Abstract
Recognition of virus presence via RIG-I (retinoic acid inducible gene I) and/or MDA5 (melanoma differentiation-associated protein 5) initiates a signaling cascade that culminates in transcription of innate response genes such as those encoding the alpha/beta interferon (IFN-α/β) cytokines. It is generally assumed that MDA5 is activated by long molecules of double-stranded RNA (dsRNA) produced by annealing of complementary RNAs generated during viral infection. Here, we used an antibody to dsRNA to show that the presence of immunoreactivity in virus-infected cells does indeed correlate with the ability of RNA extracted from these cells to activate MDA5. Furthermore, RNA from cells infected with encephalomyocarditis virus or with vaccinia virus and precipitated with the anti-dsRNA antibody can bind to MDA5 and induce MDA5-dependent IFN-α/β production upon transfection into indicator cells. However, a prominent band of dsRNA apparent in cells infected with either virus does not stimulate IFN-α/β production. Instead, stimulatory activity resides in higher-order structured RNA that contains single-stranded RNA and dsRNA. These results suggest that MDA5 activation requires an RNA web rather than simply long molecules of dsRNA.
The innate immune response to virus infection is largely dependent on type I (alpha/beta) interferons (IFN-α/β). IFN-α/β induces expression of IFN-stimulated genes that have diverse antiviral properties, including sequestration of virus proteins, blocking of cellular translation, and degradation of viral and cellular RNA (12, 13, 21). It is believed that viral genomes and replication products are the main triggers of the key pattern recognition receptors (PRRs) that sense virus infection and that signal for IFN-α/β induction. PRRs known to induce IFN-α/β in response to viruses include Toll-like receptor 3 (TLR-3), TLR-7/TLR-8, and TLR-9. These TLRs are restricted in distribution to immune cells and a few nonimmune cell types and are activated by double-stranded RNA (dsRNA), single-stranded RNA (ssRNA), and DNA delivered into endosomes during the infection process (8). Most cells rely on another set of PRRs, the retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs), to sense RNA that accumulates in the cytoplasm during infection with many viruses (21). Two RLR members are known to signal for IFN-α/β induction: RIG-I and MDA5 (melanoma differentiation-associated protein 5) (4, 11, 32). Both proteins contain an RNA binding DEAD-box helicase domain and tandem caspase recruitment domains. The caspase recruitment domains are necessary for downstream signaling via shared adaptor MAVS (the mitochondrial antiviral signaling protein; also called CARDIF, IPS-1, or VISA) (20). Notably, some viruses such as Dengue virus and West Nile virus are sensed by both RIG-I and MDA5 such that loss of either RLR is redundant for IFN-α/β responses (24). However, RIG-I is nonredundant for responses to many negative-strand RNA viruses such as influenza virus and Sendai virus and some positive-strand RNA viruses such as Japanese encephalitis virus (11). In contrast, MDA5 is essential for responses to picornaviruses (4, 11). These data suggest that although RIG-I and MDA5 are similar in sequence and signal via a conserved pathway, they are activated by distinct RNA species. Indeed, we along with others could show that RIG-I but not MDA5 is activated by 5′ triphosphorylated RNA such as that present in the genomes of influenza virus and other negative-strand RNA viruses (7, 22). Interestingly, picornaviruses do not have triphosphorylated RNA genomes (23), which may explain why they do not activate RIG-I. However, the picornavirus-derived agonist for MDA5 has not been defined, and it is unclear why MDA5 agonists are generated during infection with picornaviruses but not influenza A virus and some other RNA viruses. One possible explanation is that MDA5 is activated by long dsRNA, which is made during infection with positive-strand RNA viruses (including picornaviruses) and DNA viruses but not with negative-strand RNA viruses such as influenza virus (22, 28). Consistent with this notion, MDA5 is activated by poly(I:C), a synthetic RNA that is often described as an equivalent of long dsRNA. Notably, Kato et al. recently showed that MDA5 can be activated by long dsRNA from the genome of reoviruses (ReoVs) or made by annealing sense and antisense strands of in vitro transcribed RNA (10). Therefore, it has come to be believed that the physiological agonist for MDA5 is simply long molecules of dsRNA.
Here, we investigated the nature of MDA5 agonists that are generated during viral infection. We show that the presence of immunodetectable dsRNA in cells infected with picornaviruses, alphaviruses, ReoV, and, notably, vaccinia virus (VV), correlates with generation of MDA5 agonists and that a dsRNA-specific antibody can immunoprecipitate RNA/MDA5 complexes containing stimulatory RNA from infected cells. However, we find that infected cells contain not only dsRNA but also RNA of high molecular weight (HMW) bearing both dsRNA and ssRNA regions and show that only the HMW fraction contains stimulatory activity. Our data suggest that MDA5 may be activated by branches of RNA rather than simply by long stretches of dsRNA.
MATERIALS AND METHODS
Reagents.
IFN-A/D, a human/mouse hybrid IFN, was a gift from Ian Kerr (Cancer Research UK). Anti-dsRNA antibody clone K1 (26) was from English and Scientific Consulting Bt. The goat anti-mouse antibody and isotype control antibody immunoglobulin G1 (IgG1) was purchased from ZyMed. Goat polyclonal anti-influenza A virus (H1N1) was from Europa Bioproducts Ltd. Anti-hemagglutinin (HA) antibody (clone HA7) conjugated to horseradish peroxidase and anti-FLAG (clone M2) was from Sigma. Calf intestinal phosphatase (CIP) was from New England Biolabs. Acridine orange, ribavirin (final concentration, 400 μM), and cycloheximide (CHX; final concentration, 10 μg/ml) were purchased from Sigma. RNase A, RNase T1, and RNase V1 were purchased from Ambion. The pGEM-T vector system was bought from Promega. pHA-MDA5 was described previously (22). The FLAG-MDA5 plasmid was generated by PCR amplification using following primers: 5′-GACAATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGTCGAATGGGTATTCCACAGACGAGAATTT-3′ and 5′-GACACTAATCCTCATCACTAAATAAACAGCATTCTGAAT-3′, followed by cloning into pCDNA3.1/V5-His-TOPO (Invitrogen). T3-tagged random hexamers had the following sequence: 5′-ATTAACCCTCACTAAAGGGANNNNN-3′. pBABE-puro-LargeT was from a kind gift of Gordon Peters (Cancer Research UK).
Cells and virus titrations.
