Abstract
We wanted to develop a therapeutic approach against rabies disease by targeting the lyssavirus transcription/replication complex. Because this complex (nucleoprotein N-RNA template processed by the L polymerase and its cofactor, the phosphoprotein P) is similar to that of other negative-strand RNA viruses, we aimed to design broad-spectrum antiviral drugs that could be used as a complement to postexposure vaccination and immunotherapy. Recent progress in understanding the structure/function of the rabies virus P, N, and L proteins predicts that the amino-terminal end of P is an excellent target for destabilizing the replication complex because it interacts with both L (for positioning onto the N-RNA template) and N (for keeping N soluble, as needed for viral RNA encapsidation). Thus, peptides mimicking various lengths of the amino-terminal end of P have been evaluated, as follows: (i) for binding properties to the N-P-L partners by the two-hybrid method; (ii) for their capacity to inhibit the transcription/replication of a rabies virus minigenome encoding luciferase in BHK-21-T7 cells; and (iii) for their capacity to inhibit rabies virus infection of BHK-21-T7 cells and of two derivatives of the neuronal SK-N-SH cell line. Peptides P60 and P57 (the first 60 and first 57 NH2 residues of P, respectively) exhibited a rapid, strong, and long-lasting inhibitory potential on luciferase expression (>95% from 24 h to 55 h). P42 was less efficient in its inhibition level (75% for 18 to 30 h) and duration (40% after 48 h). The most promising peptides were synthesized in tandem with the Tat sequence, allowing cell penetration. Their inhibitory effects were observed on BHK-21-T7 cells infected with rabies virus and Lagos bat virus but not with vesicular stomatitis virus. In neuronal cells, a significant inhibition of both nucleocapsid inclusions and rabies virus release was observed.
The etiologic agents of rabies disease belong to the Lyssavirus genus, Rhabdoviridae family, Mononegavirales order (45). Rabies encephalitis remains a serious burden for both public health and the global economy, with 55,000 human deaths annually, corresponding to 1.74 million disability-adjusted life-years worldwide (24). Even though an estimated 10 million people per year receive the optimal postexposure treatment recommended by the World Health Organization (51), there is still an insufficient availability and/or access to modern vaccines and immunoglobulins, particularly in rural regions of Asia and Africa that pay the heaviest tribute to the disease. Besides improving accessibility and compliance to existing treatments, the development of antivirals is a possible alternative which is particularly attractive for a disease with a rather long incubation period (2 months on average), even considering that rabies virus (RABV) is neurotropic and not readily accessible to drugs. An experimental antiviral treatment has recently been applied to a young symptomatic woman 1 month after she was bitten by a bat (50). It consisted of the following: (i) inducing a deep coma to limit neuronal dysfunctions and to provide time for the maturation of the native immune response and (ii) applying an empirical combination of drugs (ketamine, midazolam, ribavirin, and amantadine) that had shown some effect against RABV when tested individually in vitro or in animal models (20, 21, 50). One could speculate as to whether this original protocol was successful since repetitions with some variations in various parts of the world have unfortunately failed to help other symptomatic patients to recover (18). However, it has obviously provoked an increase in interest in antirabies antiviral approaches.
The transcription/replication complex is an attractive antiviral target. It is composed of the RNA genome permanently encapsidated by the nucleoprotein N, resulting in a helical ribonucleocapsid (RNP). The ultrastructure of the RABV RNP has been studied by electron microscopy (40), and the crystal structure of RNP-like rings, formed by 11 protomers of recombinant N proteins encapsidating RNA, has been solved (1, 19). The RNP constitutes the template for the RNA-dependent RNA polymerase (L protein) and its cofactor, the phosphoprotein P. This complex is typical of negative-stranded RNA viruses, and this feature is favorable for developing broad-spectrum antiviral drugs in the absence of (or with low) cellular toxicity. Recent structural data obtained from elements of the transcription/replication complex indicate a great similarity between viruses despite substantial differences in the primary sequence of their proteins (30, 31, 33, 39, 44).
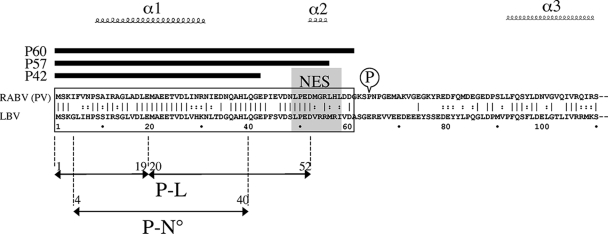
The RABV phosphoprotein P is an essential intermediate of the complex: (i) it mediates the correct positioning of the L polymerase on the N-RNA template, and (ii) it acts as a chaperone by forming N°-P (where N° is soluble N protein with no viral RNA) complexes that prevent N from binding to cellular RNA and favors its delivery to the viral RNA for encapsidation into new RNPs. Numerous studies have addressed the structure/function relationships of the P protein (Fig. 1). Coimmunoprecipitation and two-hybrid experiments have shown the presence of two independent N-binding domains (9, 12, 22). The stronger one, located at the C-terminal side, binds to the nucleoprotein N in the N-RNA complex. Its crystal structure has been solved, and the residues important for interaction have been precisely mapped (33). The weaker N-binding site, located at the N terminus of P, is responsible for the chaperone function by binding of the newly produced N° (29, 32). Using proteolysis analysis of purified N°-P complexes, residues 4 to 40 of P have been shown to directly bind N° (34). The N terminus of P is also involved in binding to the L polymerase (Fig. 1). By using coimmunoprecipitation of deletion mutants of P and L coexpressed in mammalian cells, residues 1 to 19 of P have been shown to interact with the 566 residues at the C terminus of L. This interaction may be stabilized when the amino-terminal peptide of P is enlarged up to residue 52 (10). In between the two N°/L and N-RNA binding sites at the NH2 and COOH sides, respectively, the P protein forms dimers through its central region (14, 15, 22). Interestingly, P mRNA directs the synthesis of four N-terminally truncated P products (P2 to P5) by a leaky ribosome scanning mechanism (4, 8) (Fig. 1), which allows uncoupling of binding sites to the different RNP partners.
FIG. 1.
Sequence alignment of the N-terminal half of P proteins of RABV (PV strain; Lyssavirus genotype 1 [GT1]) and LBV (isolate V267; GT2). The secondary structure prediction for the N-terminal end of the RABV P protein is from Mavrakis et al. (32). The NES of the RABV P protein (36) is outlined in gray, and the interaction domains of the RABV P protein, with the polymerase L (P-L) (10) and with the nucleoprotein N° (P-N°) (32) are indicated. The peptides P42, P57, and P60 are represented. The serine in position 63 outlines one of the two serine residues that are phosphorylated on the RABV P protein (CVS strain) by the RVPK (15). The second serine residue (position 64) is changed into a proline in the RABV PV strain.
In cells, the P (Met 1) and P2 (Met 20) products stay in the cytoplasm due to the presence of a CRM1-dependent nuclear export signal (NES) at position 48 to 59, whereas P3 (Met 53), P4 (Met 69), and P5 (Met 83) are routed to the nucleus due to a nuclear localization signal at position 172 to 297 (38). When both NES and nuclear localization signal are present, NES is dominant (38). A second NES, dependent on the phosphorylation state of the protein, has also been located at the C-terminal side of P (36). Five phosphorylation sites have been located along the P protein, targeted by two distinct cellular kinases: protein kinase C for the serine 162/210/271 sites and RABV protein kinase (RVPK) for the serine 63/64 sites (16). Variability in phosphorylation patterns, protein length, and cellular distribution provide P with a significant variability to regulate its interactions with both viral and cellular factors. For example, the P protein is the main element that hijacks the interferon (IFN) pathway (7, 41). By interacting with the TBK-1 kinase, P inhibits IFN regulatory factor 3 phosphorylation and IFN induction (6). IFN signaling is blocked by an interaction between P and STAT1 that prevents nuclear translocation of STAT1 (5, 46). P3 competes with the DNA binding domain of STAT1 in the nucleus and inhibits the expression of IFN antiviral genes (47). Finally, P and P3 interact with PML and probably abrogate its antiviral effect (4).
Because of its strong interactivity within the transcription/replication complex, the P protein is a target of choice for an antiviral strategy aiming to destabilize the complex and inhibit viral replication. The N terminus is particularly promising as a target because of its double interaction with both N° and L. Based on this observation, we have evaluated the antiviral potential of peptides encompassing different lengths at the N terminus of P as competitive inhibitors for the transcription/replication machinery. P42 (the first 42 residues) corresponds almost exactly to the N° binding site (34). P60 (the first 60 residues) encompasses the region conserved between all lyssavirus P proteins (37). A preliminary study showed that both P60 and P42 bind to N°, with P60 binding as strongly as P itself while P42 binds less efficiently (34). In the present work, the interaction map of these two peptides with both N and L has been refined, and their inhibitory potential concerning RABV replication was evaluated on RABV minireplicons as well as in infected cells. Finally, the spectrum of their antiviral effect has been estimated on a distantly related lyssavirus as well as on a vesiculovirus.
MATERIALS AND METHODS
Cells and viruses.
Baby hamster kidney cells (BHK-21-T7) were grown in Glasgow minimal essential medium supplemented with 8% fetal calf serum (FCS), 10% tryptose phosphate, and 10 mM HEPES. BHK-21-T7 is a BHK-21 cell clone that stably expresses the T7 RNA polymerase.
BSR cells (a clone of BHK cells) were grown in Dulbecco's modified Eagle's medium (DMEM)-Glutamax I (Gibco) supplemented with 8% FCS and gentamicin (40 mg/liter).
