stability and the size of core-associated DNA. (vi) The 3.5-kb pgRNA encapsidated by mutant 164 was more micrococcal nuclease sensitive than the 2.2-kb RNA encapsidated in the same mutant capsids (19, 22). These results suggest that excessive negative charge of RNA can cause structural alteration of capsids leading to increased micrococcal nuclease sensitivity. Indeed, nucleocapsids of duck hepatitis B virus class II mutants, which lack about half of the arginine-rich C terminus, disintegrated upon viral DNA maturation (18), suggesting again increased capsid instability due to increased charge imbalance. (vii) The experimental results in Fig. 1 and 2 suggest the existence of charge repulsion. (viii) Non-nucleic acid polyanions can also induce capsid reassembly (Fig. 3 and 4). (ix) The experimental results in Fig. 6 mapped the majority of charge repulsion activity to ARD-III and ARD-IV. (x) Relative to the intracellular capsids, virion-associated mature capsids are hypophosphorylated or dephosphorylated (29, 30). These results suggest that dephosphorylation of capsids occurs during capsid maturation, probably after the initiation of plus-strand DNA synthesis (3). Dephosphorylation of HBc can reduce the negative charge and serve as a counteracting mechanism for viral capsids to maintain adequate electrostatic interaction, particularly when the plus-strand DNA synthesis gradually builds up excessive negative charges (22). (xi) The replication defect of an arginine-deficient HBc mutant, 173GG, can be rescued by decreasing the negative charge content by E-to-A mutations at specific positions of HBc (Chua et al., submitted). This result suggests that, for HBV DNA synthesis, the relative amount of positive and negative charge per se is more important than the absolute amount of positive or negative charge.
A puzzle of empty particles in patients.
Predominant empty Dane-like particles are found to be prevalent in HBV-infected patients (1, 10, 32). Based on positive staining and EM, empty particles do not seem to contain any nucleic acid material. If charge repulsion between HBc subunits does exist according to our hypothesis, then how can these empty capsids be stable in natural infection? Recently, Kimura et al. (17) demonstrated that the capsids of empty particles consist of a 22-kDa core-related antigen, which lacks the entire ARD at the C terminus. Therefore, like HBc 149 capsids (Fig. 1), these empty capsids are stable without encapsidated nucleic acids, due to the lack of ARD and charge repulsion.
In summary, we established a novel in vitro disassembly/reassembly system for full-length HBV capsids. We proposed the existence of potential charge repulsion between HBc subunits in close contact in icosahedral particles. Such a positive charge repulsion force can be reduced or eliminated by electrostatic interactions with the negative charge from polyanions. The in vitro capsid reassembly system, using full-length HBc protein and a more physiological salt concentration, should provide us an opportunity to study the stoichiometry, biochemistry, and structure-function relationship of RNA encapsidation, genome maturation, and capsid stability in the future. It remains to be investigated whether this hypothesis of HBV could be applicable to other non-HBV viruses.
Acknowledgments
This work was supported by NIH RO1 grants; a UTMB John Sealy Foundation grant; the Summit Project of Academia Sinica, Taiwan; and a grant from the National Science Council of the Republic of China (NSC 96-3112-B-001-024) to C.S. P.K.C. was supported by a McLaughlin postdoctoral fellowship.
We thank Michael S. Wong for providing four chemicals, T. S. Su for providing the plasmid pCH7-8T, Robert Wang for his assistance with the construction of HBc mutants, and the EM Core Facility at UTMB, Texas, and Academia Sinica, Taipei, Taiwan.
Footnotes
Published ahead of print on 5 August 2009.
