Abstract
Cellular tropism of vaccinia virus (VACV) is regulated by host range genes, including K1L, C7L, and E3L. While E3L is known to support viral replication by antagonizing interferon (IFN) effectors, including PKR, the exact functions of K1L and C7L are unclear. Here, we show that K1L and C7L can also inhibit antiviral effectors induced by type I IFN. In human Huh7 and MCF-7 cells, a VACV mutant lacking both K1L and C7L (vK1L−C7L−) replicated as efficiently as wild-type (WT) VACV, even in the presence of IFN. However, pretreating the cells with type I IFN, while having very little effect on WT VACV, blocked the replication of vK1L−C7L− at the step of intermediate viral gene translation. Restoring either K1L or C7L to vK1L−C7L− fully restored the IFN resistance phenotype. The deletion of K1L and C7L from VACV did not affect the ability of the virus to inhibit IFN signaling or its ability to inhibit the phosphorylation of PKR and the α subunit of eukaryotic initiation factor 2, indicating that K1L and C7L function by antagonizing an IFN effector(s) but with a mechanism that is different from those of IFN antagonists previously identified for VACV. Mutations of K1L that inactivate the host range function also rendered K1L unable to antagonize IFN, suggesting that K1L supports VACV replication in mammalian cells by antagonizing the same antiviral factor(s) that is induced by IFN in Huh7 cells.
Vaccinia virus (VACV) is the prototypical member of the poxvirus family of large, complex, double-stranded DNA viruses (21). VACV has a very broad host range and is capable of infecting many vertebrate animal species. Its host range, however, can be significantly narrowed by deleting from its genome some of the so-called host range genes, the most important of which are E3L, K1L, and C7L (17). VACV mutants deleted of E3L (ΔE3L) or both K1L and C7L (ΔK1LΔC7L) replicate abortively and express only a subset of viral genes in most mammalian cell lines (3, 24). These mutants are highly attenuated in animal hosts but are capable of eliciting immune responses, making them attractive vaccine vectors for infectious diseases and cancers (27, 28). NYVAC, a VACV strain derived through deletion of 18 genes, including both K1L and C7L (27), has been used as the vector for an AIDS vaccine (2).
The functions of E3L and the host factors that restrict the replication of the ΔE3L mutant have been studied extensively. E3L encodes a 20-kDa and a 25-kDa protein that bind double-stranded RNA (dsRNA) and Z form DNA (6, 15). The E3 proteins antagonize the dsRNA-dependent protein kinase PKR (5), which exists as an inactive form in the cells and undergoes autophosphorylation and activation upon binding to dsRNA. The activated PKR phosphorylates the α subunit of eukaryotic initiation factor 2 (eIF2α), resulting in a block in protein translation at the initiation step. The infection of most mammalian cells by the ΔE3L mutant leads to the activation of PKR and a block in translating viral mRNAs (16). The replication of the ΔE3L mutant in nonpermissive HeLa cells can be rescued by silencing PKR expression (32), while its replication in permissive Huh7 (human hepatoma) cells can be blocked by upregulating PKR expression with interferon (IFN) treatment (1). In addition to affecting PKR, E3 has also been shown to inactivate IFN-stimulated gene 15 (ISG15) (14), another IFN effector that plays a role in host defense against VACV.
Like the replication of the ΔE3L mutant, the replication of the ΔK1LΔC7L mutant in nonpermissive HeLa cells is blocked at the translation of viral mRNA. However, the host factors that restrict the replication of the ΔK1LΔC7L mutant and the molecular functions of K1L or C7L remain a mystery. K1 and C7 share no amino acid sequence homology, but either K1L or C7L can complement the replication defect of the ΔK1LΔC7L mutant in most cell lines. The exception is rabbit RK13 cells, where K1L but not C7L can complement (24). K1L is present in only a few orthopoxviruses, while C7L or a functional homologue of C7L is present in almost all mammalian poxviruses (18). K1 comprises multiple ankyrin repeats, a protein motif that is involved in protein-ligand interaction. It was shown to prevent the degradation of IκBα and to thus inhibit host NF-κB activation in RK13 cells (25). C7L has no homologue outside the poxvirus family, and its molecular function remains unknown. It may play a role in inhibiting cellular apoptosis in response to VACV infection (23).