Cells were infected with the vTF7-3 T7 recombinant vaccinia virus (vT7 cells), the PV strain of RABV, Lagos bat virus (LBV), or vesicular stomatitis virus (VSV).
SK-N-SH CP is a subclone of the human neuroblastoma cell line SK-N-SH (ATCC HTB11) (26). SK-N-SH and SK-N-SH CP were grown in DMEM-Glutamax I (Gibco) high glucose (4.5 g/liter)-Na Pyr, supplemented with 10% fetal bovine serum (Gibco/BRL) plus penicillin-streptomycin (50 mg/liter).
Yeast two-hybrid method.
Various segments of the RABV (PV strain) P protein were checked by the yeast two-hybrid technique for their ability to interact with the homologous full-length N protein (450 amino acids [aa]) or with the homologous carboxy-terminal extremity of the L protein (504 aa between positions 1624 and 2127). All P-segment-encoding constructs were amplified by standard PCR using high-fidelity EX Taq (Takara) and pairs of primers 68 nucleotides long and having the following chimerical composition: the first 48 nucleotides (5′ end) were matched with the destination vector pACT2 (Forward, 5′-AACTATCTATTCGATGATGAAGATACCCCACCAAACCCAAAAAA AGAG-3′; Reverse, 5′-ATGCACAGTTGAAGTGAACTTGCGGGGTTTTTCAGTATCTACGATTCA-3′), and the remaining 20 nucleotides (3′ end) were specific to the region to be amplified.
In pACT2 these peptides are expressed fused to the COOH side of the GAL4 transcription factor-activating domain. Cloning was performed following a gap-repair procedure using corresponding reagents and media (48). Briefly, yeast cells (Saccharomyces cerevisiae AH109) were shaken at 30°C in a rich YEPD (yeast extract, peptone, and dextrose) medium until an optical density at 600 nm of 0.4 to 0.5 was reached. Then the cells were cotransformed by the lithium acetate method (48) with 250 ng of linearized pACT2 empty vector (SmaI) and 500 ng of the linear PCR product in the presence of 2 μl (11 mg/ml) of boiled salmon sperm DNA (Sigma). After incubation for 30 min at 30°C, the yeast cells were heat shocked for 15 min at 42°C and then left to grow in petri dishes for 1 week at 30°C in a synthetic medium lacking leucine to select only cells that had been transformed with recircularized pACT2 repaired by double recombination. Several clones were sequenced from each transformation in order to obtain a clone with the correct sequence and reading frame. The pACT2 vector containing the peptide was then recovered according to the procedure described by Soni and Murray (42). All plasmids were transformed and amplified in the Escherichia coli DH5α strain.
For cloning of the full-length N protein (450 aa) or the carboxy-terminal extremity of the L protein into the pGBKT7 plasmid, another recombinatorial cloning system was used (Gateway; Invitrogen). The corresponding N and L sequences were first amplified by PCR using pairs of chimerical primers. In this case, the 5′ end of the forward and reverse primers started with the attB1.1 and the attB2.1 recombination sequences, respectively, and were followed by the targeted N- or L-specific sequences: attB1.1, 5′-GGGGACAACTTTGTACAAAAAAGTTGGCATG-3′; attB2.1, 5′-GGGGACAACTTTGTACAAGAAAGTTGGTTA-3′.
This allowed the insertion of the corresponding sequence into pDONOR 207 (Invitrogen) before further transfer by double recombination into pGBKT7 in which the N protein and the C-terminal part of the L protein (L-Cter) protein had been cloned fused to the COOH side of the GAL4 transcription factor binding domain. Plasmids were amplified in E. coli DH5α strain.
For two-hybrid experiments, yeast cells were cotransformed with pGBKT7 encoding either the N or the L-Cter proteins, with pACT2 encoding one of the different P peptides. The cotransformed yeast cells were grown in medium lacking leucine, tryptophan, and histidine to allow only the growth of yeast containing both plasmids (pGBKT7 and pACT2 encode tryptophan and leucine, respectively) and to evaluate the interaction between the fused proteins (histidine under the control of the reconstituted GAL4 promoter). The addition of low concentrations of 3-amino-1,2,4-triazole (3-AT; Sigma-Aldrich) allowed us to evaluate the strength of the interaction and to eliminate false-positive results.
Plasmid constructions.
Standard manufacturer's protocols were followed for RNA extraction using Trizol reagent (Life Technologies) and Expand Reverse Transcriptase (Roche) for reverse transcription (RT) and an Expand high-fidelity PCR system (Roche) for PCR. For T7-driven expression of RABV (PV strain) proteins, pBluescript II KS (+/−) plasmids pT7-N, pT7-P, pT7-M, and pT7-G (Stratagene) encoding the full-length N (450 aa), P (297 aa), matrix (M; 202 aa), and glyco- (G; 505 aa) proteins, respectively, and pT7-L (pBluescript SK) encoding the full-length L protein (2,127 aa) were described earlier (22, 28). The RABV (PV strain) minigenome was transcribed by the T7 RNA polymerase from pDI.mut plasmids as described earlier (28). The RABV (PV strain) minigenome is composed (from 5′ to 3′) of the trailer, the L stop transcription signal, the luciferase gene in the negative sense, the N start transcription signal, and the leader sequence. This allows luciferase mRNA transcription to be controlled by N start and L stop RABV transcription signals. To perfectly mimic the extremities of the wild-type viral genome, the minigenome RNA is flanked by a hammerhead and a hepatitis-delta virus genomic ribozyme at the 5′ and 3′ ends, respectively (28).
Peptides P42, P57, and P60 were cloned into (i) either pCINeo3Flag plasmids (Sigma modified with a gateway cassette), which added three Flag (3Flag) sequences (3×DYKDDDDK) to the NH2 terminus of the peptide (22), or (ii) into peGFPC1 plasmids (Clontech), which added enhanced green fluorescent protein (eGFP) sequences to the NH2 terminus of the peptide.
Cloning junctions and the complete open reading frame of the PCR-generated fusions peptides or proteins were verified by sequencing on an ABI 377 automatic sequencer (Applied Biosystems). All plasmids were amplified in E. coli strain DH5α and purified by chromatography on Qiagen columns.
Minireplicon system assay for inhibition test.
The minireplicon system assay was slightly modified from Le Mercier et al. (28). BSR cells were plated in a 96-well plate (3 × 104 cells per well) in DMEM-Glutamax I (Gibco) supplemented with 5% FCS. After 24 h at 37°C (in 5% CO2), the medium was removed, and the cells were infected for 30 min at 37°C with vTF7-3 T7 recombinant vaccinia virus (multiplicity of infection [MOI] of 10 PFU/cell) in DMEM to allow cytoplasmic expression of the T7 RNA polymerase.
For the reconstitution of a minireplicon in the cytoplasm, the medium was removed after 30 min, and the cells in each well were transfected with pT7-N (0.1 μg), pT7-P (0.1 μg), pT7-L (0.02 μg), and pDImut (0.5 μg) using the FuGENE 6 reagent (catalog number 11814443001; Roche). Briefly, for each well, the necessary volume of FuGENE (3 μl/μg DNA) was diluted in 100 μl of serum-free DMEM; DNA was added, mixed, and incubated for at least 15 min at room temperature, and then the mix was added to the cells. After 2 h, the transfection medium was removed, fresh DMEM supplemented with 5% FCS was added, and the cells were incubated for various times at 37°C. The N, P, and L proteins and the RNA minigenome formed a functional RNP template resulting in luciferase gene transcription, and thus the amount of luciferase was related to the transcriptional activity of the reconstituted RNP. The inhibitory effect of P42, P57, or P60 on luciferase expression by the RNP was evaluated by cotransfection of 0.1 μg of plasmid pCINeo3Flag-P42, pCINeo3Flag-P57, pCINeo3Flag-P60, or pCINeo3Flag alone in vT7-BSR cells.
Pseudovirus production assay.
vT7-BSR cells were transfected with pT7-N (0.1 μg), pT7-P (0.1 μg), pT7-L (0.02 μg), and pDImut (0.5 μg) in the presence of 0.1 μg of pCINeo3Flag-P42, pCINeo3Flag-P57, pCINeo3Flag-P60, or pCINeo3Flag. In addition, pT7-M (0.1 μg) and pT7-G (0.1 μg) plasmids were added to allow release of pseudoviruses (encoding only luciferase) in the cell supernatant. Transfection was performed under the same conditions as in the minireplicon system, using FuGENE in proportional amounts to transfected DNA. The cell supernatant was transferred 48 h posttransfection to fresh vT7-BSR cells previously transfected with pT7-N (0.1 μg), pT7-P (0.1 μg), and pT7-L (0.02 μg) to enhance the luciferase signal from the infectious pseudovirus. The luciferase signal was measured 24 h later. It resulted exclusively from the transcription of incoming RNP and was thus proportional to pseudovirus production.
Luciferase assay.
At determined times posttransfection, each BSR cell well was washed with phosphate-buffered saline (PBS) and overlaid with 100 μl of passive lysis buffer (Promega) for 15 min at room temperature. Then, 50 μl per well was transferred into a new well of white 96-well plates (catalog number 675083; Greiner Bio-One), and luciferase expression was measured using a Berthold plate luminometer (Centro LB 960) by injecting 25 μl/well of the luciferase assay reagent (catalog number E1501; Promega) and counting for 5 s. Luciferase activity was measured at least in triplicate, and transfection was normalized by cotransfecting plasmid pGL4.74 hRluc/TK encoding Renilla luciferase (Promega).
