Abstract
Understanding the mechanisms underlying potential altered susceptibility to human immunodeficiency virus type 1 (HIV-1) infection in highly exposed seronegative (ES) individuals and the later clinical consequences of breakthrough infection can provide insight into strategies to control HIV-1 with an effective vaccine. From our Seattle ES cohort, we identified one individual (LSC63) who seroconverted after over 2 years of repeated unprotected sexual contact with his HIV-1-infected partner (P63) and other sexual partners of unknown HIV-1 serostatus. The HIV-1 variants infecting LSC63 were genetically unrelated to those sequenced from P63. This may not be surprising, since viral load measurements in P63 were repeatedly below 50 copies/ml, making him an unlikely transmitter. However, broad HIV-1-specific cytotoxic T-lymphocyte (CTL) responses were detected in LSC63 before seroconversion. Compared to those detected after seroconversion, these responses were of lower magnitude and half of them targeted different regions of the viral proteome. Strong HLA-B27-restricted CTLs, which have been associated with disease control, were detected in LSC63 after but not before seroconversion. Furthermore, for the majority of the protein-coding regions of the HIV-1 variants in LSC63 (except gp41, nef, and the 3′ half of pol), the genetic distances between the infecting viruses and the viruses to which he was exposed through P63 (termed the exposed virus) were comparable to the distances between random subtype B HIV-1 sequences and the exposed viruses. These results suggest that broad preinfection immune responses were not able to prevent the acquisition of HIV-1 infection in LSC63, even though the infecting viruses were not particularly distant from the viruses that may have elicited these responses.
Understanding the mechanisms of altered susceptibility or control of human immunodeficiency virus type 1 (HIV-1) infection in highly exposed seronegative (ES) persons may provide invaluable information aiding the design of HIV-1 vaccines and therapy (9, 14, 15, 33, 45, 57, 58). In a cohort of female commercial sex workers in Nairobi, Kenya, a small proportion of individuals remained seronegative for over 3 years despite the continued practice of unprotected sex (12, 28, 55, 56). Similarly, resistance to HIV-1 infection has been reported in homosexual men who frequently practiced unprotected sex with infected partners (1, 15, 17, 21, 61). Multiple factors have been associated with the resistance to HIV-1 infection in ES individuals (32), including host genetic factors (8, 16, 20, 37-39, 44, 46, 47, 49, 59, 63), such as certain HLA class I and II alleles (41), as well as cellular (1, 15, 26, 55, 56), humoral (25, 29), and innate immune responses (22, 35).
Seroconversion in previously HIV-resistant Nairobi female commercial sex workers, despite preexisting HIV-specific cytotoxic T-lymphocyte (CTL) responses, has been reported (27). Similarly, 13 of 125 ES enrollees in our Seattle ES cohort (1, 15, 17) have become late seroconverters (H. Zhu, T. Andrus, Y. Liu, and T. Zhu, unpublished observations). Here, we analyze the virology, genetics, and immune responses of HIV-1 infection in one of the later seroconverting subjects, LSC63, who had developed broad CTL responses before seroconversion.
MATERIALS AND METHODS
Study subjects.
The Seattle MSM (men having sex with men) ES cohort was initiated in 1995. Detailed information about this cohort, including procedures for subject recruitment, has been published previously (1, 15, 17, 61, 66). This cohort enrolled HIV-1-seronegative subjects who repeatedly practiced unprotected sex with known HIV-1-infected partners; some also reported sexual partners with unknown HIV-1 serostatus (1, 15, 17). All ES enrollees completed a questionnaire concerning risk behavior and provided blood during visits scheduled monthly during the initial 3 months and every 3 to 6 months thereafter.
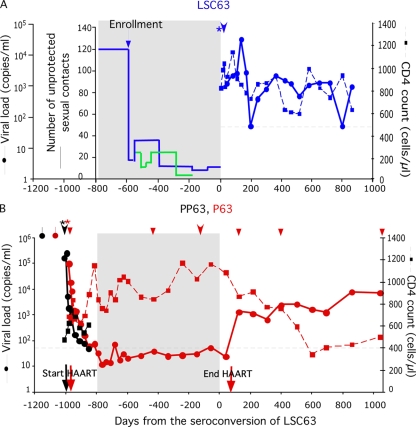
As shown in Fig. 1A, LSC63 had frequent unprotected anal insertive and receptive sex with his HIV-1-infected partner P63 for more than 6 months before enrollment into the Seattle ES cohort. The seronegative status of LSC63 upon enrollment was confirmed by HIV-1/HIV-2 enzyme-linked immunosorbent assay and Western blot assay (60), HIV-1 plasma RNA reverse transcription-PCR (Amplicor HIV-1 monitor; Roche, Branchburg, NJ), and peripheral blood mononuclear cell (PBMC) DNA PCR (52). The HIV-1/HIV-2 enzyme-linked immunosorbent assay and Western blot assay were repeated every 3 months. HIV-1 PBMC DNA PCR was performed again at 4 and 8 weeks after enrollment and then repeated yearly. LSC63 seroconverted 582 days after enrollment, after which the relationship with P63 ended. LSC63 reported engaging in unprotected anal insertive and receptive sex with four HIV-1-infected partners other than P63 and two partners with unknown HIV-1 serostatus during the period 6 to 12 months prior to seroconversion (Fig. 1A) (there is no information as to whether or not these HIV-positive partners were on antiretroviral medications). However, LSC63 engaged in sex only with P63 during the last 6 months prior to seroconversion. No other sexually transmitted diseases were reported for LSC63, nor did he receive antiretroviral treatment during the 2.5 years of follow-up after seroconversion (Fig. 1A).
FIG. 1.
Clinical, virological, and behavioral data for LSC63, P63, and PP63. (A) Numbers of unprotected sexual contacts, viral loads, and CD4+ T-cell counts of LSC63. For the numbers of unprotected sexual contacts, the blue line shows reported sexual contacts with P63, the green line shows reported sexual contacts with other partners, and the horizontal line sections represent the total number of unprotected sexual contacts reported by LSC63 during the periods encompassed. (B) Viral loads and CD4+ T-cell counts of subjects PP63 and P63. For both panels, the areas shaded gray represent the period in which LSC63 had frequent unprotected sex with P63 and the black dashed lines indicate a viral load level of 50 copies/ml., onset of acute symptoms of viral infection; *, seroconversion;, viral sequences covering nearly the entire HIV-1 coding region were obtained; ▾, viral sequences of env C2-V5 were obtained.
Before his relationship with LSC63, P63 had a sexual relationship with subject PP63 (who reported having acute symptoms of HIV-1 infection 3 months before the relationship) and subsequently developed acute symptoms. Both PP63 and P63 were enrolled in the University of Washington Primary Infection Clinic. As shown in Fig. 1B, PP63 and P63 were found to be HIV-1 seropositive 2.8 years and 2.7 years, respectively, before the seroconversion of LSC63.
Amplification and sequencing of viral genes.
Viral sequences that covered nearly the entire HIV-1 coding region (Table 1) were amplified from cellular genomic DNA of LSC63 24 days after seroconversion, from P63 on day 831 after seroconversion (−148 days from the seroconversion of LSC63), and from PP63 on day 14 after seroconversion (−994 days from the seroconversion of LSC63), after which PP63 started highly active antiretroviral therapy (HAART). In addition, HIV-1 sequences corresponding to the env C2-V5 region were obtained from P63 on days 12, 530, 831, 1104, 1377, and 2037 after his seroconversion (−967, −449, −148, 125, 398, and 1058 days from the seroconversion of LSC63, respectively) (40). P63 received HAART from day 21 to 1052 after his seroconversion (−2.7 years to 2 months relative to the seroconversion of LSC63).
TABLE 1.