As E3L supports VACV replication by antagonizing innate antiviral pathways that are inducible by IFNs, we hypothesize that K1L and C7L might function similarly by antagonizing IFN effectors. In the current study, we tested the hypothesis and found that both K1L and C7L can antagonize antiviral activities induced by type I IFNs. We found that K1L and C7L do not antagonize IFN by inhibiting IFN signaling or PKR activation, demonstrating that K1L and C7L are novel IFN antagonists functioning differently than previously identified IFN antagonists in VACV. Furthermore, we tested a panel of K1L mutant viruses and showed that K1L's functions of regulating VACV host range and of antagonizing IFN are genetically nonsegregable, suggesting that K1L supports VACV replication in mammalian cells by antagonizing the same antiviral factor(s) that is induced by IFN in Huh7 cells.
MATERIALS AND METHODS
Cells and viruses.
Huh7, MCF-7 (breast adenocarcinoma), and BHK21 cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) with 10% fetal bovine serum. Vero cells were cultured in minimum essential medium with Earle's balanced salts (Invitrogen) supplemented with 10% fetal bovine serum. Wild-type (WT) VACV strain WR and all other recombinant viruses except the E3L deletion mutant were propagated on Vero cells. The E3L deletion mutant was propagated on BHK21 cells.
Construction of mutant viruses. (i) vK1L−C7L−/GFP+.
vK1L−, a K1L deletion mutant of WR VACV, was described previously (19). vK1L−C7L−/GFP+ was constructed by replacing the C7L gene of vK1L− with the green fluorescent protein (GFP) gene under the control of the late promoter P11. The DNA fragment containing the GFP gene under the control of the P11 promoter was amplified from pYW31 (19) by PCR (Pfu; Stratagene) with primer pair 5′-TTACTTGTACAGCTCGTGC-3′ and 5′-GGCGCGCCTTTCATTTTG-3′. Approximately 300 bp each of the left and right C7L flanking sequences were amplified from VACV DNA by PCR with primer pair 5′-GCACGAGCTGTACAAGTAATATGAGTATAGTGTTAAATGACAC-3′ and 5′-ATAAACGGAGCACACCATTTAG-3′ and primer pair 5′-CAAAATGAAAGGCGCGCCCATATTGGGTACTAGTTTTACTATCAT-3′ and 5′-GTCGTAGATATTAACAAAGGTTGTG-3′. The underlined sequences in the primers denote the sequences that are not present in the VACV genome but were added to overlap the PCR product containing the GFP cassette. All three PCR products were assembled together by recombinant PCR, and the final PCR product was transfected into Vero cells infected with vK1L− at a multiplicity of infection (MOI) of 1 PFU per cell. The cell lysates were harvested 2 days later and applied to Vero cell monolayers. Plaques containing recombinant viruses expressing the GFP gene were picked under the fluorescence microscope and purified by four rounds of plaque purification.
(ii) WT-GFP+.
WT-GFP+ was constructed by inserting a GFP expression cassette next to the K1L gene of WR VACV. The transfer plasmid pK1L-V5-GFP, which contains a GFP expression cassette between the K1L open reading frame and its flanking sequence, was described previously (19). It was transfected into Vero cells infected with WT WR. Plaques containing recombinant viruses expressing the GFP gene were picked and purified as described above.
(iii) vE3L−.
vE3L− was constructed by replacing the E3L gene of WT WR with the GFP under the control of an early/late promoter. The left and right flanking regions of the WR E3L gene were PCR amplified from WR genomic DNA with primer pair 5′-TTGGTACCTCTCTTATGAATCGTATATCATCAT-3′and 5′-AGATCTACTACATATGAGTCGACGAATGCTAGCTATTCGATAAGGCAGATGGAAAATCTA-3′ and primer pair 5′-GCTAGCATTCGTCGACTCATATGTAGTAGATCTAATCTCTGCGTTAGAACGCTCGTCGA-3′ and 5′-TAGAGCTCAATAAACCGTCTATTGCCACAAATT-3′.