Quantitative RT-PCR (qRT-PCR).
Total RNAs were extracted 48 h posttransfection from vT7-BSR cells with the different elements of the minireplicon system using an RNeasy minikit (Qiagen), and two treatments with Turbo DNase (catalog number 1907; Ambion) were added to completely remove residual plasmid and genomic DNA, following the manufacturer's instructions. Positive-strand RABV cDNAs were first obtained using a specific forward primer (5′-ATGGCGCCGGGCCTT-3′) and the Superscript II (Invitrogen) reverse transcriptase in a 20-μl final volume, according to the manufacturer's instructions. In a second step, the addition of the reverse primer 5′-ACGCTTAACAACCAG-3′ allowed specific amplification of the cDNA of the positive-strand minigenome RNA resulting from the replication of the reconstituted RNP. A real-time PCR assay was performed using LC FastStart DNA Master Plus SYBR Green I (Roche) and a LightCycler (Roche), following the manufacturer's instructions. Briefly, 5 μl of cDNA solution diluted fivefold was amplified in a final volume of 15 μl with both forward and reverse primers. Cycling parameters were as follows: denaturation at 95°C for 10 min, followed by 50 cycles of denaturation at 95°C for 10 s, annealing at 48°C for 20 s, and extension at 72°C for 15 s. The melting curve was obtained with 15 s at 55°C, followed by a cooling step of 30 s at 40°C.
Western blotting.
Cell monolayers were treated with passive cell lysis buffer (Promega). Lysates were separated for 1 h at 100 V on a 4 to 12% NuPAGE Bis-Tris gel and an morpholinethanesulfonic acid migration buffer (Invitrogen) and were then electrotransferred for 1 h at 75 V onto a nitrocellulose membrane (Hybond ECL; Amersham Bioscience). The membrane was saturated overnight at 4°C in 3% bovine serum albumin (BSA)-PBS, washed three times in 0.2% Tween 20-PBS, and then subjected to sequential incubations, as follows: (i) with a rabbit anti-RNP polyclonal antibody (dilution 1/5,000) or with a mouse anti-Flag monoclonal antibody (Sigma; dilution 1/800) in 3% BSA-PBS for 1 h at room temperature, followed by three washes in 0.2% Tween 20-PBS; (ii) with a goat anti-rabbit or a goat anti-mouse antibody conjugated to horseradish peroxidase (dilution 1/10,000; Vector) in 3% BSA-PBS for 1 h at room temperature, followed by three washes in 0.2% Tween 20-PBS. Development was carried out on Hyperfilm MP (Amersham Bioscience) using an ECL SuperSignal kit (Pierce).
Determination of inhibitory effect of peptides derived from the amino terminus of P on RABV-infected cells.
BHK-21-T7 cells were cultured in Labtek chambers (35,000 cells per chamber) for 24 h at 37°C in DMEM supplemented with 5% FCS. To assess the replication inhibition with the eGFP-tagged peptide from T7-expressed plasmids, cells were transfected with 0.2 μg of plasmid peGFPC1-P42, peGFPC1-P57, peGFPC1-P60, or peGFPC1 (control) with FuGENE (Roche Diagnostics) according to the manufacturer's protocol. Five hours posttransfection and incubation at 37°C, cell monolayers were washed twice with DMEM and then infected with RABV diluted in DMEM (5 PFU/cell). After incubation for 1 h at 37°C, the medium was discarded, and cells were washed with DMEM and then incubated for 24 h at 37°C in DMEM supplemented with 5% FCS and 5% CO2.
To measure the replication inhibition with synthetic peptides, P42, P57, and P60 were obtained from New England Peptide with their carboxy-terminal extremity fused to a Tat peptide (NH2-RKKRRQRRRC-COOH) in order to allow transmembrane passage. The peptide-Tat construct ended with a C-terminal cysteine residue to fix an Alexa Fluor 488 molecule, allowing visualization in cells. BHK-21-T7, SK-N-SH, or SK-N-SH CP cell monolayers were washed twice with DMEM and then infected with RABV (5 PFU/cell). After incubation for 1 h at 37°C, cells were washed with DMEM and 10 μM concentrations of synthetic peptides were added or not (control) in DMEM supplemented with 5% FCS (BHK-21-T7) or in DMEM-Glutamax-I-high glucose-Na Pyr supplemented with 2% fetal bovine serum (SK-N-SH and SK-N-SH CP). Cells were then incubated for 14 h at 37°C in the presence of 5% CO2.
To evaluate the percentage of infection, cells were fixed with PBS-4% paraformaldehyde and then permeabilized with 80% acetone for 15 min at 0°C. The accumulation of intracellular RABV RNP was detected by incubation of fixed cells for 1 h at 37°C with rabbit anti-RNP serum, followed by incubation with Cy3 anti-rabbit conjugate under the same experimental conditions and visualized by immunofluorescence (Leica DMRB microscope). In contrast, the detection of cells containing GFP, peptide-GFP, or peptide-Alexa Fluor 488 was determined directly by fluorescence (Leica DMRB microscope).
The percentage of inhibition of infection in cells expressing P42-eGFP, P57-eGFP, or P60-eGFP was calculated as follows: [1 − (percentage of peptide-containing RABV-infected cells/percentage of control-GFP RABV-infected cells)] × 100. For each triplicate, 50 GFP-positive cells were examined for the presence of RABV RNP. The percentage of inhibition in cells expressing synthetic Tat peptides was calculated as follows: [1 − (peptide-containing RABV-infected cells/RABV-infected cells)] × 100. This approach was necessary because we did not have Tat peptide alone for a control. The MOI that was used was calculated to reach >90% of RABV nucleocapsid-positive cells. High stringency was applied during image treatments to detect very small RABV nucleocapsid aggregates, and all cells having a single small (even very small) fluorescent RNP inclusion (red point) were counted as infected. The fluorescence index was calculated with ImageJ software (Wayne Rasband, NIH), and statistical analysis was performed with GraphPad VS software.
RESULTS
Interaction map of P assessed by yeast two-hybrid analysis.
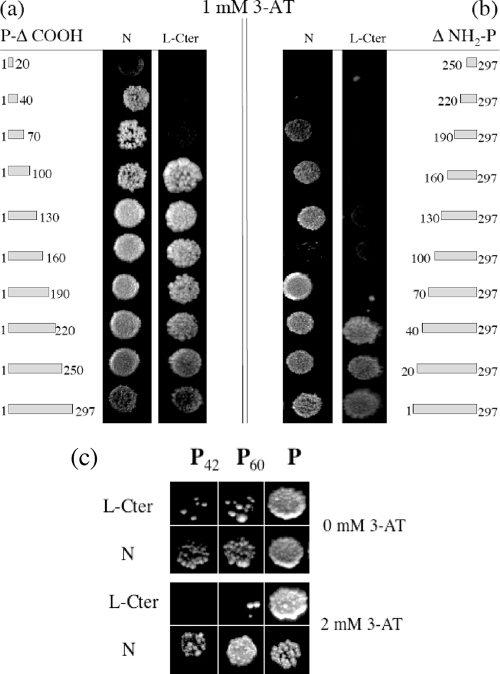
Various approaches have shown that the amino-terminal extremity of the P protein is a key element in the RNP complex by its interaction with both the N° nucleoprotein (34) and the L polymerase (10). We first completed the interaction map of P by a systematic two-hybrid analysis of progressive NH2 and COOH truncations of P with N and with the L-Cter protein that contains the P interaction site (10). As shown in Fig. 2, the interaction between N and P requires at least either one domain included in the first 40 residues of P (Fig. 2a) or one domain included in its last 108 residues (Fig. 2b). This confirms the existence of two independent N binding sites previously described at the two extremities of P: one binding to the N-RNA template at the COOH side (33) and the other to N° at the NH2 side (34). The absence of an interaction between N and the P protein with a deletion of its 99 amino-terminal residues (Fig. 2b, ΔNH2-P 100-297) is probably due to an abnormal folding of the deleted protein. With respect to the interaction of the P protein with L-Cter (encompassing the last 504 residues of L), the first 100 residues of P are required for a strong interaction (Fig. 2a). On the other hand, the interaction between P and L-Cter is not affected by a deletion of the first 40 residues of the P protein, whereas deletion of the first 70 residues totally inhibits the interaction (Fig. 2b). Thus, the domain of residues 40 to 70 of P appears crucial for the interaction with L-Cter (Fig. 2b) although the following 30 residues (up to 100) seem to stabilize the interaction (Fig. 2a).
FIG. 2.
Systematic two-hybrid analysis of progressive truncations of the carboxy-terminal extremity (a) and the amino-terminal extremity (b) of the P protein with the N protein or with the last 504 aa of the L protein (L-Cter; residues 1644 to 2127) in the presence of 1 mM 3-AT. (c) Two-hybrid experiment showing the interaction of P42, P60, and intact P with L-Cter and N at 0 mM or 2 mM 3-AT.
Taken together, our two-hybrid results obtained using NH2- and COOH-truncated P highlight the functional importance of the amino-terminal end in the transcription/replication complex since (i) the first 40 residues are sufficient for the interaction with N° and (ii) the domain 40 to 100 is essential to bind L-Cter. In the perspective of an antiviral approach, we investigated whether peptides of various lengths mimicking various parts of the amino-terminal end of P were able to destabilize the transcription/replication complex and interfere with RABV replication. We compared two the P42 and P60 NH2 residues of P. P42 corresponds almost exactly to the N° binding site (34). P60 encompasses the NH2 domain conserved between P proteins of the most divergent lyssaviruses, such as RABV, LBV, and Mokola virus (22) (Fig. 1). In addition, a previous scanning of the P protein with software predicted that P42 hosts an α-helix, possibly involved in a coiled-coil structure, and that its folding could be stabilized when increased up to 60 residues (13, 34).