PCR primers used in this study
| Gene amplified and PCR round | Forward primer | Sequence (position in HXB2) | Reverse primer | Sequence (position in HXB2) |
|---|---|---|---|---|
| C2-V5 | ||||
| 1st | P5-3 | ATCCTTTGAGCCAATTCCCATACATTATTG (6848-6877) | P2 | GACGTGCAGCCCATAGTGCTTCCTGCTGCT (7825-7800) |
| 2nd | P5 | ACACATGGAATTCGGCCAGTAGT (6966-6988) | P120 | AAGAACCCAAGGAACATAGCTCC (7792-7770) |
| gag | ||||
| 1st | PG3 | TGACTAGCGGAGGCTAGAAGGAGAGAG (763-789) | PG4 | ACCTCCAATTCCCCCTATCATTTTTGG (2408-2382) |
| 2nd | PG1 | ATGGGTGCGAGAGCGTCGGTATTAA (790-814) | PG2 | TTCTTCTAATACTGTATCATCTGCTCCTGT (2357-2328) |
| polA | ||||
| 1st | POL3 | GCAGGGCCCCTAGGAAAAAGGGCTGTT (2003-2029) | P58 | GACAAACTCCCACTCAGGAATCCA (3800-3777) |
| 2nd | POL1 | GAAATGTGGAAAGGAAGGACACCAAAT (2031-2057) | P56 | TGTCCACCATGCTTCCCATGTTTCCTTTTGTATG (3758-3725) |
| polB | ||||
| 1st | POL9-1 | AGTACCACTAACTGAAGAAGCAGA (3425-3448) | POL8 | CTGCTAGGTCAGGGTCTACTTGTGTG (5350-5325) |
| 2nd | POL7-1 | GACTTAATAGCAGAAATACAGAA (3519-3541) | POL6 | CCCTAGTGGGATGTGTACTTCTGAAC (5220-5195) |
| vif-vpr-vpu | ||||
| 1st | P11 | TGGAAAGGTGAAGGGGCAGTAGTAATA (4956-4982) | HY-P3R | CATGGCTTTAGGCTTTGATCCCAT (6580-6557) |
| 2nd | PF1 | ATGGAAAACAGATGGCAGGTGATGATTGTGTGG (5041-5073) | POR2 | GCTTTAGCATCTGATGCACAAAA (6403-6381) |
| gp120 | ||||
| 1st | PE0 | GCCTTAGGCATCTCCTATGGCAGGAAG (5954-5980) | PE3 | GGTGCAAATGAGTTTTCCAGAGCAACCCCA (8039-8010) |
| 2nd | PE1 | TAGAAAGAGCAGAAGACAGTGGCAATGA (6201-6228) | PE2 | GCCTGGAGCTGTTTGATGCCCCA (7957-7935) |
| gp41-nef | ||||
| 1st | PE11 | ATGAGGGACAATTGGAGAAGTGAATTA (7647-7673) | Nef4 | TGGTACTAGCTTGAAGCACCATCCAAA (9237-9211) |
| 2nd | P31 | TAGGAGTAGCACCCACCAAGGCAAAGAGAAGAG (7705-7737) | Nef2 | CCAATCAGGGAAGTAGCCTTGTGT (9168-9145) |
Genomic DNA was extracted from PBMC, purified CD4+ T cells, and CD14+ monocytes by using a QIAmp DNA mini kit (Qiagen, Valencia, CA) (13, 40). HIV-1 genome fragments were amplified by nested PCR using the primers listed in Table 1. PCR conditions for env C2-V5 have been described previously (13, 36, 40). The conditions for amplification of fragments that covered nearly the entire coding region were as follows for first-round PCR: 3 min at 95°C; 30 cycles of 40 s at 95°C, 40 s at 55°C, and 2.5 min at 72°C; and 7 min at 72°C. The conditions for second-round PCR were as follows: (i) for gag, 2 min at 95°C; 35 cycles of 15 s at 95°C, 30 s at 58°C, and 2 min at 72°C; and 7 min at 72°C; (ii) for polA, vif-vpr-vpu, and gp120, 2 min at 95°C; 35 cycles of 15 s at 95°C, 30 s at 55°C, and 2 min at 72°C; and 7 min at 72°C; (iii) for polB, 2 min at 95°C; 35 cycles of 15 s at 95°C, 30 s at 61°C, and 2 min at 72°C; and 7 min at 72°C; and (iv) for gp41-nef, 2 min at 95°C; 35 cycles of 15 s at 95°C, 30 s at 60°C, and 2 min at 72°C; and 7 min at 72°C. All PCR products were gel purified and cloned using a pcR2.1 TOPO TA cloning kit (Invitrogen, Carlsbad, CA). Sequencing was performed with a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) as previously described (13, 40).
Host genetic polymorphisms associated with HIV-1 transmission.
A multiplex PCR strategy was used for genotyping of CCR5-Δ32, CCR5 promoter −2459A/G, CCR2-64I, and SDF-1-3′A, and PCR-restriction fragment length polymorphism analysis was used for genotyping of the RANTES promoter, RANTES-403 and -28, DC-SIGN, and DC-SIGNR, as previously described (36, 38).
Humoral immunity to HSV-1 and HSV-2.
Antibodies to herpes simplex virus type 1 (HSV-1) and HSV-2 were detected by Western blot assays using plasma from LSC63 obtained at four time points (1.6 and 1.3 years before and 0 and 3 days after seroconversion) (3, 4).
ELISPOT assays.
CTL responses were measured in gamma interferon enzyme-linked immunospot (ELISPOT) assays, using cryopreserved PBMC as described previously (15, 17, 61). Fifteen-mer peptides, overlapping by 11 amino acids, were synthesized by the Biotechnology Center at Fred Hutchinson Cancer Research Center or by Synpep (Dublin, CA) or were kindly provided by the National Institutes of Health AIDS Research and Reference Reagent Program. These peptides spanned the entire HIV-1 HXB2 (subtype B) Gag, Env, Pol, and Nef proteins, with Vpr, Rev, Vif, and Vpu proteins corresponding to the 2001 HIV-1 subtype B consensus sequence. Predefined optimal epitopes restricted by LSC63's HLA alleles were also tested. Spot-forming cells (SFC) were counted using an automated ELISPOT reader (Immunospot; Cellular Technology, Cleveland, OH).
Phylogenetics, genetic distance, and statistical analysis.
Gene sequences were aligned using the program MacClade (42). Neighbor-joining trees were constructed, and average nucleotide pairwise distances were calculated using the HKY85 evolutionary model with the program PAUP* (64). Synonymous and nonsynonymous distances were calculated using a modified version of SNAP (www.hiv.lanl.gov) (30). For each sequenced region, we compared the genetic distances between the viral sequences from LSC63 (at 24 days after seroconversion) and viral sequences from P63 (at −148 days from seroconversion of LSC63) to the genetic distances between 142 random subtype B whole-genome sequences and viral sequences from P63. For a given region, the P value reported is the fraction of random sequences that have an average distance to P63 viruses greater than the average distance between viruses from LSC63 and P63. A P value of <0.05 was taken to indicate that the distances between viral sequences from LSC63 and P63 were significantly greater than the distances between random sequences and those from P63.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been submitted to GenBank and were given accession numbers GQ859273 to GQ859460.
RESULTS
Transmission partnerships among LSC63, P63, and PP63.