3T3, 293T, HeLa S3, and Vero cells were from Cancer Research UK. HEK293 cells were a gift from Friedemann Weber (Freiburg, Germany). LL171 cells (L929 containing a stable IFN-stimulated response element-luciferase reporter plasmid [ISRE-Luc]) were a kind gift from Mireia Pelegrin (Montpellier, France). Mouse embryonic fibroblasts (MEFs) from MDA5−/−, RIG-I−/−, and wild-type littermate controls were generated as described previously (11) and were immortalized with simian virus 40 large T antigen by infection with retrovirus prepared from 48-h supernatants of Phoenix cells transfected with pBABE-puro-LargeT and a vesicular stomatitis virus G protein expression plasmid (pVSV-G; Clontech). Immortalized MEFs were selected on puromycin (final concentration, 2 μg/ml) for 2 weeks. All cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, 2 mM glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Encephalomyocarditis virus (EMCV) titration was done by determining the 50% tissue culture infective dose using Vero cells.
Viruses, stimuli, and cytokine induction assays.
Influenza A/PR/8/34 virus and ΔNS1 virus were a gift from Thomas Muster (Vienna, Austria). Semliki Forest virus (SFV), EMCV, Sindbis virus (SiV), and ReoV were a gift from Ian Kerr. Theiler's murine encephalomyelitis virus (TMEV) was kindly provided by Thomas Michiels (Leuven, Belgium). VV was propagated on HeLa cells. 5′ Triphosphate-containing RNA (PPP-RNA; 7SK-antisense RNA) was described earlier (22). To generate stimulatory RNA preparations, Vero or HeLa cells were infected overnight at a multiplicity of infection (MOI) of 0.1, unless indicated otherwise. RNA from cells and immunoprecipitates was isolated by Trizol reagent (Invitrogen) according to the protocol provided by the manufacturer.
For stimulation, cells were seeded in 24 plates at 2 ×105 cells per ml and transfected with RNA for 12 to 15 h. IFN-α was measured by sandwich enzyme-linked immunosorbent assay (ELISA) as described previously (22). Total IFN-α/β was measured by titration on LL171 cells and compared to recombinant IFN-A/D used as a cytokine standard.
Confocal microscopy.
HeLa or Vero cells were grown overnight on coverslips and infected with viruses for the times specified in the figure legends. Cells were fixed in 4% paraformaldehyde, blocked with phosphate-buffered saline (PBS) containing 1% bovine serum albumin, permeabilized in 0.1% Triton X-100, and stained with K1 antibody or goat polyclonal anti-influenza A virus, followed by appropriate secondary antibodies including a goat anti-mouse antibody, Alexa Fluor 488-conjugated anti-mouse (Molecular Probes), and DRAQ5. Coverslips were mounted on a slide, and images were acquired with a laser scanning confocal microscope (LSM 510; Zeiss).
Western blotting and immunoprecipitation.
293T cells were transfected with pHA-MDA5 and 48 h later lysed in PBS containing 0.2% Triton X-100, protease inhibitors (Complete, Roche), and RNasin (Promega). Equal amounts of cell lysate or RNA of EMCV-infected Vero cells (Vero-EMCV RNA) or of VV-infected HeLa cells (HeLa-VV RNA) were incubated with Sepharose beads (Gammabind plus; Pierce) that had been loaded with 2 μg of anti-dsRNA K1 antibody or isotype control IgG1. Lysates or Sepharose beads were mixed with sample buffer and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, blotted onto an Immobilon-P membrane (Millipore), probed with anti-HA conjugated to horseradish peroxidase, washed four times in PBS-Tween (0.05%), and detected using the Supersignal West Pico developing reagent (Pierce).
RNA analysis.
RNA from uninfected or infected cells was isolated using Trizol, and the amount of RNA indicated in the figure legends was separated on a 1% agarose gel. Gels were stained for 15 min with 1 mM acridine orange in Tris-acetate EDTA buffer. Thereafter, the gel was destained in deionized water for 1 to 3 h until clear bands were visible. Pictures were taken using a UV transilluminator and a Dimage Xt digital camera (Minolta) mounted on a stand. The blue channel (showing the UV lamp) was removed using Photoshop CS (Adobe). To isolate RNA from agarose gels, low-melting-point agarose (0.7%) was used followed by phenol-chloroform extraction.
For RNase digestion, 1 μg of RNA was diluted in RNA structure buffer (Ambion) and incubated for 1 h with 10-fold dilutions of a mix of RNase A (0.1 μg/μl) and RNase T1 (0.1 U/μl) or RNase V1 (0.01 U/μl). RNA was loaded on an agarose gel containing 0.05 μg/ml ethidium bromide or directly used for transfection into 3T3 cells.
To quantify IFN-β mRNA and EMCV viral RNA, total RNA was isolated from infected or uninfected cells using an RNeasy kit (Qiagen) combined with a DNA digestion step (DNase set; Qiagen). Single-stranded cDNA was synthesized using SuperScript II (Invitrogen) and random hexamer primers. Real-time PCR amplification for IFN-β was carried out using TaqMan universal master mix (Applera) and predeveloped TaqMan assay reagents (containing primers and fluorescent probe) for murine IFN-β and 18S rRNA (Applera). EMCV RNA was quantified by SYBR Green (Invitrogen) reverse transcription-PCR using the following primers: 5′-TTGAAAGCCGGGGGTGGGAGATCC-3′ and 5′-TCTGTTGTTATTTTGGGGTGGC-3′. PCRs were analyzed on an ABI 7900HT thermal cycler (Applera).
RESULTS
Activation of MDA5 and RIG-I by RNA isolated from cells infected with RNA viruses.
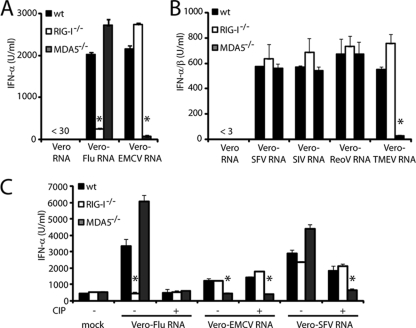
To characterize RNA species that might act as RLR agonists in virus-infected cells, we isolated RNA from uninfected Vero cells (Vero-RNA) or Vero cells that had been infected with influenza virus (Vero-Flu RNA) or Vero-EMCV RNA and tested its IFN-α/β-inducing ability. Vero-RNA did not elicit IFN-α production (Fig. 1A) or induce activation of an IFN-β reporter construct (data not shown) when transfected into MEFs or human fibroblast cell lines. In contrast, total RNA extracted from virus-infected cells was highly stimulatory in the same assays (Fig. 1A and data not shown). Use of the stimulatory RNA faithfully reproduced the pattern of RLR dependence seen with intact virus: RIG-I-deficient MEFs were selectively unresponsive to Vero-Flu RNA whereas MDA5-deficient MEFs did not respond to Vero-EMCV RNA (Fig. 1A), as reported for intact influenza virus and EMCV (4, 11).