The left and right flanking sequences were assembled together by recombinant PCR and inserted into cloning vector pBS-KS+ (Stratagene) between the KpnI and SacI sites, resulting in plasmid pΔE3L. The gpt gene driven by an early and late promoter, p7.5, was subcloned into pΔE3L at the BglII site. A cassette containing the enhanced GFP (EGFP) gene, driven by an early and a late promoter, was cloned between the NheI and NdeI sites, resulting in pΔE3L/EGFP. Two micrograms of pΔE3L/EGFP was transfected into BHK21 cells infected with WT WR. Recombinant virus was grown in selective medium with mycophenolic acid (12) and screened for GFP expression.
vK1L−C7L−, its revertants, and the panel of K1L mutants were described previously (18, 19). The K1L revertant (K1L-rev) of vK1L−C7L− was previously referred to as vK1L-WT (19), while the C7L revertant (C7L-rev) of vK1L−C7L− was previously referred to as vVACV-C7L (18).
The purity of the mutants was confirmed by PCR with primers flanking the deleted gene.
IFN sensitivity assays.
Huh7 or MCF-7 cells were grown to 80% confluence in 12-well culture plates; treated with 0, 2, 20, 200, or 2,000 U/ml of human IFN-α, IFN-β, or IFN-γ (PBL Biomedical Laboratories) for 24 h; and then infected with WT or mutant VACV at an MOI of 0.05 or 5. One hour postinfection (hpi), the media were removed, the cells were washed twice with Dulbecco's phosphate-buffered saline, and fresh medium containing no or the same concentration of IFN was added. At 24 hpi, the infected cells expressing GFP were visualized under an inverted fluorescence microscope using a 10× objective and photographed with a digital camera. Sets of cells were also harvested at 0, 24, or 48 hpi, and the viral titers were determined by duplicate plaque assays on Vero cells.
Western blot analysis.
Huh7 cells were left untreated or treated with 200 U/ml of IFN-β and infected with VACV at an MOI of 5 for 8 h. The cells were then lysed for Western blot analysis as described previously (19). The detection antibodies were as follows: mouse monoclonal antibodies against PKR (Santa Cruz Biotechnology), Hsp70 (Santa Cruz), GFP (Santa Cruz), VACV E3 (a gift of Stuart Isaacs) (30), and WR148 (18); goat polyclonal antibody against phospho-STAT1 (Santa Cruz); rabbit monoclonal antibodies against phospho-eIF2α (Ser51) (cell signaling; catalog no. 3597S); and rabbit polyclonal antibodies against phospho-PKR (Thr446) (cell signaling; catalog no. 3076S), eIF2α (Santa Cruz), STAT1 (Upstate), VACV D12, and G8 (1).
RT-PCR.
Huh7 cells were left untreated or treated with 200 U/ml of IFN-β and infected with VACV at an MOI of 5 for 8 h. The cells were then lysed, and total RNA was extracted with Trizol. Reverse transcription-PCR (RT-PCR) was performed with an iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions. The following PCR primers were used: ISG15 forward primer 5′-CCGTGAAGATGCTGGCG-3′, ISG15 reverse primer 5′-CGAAGGTCAGCCAGAAC-3′, MxA forward primer 5′-AAGGTCAGTTACCAGGACTACGAGA-3′ and reverse primer 5′-ACAATCATGTAACCCTTCTTCAGGT-3, human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward primer 5′-AAGGTGAAGGTCGGAGTCAACGGA-3′ and reverse primer 5′-TTACTCCTTGGAGGCCATGT-3′, vvD12L forward primer 5′-ATGGATGAAATTGTAAAAAATATCCGGGA-3′ and reverse primer 5′-TCACAGCAGTAGTTTAACTAGTCT-3′, and vvG8R forward primer 5′-AATGTAGACTCGACGGATGAGTTA-3′ and reverse primer 5′-TCGTCATTATCCATTACGATTCTAGTT-3′.
PKR silencing.