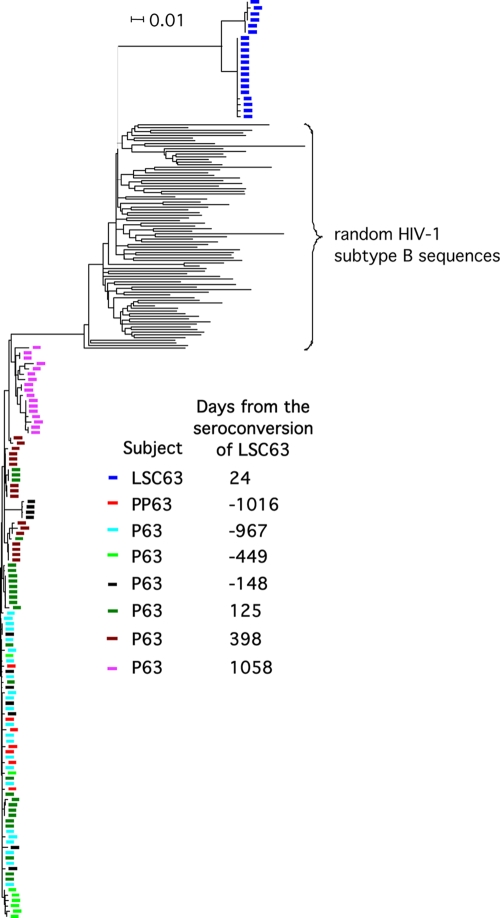
Nucleotide sequences of env C2-V5 from P63 and PP63 around the time of their seroconversions were homogeneous and clustered (Fig. 2), confirming the reported transmission partnership. LSC63 seroconverted after more than 25 months and more than 200 reported instances of unprotected sex with P63, as well as more than 80 reported unprotected sexual contacts with other partners (Fig. 1A). However, nucleotide sequences of env C2-V5 from LSC63 formed two related clusters that were phylogenetically unlinked to sequences from P63 (Fig. 2), indicating infection of LSC63 by a source other than P63. LSC63, receiving no HAART during the 2.5-year study, maintained a plasma viral load of ∼103 copies/ml and a CD4+ T-cell count of more than 500 cells/μl (Fig. 1A).
FIG. 2.
Unrooted neighbor-joining tree of env C2-V5 nucleotide sequences from PP63, P63, and LSC63. To assess possible infecting-partner-to-recipient relationships, env C2-V5 nucleotide sequences from each subject were aligned with randomly chosen HIV-1-B sequences from the Los Alamos HIV Sequence Database.
During his relationship with LSC63, P63 received HAART, which effectively suppressed plasma viremia to a level below 50 copies/ml (Fig. 1B) and resulted in a low level of genetic diversification [0.047% per nucleotide per year in diversity increase in the C2-V5 region of env compared to an average of 1% per nucleotide per year in untreated subtype B-infected subjects (62)] (Fig. 2). The viral load in P63 increased to more than 103 copies/ml after the termination of HAART. LSC63 was found to be heterozygous for CCR5-Δ32 and CCR5 promoter −2459A/G mutations (Table 2). This host gene combination has been shown to offer an advantage in resisting sexual HIV-1 transmission (8, 18, 19, 31, 43, 47, 59, 63). Although symptoms of HSV primary infection were not observed during the course of the study, we also examined HSV-1 and HSV-2 infection in LSC63, since HSV-2 is associated with a threefold-increased risk for HIV acquisition (6, 7). LSC63 was both HSV-1 and HSV-2 positive by Western blot assay in all four samples examined between 1.6 years before and 3 days after seroconversion.
TABLE 2.
Genetic polymorphisms of LSC63 at sites associated with increased resistance against HIV-1 infection
| Polymorphism | Genotype in LSC63 |
|---|---|
| CCR5-Δ32 | Heterozygous |
| CCR5-P-2459A/G | Heterozygous |
| CCR2-64I | Wild type |
| SDF-1-3′A | Wild type |
| RANTES-403 | Heterozygous |
| RANTES-28 | Wild type |
| DC-SIGNR | 7/5a |
| DC-SIGN | 7/7a |
Number of repeats of the 23-amino-acid sequence in the neck region of DC-SIGNR or DC-SIGN.
CTL responses in LSC63 before and after seroconversion.
In a previous study, broad CTL responses were detected using peptides derived from consensus or reference strain sequences (as described in Materials and Methods) in LSC63 before seroconversion (17). At day −172, 13 peptide pools, including 9/10 pools of Pol peptides, 2/9 pools of Env peptides, and 2/2 pools of Nef peptides, were recognized by CD8+ T cells, with a median magnitude of 198 SFC/106 PBMC (range, 49 to 380) (17). However, no viral DNA was detected by PCR at days −286, −161, and −90, even when 100 million or more PBMC were tested at each time point using an ultrasensitive PCR assay designed for the detection of low levels of HIV-1 infection (66). Three days after seroconversion, CTL recognition of 13 peptides, including five optimally defined epitopes (Table 3), was detected, with a median magnitude of 164 SFC/106 PBMC (range, 57 to 1543). Of these peptides, two were located in Gag, three in Pol, two in Vpr (pools of Vpr peptides were not examined before seroconversion), and six in Env.
TABLE 3.
Peptides recognized after seroconversion and corresponding viral sequences in P63 and LSC63
| Peptide and status of poola | Patient | Sequenceb | Protein (position in HXB2) | SFC/106 PBMC | HLA restrictionc |
|---|---|---|---|---|---|
| Pool was recognized | |||||
| PW10 | PIQKETWETW | Pol RT (392-401) | 105 | A32 | |
| P63 | --------A- | ||||
| LSC63 | --------A- | ||||
| KI15 | KRKGGIGGYSAGERI | Pol INT (186-196) | 164 | NA | |
| P63 | -------R------- | ||||
| LSC63 | --------------- | ||||
| GI15 | GIGGYSAGERIVDII | Pol INT (190-204) | 102 | NA | |
| P63 | ---R----------- | ||||
| LSC63 | ----------I---- | ||||
| CF15 | CNTSVITQACPKISF | Env gp120 (196-210) | 215 | NA | |
| P63 | ------------V-- | ||||
| LSC63 | ------------V-- | ||||
| YL10 | YCAPAGFAIL* | Env gp120 (218-226) | 274 | Cw1 | |
| P63 | ---------- | ||||
| LSC63 | ---------- | ||||
| KQ15 | KFSGKGSCKNVSTVQ | Env gp120 (232-246) | 276 | NA | |
| P63 | T-N-T-P-T------ | ||||
| LSC63 | --N-T-A-T------ | ||||
| Pool was not recognized | |||||
| KK10 | KRWIILGLNK | Gag p24 (131-140) | 1,264 | B27 | |
| P63 | ---------- | ||||
| LSC63 | ---------- | ||||
| GC15 | GNFRNQRKIVKCFNC | Gag p7 (4-18) | 57 | NA | |
| P63 | --------T------ | ||||
| LSC63 | --------NI----- | ||||
| HG11 | HFPRIWLHGLG* | Vpr (33-43) | 72 | NA | |
| P63 | ----P------ | ||||
| LSC63 | ----P------ | ||||
| RV11 | RIGCRHSRIGV | Vpr (73-83) | 75 | NA | |
| P63 | -L--------I | ||||
| LSC63 | ----H-----I | ||||
| RW9 | RIKQIINMW | Env gp120 (419-427) | 73 | A32 | |
| P63 | -----V--- | ||||
| LSC63 | --------- | ||||
| IP11 | IVNRVRQGYSP* | Env gp41 (193-203) | 1,152 | NA | |
| P63 | ----------- | ||||
| LSC63 | ----------- | ||||
| GY10 | GRRGWEVLKY | Env gp41 (275-284) | 1,543 | B27 | |
| P63 | ---------- | ||||
| LSC63 | ------I--I |
Peptides were located or were not located in pools that were recognized before seroconversion of LSC63.
Viral sequences refer to the consensus sequences of P63 obtained at day −148 from the seroconversion of LSC63 and the consensus sequences of LSC63 obtained at day 24 from his seroconversion. −, amino acids in the viral sequences identical to the amino acid in the corresponding test peptides; those different from the test peptides are singled out. Peptides labeled with an asterisk have corresponding viral sequences in P63 and LSC63 that differ within the 5-amino-acid flanking regions.