FIG. 1.
RNAs from virus-infected cells act as RLR agonists. (A and B) MEFs of the indicated genotype were transfected with 0.2 μg of the indicated RNA, and accumulation of IFN-α in supernatants was measured by ELISA after overnight incubation. (B) MEFs of the indicated genotype were transfected with 0.2 μg of RNA from virus-infected cells. After overnight incubation, IFN-α/β in the supernatant was measured on indicator cells containing an ISRE-Luc reporter. (C) RNA from infected cells was treated with CIP (+) or buffer only (−) before transfection. IFN-α was measured by ELISA. Graphs show one of at least three representative experiments. Error bars show standard deviation of triplicate (A and C) or quadruplicate (B) measurements. Asterisks indicate a P value of <0.01 compared to wild-type (wt) control MEFs.
We extended our study to RNA isolated from cells infected with other RNA viruses (Table 1), including SFV, SiV, ReoV, and TMEV. RNA isolated from TMEV-infected cells induced IFN-α/β in an MDA5-dependent but RIG-I-independent manner (Fig. 1B), consistent with the observation that picornaviruses selectively activate MDA5 (4, 11). In contrast, IFN-α/β production by cells transfected with Vero-SFV RNA, Vero-ReoV RNA, and Vero-SiV RNA was unaffected by deficiency in either RIG-I or MDA5 (Fig. 1B). This suggested that infection with these viruses can generate both RIG-I and MDA5 agonists, leading to RLR redundancy (6, 11, 14). To test this possibility, we treated RNA from SFV-infected cells with CIP to destroy putative RIG-I agonists bearing 5′ triphosphates. For controls, we confirmed that CIP treatment of Vero-Flu RNA rendered it nonstimulatory but did not affect the stimulatory ability of Vero-EMCV RNA (Fig. 1C). Notably, CIP treatment rendered Vero-SFV RNA strictly dependent on MDA5 for its stimulatory ability (Fig. 1C). We tentatively conclude that some viruses (e.g., SFV) generate 5′ phosphate-bearing stimulatory RNAs that activate RIG-I, as well as 5′ phosphate-independent RNAs that act as agonists for MDA5 in infected cells.
TABLE 1.
Viruses used in this study
| Virus (abbreviation) | Genome type |
|---|---|
| Influenza A virus (Flu) | (−) ssRNA |
| Encephalomyocarditis virus (EMCV) | (+) ssRNA |
| Theiler's murine encephalitis virus (TMEV) | (+) ssRNA |
| Semliki Forest virus (SFV) | (+) ssRNA |
| Sindbis virus (SiV) | (+) ssRNA |
| Reovirus (ReoV) | dsRNA |
| Vaccinia virus (VV) | DNA |
One caveat with the above interpretation is that the IFN-stimulatory activity assayed in these experiments could in theory result from de novo virus replication arising from the presence of viral mRNA and viral genomes in the transfected RNA. Indeed, we found that 3T3 cells transfected with Vero-EMCV RNA produced detectable amounts of virus (Fig. 2A) and accumulated viral RNA (Fig. 2C) although at much lower levels than when cells were infected with live EMCV (Fig. 2A and C). To test whether IFN-α/β induction is dependent on virus production, we pretreated 3T3 cells with the antiviral drug ribavirin. This reduced the amount of progeny virus after EMCV infection by 4 to 5 log10 and abrogated virus production following Vero-EMCV RNA transfection (Fig. 2A). Importantly, this treatment did not decrease IFN-α production in response to transfected Vero-EMCV RNA (Fig. 2B). Similarly, the accumulation of EMCV RNA after transfection with Vero-EMCV RNA was reduced more than 1,000-fold by pretreatment with CHX, which blocks cellular translation and therefore expression of proteins essential for viral replication (Fig. 2C), yet the accumulation of IFN-β mRNA was unaffected (Fig. 2D). Furthermore, despite the fact that direct infection with EMCV led to greater accumulation of viral RNA (Fig. 2C), IFN-β induction was superior when RNA from virus-infected cells was transfected (Fig. 2D). Similar effects were seen in experiments using RNA extracted from cells infected with other RNA viruses (data not shown). Therefore, we conclude that RNA extracted from cells infected with RNA viruses contains preformed stimulatory species that act as agonists for RLRs independently of further virus replication.
FIG. 2.
Transfection of RNA from virus-infected cells generates progeny virus. (A and B) 3T3 cells were left untreated or were pretreated with ribavirin for 1 h and transfected with Vero-EMCV RNA (0.2 μg in A; 1 and 0.2 μg in B) or infected with EMCV (MOI of 1). (A) Accumulation of infectious virus particles quantified by 50% tissue culture infective dose. (B) IFN-α was measured after overnight incubation. ELISA shows average of triplicate measurements + standard deviation. (C and D) Quantitative reverse transcription-PCR analysis for EMCV RNA (C) and IFN-β mRNA (D) in cells treated with CHX for 30 min and subsequently infected with EMCV or transfected with 0.2 μg of Vero-EMCV RNA for 4 h. Bars show averages of duplicate measurements + standard deviations. Graphs show one of two (A and B) or three representative experiments (C and D). Error bars show standard deviations of quadruplicate (A), triplicate (B), or duplicate (C and D) measurements. Asterisks indicate a P value of <0.001 compared to dimethyl sulfoxide (DMSO) treatment.
VV generates dsRNA that activates MDA5.