Using two-hybrid experiments and coimmunoprecipitation in BSR cells, we have previously shown that P42 and P60 are able to bind N although P42 does not do so as strongly as P60 or P (34). We further verified that these interactions were maintained under more stringent conditions by adding 2 mM 3-AT (Fig. 2c). The results shown in Fig. 2c also indicate that the interaction of P42 and P60 with L-Cter is weak in the absence of 3-AT and disappears with 2 mM 3-AT. This result is coherent with the absence of an interaction of the first 40 and first 70 NH2 amino acids of P with L-Cter in the presence of 1 mM 3-AT (Fig. 2a). In summary, P42 and P60 clearly interact with N and very weakly with L-Cter, and P60 is better than P42 in both cases.
Inhibition by P42 and P60 of a RABV minireplicon.
To evaluate whether P42 and P60 were able to interfere with RABV replication, we set up a minireplicon assay based on plasmids under the control of the T7 promoter in BSR cells infected by recombinant vT7-vaccinia virus. This system reconstitutes a functional RABV RNP in the cytoplasm upon transfection of four plasmids: one plasmid encoding a minigenome composed of the leader/trailer genomic regions flanking a luciferase reporter gene under the control of RABV transcription signals and three plasmids encoding the N, P, and L proteins. The production of luciferase in this system combines successively the encapsidation of the minigenome into RNP and its transcription by the L/P complex. In addition, the presence of large amounts of N probably stimulates RNP replication, thus increasing the number of templates available for luciferase expression.
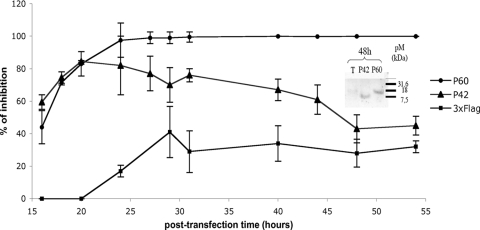
To investigate the inhibitory potential of P42 and P60 on transcription/replication, the kinetics of luciferase expression was followed for 55 h after cotransfection of plasmids encoding P42-3Flag, P60-3Flag, or 3Flag alone, together with the minireplicon system. Both P42-3Flag and P60-3Flag exhibited an inhibitory effect significantly higher than 3Flag alone (Fig. 3). This indicated that the inhibition was P peptide specific even if the 3Flag tag somehow contributed to the effect, as was shown by coupling different tags, such as cMyc or GFP, to P42 and P60 (data not shown). It should be noted that the luciferase signal constantly increased all the time but that this increase was very limited in the presence of P60-3Flag (stable around 104 units/luciferase after 55 h), whereas it was less controlled in the presence of P42-3Flag and still less with 3Flag alone (reaching 106 relative luciferase units (RLU) after 55 h) (data not shown). Thus, the inhibitory effect of the peptides was clearly measured on transcriptionally active reconstituted RNPs. The kinetics of inhibition by P42-3Flag and P60-3Flag evolved differently (Fig. 3). While the two peptides inhibited the luciferase signal during the first 20 h after transfection equally efficiently (70 to 75%), P60-3Flag reached a very strong inhibitory effect (>98%) after 24 h that was maintained up to 55 h. In contrast, the inhibitory effect of P42-3Flag progressively decreased after 24 h and stabilized at 43% after 48 h, a value significantly higher than 3Flag alone (28%). This decrease in inhibition was not due to a more specific sensitivity of P42-3Flag to degradation since almost the same quantities of P42-3Flag and P60-3Flag were observed 48 h after transfection (Fig. 3, inset).
FIG. 3.
Kinetics of inhibition of luciferase expression from an RABV minireplicon by plasmids encoding P42 or P60. BSR cells were first infected by a recombinant vT7-vaccinia virus and then transfected with plasmids encoding the RABV minigenome; the N, P, and L proteins; and pCINeo plasmids encoding P42-3Flag (▴), P60-3Flag (•), or 3Flag alone (▪). Their inhibitory effect on luciferase expression was evaluated with time (each point was carried out in triplicate). The amount of P42-3Flag or P60-3Flag in the cells was evaluated by Western blotting at 48 h posttransfection using an anti-Flag antibody (inset). The results are summarized in Table 1.
The inhibitory effect could be due to interference of P42 and P60 with both minigenome encapsidation (interference with the P-N° interaction) and transcription (interference with the P-L interaction). However, several factors suggest that their inhibitory effect is most likely mediated by the interaction of P42 or P60 with N° rather than with L: (i) the 5N/1L ratio used in the minireplicon system, (ii) the better affinity of P42 and P60 for N than for L (Fig. 2c), and (iii) the partial overlapping of the N° and L binding sites on P42 and P60 (10, 34). Thus, we hypothesized that the greater inhibition of P60 over time was due to its better capacity to compete with P for N° binding and, consequently, to interfere with genome encapsidation. This would limit the amount of newly formed RNPs in the minireplicon system while P42 would not be able to control this as efficiently.
This hypothesis was supported by three additional experiments performed at 48 h posttransfection, i.e., when the difference in the inhibitory effects between the two peptides is at its maximum. (i) qRT-PCR indicated that the cytoplasmic amount of plus-strand minigenome, which results from the replication of encapsidated minus-strand RNA (RNP), was significantly more reduced in the presence of P60-3Flag (46%) than in the presence of P42-3Flag (only 10%) (Table 1). (ii) We further evaluated the amounts of pseudovirus released by vT7-BSR cells by transferring supernatants to fresh cells and measuring the luciferase signal 24 h later. Recipient cells were previously transfected with pT7-N, pT7-P, and pT7-L (but not with pDImut) in order to enhance the luciferase signal resulting exclusively from incoming RNP embedded into pseudoviruses. Signals produced by supernatants obtained in the presence of P42-3Flag were 75% lower while those obtained in the presence of P60-3Flag or P57-3Flag were 92.5% and >99% lower, respectively, than those obtained in the presence of 3Flag alone (Fig. 4). (iii) Finally, a dose-dependent inhibitory assay with the minireplicon system was performed after transfection by plasmids encoding P42-3Flag or P60-3Flag. The specific inhibitory potential was calculated in comparison with the plasmid encoding 3Flag alone (Fig. 5). Both P42-3Flag and P60-3Flag showed a dose-dependent specific inhibitory effect. However, compared to the fixed quantity of P-encoding plasmid (100 ng) used in this assay, the addition of only 100 ng of the P60-encoding plasmid already resulted in 72% inhibition. In contrast, 500 ng of the P42-encoding plasmid was necessary to reach a similar level of inhibition (82%). Taken together, these results are compatible with the interference potential of peptides for encapsidation/replication via a strong competitive effect of P60 and P57 with P for N° while P42 appears less efficient in this respect.
TABLE 1.
Summary of the percentage of inhibition by P42, P60, and P57 in different experimental setups
| Peptide delivery (cell line) | Experimental setup
|
Inhibitory potential (%)b
|
|||||
|---|---|---|---|---|---|---|---|
| System | Parameter | Time (hpt)a | P42 | P60 | P57 | P1-20/P20-40/ P40-60 | |
| cDNA peptide-3Flag (vT7-BSR) | Minireplicon | Luciferase expression | 20 | 74 | 71.5 | 74 | ND |
| Minireplicon | Luciferase expression | 48 | 42.6 | 99.5 | 99.6 | ND | |
| Pseudovirus | Luciferase expression | 48 | 75 | 92.5 | 99 | ND | |
| qRT-PCR | RNP replication | 48 | 10 | 46 | ND | ND | |
| cDNA peptide-eGFP (BHK-21-T7) | Infection | Immunofluorescence | 20 | 71 | 70 | 75 | ND |
| Synthetic peptide-Tat (BHK-21-T7) | Infection | Immunofluorescence | 15 | 70 | 90 | ND | ND |
| Synthetic peptide-Tat (SK-N-SH) | Infection | Immunofluorescence | 15 | 79 | c | ND | 54 |
| Infection | Virus titer | 15 | 56 | c | ND | 58 | |
| Synthetic peptide-Tat (SK-N-SH CP) | Infection | Immunofluorescence | 15 | 10 | c | ND | 95 |
| Infection | Virus titer | 15 | 35 | c | ND | 66 | |
hpt, hours posttransfection.
In the minireplicon system, the values for the 3Flag controls were 0% at 20 h posttransfection and 28% at 48 h posttransfection. ND, not done.
P60 does not penetrate SK-N-SH or SK-N-SH CP neuronal cells.
FIG. 4.
Inhibition of RABV pseudovirus production by plasmids encoding P42, P57, or P60. BSR cells were infected by a recombinant vT7-vaccinia virus (vT7-BSR) and then transfected with plasmids encoding the RABV minigenome (DI); the N, P, L, M and G proteins; and pCINeo plasmids encoding P42-3Flag, P57-3Flag, P60-3Flag, or 3Flag alone. The inhibitory effect on pseudovirus production was evaluated at 48 h posttransfection (each point was carried out in triplicate) by transferring the cell supernatant to new vT7-BSR cells transfected with plasmids encoding the N, P, and L proteins in order to enhance the luciferase signal from incoming pseudovirus. This signal, evaluated at 24 h postinfection, is proportional to the amount of incoming pseudovirus.