Peptides with HLA restriction listed are optimally defined epitopes. NA, not applicable.
Six of the 13 peptides recognized after seroconversion were located in pools recognized prior to seroconversion; however, the corresponding CTL responses were of low magnitude (105 to 276 SFC/106 PBMC) (Table 3). The three peptides (Env IP11 and the HLA-B27-restricted Gag KK10 and Env GY10) associated with the highest-magnitude responses (1,152 to 1,264 SFC/106 PBMC) at day 3 were not located in the pools recognized before seroconversion. To determine whether sequence discrepancy between the infecting viruses in LSC63 and the viruses to which he was exposed through P63 (termed the exposed virus) contributed to the discordant CTL recognition, we compared their sequences within the peptides recognized after seroconversion and extending 5 amino acids into the flanking regions.
Of the six peptides in peptide pools recognized before seroconversion, viral sequences from P63 and LSC63 were identical to each other within the peptides and the flanking regions of PW10 and CF15 but different from the tested peptide sequence (Tables 3 and 4). Peptide YL10 was found in both P63 and LSC63, with flanking sequences differing. Peptide KI15 was found in LSC63 but not in P63. Peptides GI15 and KQ15 were not found in LSC63 or P63, and the corresponding sequences differed between the two subjects. Due to a lack of available PBMC for CTL recognition testing, we do not know whether these six peptides were recognized in LSC63 before seroconversion.
TABLE 4.
Relationships of viral sequences in P63 and LSC63 encoding peptides recognized after seroconversion of LSC63
| Sequence relationship within the tested peptide (and within in the 5 amino acids flanking the tested peptide)a | No. of peptides (no. of HLA-B27-restricted peptides) that were:
|
|
|---|---|---|
| Located in pools recognized preseroconversion | Not located in pools recognized preseroconversion | |
| P63 = LSC63 = peptide (P63 = LSC63) | 0 | 1 (1) |
| P63 = LSC63 = peptide (P63 ≠ LSC63) | 1 (0) | 2 (0) |
| P63 = LSC63 ≠ peptide (P63 = LSC63) | 2 (0) | 0 |
| P63 = peptide ≠ LSC63 | 0 | 1 (1) |
| LSC63 = peptide ≠ P63 | 1 (0) | 1 (0) |
| P63≠LSC63 ≠ peptide | 2 (0) | 2 |
| Total | 6 (0) | 7 (2) |
Peptide, P63, and LSC63 represent the synthesized peptides (see Materials and Methods) and the corresponding viral sequences from patients.
Of the seven peptides that were not recognized before seroconversion, the sequences of peptides KK10, GH11, and IP11 were found in both P63 and LSC63, with flanking sequences differing for GH11 and IP11 (Tables 3 and 4). We predicted epitope processing of GH11 and IP11 in silico by using the NetChoP 3.0 server (48) and found that the differences in the flanking regions should not affect the processing of the two epitopes (data not shown). Viral sequences corresponding to the other four peptides were different in LSC63 and P63. The RW9 peptide was present only in LSC63, while the GY10 peptide was found only in P63. Sequences corresponding to peptides GC15 and RV11 were not found in either subject. Hence, the lack of preseroconversion recognition for six of the seven peptides recognized after seroconversion was associated with differences between viral epitopes or their immediate flanking regions in the exposed versus infecting viruses of LSC63.
CTL responses to HLA-B27-restricted epitopes.
The class I HLA alleles expressed by LSC63 were A26, A32, B27, Cw01, and Cw02. HLA-B27 has been associated with control of HIV infection (23, 50). Two B27-restricted epitopes were recognized in LSC63 after seroconversion (Table 3 and 4). KK10 in Gag, the most commonly recognized HLA-B27 epitope (2), was recognized with the second-highest magnitude (1,264 SFC/106 PBMC) in LSC63 after seroconversion. However, this epitope was not recognized before seroconversion despite the fact that the sequences of the exposed and the infecting viruses were identical at the epitope and the flanking regions. GY10 (GRRGWEVLKY) of Env-gp41 was the epitope recognized with the highest magnitude (1,543 SFC/106 PBMC) after seroconversion, despite the fact that the viral sequences corresponding to GY10 in LSC63 were different at two amino acids (GRRGWEVIKI). The viral sequences in P63 were identical to the testing peptide, and it is possible that low-level CTL responses to GRRGWEVLKY were elicited by the exposure to P63 viruses and the infecting viruses of LSC63 were escape mutants; however, recognition of the pool containing GY10 was not detected in LSC63 before seroconversion. It is also possible that the infecting viruses of LSC63 initially had the sequence GRRGWEVLKY, which elicited strong responses to GY10, and were replaced by the escape mutant GRRGWEVIKI. The third possibility is that the strong responses to GY10 after seroconversion of LSC63 were from cross-reactivity of CTL responses elicited by GRRGWEVIKI.
Genetic distances between viruses from LSC63 and P63.
To further examine whether there was protection from the P63 viruses due to prior exposure, we analyzed the genetic differences between the exposed and the infecting viruses. If genetic distances, especially nonsynonymous distances, between viruses of P63 and LSC63 were significantly greater than those between viruses of P63 and random subjects, exposure to P63 viruses might have provided LSC63 some degree of protection against infection by relatively closely related viruses.
Using 142 random subtype B whole-genome sequences as controls, we found that the nucleotide pairwise distances between P63 and LSC63 were significantly greater than the distances between P63 and random subtype B sequences in gp41 and nef (P = 0.035 and P = 0.042, respectively) (Table 5). These differences were mostly due to greater nonsynonymous distances between the two subjects (P = 0.021 and P = 0.049 in gp41 and nef, respectively). Nonsynonymous distances between viruses in the two subjects were also significantly greater in the polB region, which encodes the 3′ half of the Pol protein (P = 0.014). Before seroconversion of LSC63, no CTL responses to pools containing peptides encoded by gp41 were detected, whereas responses to five pools containing peptides encoded by polB (49 to 241 SFC/106 PBMC) and two pools containing peptides encoded by nef were detected (86 and 198 SFC/106 PBMC). After seroconversion, CTL recognition of Nef was not detected, while Pol peptides (PW10, KI15, and GI15) (Table 4) that were recognized were encoded by polB and located in pools recognized before seroconversion. Synonymous distances between the two subjects were not significantly different from those between P63 and random subtype B sequences at most regions, except for vif (P = 0.042). Interestingly, five pools containing peptides encoded by polA (encoding the 5′ half of the Pol protein) were recognized before seroconversion, and yet the pairwise total or nonsynonymous distances in polA between the two subjects were not significantly different from those between P63 and random subtype B sequences. It should be noted that the differences observed above became nonsignificant after application of the conservative Bonferroni correction for multiple testing.
TABLE 5.
Genetic distances between HIV-1 viruses from LSC63 and P63a
| Gene or region | Pairwise nucleotide distance | P value | Synonymous distance | P value | Nonsynonymous distance | P value |
|---|---|---|---|---|---|---|
| gag | 0.074 | 0.380 | 0.199 | 0.422 | 0.043 | 0.324 |
| polA | 0.061 | 0.338 | 0.163 | 0.570 | 0.035 | 0.127 |
| polB | 0.072 | 0.092 | 0.166 | 0.380 | 0.047 | 0.014 |
| Central region | 0.110 | 0.246 | ||||
| vif | 0.196 | 0.042 | 0.065 | 0.662 | ||
| vpr | 0.168 | 0.788 | 0.067 | 0.451 | ||
| vpu | 0.225 | 0.317 | 0.121 | 0.261 | ||
| gp120 | 0.152 | 0.120 | 0.207 | 0.282 | 0.136 | 0.099 |
| gp41 | 0.126 | 0.035 | 0.199 | 0.239 | 0.105 | 0.021 |
| nef | 0.150 | 0.042 | 0.249 | 0.465 | 0.124 | 0.049 |
Genetic distances (per nucleotide) between the exposed viruses and the infecting viruses of LSC63 were calculated. The control group was comprised of 142 random subtype B whole-genome sequences. The P value is the percentage of random sequences whose distances to viruses of P63 were greater than the distance between the viruses of LSC63 and P63. P values of <0.05 are in bold font.