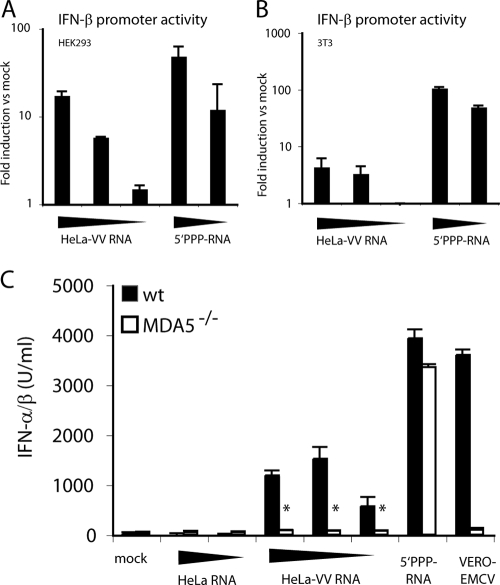
The presence of infectious RNAs in preparations extracted from cells infected with RNA viruses complicated our attempts to characterize agonists for RLRs. We therefore sought to establish an additional virus model in which RNA extracted from infected cells would be noninfectious. DNA viruses have been reported to generate dsRNA in infected cells (1, 9, 28), but, by definition, such RNA cannot be infectious. Notably, HeLa-VV RNA activated the IFN-β promoter upon transfection into HEK293 (Fig. 3A) or 3T3 cells (Fig. 3B) and induced secretion of IFN-α/β protein after transfection in MEFs (Fig. 3C). The stimulatory activity of HeLa-VV RNA was appreciably lower than that of Vero-EMCV RNA or of a control 5′ PPP-RNA (Fig. 3A to C). Nevertheless, it was completely MDA5 dependent as MEFs that lacked MDA5 were totally unresponsive to HeLa-VV RNA (Fig. 3C). Therefore, RNA from VV-infected cells can be used as a tool to dissect MDA5-dependent recognition processes without the added complication of infectious RNA and de novo virus replication.
FIG. 3.
RNA from VV-infected cells stimulates MDA5. HEK293 (A) or 3T3 (B) cells were transfected with reporter constructs for IFN-β-luciferase and control plasmid pRL-TK and subsequently stimulated with HeLa-VV RNA (1, 0.2, and 0.04 μg) or control 5′ PPP-RNA (0.2 and 0.04 μg), respectively. Graphs show relative activation of the IFN-β promoter. Data are the average of duplicate measurements ± standard deviation. (C) MEFs of the indicated genotype were transfected with HeLa-RNA (1 and 0.2 μg), HeLa-VV RNA (1, 0.2, and 0.04 μg), Vero-EMCV RNA (0.2 μg), or with control 5′ PPP-RNA (0.2 μg). The graph shows the accumulation of IFN-α/β in supernatants after overnight incubation. *, P < 0.001 compared to wild-type (wt) MEFs. Data are average ± standard deviations of duplicate (A and B) or quadruplicate (C) measurements from one experiment repeated three times.
The dsRNA-specific K1 antibody recognizes an MDA5 agonist.
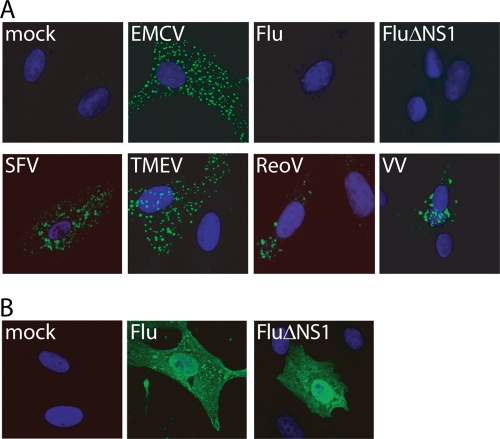
VV, EMCV, and SFV have all previously been shown to generate dsRNA that can be detected by staining cells with the dsRNA-specific K1 monoclonal antibody (MAb) (27, 28). Because all three viruses activate MDA5 (see above), we tested for K1 immunoreactivity upon infection with all MDA5-activating viruses. K1 stained cells infected with EMCV, TMEV, SFV, ReoV, SiV, and VV but did not stain cells infected with influenza virus or influenza virus lacking the NS1 protein (FluΔNS1) (Fig. 4A), despite the fact that the latter were clearly infected (Fig. 4B). Given this correlation between MDA5-stimulatory ability and generation of K1 ligands, we next asked whether RNA bound to the dsRNA-specific K1 antibody can also bind to MDA5. We transfected 293T cells with HA-tagged MDA5 and infected these cells with EMCV (MOI of 10 for 8 h) or left them uninfected. The dsRNA antibody did not coprecipitate MDA5 when the cells were not infected, as expected (Fig. 5A). However, infection with EMCV promoted formation of a complex between MDA5 and K1. Notably, this complex could also be formed after cell lysis by the addition to the lysates of poly(I:C), which binds strongly to both K1 (26) and to MDA5 (11), or of Vero-EMCV RNA (Fig. 5A).
FIG. 4.
MDA5-activating viruses generate immunodetectable dsRNA. (A) Vero cells were infected with the indicated virus (MOI of 0.5 to 1) for 8 h (EMCV, TMEV, and SFV) or 16 h (influenza virus (Flu), FluΔNS1, and ReoV). HeLa cells were infected with VV (MOI of 0.5) for 16 h. Cells were fixed and stained with the K1 (dsRNA) MAb (green) and DAPI (blue). (B) Influenza virus-infected cells stained with anti-influenza (Flu) antibodies (green) and DAPI (blue).
FIG. 5.
K1 and MDA5 bind to the same type of RNA. (A) 293T cells were transfected with HA-tagged MDA5 and 40 h later infected with EMCV (if EMCV) or left uninfected. Eight hours later, cell lysates were prepared and used for coimmunoprecipitation experiments. Uninfected cells (lanes 5, 6, and 8) or infected cells (lane 7) were used for immunoprecipitation with the K1 (dsRNA) antibody. Poly(I:C) and Vero-EMCV RNA were added to the lysates in lanes 6 and 8, respectively. HA-MDA5 in total cell lysates (lanes 1 to 3) or K1 immunoprecipitates (lanes 4 to 8) was visualized by Western blotting (WB) with antibody against the HA epitope. (B and C) The K1 antibody or a control irrelevant antibody was used to precipitate RNA extracted from EMCV-infected Vero cells (B) or VV-infected HeLa cells (C). The RNA in the precipitated fraction was extracted with Trizol and used to transfect 3T3 cells. Graphs show average (+ standard deviation) accumulation of IFN-α/β in supernatants (measured in quadruplicate) after 16 h. One representative experiment of three is shown.
To test whether RNA bound to the K1 antibody has stimulatory activity, we precipitated RNA extracted from EMCV-infected cells and tested the bound fractions for IFN-α/β induction. As expected, control Sepharose beads coated with IgG did not precipitate stimulatory activity from Vero-EMCV RNA (Fig. 5B). However, beads coupled to the K1 antibody precipitated Vero-EMCV RNA that induced large amounts of IFN-α/β upon transfection into indicator cells (Fig. 5B). Similar to Vero-EMCV RNA, the fraction of HeLa-VV RNA precipitated by K1 contained stimulatory activity, whereas control IgG-precipitated RNA was inactive (Fig. 5C). Notably, the amount of RNA isolated from K1-precipitated fractions was dramatically reduced (>100-fold) compared to the amount isolated from unbound fractions (data not shown), indicating that the dsRNA recognizing MAb bound selected RNA species. Collectively, these experiments suggest that the K1 antibody can bind to MDA5 agonist(s) present in virus-infected cells and indicate that MDA5 and the K1 antibody bind to similar RNAs.