Experiments were performed essentially as described previously (1). Briefly, Huh7 cells were transfected with 100 nM of small interfering RNA (siRNA) specific to human PKR 5′-P-ACUUUGUCUAGUUUCUCGCUU (Dharmacon RNA Technologies) and Silencer Cy3-labeled negative control 1 (catalog no. 4621; Ambion, Inc.). Forty-eight hours later, cells were treated with 200 U/ml of IFN-β for 24 h and then infected with VACV at an MOI of 5. Virus titers were determined by plaque assay. PKR silencing was confirmed by Western blotting.
RESULTS
VACV mutants deleted of both K1L and C7L replicate as efficiently as WT VACV in human Huh7 and MCF-7 cells.
In many human cell lines, VACV mutants deleted of both K1L and C7L replicate abortively and express only a subset of viral genes (24). This presents a problem for deducing the functions of K1L and C7L from the phenotypes of the mutants in human cells, as it is difficult to determine whether the phenotypes are due to the deletion of K1L and C7L or to reduced expression of other viral proteins. To circumvent this problem in our current study, we first identified human cell lines that were fully permissive for the replication of a K1L and C7L deletion mutant. We infected several human cell lines with vK1L−C7L−/GFP+, a VACV mutant deleted of both K1L and C7L and encoding a GFP open reading frame under the control of VACV late promoter P11. In A431 (epidermoid carcinoma), HT-3 (cervical carcinoma), Ca Ski (cervical carcinoma), and SKOV-3 (ovary adenocarcinoma) cells, vK1L−C7L−/GFP+ replicated abortively, as indicated by the lack of GFP expression (data not shown). However, in both Huh7 (human hepatoma) and MCF-7 (breast adenocarcinoma) cells, vK1L−C7L−/GFP+ expressed late viral proteins, as indicated by GFP expression, and formed plaques that were similar in size to those formed by WT-GFP+, a WR VACV with a similar GFP cassette (Fig. 1A). The growth rate of vK1L−C7L−/GFP+ in Huh7 and MCF-7 cells was very similar to that of WT-GFP+ at both low and high MOIs (Fig. 1B and data not shown).
FIG. 1.
VACV mutant deleted of both K1L and C7L replicates as efficiently as WT VACV in human Huh7 and MCF-7 cells. (A) Plaque morphology of WT-GFP+ and vK1L−C7L−/GFP+ on Huh7 and MCF-7 cells. WT-GFP+ was constructed by inserting a GFP expression cassette into the WT WR strain. vK1L−C7L−/GFP+ was constructed from the WT WR strain by deleting the K1L gene and replacing the C7L gene with a GFP expression cassette. The cells were infected at an MOI of 0.05 PFU/cell, and the GFP-expressing cells (shown as white) were photographed at 36 hpi. (B) Growth curves of WT-GFP+ and vK1L−C7L−/GFP+ at high (5 PFU/cell) and low (0.05 PFU/cell) MOIs. Virus yields at 0, 24, and 48 hpi were determined by duplicate plaque assays on Vero cells. The average titers are shown with standard deviations indicated by vertical bars.
vK1L−C7L−/GFP+ is sensitive to type I IFN in permissive human cells.
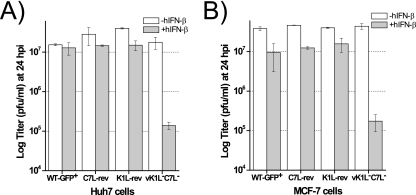
To determine whether K1L and C7L are required for VACV to antagonize IFN activities, we examined whether the deletion of K1L and C7L from VACV rendered the virus sensitive to IFNs. We pretreated Huh7 cells for 24 h with increasing concentrations of human IFN-β (hIFN-β), infected the cells with vK1L−C7L−/GFP+ or WT-GFP+ at an MOI of 5 in the absence of hIFN-β, and measured the viral yields at 24 hpi. Treatment with up to 200 U/ml of hIFN-β had very little effect on the replication of WT-GFP+, and in comparison with the control receiving no hIFN-β treatment, treatment with 2,000 U/ml of hIFN-β reduced the 24-h yield of WT-GFP+ less than 10-fold (Fig. 2A). In contrast, hIFN-β reduced the yield of vK1L−C7L−/GFP+ in a dose-dependent manner. The greatest difference between the yields of vK1L−C7L−/GFP+ and WT-GFP+ was observed when the cells were treated with 200 U/ml of hIFN-β, which reduced the yield of vK1L−C7L−/GFP+ approximately 100-fold.