FIG. 5.
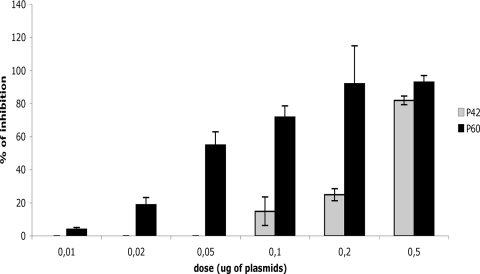
Dose-dependent assay of inhibition of luciferase expression from an RABV minireplicon by plasmids encoding P42 or P60. BSR cells were infected by a recombinant vT7-vaccinia virus and then transfected with plasmids encoding the RABV minigenome; the N, P, and L proteins; and increasing amounts of T7-expressed pCINeo plasmids encoding P42-3Flag (gray) or P60-3Flag (black). The inhibitory effect on luciferase expression was evaluated at 48 h posttransfection by comparison with cells transfected with a plasmid encoding 3Flag alone (each point was carried out in triplicate).
Inhibitory potential of P42 and P60 on the infection of BHK-21-T7 cells by RABV.
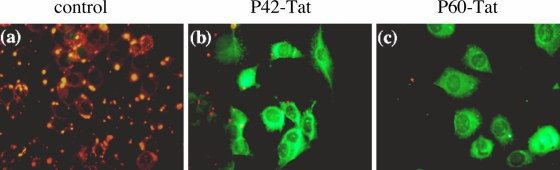
We further analyzed the inhibitory potential of P42 and P60 in BHK-21-T7 cells infected with RABV at an MOI allowing 90% infection. In a first step, we delivered peptides from plasmids peGFPC1-P42/P60, encoding P42-eGFP or P60-eGFP, transfected immediately prior to infection (Fig. 6). The eGFP protein was linked to the carboxy-terminal extremity of each peptide to identify cells expressing P42 or P60. At 20 h posttransfection, the percentage of infection in cells expressing the eGFP-tagged peptide was compared to the percentage of infection in cells expressing eGFP alone. Both P42-eGFP and P60-eGFP were found to inhibit RABV infection at very similar levels, i.e., 70% and 71% of cells, respectively (Table 1). These values were similar to the percentage of inhibition observed in the minireplicon system at this time (Fig. 3). In a second step, P42 and P60 were synthesized in tandem with a Tat sequence at their carboxy terminal extremities to allow transmembrane passage. An Alexa Fluor 488 molecule was linked at the carboxy-terminal end to identify cells having engulfed peptides. At 1 h postinfection, cells were washed, and a 10 μm concentration of P42-Tat or P60-Tat was added to the medium. The uptake of both peptides by cells was completed within the first 2 h (data not shown), and the effect on infection was observed 15 h later (Fig. 7). As reported in the literature (23), no cytotoxic effect was observed due to the Tat peptide at similar concentrations. While almost 100% of control BHK-21-T7 cells were infected in the absence of peptides (Fig. 7a), the presence of P60-Tat in cells protected >90% of them from infection (Fig. 7c and Table 1). P42-Tat was slightly less efficient, protecting 70% of cells from infection (Fig. 7b and Table 1). In conclusion, P42 and, particularly, P60 given 1 h postexposure to virus were found to be excellent candidates to inhibit RABV infection in BHK-21-T7 cells.
FIG. 6.
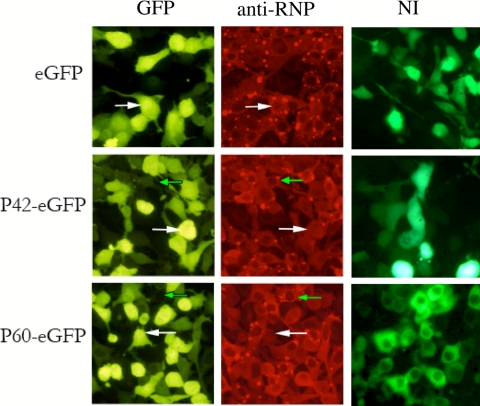
Inhibition of RABV infection by plasmids encoding P42 or P60. BHK-21-T7 cells were infected with RABV at an MOI allowing 90% infection and immediately transfected with T7-expressed peGFPC1 plasmids encoding P42-eGFP, P60-eGFP, or eGFP alone. Twenty hours later, cells were visualized by fluorescence. Infection was revealed by an anti-RNP polyclonal antibody (in red) and transfected cells expressing eGFP-tagged peptides or eGFP alone (green). The percentage of infection in cells expressing the peptide-eGFP was compared to the percentage of cells expressing eGFP alone. Green and white arrows indicate examples of peptide-expressing and nonexpressing cells, respectively. Those expressing P42-eGFP or P60-eGFP are mostly noninfected while those expressing eGFP alone are mostly infected (red punctation). The results are summarized in Table 1. The right-hand column shows images at lower exposure of noninfected (NI) cells transfected with peptide-eGFP and eGFP alone in order to observe the cellular localization of the peptides.
FIG. 7.
Inhibition of RABV infection by synthetic peptides P42-Tat or P60-Tat. Synthetic peptide (10 μM) P60 or P42 fused to a Tat peptide (P60-Tat and P42-Tat, respectively) marked with an Alexa Fluor 488 molecule at the carboxy-terminal extremity was added, or not (control), at 1 h postinfection, to the medium of BHK-21-T7 cells infected with RABV at an MOI allowing 90% infection. Fifteen hours later, cells were revealed by fluorescence. Infection was detected by an anti-RNP polyclonal antibody (red), and cells carrying the peptide-Tat fusion are shown in green. The percentage of infection in cells that received the Tat peptides was compared to that of cells without peptide. The results are summarized in Table 1.
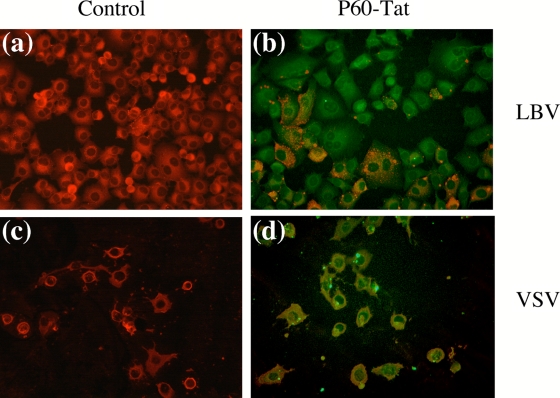
Since P60-Tat was the best candidate for postexposure inhibition, it was further tested for the broadness of its spectrum of inhibition against two other members of the Rhabdoviridae family: LBV, which belongs to phylogroup II of the Lyssavirus genus, only distantly related to RABV (2), and VSV, which is the prototype of the Vesiculovirus genus, even more distantly related to RABV. BHK-21-T7 cells were infected with LBV and VSV to obtain almost 100% infection. As shown in Fig. 8, the anti-RNP antibodies used to reveal infection gave a diffuse staining with VSV (Fig. 8c) compared to the more punctuated staining with LBV (Fig. 8a). The latter staining is classical for lyssavirus infection (Fig. 7a) that proceeds by intracellular accumulation of RNP into Negri bodies (25, 27, 35). When cells were treated 1 h postinfection with P60-Tat and the infection was revealed 15 h later, no inhibition was observed against VSV (Fig. 8d), and cells exhibited the classical cell-rounding effect (3). However, a partial level of protection (estimated at 50% of the cells) was observed against LBV (Fig. 8b) with a slight dose-dependent effect; i.e., the higher the P60-Tat peptide concentration in the cells, the less they were infected. The moderate inhibitory effect of P60-Tat against LBV compared to RABV is logical, given the divergence between the two P proteins (Fig. 1). The region of residues 1 to 60 nevertheless remains relatively conserved among lyssavirus P proteins and is thus an attractive target to set up an antilyssavirus strategy.
FIG. 8.
Inhibition of LBV and VSV infection by synthetic peptide P60-Tat. Synthetic peptide P60 (10 μM) fused to a Tat peptide (P60-Tat) marked with an Alexa Fluor 488 molecule at the C-terminal extremity was added, or not (control), 1 h postinfection to the medium of BHK-21-T7 cells infected with LBV or with VSV virus at an MOI allowing 90% infection. Fifteen hours later, cells were visualized by fluorescence. Infected cells were revealed by an anti-RNP polyclonal antibody (red), and cells having peptide-Tat were visualized in green. The infection level in cells that received the Tat-peptide was compared to that of cells without peptide.
Inhibitory potential of P42 and P60 on the infection of neuronal cells by RABV.