Lack of specific sequence motifs in stimulatory RNA fractions from VV-infected cells.
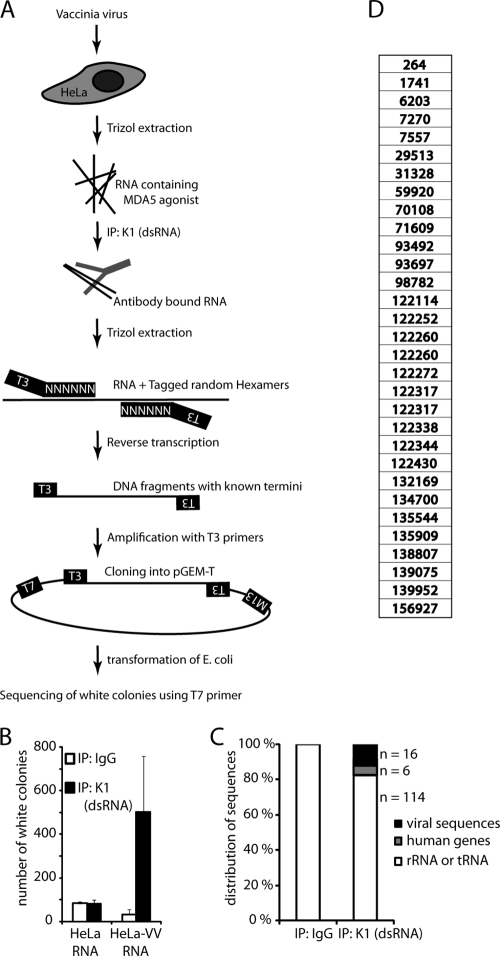
We attempted to define specific sequences in the stimulatory RNA from VV-infected cells. As we did not know whether terminal OH groups were present in MDA5-agonistic RNA and whether sequences necessary for activating MDA5 were at the end of the putative RNA sequence, we resorted to an approach that involved tagged random hexamer priming (29) (Fig. 6A gives a schematic representation). RNA from virus-infected or uninfected cells was used for immunoprecipitation with K1 or control antibodies. RNA was reverse transcribed and amplified by PCR for 5 to 10 cycles, and the resulting DNA fragments were ligated into pGEM-T (Promega). Colonies were screened for β-galactosidase activity, and white colonies were selected for sequencing. K1 immunoprecipitates of HeLa-VV RNA yielded more colonies than control precipitations (IgG precipitates from HeLa-VV RNA or K1 precipitations of RNA from uninfected cells) (Fig. 6B). This was encouraging as it suggested enrichment of specific stimulatory RNAs with the K1 antibody. However, sequencing revealed mostly rRNA, a few viral sequences, and a few sequences from human genes (Fig. 6C shows a typical example of one experiment). In total, we identified 31 clones containing viral sequences, and in one of three experiments, there was some enrichment in the A11R gene locus (bp 121859 to 122815) (Fig. 6D). However, overall we failed to identify any particular sequence or sequence motif or even clustering when sequences were aligned to the virus genome. Thus, the data obtained by this method do not reveal any particular sequence preference for MDA5-stimulatory RNA.
FIG. 6.
Lack of specific sequence motifs in MDA5-agonistic RNA. (A) Experimental outline. RNA from cells infected with VV (13 or 24 h) was immunoprecipitated with K1 or IgG control antibodies. RNA was isolated and reverse transcribed using random hexamer primers with an attached T3 promoter site. cDNA was amplified in a PCR step (5 to 10 cycles) with T3 primers, and the resulting DNA was cloned into pGEM-T (Promega). Bacterial colonies were screened for β-galactosidase activity, and white colonies that contained an insert were sequenced using T7 primers. (B) Average number of white CFU from a typical experiment. Error bars represent standard deviations from two independent transformations. (C) Distribution of identified sequences from IgG immunoprecipitation (n = 21) and K1 (dsRNA) immunoprecipitation (n = 136). The average recovered sequence length was 369 bp. One representative experiment of three is shown. (D) Identified sequences from 31 identified VV gene products. The number shows start of the sequence in the VV genome (VV NCBI accession number AY243312). IP, immunoprecipitation.
HMW RNA but not dsRNA stimulates MDA5.
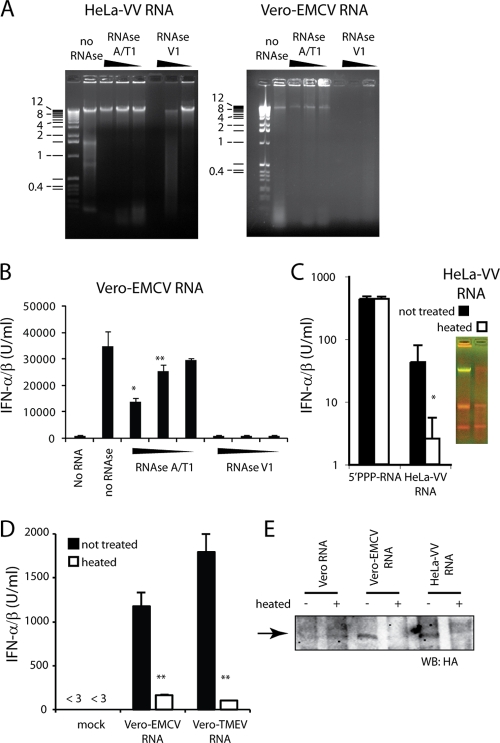
The association between MDA5-agonistic activity and recognition by K1, a dsRNA-specific MAb, prompted us to analyze RNA preparations from virus-infected cells for the presence of dsRNA. Double-strandedness of nucleotides can be visualized by staining with acridine orange, a metachromatic stain that stains single-stranded polynucleotides red and double-stranded polynucleotides green (18). In the past, this dye has been used extensively to categorize viruses according to the nature of their genome (15-17). As expected, a double-stranded DNA marker appears green on an acridine orange-stained gel, whereas in vitro transcribed ssRNA appears red (Fig. 7A). RNA isolated from uninfected HeLa cells (HeLa-RNA) or Vero-RNA showed two prominent red bands of rRNA (Fig. 7A to C). Consistent with the fact that EMCV degrades rRNA (30), the rRNA bands were faint in Vero-EMCV RNA, which contained a prominent dsRNA band of 11,000 or more bp (Fig. 7A and C). Similarly, HeLa-VV RNA contained a dsRNA band that was absent from uninfected cells (Fig. 7B and C). In addition, RNA from both EMCV- and VV-infected cells contained a very-high-molecular-weight (HMW) fraction that did not enter the agarose gel and remained trapped in the well and variably stained green and red with acridine orange (Fig. 7A to C). Similarly, well-trapped HMW RNA ranging in staining from green to red was detected in all RNA preparations from virus-infected cells able to stimulate MDA5 but not in Vero-Flu RNA, which exclusively activates RIG-I (Fig. 7C). We isolated the dsRNA band and the HMW well-trapped fraction from Vero-EMCV cells and tested them for stimulatory activity (Fig. 7D, inset). To our surprise, only the HMW RNA and not the dsRNA fraction elicited IFN-α/β when transfected into 3T3 cells (Fig. 7D). Similarly, HMW RNA from VV-infected cells contained stimulatory potential, whereas the dsRNA fraction did not generate measurable IFN-α/β (Fig. 7E). For both viruses, HMW RNA allowed association of MDA5 to agarose beads coupled to the dsRNA antibody (Fig. 7F). Thus, we conclude that HMW RNA is generated by some viruses and that this RNA has the ability to bind to and activate MDA5. Interestingly, although dsRNA arising from virus infection can bind to MDA5, it is not necessarily an agonist for the helicase.