DISCUSSION
In this study, we examined the genetics of breakthrough HIV-1 in an ES subject (LSC63) who had been seronegative for more than 2 years despite repeated unprotected sexual contacts with an HIV-1-positive partner (P63), as well as the HIV-1-specific CTL responses before and after LSC63's seroconversion. Genetic analyses of HIV-1 sequences of LSC63, P63, and P63's infecting source (PP63) indicate that subject LSC63 was not infected by P63 viruses. Low viral load (<50 copies/ml) in P63 during the sexual relationship with LSC63 may have prevented the transmission of P63's HIV-1 to LSC63. However, broad CTL responses were detected in LSC63 before seroconversion. Compared to those detected after seroconversion, these CTL responses were of low magnitude and half of them targeted different regions of the viral proteome. Furthermore, for the majority of the coding regions of HIV-1 (with the possible exceptions of polB, gp41, and nef), the genetic distances between the infecting and the exposed viruses of LSC63 were comparable to the distances between random subtype B HIV-1 sequences and the exposed viruses. These results suggest that repeated sexual exposure to low levels of HIV-1 from a seropositive partner was able to elicit HIV-1-specific CTL responses in LSC63. However, these responses were not able to prevent HIV infection in the subject, even though the infecting viral strain was not significantly dissimilar from random HIV-1 strains.
Viral loads in HIV-1-positive partners are a strong predictor of heterosexual transmission (51, 53, 54). It has also been reported that the HIV-1-positive partners in monogamous homosexual relationships between whom HIV-1 transmission had not occurred had significantly lower cellular infectious viral loads than HIV-1-transmitting donors in monogamous homosexual relationships (5). In addition, heterozygous CCR5-Δ32 and CCR5 promoter mutations associated with resistance to HIV-1 infection (18) were identified in LSC63 and may have also contributed to the delay in HIV-1 infection in LSC63. LSC63 was HSV-2 positive 1.6 years before his HIV-1 seroconversion. Thus, HSV-2 infection might have played a minor role in his acquisition of HIV-1.
Consistent with findings for the late seroconverters from the Nairobi commercial sex worker cohort (24), we found that CTL responses in LSC63 before seroconversion were of low magnitudes and that CTL targets before and after seroconversion were different. Of the 13 peptides recognized after seroconversion, 6 were located in pools recognized before seroconversion. In only 3/13 peptides were the sequences of the exposed and the infecting viruses identical over both the targeted peptides and the immediate 5-amino-acid flanking regions, and two of them were located in pools recognized before seroconversion. It has been reported that mutations flanking CTL epitopes at least as far as the fifth position downstream can prevent appropriate epitope processing and presentation (67) and lead to escape (10, 65). Although our in silico analysis did not predict alteration of epitope processing by sequence differences at flanking regions of the epitopes between the exposed and the infecting viruses, in vitro digestion analyses of peptides containing those epitopes and flanking regions will be required (34) to further determine the impact of the sequence variation observed in the flanking regions. Therefore, discordant CTL responses before and after seroconversion might reflect the sequence differences between the exposed and the infecting viruses.
HIV-1-specific CTL activities have been associated with resistance to HIV-1 infection (1, 11, 15, 58). However, in the study of HIV-1-resistant female commercial sex workers in Nairobi (27, 28), most of the peptides recognized before seroconversion were present in the later-infecting viruses, indicating a minor role of viral escape in late seroconversion in these individuals. We were not able to identify the individual CTL peptides recognized before seroconversion in LSC63 or examine the functionality of CTL responses before seroconversion due to a lack of specimens. However, our genetic analyses suggested that, when compared to random HIV-1 subtype B sequences, the viruses establishing infection in LSC63 had distances similar to those of the exposed viruses over most of the coding regions of HIV-1. Only for Nef and the 3′ half of Pol were CTL responses detected before seroconversion and significantly greater genetic distances observed between the exposed and the infecting viruses and random strain sequences. We note that the choice of random control sequences might affect these interpretations (D. C. Nickle, Y. Liu, K. Davis, C. Celum, G. H. Learn, and J. I. Mullins, unpublished data). The ideal control sequences for this study would be random sequences from subjects that had been infected for a period of time comparable to P63 and had effective HAART shortly after HIV-1 infection. Nevertheless, our results indicate that although the viruses infecting LSC63 might contain sequences that could have escaped some specific CTL responses elicited before seroconversion, the majority of the protein-coding regions of HIV-1 variants in LSC63 (except gp41, nef, and the 3′ half of pol) were not unusually distant from the viruses to which LSC63 had been previously regularly exposed.
Without antiviral treatment, LSC63 maintained a viral load of ∼103 copies/ml and a CD4+ T-cell count of over 500 cells/μl during the 2.5 years of study after seroconversion. LSC63 had the protective B27 allele (23, 50); developed strong CTL responses after seroconversion, especially B27-restricted responses; and was CCR5-Δ32 and CCR5 promoter heterozygous. All of these factors might have contributed to the initial relatively benign clinical outcome. Before seroconversion, however, in spite of broad CTL responses, no B27-restricted CTL responses were detected.
In summary, we found that repeated exposure to low-level HIV-1 was able to elicit multiple, although low-magnitude, HIV-1-specific CTL responses in LSC63. However, only marginal evidence was obtained for these responses being able to restrict infection with related viruses. Further study of ES individuals who seroconvert and those who fail to seroconvert may assist in the identification of protective CTL responses.
Acknowledgments
This work was supported by National Institutes of Health grants AI45402, AI55336, AI49109, and AI56994 to T.Z.; AI35605 and AI047086 to M.J.M.; AI57005 to J.I.M.; and AI27757 to the Computational Biology Core of the University of Washington Center for AIDS Research.
We thank Blythe Adams and Leah Hampson for technical assistance; Julie Czartoski, Claire Stevens, Janine Maenza, Kurt Diem, and Ann Collier for recruitment and coordination of study subjects and sampling; and the study participants for their time and effort.
Footnotes
Published ahead of print on 12 August 2009.