REFERENCES
- 1.Alberti, A., S. Diana, G. H. Scullard, W. F. Eddleston, and R. Williams. 1978. Full and empty Dane particles in chronic hepatitis B virus infection: relation to hepatitis B e antigen and presence of liver damage. Gastroenterology 75:869-874. [PubMed] [Google Scholar]
- 2.Bartenschlager, R., M. Junker-Niepmann, and H. Schaller. 1990. The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J. Virol. 64:5324-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basagoudanavar, S. H., D. H. Perlman, and J. Hu. 2007. Regulation of hepadnavirus reverse transcription by dynamic nucleocapsid phosphorylation. J. Virol. 81:1641-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beames, B., and R. E. Lanford. 1993. Carboxy-terminal truncations of the HBV core protein affect capsid formation and the apparent size of encapsidated HBV RNA. Virology 194:597-607. [DOI] [PubMed] [Google Scholar]
- 5.Birnbaum, F., and M. Nassal. 1990. Hepatitis B virus nucleocapsid assembly: primary structure requirements in the core protein. J. Virol. 64:3319-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blumberg, B. S. 1997. Hepatitis B virus, the vaccine, and the control of primary cancer of the liver. Proc. Natl. Acad. Sci. USA 94:7121-7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, B. J., and J. E. Richmond. 1982. Electron microscopy of hepatitis B core antigen synthesized in E. coli. Nature 296:677-679. [DOI] [PubMed] [Google Scholar]
- 8.Gallina, A., F. Bonelli, L. Zentilin, G. Rindi, M. Muttini, and G. Milanesi. 1989. A recombinant hepatitis B core antigen polypeptide with the protamine-like domain deleted self-assembles into capsid particles but fails to bind nucleic acids. J. Virol. 63:4645-4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gazina, E. V., J. E. Fielding, B. Lin, and D. A. Anderson. 2000. Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and duck hepatitis B viruses. J. Virol. 74:4721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerin, J. L., E. C. Ford, and R. H. Purcell. 1975. Biochemical characterization of Australia antigen. Evidence for defective particles of hepatitis B virus. Am. J. Pathol. 81:651-668. [PMC free article] [PubMed] [Google Scholar]
- 11.Hatton, T., S. Zhou, and D. N. Standring. 1992. RNA- and DNA-binding activities in hepatitis B virus capsid protein: a model for their roles in viral replication. J. Virol. 66:5232-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirsch, R. C., J. E. Lavine, L. J. Chang, H. E. Varmus, and D. Ganem. 1990. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature 344:552-555. [DOI] [PubMed] [Google Scholar]
- 13.Hui, E. K., K. L. Chen, and S. J. Lo. 1999. Hepatitis B virus maturation is affected by the incorporation of core proteins having a C-terminal substitution of arginine or lysine stretches. J. Gen. Virol. 80:2661-2671. [DOI] [PubMed] [Google Scholar]
- 14.Hunter, G. J., D. H. Rowitch, and R. N. Perham. 1987. Interactions between DNA and coat protein in the structure and assembly of filamentous bacteriophage fd. Nature 327:252-254. [DOI] [PubMed] [Google Scholar]
- 15.Junker-Niepmann, M., R. Bartenschlager, and H. Schaller. 1990. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packing of foreign RNA. EMBO J. 9:3389-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kann, M., and W. H. Gerlich. 1994. Effect of core protein phosphorylation by protein kinase C on encapsidation of RNA within core particles of hepatitis B virus. J. Virol. 68:7993-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kimura, T., N. Ohno, N. Terada, A. Rokuhara, A. Matsumoto, S. Yagi, E. Tanaka, K. Kiyosawa, S. Ohno, and N. Maki. 2005. Hepatitis B virus DNA-negative Dane particles lack core protein but contain a 22-kDa precore protein without C-terminal arginine-rich domain. J. Biol. Chem. 280:21713-21719. [DOI] [PubMed] [Google Scholar]
- 18.Kock, J., S. Wieland, H. E. Blum, and F. von Weizsacker. 1998. Duck hepatitis B virus nucleocapsids formed by N-terminally extended or C-terminally truncated core proteins disintegrate during viral DNA maturation. J. Virol. 72:9116-9120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kock, J., M. Nassal, K. Deres, H. E. Blum, and F. von Weizsacker. 2004. Hepatitis B virus nucleocapsids formed by carboxy-terminally mutated core proteins contain spliced viral genomes but lack full-size DNA. J. Virol. 78:13812-13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konig, S., G. Beterams, and M. Nassal. 1998. Mapping of homologous interaction sites in the hepatitis B virus core protein. J. Virol. 72:4997-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lan, Y. T., J. Li, W. Liao, and J. Ou. 1999. Roles of the three major phosphorylation sites of hepatitis B virus core protein in viral replication. Virology 259:342-348. [DOI] [PubMed] [Google Scholar]
- 22.Le Pogam, S., P. K. Chua, M. Newman, and C. Shih. 2005. Exposure of RNA templates and encapsidation of spliced viral RNA are influenced by the arginine-rich domain of human hepatitis B virus core antigen (HBcAg 165-173). J. Virol. 79:1871-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lingappa, J. R., M. A. Newman, K. C. Klein, and J. E. Dooher. 2005. Comparing capsid assembly of primate lentiviruses and hepatitis B virus using cell-free systems. Virology 333:114-123. [DOI] [PubMed] [Google Scholar]
- 24.Lott, L., B. Beames, L. Notvall, and R. E. Lanford. 2000. Interaction between hepatitis B virus core protein and reverse transcriptase. J. Virol. 74:11479-11489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melegari, M., V. Bruss, and W. H. Gerlich. 1991. The arginine-rich carboxyterminal domain is necessary for RNA packaging by hepatitis B core protein. p. 164-168. In F. B. Hollinger, S. M. Lemon, and H. S. Margolis (ed.), Viral hepatitis and liver disease. Williams & Wilkins, Baltimore, MD.