FIG. 7.

HMW RNA activates MDA5. (A) Amounts of 1 μg and 0.2 μg of the indicated nucleic acid were electrophoretically separated on a 1% agarose gel and subsequently stained with acridine orange. Double-stranded nucleic acid stains green; single-stranded nucleic acid stains red. (B) A 1-μg and 0.2-μg DNA ladder and 1 μg of HeLa-VV RNA were electrophoretically separated at the indicated time points on an agarose gel and stained with acridine orange. (C) A 1-μg DNA ladder and 1 μg of the indicated RNA preparation were electrophoretically separated on an agarose gel and stained with acridine orange. Numbers to the left of the DNA ladders (A to C) show sizes of DNA markers. (D and E) The indicated RNA fraction (see inset of acridine orange gel) was isolated from the agarose gel, and 0.5 or 0.1 μg was used to transfect 3T3 cells. In panel D 3T3 cells were treated with ribavirin to prevent EMCV replication. Graphs show average accumulation + standard deviation of IFN-α/β after overnight culture measured in quadruplicate. (F) Lysate of 293T cells transfected with FLAG-MDA5 for 48 h was used for immunoprecipitation with the K1 (dsRNA) antibody in the absence or presence of 1 μg of the indicated RNA. K1 immunoprecipitates were visualized by Western blotting (WB) with antibody against the FLAG epitope. The arrow points to FLAG-tagged MDA5.
HMW RNA contained both ssRNA and dsRNA regions as it stained red and green on acridine orange gels, with some variability between experiments (Fig. 7A to C). The presence and functional importance of dsRNA and ssRNA regions in HMW RNA were further suggested by experiments using nuclease digestion with the ssRNA-specific nucleases RNase A and RNase T1 and the dsRNA-specific RNase V1 (Fig. 8A and B). Therefore, we envisaged that HMW RNA may resemble an RNA web rather than perfectly base-paired long molecules of dsRNA. We hypothesized that destroying the secondary and tertiary structure of this HMW RNA would diminish its potential to stimulate MDA5. Indeed, when we heat denatured and flash froze HeLa-VV RNA, both the dsRNA fraction and the HMW RNA fraction disappeared (Fig. 8C, insert), together with the ability to stimulate IFN-α/β (Fig. 8C). In contrast, heating and flash freezing of an in vitro transcribed control 5′ PPP-RNA did not result in a change of IFN induction (Fig. 8C). The stimulatory activity of RNA preparations from cells infected with picornaviruses was similarly sensitive to heat denaturation followed by rapid freezing (Fig. 8D). Finally, the association between MDA5 and beads coated with the dsRNA antibody was lost when RNA denatured by heating and freezing was used (Fig. 8E). We conclude that the ability to stimulate MDA5 requires highly structured RNA.
FIG. 8.
Destroying higher-order RNA structures reduces MDA5 activation. (A and B) One microgram of HeLa-VV RNA or Vero-EMCV RNA was digested with 10-fold serial dilutions of the ssRNA-specific RNase A and RNase T1 or with the dsRNA-specific RNase V1 for 1 h. RNA was visualized on an agarose gel (A) or transfected into 3T3 cells for measuring IFN-α/β production (B). Numbers to the left of the DNA ladder (A) show sizes of DNA markers. (C) Two micrograms of HeLa-VV RNA was heated to 99°C for 5 min and flash frozen on dry ice. After the RNA was thawed, 1 μg was separated on an acridine orange gel (inset). In addition, 0.2 μg of the indicated RNA was used to transfect 3T3 cells, and IFN-α/β accumulation in supernatants was measured after overnight culture. (D) Vero-EMCV RNA and Vero-TMEV RNA were heated and flash frozen on dry ice, and after the RNA was thawed, 0.2 μg was transfected into 3T3 cells. The graph shows IFN-α/β accumulation in supernatants measured after overnight incubation. Asterisks in panels C and D indicate statistical significance between untreated and treated samples (*, P < 0.001; **, P < 0.05). (E) Lysate from 293T cells transfected with HA-MDA5 was used for immunoprecipitation with the K1 antibody in the presence of Vero-RNA, Vero-EMCV RNA, or HeLa-VV RNA that had been left untreated (−) or heated and flash frozen (+). Immunoprecipitates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and subjected to Western blotting (WB) with an anti-HA antibody. The arrow points to HA-tagged MDA5.