REFERENCES
- 1.Akridge, R., F. Hladik, J. Markee, C. Alef, H. Kelley, A. Collier, A. Collier, and M. J. McElrath. 1999. Cellular immunity and target cell susceptibility in persons with repeated HIV-1 exposure. Immunol. Lett. 66:15-19. [DOI] [PubMed] [Google Scholar]
- 2.Altfeld, M., E. T. Kalife, Y. Qi, H. Streeck, M. Lichterfeld, M. N. Johnston, N. Burgett, M. E. Swartz, A. Yang, G. Alter, X. G. Yu, A. Meier, J. K. Rockstroh, T. M. Allen, H. Jessen, E. S. Rosenberg, M. Carrington, and B. D. Walker. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8(+) T cell response against HIV-1. PLoS Med. 3:e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashley, R., A. Cent, V. Maggs, A. Nahmias, and L. Corey. 1991. Inability of enzyme immunoassays to discriminate between infections with herpes simplex virus types 1 and 2. Ann. Intern. Med. 115:520-526. [DOI] [PubMed] [Google Scholar]
- 4.Ashley, R. L., J. Militoni, F. Lee, A. Nahmias, and L. Corey. 1988. Comparison of Western blot (immunoblot) and glycoprotein G-specific immunodot enzyme assay for detecting antibodies to herpes simplex virus types 1 and 2 in human sera. J. Clin. Microbiol. 26:662-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaak, H., A. B. van't Wout, M. Brouwer, M. Cornelissen, N. A. Kootstra, N. Albrecht-van Lent, R. P. Keet, J. Goudsmit, R. A. Coutinho, and H. Schuitemaker. 1998. Infectious cellular load in human immunodeficiency virus type 1 (HIV-1)-infected individuals and susceptibility of peripheral blood mononuclear cells from their exposed partners to non-syncytium-inducing HIV-1 as major determinants for HIV-1 transmission in homosexual couples. J. Virol. 72:218-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corey, L., and H. H. Handsfield. 2000. Genital herpes and public health: addressing a global problem. JAMA 283:791-794. [DOI] [PubMed] [Google Scholar]
- 7.Corey, L., A. Wald, C. L. Celum, and T. C. Quinn. 2004. The effects of herpes simplex virus-2 on HIV-1 acquisition and transmission: a review of two overlapping epidemics. J. Acquir. Immune Defic. Syndr. 35:435-445. [DOI] [PubMed] [Google Scholar]
- 8.Dean, M., M. Carrington, C. Winkler, G. A. Huttley, M. W. Smith, R. Allikmets, J. J. Goedert, S. P. Buchbinder, E. Vittinghoff, E. Gomperts, S. Donfield, D. Vlahov, R. Kaslow, A. Saah, C. Rinaldo, R. Detels, S. J. O'Brien, et al. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science 273:1856-1862. [DOI] [PubMed] [Google Scholar]
- 9.De Maria, A., C. Cirillo, and L. Moretta. 1994. Occurrence of human immunodeficiency virus type 1 (HIV-1)-specific cytolytic T cell activity in apparently uninfected children born to HIV-1-infected mothers. J. Infect. Dis. 170:1296-1299. [DOI] [PubMed] [Google Scholar]
- 10.Draenert, R., S. Le Gall, K. J. Pfafferott, A. J. Leslie, P. Chetty, C. Brander, E. C. Holmes, S. C. Chang, M. E. Feeney, M. M. Addo, L. Ruiz, D. Ramduth, P. Jeena, M. Altfeld, S. Thomas, Y. Tang, C. L. Verrill, C. Dixon, J. G. Prado, P. Kiepiela, J. Martinez-Picado, B. D. Walker, and P. J. Goulder. 2004. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J. Exp. Med. 199:905-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fowke, K. R., R. Kaul, K. L. Rosenthal, J. Oyugi, J. Kimani, W. J. Rutherford, N. J. Nagelkerke, T. B. Ball, J. J. Bwayo, J. N. Simonsen, G. M. Shearer, and F. A. Plummer. 2000. HIV-1-specific cellular immune responses among HIV-1-resistant sex workers. Immunol. Cell Biol. 78:586-595. [DOI] [PubMed] [Google Scholar]
- 12.Fowke, K. R., N. J. Nagelkerke, J. Kimani, J. N. Simonsen, A. O. Anzala, J. J. Bwayo, K. S. MacDonald, E. N. Ngugi, and F. A. Plummer. 1996. Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet 348:1347-1351. [DOI] [PubMed] [Google Scholar]
- 13.Fulcher, J. A., Y. Hwangbo, R. Zioni, D. Nickle, X. Lin, L. Heath, J. I. Mullins, L. Corey, and T. Zhu. 2004. Compartmentalization of human immunodeficiency virus type 1 between blood monocytes and CD4+ T cells during infection. J. Virol. 78:7883-7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibbons, J., J. M. Cory, I. K. Hewlett, J. S. Epstein, and M. E. Eyster. 1990. Silent infections with human immunodeficiency virus type 1 are highly unlikely in multitransfused seronegative hemophiliacs. Blood 76:1624-1626. [PubMed] [Google Scholar]
- 15.Goh, W. C., J. Markee, R. E. Akridge, M. Meldorf, L. Musey, T. Karchmer, M. Krone, A. Collier, L. Corey, M. Emerman, and M. J. McElrath. 1999. Protection against human immunodeficiency virus type 1 infection in persons with repeated exposure: evidence for T cell immunity in the absence of inherited CCR5 coreceptor defects. J. Infect. Dis. 179:548-557. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez, E., R. Dhanda, M. Bamshad, S. Mummidi, R. Geevarghese, G. Catano, S. A. Anderson, E. A. Walter, K. T. Stephan, M. F. Hammer, A. Mangano, L. Sen, R. A. Clark, S. S. Ahuja, M. J. Dolan, and S. K. Ahuja. 2001. Global survey of genetic variation in CCR5, RANTES, and MIP-1alpha: impact on the epidemiology of the HIV-1 pandemic. Proc. Natl. Acad. Sci. USA 98:5199-5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hladik, F., A. Desbien, J. Lang, L. Wang, Y. Ding, S. Holte, A. Wilson, Y. Xu, M. Moerbe, S. Schmechel, and M. J. McElrath. 2003. Most highly exposed seronegative men lack HIV-1-specific, IFN-gamma-secreting T cells. J. Immunol. 171:2671-2683. [DOI] [PubMed] [Google Scholar]
- 18.Hladik, F., H. Liu, E. Speelmon, D. Livingston-Rosanoff, S. Wilson, P. Sakchalathorn, Y. Hwangbo, B. Greene, T. Zhu, and M. J. McElrath. 2005. Combined effect of CCR5-Δ32 heterozygosity and the CCR5 promoter polymorphism −2459 A/G on CCR5 expression and resistance to human immunodeficiency virus type 1 transmission. J. Virol. 79:11677-11684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffman, T. L., R. R. MacGregor, H. Burger, R. Mick, R. W. Doms, and R. G. Collman. 1997. CCR5 genotypes in sexually active couples discordant for human immunodeficiency virus type 1 infection status. J. Infect. Dis. 176:1093-1096. [DOI] [PubMed] [Google Scholar]
- 20.Huang, Y., W. A. Paxton, S. M. Wolinsky, A. U. Neumann, L. Zhang, T. He, S. Kang, D. Ceradini, Z. Jin, K. Yazdanbakhsh, K. Kunstman, D. Erickson, E. Dragon, N. R. Landau, J. Phair, D. D. Ho, and R. A. Koup. 1996. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med. 2:1240-1243. [DOI] [PubMed] [Google Scholar]
- 21.Imagawa, D. T., M. H. Lee, S. M. Wolinsky, K. Sano, F. Morales, S. Kwok, J. J. Sninsky, P. G. Nishanian, J. Giorgi, J. L. Fahey, et al. 1989. Human immunodeficiency virus type 1 infection in homosexual men who remain seronegative for prolonged periods. N. Engl. J. Med. 320:1458-1462. [DOI] [PubMed] [Google Scholar]