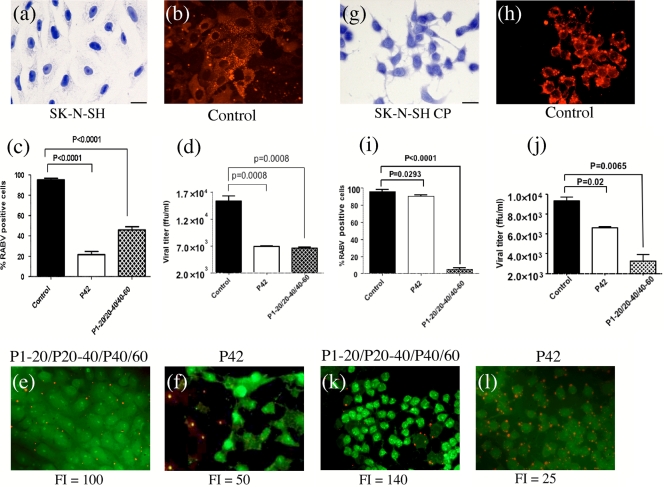
The inhibitory effect of P60-Tat or P42-Tat was then tested in neuronal cells, which are the target cells for neurotropic viruses like RABV. Two different neuroblastoma cell lines, SK-N-SH (Fig. 9a) and a subclone SK-N-SH CP (Fig. 9g) were tested under the same conditions as those described for the BHK-21-T7 cells. SK-N-SH-CP exhibits a clear neuron-like morphology with long neurites (Fig. 9g) while the parental SK-N-SH exhibits an epithelial-like morphology (Fig. 9a). Moreover, SK-N-SH-CP cells are disabled for expression of IFN-β and possess a modified form of Toll-like receptor 3 (26). It is well known that delivery of proteins/peptides or other macromolecules into neurons can be difficult, possibly in relation to their differentiated state (17, 49). Accordingly, P60-Tat uptake was observed neither in SK-N-SH nor in SK-N-SH CP cells (data not shown). In contrast, uptake of P42-Tat by both cell types was observed within 2 h, and the effect on RABV infection was evaluated 15 h later. This timing was chosen to be homogeneous with the experiments performed in BHK-21-T7 cells although human neuroblastoma cells are generally slower (20 h to 24 h) to reach the plateau of virus production. Ten micrograms of P42-Tat showed a better fluorescence index (combining penetration and intracellular stability) in SK-N-SH (fluorescence index of 50) (Fig. 9f) than in SK-N-SH CP (fluorescence index of 25) (Fig. 9l) cells. This correlated with an inhibitory effect on intracellular RNP of 79% (Fig. 9c and f) and 10% (Fig. 9i and l), respectively, whereas more than 95% of nontreated cells contained intracellular RNP (Fig. 9b, c, h, and i).
FIG. 9.
Inhibition of RABV infection in neuroblastoma cell lines by synthetic peptides P42-Tat or by the peptide cocktail P1-20-Tat/P20-40-Tat/P40-60-Tat. SK-N-SH (a) and SK-N-SH CP (g) neuronal cells were infected with RABV at an MOI allowing at least 90% infection (panels b and h, respectively). At 1 h postinfection, a 10 μM concentration of peptide P42-Tat (f and l) or of each peptide in the cocktail P1-20-Tat/P20-40-Tat/P40-60-Tat (e and k), each marked with an Alexa Fluor 488 molecule at the carboxy-terminal extremity, was added to the culture medium. Fifteen hours later, RABV nucleocapsids were revealed by fluorescence (in red), and cells having engulfed peptide-Tat were visible in green. The fluorescence index (FI) indicates the efficacy of penetration/stability of the peptides in cells. The percentage of cells exhibiting RABV nucleocapsid fluorescent inclusions was evaluated for each condition (c and i). The virus titer in cell supernatants 15 h postinfection is indicated (d and j). FFU, focus-forming units.
As size appears to limit the efficient penetration into neuronal cells, we cleaved P60-Tat into three juxtaposed smaller peptides (residues 1 to 20 of P fused to Tat [P1-20-Tat] or residues 20 to 40 [P20-40-Tat] or 40 to 60 [P40-60-Tat]) and tested the inhibitory effect of this cocktail under similar conditions, using 10 μM for each peptide. In contrast to P60-Tat, the cocktail gave a very good fluorescence index in the two cell lines, which was slightly better in SH-N-SH CP (fluorescence index of 140) (Fig. 9k) than in SH-N-SH (fluorescence index of 100) (Fig. 9e) cells. Here, again, the inhibitory effect on intracellular RNP correlated with the penetration/stability capacity since >95% inhibition was observed in SH-N-SH CP cells (Fig. 9i and k) compared to 54% in SH-N-SH cells (Fig. 9c and e). All together, these results show that the potential for intracellular RNP inhibition is related to the penetration/stability of the peptide into the cell.
Finally, we checked whether the inhibitory effect observed on intracellular RNP was correlated with the viral production by the infected cell (Fig. 9d and j). In SK-N-SH cells, the strong inhibition of P42-Tat and of the peptide cocktail (P1-20-Tat/P20-40-Tat/P40-60-Tat) on intracellular RNP (79% and 54%, respectively) (Fig. 9c) corresponded to a strong drop in virus production (56% and 58%, respectively) (Fig. 9d). In SK-N-SH CP, the same correlation existed with the P1-20-Tat/P20-40-Tat/P40-60-Tat cocktail: 95% inhibition of intracellular RNP (Fig. 9i) and 66% of virus production (Fig. 9j). On the other hand, P42-Tat, although showing a limited inhibition on intracellular RNP (10%) (Fig. 9i), had a significant effect on virus release (35%) (Fig. 9j).
Localization of P42 and P60 in transfected cells.
An obvious difference between P60 and P42 is a CRM1-dependent NES located between residues 49 and 58 of P and consequently present in P60 but absent from P42. By transfecting plasmids encoding P60-eGFP or P42-eGFP proteins into BHK-21-T7 cells, we first confirmed the functionality of the NES since P60-eGFP was clearly confined to the cytoplasm, whereas P42-eGFP was equally distributed between the cytoplasm and the nucleus, as was eGFP alone (Fig. 6, noninfected cells). To elucidate whether the cytoplasmic targeting of P60 could explain its better inhibitory potential than P42 on RABV replication, we generated a plasmid encoding P57, which is the largest peptide that can be designed for P that lacks the cytoplasmic targeting signal. In addition, P57 is predicted to constitute a structured region at the amino-terminal end of P, whereas the following segment of residues 56 to 91 is predicted to be disordered (13). As expected, P57-eGFP had lost the complete NES and was equally distributed in the cytoplasm and in the nucleus, as was P42 (data not shown). However, the inhibitory effect of P57 was as good as that of P60, with almost identical kinetics of inhibition in the minireplicon system, attaining >99% at 48 h posttransfection (Table 1). These results indicate that the difference in the inhibitory potential of P42 and P60 is not due either to their cellular localization or to their stability in the cell but, rather, to their different affinities for N° and L, allowing better competition with P in the RABV replication complex.
DISCUSSION
Developing an antiviral approach against rabies disease is confronted with two main difficulties: the virus travels and replicates in neurons, which impairs the delivery of candidate drugs, and the disease burden is mainly in developing countries (55,000 deaths/year) in which resources are limited (24). However, rabies disease also offers three specific advantages: (i) the contaminating event is generally obvious (a bite); (ii) the incubation period is rather long (2 months on average), leaving time to apply a therapy; and (iii) the replication complex is well documented (1) and similar to that of other Mononegavirales (30, 31), making the RABV a possible model for developing antiviral strategies applicable to other viruses. In this paper, we have explored the inhibitory potential of P-derived peptides competing with critical domains of the replication complex. These are not only promising antiviral compounds but also excellent tools to increase our understanding of the functional relationships within the complex itself.
Previous contributions have outlined the critical role of the N-terminal extremity of P in the transcription/replication complex as being able to bind both N° (34) and L (10). Our systematic two-hybrid scanning analysis of P by successive truncations of both the NH2 and COOH ends confirmed the importance of the first 40 residues for binding to N (the two-hybrid experiments do not distinguish between N-RNA and N°). With respect to the L-binding domain, we have demonstrated a critical involvement of amino acids 40 to 100 and, particularly, the stretch of amino acids 40 to 70. This result is not in agreement with previous coimmunoprecipitation data concerning deletion mutants of P and L proteins of RABV (10) that mapped the L-Cter binding domain closer to the NH2 end of P, within the first 19 NH2 residues, although the following residues up to 52 (1 to 52) stabilized the interaction. In our hands, the first 42 or 60 residues of P interacted poorly with L. This discrepancy between results from different laboratories could be explained by experimental differences between the study of Chenik et al. (10) and the present study: (i) the method used (coimmunoprecipitation versus the yeast two-hybrid assay here); (ii) the viral strains used (P-CVS and L-Sad versus P-PV and L-PV here); (iii) the position of the fused gene (GFP at the COOH side of the P fragments versus the GAL4 activation domain at the NH2 side of the P fragments here); (iv) the last 566 residues of L-Cter versus the last 504 residues here). It is of note, however, that the present work offers a new perspective in proposing that the two most important functional domains at the NH2 end of P would not be overlapping as previously reported (residues 4 to 40 for N° and 1 to 19/52 for L) but juxtaposed (residues 4 to 40 for N° and 40 to 70 for L). The fact that the main L binding domain (residues 40 to 70) would then mostly correspond to a predicted disordered region of the P protein (mapped as residues 56 to 91) (13) could be a guarantee for flexibility, and N° bound to the NH2 end of P could serve as a chaperone for the L binding site. Finally, the L binding site in position 40 to 70/100 on P is consistent with the observation that the domain of residues 79 to 123 of the P protein of VSV is critical for binding to L (11), possibly through phosphorylation of Ser and Thr residues between residues 49 to 64 (43). Interestingly, Ser 63 to 64 of the RABV CVS strain located in a related position (Fig. 1) are also phosphorylated by the cellular RABV protein kinase RVPK (16).
Despite the remaining uncertainty about the precise mapping of the L binding site, it is clear that the NH2 end of P interacts with both N and L within the replication complex. We thus decided to evaluate the inhibition potential of two peptides covering the first 42 (P42) and 60 (P60) NH2-terminal residues of the RABV P protein. Two-hybrid experiments showed the following: (i) the two peptides interact with both N° and L-Cter, indicating that they preserve a fold similar to that in the intact P protein; (ii) their interaction with N° is much more efficient than that with L-Cter, which is very weak; and (iii) P60 interacts better than P42 with both N and L-Cter, suggesting that the region of residues 43 to 60, in which a smaller α-helix is predicted (Fig. 1), could somehow stabilize the interactions (34).