- 26.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nassal, M. 1992. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J. Virol. 66:4107-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman, M., F.-M. Suk, M. Cajimat, P. K. Chua, and C. Shih. 2003. Stability and morphology comparisons of self-assembled virus-like particles from wild-type and mutant human hepatitis B virus capsid proteins. J. Virol. 77:12950-12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perlman, D. H., E. A. Berg, P. B. O'connor, C. E. Costello, and J. Hu. 2005. Reverse transcription-associated dephosphorylation of hepadnavirus nucleocapsids. Proc. Natl. Acad. Sci. USA 102:9020-9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pugh, J., A. Zweidler, and J. Summers. 1989. Characterization of the major duck hepatitis B virus core particle protein. J. Virol. 63:1371-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purcell, R. H. 1994. Hepatitis viruses: changing patterns of human disease. Proc. Natl. Acad. Sci. USA 91:2401-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakamoto, Y., G. Yamada, M. Mizuno, T. Nishihara, S. Kinoyama, T. Kobayashi, T. Takahashi, and H. Nagashima. 1983. Full and empty particles of hepatitis B virus in hepatocytes from patients with HBsAg-positive chronic active hepatitis. Lab. Investig. 48:678-682. [PubMed] [Google Scholar]
- 33.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steven, A. C., J. F. Conway, N. Cheng, N. R. Watts, D. M. Belnap, A. Harris, S. J. Stahl, and P. T. Wingfield. 2005. Structure, assembly, and antigenicity of hepatitis B virus capsid proteins. Adv. Virus Res. 64:125-164. [DOI] [PubMed] [Google Scholar]
- 35.Su, T. S., C. J. Lai, J. L. Huang, L. H. Lin, Y. K. Yauk, C. M. Chang, S. J. Lo, and S. H. Han. 1989. Hepatitis B virus transcript produced by RNA splicing. J. Virol. 63:4011-4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wieland, S. F., and F. V. Chisari. 2005. Stealth and cunning: hepatitis B and hepatitis C viruses. J. Virol. 79:9369-9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wingfield, P. T., S. J. Stahl, R. W. Williams, and A. C. Steven. 1995. Hepatitis core antigen produced in Escherichia coli: subunit composition, conformational analysis, and in vitro capsid assembly. Biochemistry 34:4919-4932. [DOI] [PubMed] [Google Scholar]
- 38.Wynne, S. A., R. A. Crowther, and A. G. Leslie. 1999. The crystal structure of the human hepatitis B virus capsid. Mol. Cell 3:771-780. [DOI] [PubMed] [Google Scholar]
- 39.Yu, M., and J. Summers. 1991. A domain of the hepadnavirus capsid protein is specifically required for DNA maturation and virus assembly. J. Virol. 65:2511-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan, T. T., G. K. Sahu, W. E. Whitehead, R. Greenberg, and C. Shih. 1999. The mechanism of an “immature secretion” phenotype of a highly frequent naturally occurring missense mutation at codon 97 of human hepatitis B virus core gene. J. Virol. 73:5731-5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zlotnick, A., N. Cheng, S. J. Stahl, J. F. Conway, A. C. Steven, and P. T. Wingfield. 1997. Localization of the C terminus of the assembly domain of hepatitis B virus capsid protein: implications for morphogenesis and organization of encapsidated RNA. Proc. Natl. Acad. Sci. USA 94:9556-9561. [DOI] [PMC free article] [PubMed] [Google Scholar]