- 22.Jennes, W., S. Verheyden, C. Demanet, C. A. Adje-Toure, B. Vuylsteke, J. N. Nkengasong, and L. Kestens. 2006. Cutting edge: resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. J. Immunol. 177:6588-6592. [DOI] [PubMed] [Google Scholar]
- 23.Kaslow, R. A., M. Carrington, R. Apple, L. Park, A. Munoz, A. J. Saah, J. J. Goedert, C. Winkler, S. J. O'Brien, C. Rinaldo, R. Detels, W. Blattner, J. Phair, H. Erlich, and D. L. Mann. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405-411. [DOI] [PubMed] [Google Scholar]
- 24.Kaul, R., T. Dong, F. A. Plummer, J. Kimani, T. Rostron, P. Kiama, E. Njagi, E. Irungu, B. Farah, J. Oyugi, R. Chakraborty, K. S. MacDonald, J. B. Bwayo, A. McMichael, and S. Rowland-Jones. 2001. CD8+ lymphocytes respond to different HIV epitopes in seronegative and infected subjects. J. Clin. Investig. 107:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaul, R., F. Plummer, M. Clerici, M. Bomsel, L. Lopalco, and K. Broliden. 2001. Mucosal IgA in exposed, uninfected subjects: evidence for a role in protection against HIV infection. AIDS 15:431-432. [DOI] [PubMed] [Google Scholar]
- 26.Kaul, R., F. A. Plummer, J. Kimani, T. Dong, P. Kiama, T. Rostron, E. Njagi, K. S. MacDonald, J. J. Bwayo, A. J. McMichael, and S. L. Rowland-Jones. 2000. HIV-1-specific mucosal CD8+ lymphocyte responses in the cervix of HIV-1-resistant prostitutes in Nairobi. J. Immunol. 164:1602-1611. [DOI] [PubMed] [Google Scholar]
- 27.Kaul, R., S. L. Rowland-Jones, J. Kimani, T. Dong, H. B. Yang, P. Kiama, T. Rostron, E. Njagi, J. J. Bwayo, K. S. MacDonald, A. J. McMichael, and F. A. Plummer. 2001. Late seroconversion in HIV-resistant Nairobi prostitutes despite pre-existing HIV-specific CD8+ responses. J. Clin. Investig. 107:341-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaul, R., S. L. Rowland-Jones, J. Kimani, K. Fowke, T. Dong, P. Kiama, J. Rutherford, E. Njagi, F. Mwangi, T. Rostron, J. Onyango, J. Oyugi, K. S. MacDonald, J. J. Bwayo, and F. A. Plummer. 2001. New insights into HIV-1 specific cytotoxic T-lymphocyte responses in exposed, persistently seronegative Kenyan sex workers. Immunol. Lett. 79:3-13. [DOI] [PubMed] [Google Scholar]
- 29.Kaul, R., D. Trabattoni, J. J. Bwayo, D. Arienti, A. Zagliani, F. M. Mwangi, C. Kariuki, E. N. Ngugi, K. S. MacDonald, T. B. Ball, M. Clerici, and F. A. Plummer. 1999. HIV-1-specific mucosal IgA in a cohort of HIV-1-resistant Kenyan sex workers. AIDS 13:23-29. [DOI] [PubMed] [Google Scholar]
- 30.Korber, B. 2000. HIV signature and sequence variation analysis, p. 55-72. In A. G. Rodrigo and G. H. Learn (ed.), Computational analysis of HIV molecular sequences. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 31.Kostrikis, L. G., A. U. Neumann, B. Thomson, B. T. Korber, P. McHardy, R. Karanicolas, L. Deutsch, Y. Huang, J. F. Lew, K. McIntosh, H. Pollack, W. Borkowsky, H. M. Spiegel, P. Palumbo, J. Oleske, A. Bardeguez, K. Luzuriaga, J. Sullivan, S. M. Wolinsky, R. A. Koup, D. D. Ho, and J. P. Moore. 1999. A polymorphism in the regulatory region of the CC-chemokine receptor 5 gene influences perinatal transmission of human immunodeficiency virus type 1 to African-American infants. J. Virol. 73:10264-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kulkarni, P. S., S. T. Butera, and A. C. Duerr. 2003. Resistance to HIV-1 infection: lessons learned from studies of highly exposed persistently seronegative (HEPS) individuals. AIDS Rev. 5:87-103. [PubMed] [Google Scholar]
- 33.Langlade-Demoyen, P., N. Ngo-Giang-Huong, F. Ferchal, and E. Oksenhendler. 1994. Human immunodeficiency virus (HIV) nef-specific cytotoxic T lymphocytes in noninfected heterosexual contact of HIV-infected patients. J. Clin. Investig. 93:1293-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Gall, S., P. Stamegna, and B. D. Walker. 2007. Portable flanking sequences modulate CTL epitope processing. J. Clin. Investig. 117:3563-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehner, T. 2003. Innate and adaptive mucosal immunity in protection against HIV infection. Vaccine 21(Suppl. 2):S68-S76. [DOI] [PubMed] [Google Scholar]
- 36.Liu, H., M. Carrington, C. Wang, S. Holte, J. Lee, B. Greene, F. Hladik, D. M. Koelle, A. Wald, K. Kurosawa, C. R. Rinaldo, C. Celum, R. Detels, L. Corey, M. J. McElrath, and T. Zhu. 2006. Repeat-region polymorphisms in the gene for the dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin-related molecule: effects on HIV-1 susceptibility. J. Infect. Dis. 193:698-702. [DOI] [PubMed] [Google Scholar]
- 37.Liu, H., D. Chao, E. E. Nakayama, H. Taguchi, M. Goto, X. Xin, J. K. Takamatsu, H. Saito, Y. Ishikawa, T. Akaza, T. Juji, Y. Takebe, T. Ohishi, K. Fukutake, Y. Maruyama, S. Yashiki, S. Sonoda, T. Nakamura, Y. Nagai, A. Iwamoto, and T. Shioda. 1999. Polymorphism in RANTES chemokine promoter affects HIV-1 disease progression. Proc. Natl. Acad. Sci. USA 96:4581-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu, H., Y. Hwangbo, S. Holte, J. Lee, C. Wang, N. Kaupp, H. Zhu, C. Celum, L. Corey, M. J. McElrath, and T. Zhu. 2004. Analysis of genetic polymorphisms in CCR5, CCR2, stromal cell-derived factor-1, RANTES, and dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin in seronegative individuals repeatedly exposed to HIV-1. J. Infect. Dis. 190:1055-1058. [DOI] [PubMed] [Google Scholar]
- 39.Liu, R., W. A. Paxton, S. Choe, D. Ceradini, S. R. Martin, R. Horuk, M. E. MacDonald, H. Stuhlmann, R. A. Koup, and N. R. Landau. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367-377. [DOI] [PubMed] [Google Scholar]
- 40.Llewellyn, N., R. Zioni, H. Zhu, T. Andrus, Y. Xu, L. Corey, and T. Zhu. 2006. Continued evolution of HIV-1 circulating in blood monocytes with antiretroviral therapy: genetic analysis of HIV-1 in monocytes and CD4+ T cells of patients with discontinued therapy. J. Leukoc. Biol. 80:1118-1126. [DOI] [PubMed] [Google Scholar]
- 41.MacDonald, K. S., K. R. Fowke, J. Kimani, V. A. Dunand, N. J. Nagelkerke, T. B. Ball, J. Oyugi, E. Njagi, L. K. Gaur, R. C. Brunham, J. Wade, M. A. Luscher, P. Krausa, S. Rowland-Jones, E. Ngugi, J. J. Bwayo, and F. A. Plummer. 2000. Influence of HLA supertypes on susceptibility and resistance to human immunodeficiency virus type 1 infection. J. Infect. Dis. 181:1581-1589. [DOI] [PubMed] [Google Scholar]
- 42.Maddison, W. P., and D. R. Maddison. 2001. MacClade: analysis of phylogeny and character evolution, version 4. Sinauer Associates, Inc., Sunderland, MA.