The inhibitory potential of P42 and P60 on RABV replication was evaluated in different ways: (i) in a minireplicon system in which a functional RABV RNP expressing luciferase is reconstituted in the cytoplasm after plasmid transfection, and P42 and P60 were coexpressed from transfected plasmids; (ii) in RABV-infected cells in which P42 and P60 were either coexpressed from transfected plasmids or delivered as synthetic peptides fused to a Tat peptide to allow transmembrane passage; and (iii) in terms of the production of pseudovirus in cell supernatants in both systems. Inhibition was observed in all cases but at different levels and following different dynamics. Twenty hours after transfection with their expression plasmids, P42 and P60 induced similar levels of inhibition (70 to 75%) both in the minireplicon system (vT7-BSR) and in BHK-21-T7-infected cells (Table 1). Later, an erosion of the inhibitory effect by P42 was observed (to only 42.6% after 40 h compared to 28% for the 3Flag control) while P60 reached >95% inhibition after 24 h and maintained a long-lasting inhibitory effect at >99% up to 55 h. The difference in inhibition potential is not due to a specific instability of P42, which was still observed by Western blotting at similar levels as P60. Nor is it due to the preferential localization of P60 into the cytoplasm, where RABV replication occurs, since P57, which is the largest peptide lacking the CRM1-dependent NES, mimics the subcellular localization of P42 but the inhibition pattern of P60 (Table 1). The carrier protein fused to the COOH end of the P peptides may certainly contribute to the inhibition effect, but this contribution seems to be secondary compared to the specific P peptide inhibition.
The minireplicon system favors peptide interference through P-N° interaction (RNA encapsidation and replication) since the N-to-L ratio is 5/1 and since both P42 and P60 show much higher affinity for N than for L. The inhibition by P42 and P60 thus appears mainly due to interference with the P-N° interaction rather than with that of P-L. This is further supported by the observation by qRT-PCR of a higher inhibitory potential of P60 for the plus-strand minigenome production which results from the replication of the encapsidated negative-strand RNP. Indeed, while the efficacy of P42 to compete with native P for the N° binding site progressively decreases when larger quantities of N° proteins are produced (>20 h posttransfection), P60 maintains a large competitive capacity for the N° binding site at later times due to its higher affinity. Accordingly, P60 is better at limiting pseudovirus release in cell supernatant (92.5% after 48 h) although, in this case, P42 appears less inferior (75%) at inhibiting RNP expression and replication, suggesting that a complementary inhibition step during virus formation cannot be formally excluded. Assuming that the predicted N° binding site of residues 4 to 40 (34) is present on both peptides, it is supposed that enlarging the segment of residues 1 to 42 up to position 57 or 60 would stabilize the global peptide structure. It provides a greater affinity for the N° binding site as confirmed by the dose-dependent inhibitory assay showing that P60 inhibits at lower amounts and at better levels than P42. The improved stability could result from a better folding of the peptide itself, improving its fitting into the relevant cavity of the N° protein. With respect to P-L interference, a closer targeting of the region of residues 40 to 70, which was shown to be crucial for L-P interaction, should offer better candidates for inhibition.
Finally, the inhibitory potentials of P42 and P60 were tested in a postexposure model on two types of RABV-infected neuronal cell lines, SK-N-CH and its subclone SK-N-CH CP, that constitute relevant cells for a neurotropic virus. P60-Tat uptake was not observed, a classical phenomena for large peptides in neurons, and consequently displayed no inhibition. Globally, we observed a correlation between the penetration index of peptides into neurons and their inhibitory effect. A cocktail of three juxtaposed peptides encompassing P60 (P1-20-Tat/P20-40-Tat/P40-60-Tat) was efficient at inhibiting both neuron infection and virus production up to very significant levels (55% to 95%) (Table 1). P42-Tat also displayed a significant inhibitory effect on infection, which was better in SK-N-CH (80%) than in SK-N-CH CP (only 10%) cells. However, even in SK-N-CH CP cells, the inhibition of virus release by P42 was significantly higher (35%), further suggesting that distal steps in the release process may also be targeted. All together, these results indicate that peptides at the NH2 extremity are excellent candidates for designing antiviral drugs against RABV and even to a distant lyssavirus like LBV. Experiments are in progress to design shorter stretches with an optimized inhibitory potential, associating efficient competitors for either P-N or P-L interactions.
Acknowledgments
This work was supported by a grant from the French ANR (ANR-07-001-01 [ANRAGE]) and Lyonbiopôle. G.C. was supported by an MENRT fellowship from the French government. M.C. was supported by a fellowship of the Fondation Mérieux (Lyon, France).
We thank Francine Gerard and Marc Jamin for extensive discussions, Pauline Ménager and Claudine Rousseaux for their assistance in microscopy and qRT-PCR, respectively, and Katherine Kean for corrections of the English.
Footnotes
Published ahead of print on 12 August 2009.
REFERENCES
- 1.Albertini, A. A., A. K. Wernimont, T. Muziol, R. B. Ravelli, C. R. Clapier, G. Schoehn, W. Weissenhorn, and R. W. Ruigrok. 2006. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313:360-363. [DOI] [PubMed] [Google Scholar]
- 2.Badrane, H., C. Bahloul, P. Perrin, and N. Tordo. 2001. Evidence of two Lyssavirus phylogroups with distinct pathogenicity and immunogenicity. J. Virol. 75:3268-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blondel, D., G. G. Harminson, and M. Schubert. 1990. Role of matrix protein in cytopathogenesis of vesicular stomatis virus. J. Virol. 64:1716-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blondel, D., T. Regad, N. Poisson, B. Pavie, F. Harper, P. P. Pandolfi, H. De The, and M. K. Chelbi-Alix. 2002. Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene 21:7957-7970. [DOI] [PubMed] [Google Scholar]
- 5.Brzozka, K., S. Finke, and K. K. Conzelmann. 2005. Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J. Virol. 79:7673-7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brzozka, K., S. Finke, and K. K. Conzelmann. 2006. Inhibition of interferon signaling by rabies virus phosphoprotein P: activation-dependent binding of STAT1 and STAT2. J. Virol. 80:2675-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chelbi-Alix, M. K., A. Vidy, J. El Bougrini, and D. Blondel. 2006. Rabies viral mechanisms to escape the IFN system: the viral protein P interferes with IRF-3, Stat1, and PML nuclear bodies. J. Interferon Cytokine Res. 26:271-280. [DOI] [PubMed] [Google Scholar]
- 8.Chenik, M., K. Chebli, and D. Blondel. 1995. Translation initiation at alternate in-frame AUG codons in the rabies virus phosphoprotein mRNA is mediated by a ribosomal leaky scanning mechanism. J. Virol. 69:707-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chenik, M., K. Chebli, Y. Gaudin, and D. Blondel. 1994. In vivo interaction of rabies virus phosphoprotein (P) and nucleoprotein (N): existence of two N-binding sites on P protein. J. Gen. Virol. 75:2889-2896. [DOI] [PubMed] [Google Scholar]
- 10.Chenik, M., M. Schnell, K. K. Conzelmann, and D. Blondel. 1998. Mapping the interacting domains between the rabies virus polymerase and phosphoprotein. J. Virol. 72:1925-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emerson, S. U., and M. Schubert. 1987. Location of the binding domains for the RNA polymerase L and the ribonucleocapsid template within different halves of the NS phosphoprotein of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 84:5655-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu, Z. F., Y. Zheng, W. H. Wunner, H. Koprowski, and B. Dietzschold. 1994. Both the N- and the C-terminal domains of the nominal phosphoprotein of rabies virus are involved in binding to the nucleoprotein. Virology 200:590-597. [DOI] [PubMed] [Google Scholar]
- 13.Gerard, F. C., E. A. Ribeiro, C. Leyrat, I. Ivanov, D. Blondel, S. Longhi, R. W. Ruigrok, and M. Jamin. 2009. Modular organization of rabies virus phosphoprotein. J. Mol. Biol. 388:978-996. [DOI] [PubMed] [Google Scholar]
- 14.Gerard, F. C., A. Ribeiro Ede, Jr., A. A. Albertini, I. Gutsche, G. Zaccai, R. W. Ruigrok, and M. Jamin. 2007. Unphosphorylated Rhabdoviridae phosphoproteins form elongated dimers in solution. Biochemistry 46:10328-10338. [DOI] [PubMed] [Google Scholar]
- 15.Gigant, B., F. Iseni, Y. Gaudin, M. Knossow, and D. Blondel. 2000. Neither phosphorylation nor the amino-terminal part of rabies virus phosphoprotein is required for its oligomerization. J. Gen. Virol. 81:1757-1761. [DOI] [PubMed] [Google Scholar]