DISCUSSION
The identification of RIG-I and MDA5 as pattern recognition receptors for viruses marked a milestone in our understanding of antiviral innate responses (11, 13, 31, 32). Interestingly, genetic deletion experiments demonstrated that RIG-I and MDA5 possess distinct virus specificities and that MDA5 but not RIG-I is essential for innate responses to picornaviruses, whereas RIG-I but not MDA5 is indispensable for innate immunity to influenza A virus, paramyxoviruses, and Japanese encephalitis virus (11). The basis for differential virus recognition by the two RLRs is not entirely understood. RIG-I agonists have been variously defined as 5′ PPP-RNAs, short dsRNA duplexes bearing or not a 5′ monophosphate, or 3′ monophosphate-containing cleavage products of RNase L (7, 10, 22). In contrast, agonists for MDA5 remain less well characterized. It is generally thought that MDA5 recognizes long dsRNA stretches such as those found in poly(I:C) and in the genomes of ReoVs (10). In this study, we set out to investigate the characteristics of RNA species that can stimulate MDA5 in virus-infected cells. As others have noted (7), we found that RNA extracted from uninfected cells does not stimulate IFN production, indicating that it does not contain enough RNA species bearing, for example, 5′ triphosphate moieties and/or the necessary secondary structure to act as agonists for RLRs. In contrast, RNA extracted from cells infected with RNA viruses contains abundant stimulatory activity, and the dependence of that activity on RIG-I and MDA5 largely matches the dependence reported for infection with the intact virus (Fig. 1 and 3). In particular, we show that influenza A virus generates stimulatory RNA for RIG-I, whereas TMEV, EMCV, SiV, ReoV, and SFV all generate RNAs capable of stimulating MDA5 activity (Fig. 1). RNA isolated from cells infected with RNA viruses is infectious, which can complicate approaches to identifying the stimulatory fraction. For example, we isolated a single fraction of RNA from EMCV-infected cells which was able to induce large amounts of IFN-α/β upon transfection into reporter cells (data not shown). However, this fraction turned out to contain the intact EMCV genome, and its transfection resulted in a full infectious cycle, rendering it difficult to determine whether the IFN-inducing activity was attributable to properties of the genome itself or to RNAs made during subsequent virus replication (data not shown). Although this problem could be circumvented in part by using ribavirin, more definitive evidence for a preformed stimulatory species came from analysis of RNA extracted from cells infected with DNA viruses. A recent paper suggests that modified VV Ankara induces IFN-α/β in an MDA5- and MAVS-dependent manner (3). Here, we show that RNAs generated during VV infection indeed activate MDA5-dependent IFN-α/β responses. Therefore, DNA viruses have the potential to generate RNA that activates RLRs, and VV infection may be a useful system in which to characterize MDA5-agonistic RNAs.
All viruses that generated MDA5 agonists in infected cells also induced accumulation of dsRNA, which was detectable by staining with the dsRNA-specific K1 antibody (Fig. 4). The striking punctate pattern of this staining suggested association with specific subcellular compartments. However, we were unable to colocalize K1 staining with markers of the endoplasmic reticulum, endosomes, ribosomes or P-bodies (data not shown). Thus, the origin of the RNA bound by the K1 antibody is uncertain, and it could reflect either species generated at specific sites of virus replication or RNA being targeted for degradation in specialized compartments. As K1 binds poly(I:C) (26), a known stimulus for MDA5, we investigated the possibility that MDA5-agonistic activity overlaps with K1 immunoreactivity. We confirmed that the K1 antibody binds to an MDA5 agonist by pull-down experiments of stimulatory RNA isolated from virus-infected cells (Fig. 5B and C). RIG-I has been suggested to be preferentially activated when the terminal triphosphate group is presented in the context of certain sequence motifs (25). We therefore tried to clone and sequence the RNA that binds to K1-coated beads but failed to obtain meaningful results: we enriched for viral sequences that were expressed at the time of cell lysis but did not find any specific pattern that could help define a specific MDA5 agonist (Fig. 6). Therefore, we used alternative methods to characterize this RNA. Notably, we found that RNA isolated from cells infected with MDA5-activating viruses contained prominent dsRNA bands, as determined by acridine orange staining. These bands represented candidate agonists as Kato et al. have previously shown that a 4-kbp fragment of annealed antisense and sense in vitro transcribed RNA, as well as long segments of genomic dsRNA from ReoV, can activate MDA5 (10). However, we found that the major dsRNA band (larger than 11 kbp) isolated from cells infected with EMCV and VV bound to the helicase but, surprisingly, was unable to activate MDA5 (Fig. 7D to F). Instead, MDA5 was activated by the HMW RNA fraction that did not enter the gel. Acridine orange staining and RNase digestion suggest that this HMW RNA contains single-stranded and double-stranded regions and can bind to both MDA5 and the K1 antibody (Fig. 7 and 8A). Notably, destroying its structure through a heating and freezing cycle abrogated MDA5 binding and IFN-α/β stimulatory activity (Fig. 8C and D). Therefore, we believe that double strandedness may be necessary but is not sufficient for IFN-α/β induction and that the MDA5-stimulating RNA present in virus-infected cells is in the form of RNA aggregates consisting of single-stranded and double-stranded moieties that are generated during the replication/transcription process. Poly(I:C) binds to K1 and acts as an agonist for MDA5 signaling. Although poly(I:C) is regarded as a dsRNA homologue, it does not consist of perfectly matched dsRNA strands because poly(I:C) is generated through annealing of enzymatically produced inosine and cytidine homopolymers of undefined length (5), which results in a web-like structure. We speculate that branched RNA may be an important part in MDA5-dependent recognition. Interestingly, recognition of branched nucleic acid is a long-studied phenomenon: junction-resolving enzymes recognize Holliday junctions (branched DNA) during DNA replication (2). Binding of junction-resolving enzymes must meet specific structural requirements (DNA branches) but is independent of the exact nucleic acid sequence. Similarly, MDA5 recognition appears to require structural features (Fig. 7 and 8), but we could not identify specific sequences (Fig. 6). Reducing the size of poly(I:C) by sonication decreases its ability to activate MDA5 (10), which could be explained by the destruction of the RNA web necessary for MDA5 activation. Notably, annealing polyadenylic and polyuridylic homopolymers of undefined length to form poly(A·U) is not sufficient to generate RLR-stimulatory activity (data not shown). One possible explanation could lie in the high affinity of poly(A) for poly(U), which would make poly(A·U) quite inflexible and prevent it from adopting a loose web structure, unlike poly(I:C). Recently, Myong et al. reported that recognition of triphosphorylated RNA cannot activate RIG-I without local secondary RNA structure (19). Similarly, dsRNA or RNA aggregates may be necessary for binding of MDA5 (Fig. 7F), but this may not be sufficient for potent MDA5 activation, which may require hitherto undefined additional signals to distinguish cellular from viral RNA. We speculate that, similar to junction-resolving enzymes that recognize branched DNA during cellular replication, MDA5 may have evolved to recognize branched RNA during virus replication.
Acknowledgments
We thank the members of the immunobiology laboratory for stimulating discussions, Mike Gait and Svend Petersen-Mahrt for invaluable intellectual input, Ina Weisswange for infecting cells with VV, and Catalina Vasquez for secretarial support.
This work was supported by Cancer Research UK and a long-term EMBO fellowship to A.P. J.R. is a recipient of FEBS and ABSP long-term fellowships.
Footnotes
Published ahead of print on 5 August 2009.