- 43.Marmor, M., H. W. Sheppard, D. Donnell, S. Bozeman, C. Celum, S. Buchbinder, B. Koblin, and G. R. Seage III. 2001. Homozygous and heterozygous CCR5-delta32 genotypes are associated with resistance to HIV infection. J. Acquir. Immune Defic. Syndr. 27:472-481. [DOI] [PubMed] [Google Scholar]
- 44.Martin, M. P., M. Dean, M. W. Smith, C. Winkler, B. Gerrard, N. L. Michael, B. Lee, R. W. Doms, J. Margolick, S. Buchbinder, J. J. Goedert, T. R. O'Brien, M. W. Hilgartner, D. Vlahov, S. J. O'Brien, and M. Carrington. 1998. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science 282:1907-1911. [DOI] [PubMed] [Google Scholar]
- 45.Mazzoli, S., D. Trabattoni, S. Lo Caputo, S. Piconi, C. Ble, F. Meacci, S. Ruzzante, A. Salvi, F. Semplici, R. Longhi, M. Fusi, N. Tofani, M. Biasin, M. Villa, F. Mazzotta, and M. Clerici. 1997. HIV-specific mucosal and cellular immunity in HIV-seronegative partners of HIV-seropositive individuals. Nat. Med. 3:1250-1257. [DOI] [PubMed] [Google Scholar]
- 46.McDermott, D. H., M. J. Beecroft, C. A. Kleeberger, F. M. Al-Sharif, W. E. Ollier, P. A. Zimmerman, B. A. Boatin, S. F. Leitman, R. Detels, A. H. Hajeer, and P. M. Murphy. 2000. Chemokine RANTES promoter polymorphism affects risk of both HIV infection and disease progression in the Multicenter AIDS Cohort Study. AIDS 14:2671-2678. [DOI] [PubMed] [Google Scholar]
- 47.Michael, N. L., G. Chang, L. G. Louie, J. R. Mascola, D. Dondero, D. L. Birx, and H. W. Sheppard. 1997. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat. Med. 3:338-340. [DOI] [PubMed] [Google Scholar]
- 48.Nielsen, M., C. Lundegaard, O. Lund, and C. Kesmir. 2005. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics 57:33-41. [DOI] [PubMed] [Google Scholar]
- 49.O'Brien, S. J., and J. P. Moore. 2000. The effect of genetic variation in chemokines and their receptors on HIV transmission and progression to AIDS. Immunol. Rev. 177:99-111. [DOI] [PubMed] [Google Scholar]
- 50.O'Brien, S. J., G. W. Nelson, C. A. Winkler, and M. W. Smith. 2000. Polygenic and multifactorial disease gene association in man: lessons from AIDS. Annu. Rev. Genet. 34:563-591. [DOI] [PubMed] [Google Scholar]
- 51.Operskalski, E. A., D. O. Stram, M. P. Busch, W. Huang, M. Harris, S. L. Dietrich, E. R. Schiff, E. Donegan, J. W. Mosley, et al. 1997. Role of viral load in heterosexual transmission of human immunodeficiency virus type 1 by blood transfusion recipients. Am. J. Epidemiol. 146:655-661. [DOI] [PubMed] [Google Scholar]
- 52.Paxton, W. B., R. W. Coombs, M. J. McElrath, M. C. Keefer, J. Hughes, F. Sinangil, D. Chernoff, B. Williams, L. Demeter, and L. Corey. 1997. Longitudinal analysis of quantitative virologic measures in human immunodeficiency virus-infected subjects with ≥400 CD4 lymphocytes: implications for applying measurements to individual patients. J. Infect. Dis. 175:247-254. [DOI] [PubMed] [Google Scholar]
- 53.Pedraza, M. A., J. del Romero, F. Roldan, S. Garcia, M. C. Ayerbe, A. R. Noriega, and J. Alcami. 1999. Heterosexual transmission of HIV-1 is associated with high plasma viral load levels and a positive viral isolation in the infected partner. J. Acquir. Immune Defic. Syndr. 21:120-125. [PubMed] [Google Scholar]
- 54.Quinn, T. C., M. J. Wawer, N. Sewankambo, D. Serwadda, C. Li, F. Wabwire-Mangen, M. O. Meehan, T. Lutalo, R. H. Gray, et al. 2000. Viral load and heterosexual transmission of human immunodeficiency virus type 1. N. Engl. J. Med. 342:921-929. [DOI] [PubMed] [Google Scholar]
- 55.Rowland-Jones, S. L., T. Dong, L. Dorrell, G. Ogg, P. Hansasuta, P. Krausa, J. Kimani, S. Sabally, K. Ariyoshi, J. Oyugi, K. S. MacDonald, J. Bwayo, H. Whittle, F. A. Plummer, and A. J. McMichael. 1999. Broadly cross-reactive HIV-specific cytotoxic T-lymphocytes in highly-exposed persistently seronegative donors. Immunol. Lett. 66:9-14. [DOI] [PubMed] [Google Scholar]
- 56.Rowland-Jones, S. L., T. Dong, K. R. Fowke, J. Kimani, P. Krausa, H. Newell, T. Blanchard, K. Ariyoshi, J. Oyugi, E. Ngugi, J. Bwayo, K. S. MacDonald, A. J. McMichael, and F. A. Plummer. 1998. Cytotoxic T cell responses to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J. Clin. Investig. 102:1758-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rowland-Jones, S. L., D. F. Nixon, M. C. Aldhous, F. Gotch, K. Ariyoshi, N. Hallam, J. S. Kroll, K. Froebel, and A. McMichael. 1993. HIV-specific cytotoxic T-cell activity in an HIV-exposed but uninfected infant. Lancet 341:860-861. [DOI] [PubMed] [Google Scholar]
- 58.Rowland-Jones, S. L., J. Sutton, K. Ariyoshi, T. Dong, F. Gotch, S. McAdam, D. Whitby, S. Sabally, A. Gallimore, T. Corrah, M. Takiguchi, T. Schultz, A. McMichael, and H. Whittle. 1995. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat. Med. 1:59-64. [DOI] [PubMed] [Google Scholar]
- 59.Samson, M., F. Libert, B. J. Doranz, J. Rucker, C. Liesnard, C. M. Farber, S. Saragosti, C. Lapoumeroulie, J. Cognaux, C. Forceille, G. Muyldermans, C. Verhofstede, G. Burtonboy, M. Georges, T. Imai, S. Rana, Y. Yi, R. J. Smyth, R. G. Collman, R. W. Doms, G. Vassart, and M. Parmentier. 1996. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722-725. [DOI] [PubMed] [Google Scholar]
- 60.Schacker, T., A. C. Collier, J. Hughes, T. Shea, and L. Corey. 1996. Clinical and epidemiologic features of primary HIV infection. Ann. Intern. Med. 125:257-264. [DOI] [PubMed] [Google Scholar]
- 61.Schmechel, S. C., N. Russell, F. Hladik, J. Lang, A. Wilson, R. Ha, A. Desbien, and M. J. McElrath. 2001. Immune defence against HIV-1 infection in HIV-1-exposed seronegative persons. Immunol. Lett. 79:21-27. [DOI] [PubMed] [Google Scholar]
- 62.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith, M. W., M. Dean, M. Carrington, C. Winkler, G. A. Huttley, D. A. Lomb, J. J. Goedert, T. R. O'Brien, L. P. Jacobson, R. Kaslow, S. Buchbinder, E. Vittinghoff, D. Vlahov, K. Hoots, M. W. Hilgartner, S. J. O'Brien, et al. 1997. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Science 277:959-965. [DOI] [PubMed] [Google Scholar]
- 64.Swofford, D. L. 1999. PAUP* 4.0: Phylogenetic Analysis Using Parsimony (* and other methods), 4.0b2a ed. Sinauer Associates, Inc., Sunderland, MA.
- 65.Yokomaku, Y., H. Miura, H. Tomiyama, A. Kawana-Tachikawa, M. Takiguchi, A. Kojima, Y. Nagai, A. Iwamoto, Z. Matsuda, and K. Ariyoshi. 2004. Impaired processing and presentation of cytotoxic-T-lymphocyte (CTL) epitopes are major escape mechanisms from CTL immune pressure in human immunodeficiency virus type 1 infection. J. Virol. 78:1324-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu, T., L. Corey, Y. Hwangbo, J. M. Lee, G. H. Learn, J. I. Mullins, and M. J. McElrath. 2003. Persistence of extraordinarily low levels of genetically homogeneous human immunodeficiency virus type 1 in exposed seronegative individuals. J. Virol. 77:6108-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zimbwa, P., A. Milicic, J. Frater, T. J. Scriba, A. Willis, P. J. R. Goulder, T. Pillay, H. Gunthard, J. N. Weber, H.-T. Zhang, and R. E. Phillips. 2007. Precise identification of a human immunodeficiency virus type 1 antigen processing mutant. J. Virol. 81:2031-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]