- 16.Gupta, A. K., D. Blondel, S. Choudhary, and A. K. Banerjee. 2000. The phosphoprotein of rabies virus is phosphorylated by a unique cellular protein kinase and specific isomers of protein kinase C. J. Virol. 74:91-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasadsri, L., J. Kreuter, H. Hattori, T. Iwasaki, and J. M. George. 2009. Functional protein delivery into neurons using polymeric nanoparticles. J. Biol. Chem. 284:6972-6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hemachudha, T., B. Sunsaneewitayakul, T. Desudchit, C. Suankratay, C. Sittipunt, S. Wacharapluesadee, P. Khawplod, H. Wilde, and A. C. Jackson. 2006. Failure of therapeutic coma and ketamine for therapy of human rabies. J. Neurovirol. 12:407-409. [DOI] [PubMed] [Google Scholar]
- 19.Iseni, F., A. Barge, F. Baudin, D. Blondel, and R. W. Ruigrok. 1998. Characterization of rabies virus nucleocapsids and recombinant nucleocapsid-like structures. J. Gen. Virol. 79:2909-2919. [DOI] [PubMed] [Google Scholar]
- 20.Jackson, A. C. 2006. Rabies: new insights into pathogenesis and treatment. Curr. Opin. Neurol. 19:267-270. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, A. C., M. J. Warrell, C. E. Rupprecht, H. C. Ertl, B. Dietzschold, M. O'Reilly, R. P. Leach, Z. F. Fu, W. H. Wunner, T. P. Bleck, and H. Wilde. 2003. Management of rabies in humans. Clin. Infect. Dis. 36:60-63. [DOI] [PubMed] [Google Scholar]
- 22.Jacob, Y., E. Real, and N. Tordo. 2001. Functional interaction map of lyssavirus phosphoprotein: identification of the minimal transcription domains. J. Virol. 75:9613-9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, S. W., R. Christison, K. Bundell, C. J. Voyce, S. M. Brockbank, P. Newham, and M. A. Lindsay. 2005. Characterisation of cell-penetrating peptide-mediated peptide delivery. Br. J. Pharmacol. 145:1093-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knobel, D. L., S. Cleaveland, P. G. Coleman, E. M. Fevre, M. I. Meltzer, M. E. Miranda, A. Shaw, J. Zinsstag, and F. X. Meslin. 2005. Re-evaluating the burden of rabies in Africa and Asia. Bull. W. H. O. 83:360-368. [PMC free article] [PubMed] [Google Scholar]
- 25.Kristensson, K., D. K. Dastur, D. K. Manghani, H. Tsiang, and M. Bentivoglio. 1996. Rabies: interactions between neurons and viruses. A review of the history of Negri inclusion bodies. Neuropathol. Appl. Neurobiol. 22:179-187. [PubMed] [Google Scholar]
- 26.Lafon, M., F. Megret, S. G. Meuth, O. Simon, R. M. L. Velandia, M. Lafage, L. Chen, L. Alexopoulou, R. A. Flavell, C. Prehaud, and H. Wiendls. 2008. Detrimental contribution of the immuno-inhibitor B7-H1 to rabies virus encephalitis. J. Immunol. 180:7506-7515. [DOI] [PubMed] [Google Scholar]
- 27.Lahaye, X., A. Vidy, C. Pomier, L. Obiang, F. Harper, Y. Gaudin, and D. Blondel. 2009. Functional characterization of Negri bodies (NBs) in rabies virus-infected cells: evidence that NBs are sites of viral transcription and replication. J. Virol. 83:7948-7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Mercier, P., Y. Jacob, K. Tanner, and N. Tordo. 2002. A novel expression cassette of lyssavirus shows that the distantly related Mokola virus can rescue a defective rabies virus genome. J. Virol. 76:2024-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, P., J. Yang, X. Wu, and Z. F. Fu. 2004. Interactions amongst rabies virus nucleoprotein, phosphoprotein and genomic RNA in virus-infected and transfected cells. J. Gen. Virol. 85:3725-3734. [DOI] [PubMed] [Google Scholar]
- 30.Luo, M., T. J. Green, X. Zhang, J. Tsao, and S. Qiu. 2007. Conserved characteristics of the rhabdovirus nucleoprotein. Virus Res. 129:246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo, M., T. J. Green, X. Zhang, J. Tsao, and S. Qiu. 2007. Structural comparisons of the nucleoprotein from three negative strand RNA virus families. Virol. J. 4:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mavrakis, M., F. Iseni, C. Mazza, G. Schoehn, C. Ebel, M. Gentzel, T. Franz, and R. W. Ruigrok. 2003. Isolation and characterisation of the rabies virus N°-P complex produced in insect cells. Virology 305:406-414. [DOI] [PubMed] [Google Scholar]
- 33.Mavrakis, M., A. A. McCarthy, S. Roche, D. Blondel, and R. W. Ruigrok. 2004. Structure and function of the C-terminal domain of the polymerase cofactor of rabies virus. J. Mol. Biol. 343:819-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mavrakis, M., S. Mehouas, E. Real, F. Iseni, D. Blondel, N. Tordo, and R. W. Ruigrok. 2006. Rabies virus chaperone: identification of the phosphoprotein peptide that keeps nucleoprotein soluble and free from non-specific RNA. Virology 349:422-429. [DOI] [PubMed] [Google Scholar]
- 35.Menager, P., P. Roux, F. Megret, J. P. Bourgeois, A. M. Le Sourd, A. Danckaert, M. Lafage, C. Prehaud, and M. Lafon. 2009. Toll-like receptor 3 (TLR3) plays a major role in the formation of rabies virus Negri Bodies. PLoS Pathog. 5:e1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moseley, G. W., R. P. Filmer, M. A. DeJesus, and D. A. Jans. 2007. Nucleocytoplasmic distribution of rabies virus P-protein is regulated by phosphorylation adjacent to C-terminal nuclear import and export signals. Biochemistry 46:12053-12061. [DOI] [PubMed] [Google Scholar]
- 37.Nadin-Davis, S. A., M. Abdel-Malik, J. Armstrong, and A. I. Wandeler. 2002. Lyssavirus P gene characterisation provides insights into the phylogeny of the genus and identifies structural similarities and diversity within the encoded phosphoprotein. Virology 298:286-305. [DOI] [PubMed] [Google Scholar]
- 38.Pasdeloup, D., N. Poisson, H. Raux, Y. Gaudin, R. W. Ruigrok, and D. Blondel. 2005. Nucleocytoplasmic shuttling of the rabies virus P protein requires a nuclear localization signal and a CRM1-dependent nuclear export signal. Virology 334:284-293. [DOI] [PubMed] [Google Scholar]
- 39.Ribeiro, E. A., Jr., A. Favier, F. C. Gerard, C. Leyrat, B. Brutscher, D. Blondel, R. W. Ruigrok, M. Blackledge, and M. Jamin. 2008. Solution structure of the C-terminal nucleoprotein-RNA binding domain of the vesicular stomatitis virus phosphoprotein. J. Mol. Biol. 382:525-538. [DOI] [PubMed] [Google Scholar]
- 40.Schoehn, G., F. Iseni, M. Mavrakis, D. Blondel, and R. W. Ruigrok. 2001. Structure of recombinant rabies virus nucleoprotein-RNA complex and identification of the phosphoprotein binding site. J. Virol. 75:490-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimizu, K., N. Ito, M. Sugiyama, and N. Minamoto. 2006. Sensitivity of rabies virus to type I interferon is determined by the phosphoprotein gene. Microbiol. Immunol. 50:975-978. [DOI] [PubMed] [Google Scholar]
- 42.Soni, R., and J. A. Murray. 1992. A rapid and inexpensive method for isolation of shuttle vector DNA from yeast for the transformation of E. coli. Nucleic Acids Res. 20:5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takacs, A. M., and A. K. Banerjee. 1995. Efficient interaction of the vesicular stomatis virus P protein with the L protein or the N protein in cells expressing the recombinant proteins. Virology 208:821-826. [DOI] [PubMed] [Google Scholar]
- 44.Tarbouriech, N., J. Curran, R. W. H. Ruigrok, and W. P. Burmeister. 2000. Tetrameric coiled-coil domain of Sendai virus phosphoprotein. Nat. Struct. Biol. 7:777-781. [DOI] [PubMed] [Google Scholar]
- 45.Tordo, N., A. Benmansour, C. Calisher, R. G. Dietzgen, R. X. Fang, A. O. Jackson, G. Kurath, S. Nadin-Davis, R. R. Tesh, and P. J. Walker. 2005. Rhabdoviridae, p. 623-629. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy: classification and nomenclature of viruses. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
- 46.Vidy, A., M. Chelbi-Alix, and D. Blondel. 2005. Rabies virus P protein interacts with STAT1 and inhibits interferon signal transduction pathways. J. Virol. 79:14411-14420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vidy, A., J. El Bougrini, M. K. Chelbi-Alix, and D. Blondel. 2007. The nucleocytoplasmic rabies virus P protein counteracts interferon signaling by inhibiting both nuclear accumulation and DNA binding of STAT1. J. Virol. 81:4255-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walhout, A. J., and M. Vidal. 2001. High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24:297-306. [DOI] [PubMed] [Google Scholar]
- 49.Whittlesey, K. J., and L. D. Shea. 2004. Delivery systems for small molecule drugs, proteins, and DNA: the neuroscience/biomaterial interface. Exp. Neurol. 190:1-16. [DOI] [PubMed] [Google Scholar]
- 50.Willoughby, R. E., Jr., K. S. Tieves, G. M. Hoffman, N. S. Ghanayem, C. M. Amlie-Lefond, M. J. Schwabe, M. J. Chusid, and C. E. Rupprecht. 2005. Survival after treatment of rabies with induction of coma. N. Engl. J. Med. 352:2508-2514. [DOI] [PubMed] [Google Scholar]
- 51.World Health Organization. 1992. WHO Expert Committee on Rabies, eighth report. WHO Tech. Rep. Ser. 824:1-84. [PubMed] [Google Scholar]