REFERENCES
- 1.Colby, C., and P. H. Duesberg. 1969. Double-stranded RNA in vaccinia virus infected cells. Nature 222:940-944. [DOI] [PubMed] [Google Scholar]
- 2.Declais, A. C., and D. M. Lilley. 2008. New insight into the recognition of branched DNA structure by junction-resolving enzymes. Curr. Opin. Struct. Biol. 18:86-95. [DOI] [PubMed] [Google Scholar]
- 3.Delaloye, J., T. Roger, Q. G. Steiner-Tardivel, D. Le Roy, M. Knaup Reymond, S. Akira, V. Petrilli, C. E. Gomez, B. Perdiguero, J. Tschopp, G. Pantaleo, M. Esteban, and T. Calandra. 2009. Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS Pathog. 5:e1000480. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Gitlin, L., W. Barchet, S. Gilfillan, M. Cella, B. Beutler, R. A. Flavell, M. S. Diamond, and M. Colonna. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA 103:8459-8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grunberg-Manago, M., P. J. Ortiz, and S. Ochoa. 1956. Enzymic synthesis of polynucleotides. I. Polynucleotide phosphorylase of azotobacter vinelandii. Biochim. Biophys. Acta 20:269-285. [DOI] [PubMed] [Google Scholar]
- 6.Holm, G. H., J. Zurney, V. Tumilasci, S. Leveille, P. Danthi, J. Hiscott, B. Sherry, and T. S. Dermody. 2007. Retinoic acid-inducible gene-I and interferon-beta promoter stimulator-1 augment proapoptotic responses following mammalian reovirus infection via interferon regulatory factor-3. J. Biol. Chem. 282:21953-21961. [DOI] [PubMed] [Google Scholar]
- 7.Hornung, V., J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K. K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994-997. [DOI] [PubMed] [Google Scholar]
- 8.Iwasaki, A., and R. Medzhitov. 2004. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5:987-995. [DOI] [PubMed] [Google Scholar]
- 9.Jurale, C., J. R. Kates, and C. Colby. 1970. Isolation of double-stranded RNA from T4 phage infected cells. Nature 226:1027-1029. [DOI] [PubMed] [Google Scholar]
- 10.Kato, H., O. Takeuchi, E. Mikamo-Satoh, R. Hirai, T. Kawai, K. Matsushita, A. Hiiragi, T. S. Dermody, T. Fujita, and S. Akira. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205:1601-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101-105. [DOI] [PubMed] [Google Scholar]
- 12.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131-137. [DOI] [PubMed] [Google Scholar]
- 13.Kawai, T., and S. Akira. 2008. Toll-like receptor and RIG-I-like receptor signaling. Ann. N. Y. Acad. Sci. 1143:1-20. [DOI] [PubMed] [Google Scholar]
- 14.Loo, Y. M., J. Fornek, N. Crochet, G. Bajwa, O. Perwitasari, L. Martinez-Sobrido, S. Akira, M. A. Gill, A. Garcia-Sastre, M. G. Katze, and M. Gale, Jr. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82:335-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsunaga, Y., S. Matsuno, and J. Mukoyama. 1977. Isolation and characterization of a parvovirus of rabbits. Infect. Immun. 18:495-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayor, H. D. 1963. The nucleic acids of viruses as revealed by their reactions with fluorochrome acridine orange. Int. Rev. Exp. Pathol. 2:1-45. [PubMed] [Google Scholar]
- 17.Mayor, H. D., and A. R. Diwan. 1961. Studies on the acridine orange staining of two purified RNA viruses: poliovirus and tobacco mosaic virus. Virology 14:74-82. [DOI] [PubMed] [Google Scholar]
- 18.McMaster, G. K., and G. G. Carmichael. 1977. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc. Natl. Acad. Sci. USA 74:4835-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myong, S., S. Cui, P. V. Cornish, A. Kirchhofer, M. U. Gack, J. U. Jung, K. P. Hopfner, and T. Ha. 2009. Cytosolic viral sensor RIG-I is a 5′-triphosphate-dependent translocase on double-stranded RNA. Science 323:1070-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Onomoto, K., M. Yoneyama, and T. Fujita. 2007. Regulation of antiviral innate immune responses by RIG-I family of RNA helicases. Curr. Top. Microbiol. Immunol. 316:193-205. [DOI] [PubMed] [Google Scholar]
- 21.Pichlmair, A., and C. Reis e Sousa. 2007. Innate recognition of viruses. Immunity 27:370-383. [DOI] [PubMed] [Google Scholar]
- 22.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997-1001. [DOI] [PubMed] [Google Scholar]
- 23.Racaniello, V. R. 2001. Picornaviridae: the viruses and their replication, p. 685-722. In D. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 24.Saito, T., and M. Gale, Jr. 2007. Principles of intracellular viral recognition. Curr. Opin. Immunol. 19:17-23. [DOI] [PubMed] [Google Scholar]
- 25.Saito, T., D. M. Owen, F. Jiang, J. Marcotrigiano, and M. Gale, Jr. 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schonborn, J., J. Oberstrass, E. Breyel, J. Tittgen, J. Schumacher, and N. Lukacs. 1991. Monoclonal antibodies to double-stranded RNA as probes of RNA structure in crude nucleic acid extracts. Nucleic Acids Res. 19:2993-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulz, O., S. S. Diebold, M. Chen, T. I. Naslund, M. A. Nolte, L. Alexopoulou, Y. T. Azuma, R. A. Flavell, P. Liljestrom, and C. Reis e Sousa. 2005. Toll-like receptor 3 promotes cross-priming to virus-infected cells. Nature 433:887-892. [DOI] [PubMed] [Google Scholar]
- 28.Weber, F., V. Wagner, S. B. Rasmussen, R. Hartmann, and S. R. Paludan. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J. Virol. 80:5059-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong, K. K., L. C. Stillwell, C. A. Dockery, and J. D. Saffer. 1996. Use of tagged random hexamer amplification (TRHA) to clone and sequence minute quantities of DNA—application to a 180 kb plasmid isolated from Sphingomonas F199. Nucleic Acids Res. 24:3778-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wreschner, D. H., T. C. James, R. H. Silverman, and I. M. Kerr. 1981. Ribosomal RNA cleavage, nuclease activation and 2-5A(ppp(A2′p)nA) in interferon-treated cells. Nucleic Acids Res. 9:1571-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoneyama, M., and T. Fujita. 2007. Function of RIG-I-like receptors in antiviral innate immunity. J. Biol. Chem. 282:15315-15318. [DOI] [PubMed] [Google Scholar]
- 32.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]