FIG. 2.
Type I IFN inhibits the replication of vK1L−C7L−/GFP+. (A) Replication of vK1L−C7L−/GFP+ is inhibited by hIFN-β in a dose-dependent manner. Huh7 cells were pretreated with hIFN-β at concentrations of 0, 2, 20, 200, or 2,000 U/ml for 24 h at 37°C, infected by the indicated viruses at an MOI of 5, and harvested at 24 hpi. The viral yields at 24 hpi were determined as described in the legend to Fig. 1B. (B) Replication of vK1L−C7L−/GFP+ is inhibited by at least seven hIFN-α subtypes. The experiment was performed as described for panel A except that the cells were pretreated with 200 U/ml of hIFN-α or hIFN-γ. (C) vK1L−C7L−/GFP+ replicates abortively in hIFN-β-treated Huh7 cells. The cells were treated with 200 U/ml of hIFN-β for 24 h and infected with the indicated viruses at an MOI of 5 PFU/cell, and the GFP-expressing cells (shown as white) were photographed at 24 hpi. (D) Growth curves of WT-GFP+ and vK1L−C7L−/GFP+ in hIFN-β-treated Huh7 cells. The cells were treated with 200 U/ml of hIFN-β for 24 h and then infected at either high (5 PFU/cell) or low (0.05 PFU/cell) MOIs. Virus yields at 0, 24, and 48 hpi were determined by plaque assay.
To find out the specificity of the IFNs that vK1L−C7L−/GFP+ is sensitive to, we then tested the sensitivities of vK1L−C7L−/GFP+ and WT-GFP+ to all 12 hIFN-α subtypes (type I IFNs) and hIFN-γ (type II IFN). When the cells were pretreated with 200 U/ml of the IFNs, the 24-h yield of WT-GFP+ remained essentially the same as the yield with no pretreatment. In contrast, seven of the hIFN-α subtypes (α6, α7, α8, α10, α14, α16, and α17) reduced the 24-h yield of vK1L−C7L−/GFP+ from 10- to 40-fold (Fig. 2B). The remaining IFN-α subtypes and the IFN-γ subtype reduced the vK1L−C7L−/GFP+ yield less than 10-fold. The replication of vK1L−C7L−/GFP+ was not affected when the cells were treated with hIFN-α or hIFN-β along with the respective neutralizing antibodies (data not shown), therefore excluding the possibility that the phenomenon was due to any contaminant in commercial IFN preparations.
Since we found the greatest difference between the yields of vK1L−C7L−/GFP+ and WT-GFP+ when the cells were treated with 200 U/ml of hIFN-β, we performed all our subsequent experiments with hIFN-β at this concentration. Under such a condition, very few cells infected by vK1L−C7L−/GFP+ expressed GFP (Fig. 2C), indicating that hIFN-β blocked the replication of the mutant before the expression of VACV late proteins.
To further confirm the mutant's sensitivity to type I IFN, we then measured the growth curves of vK1L−C7L−/GFP+ and WT-GFP+ in hIFN-β-treated Huh7 cells under both low and high MOI conditions (Fig. 2D). Over a 48-h infection period, the WT-GFP+ titer continued to increase, but the vK1L−C7L−/GFP+ titer increased only slightly by 48 hpi under low MOI conditions and did not increase at all under high MOI conditions. The difference between the yields of vK1L−C7L−/GFP+ and WT-GFP+ was obvious at 24 hpi and became even more prominent at 48 hpi.
To determine the time required for hIFN-β to induce effective inhibition of the mutant replication, we added hIFN-β to Huh7 cells at various times before and/or during virus infection and then measured the viral yields at 24 hpi. While pretreating Huh7 cells with hIFN-β greatly reduced the yield of vK1L−C7L−/GFP+, adding hIFN-β during the infection had no effect on the replication of either the WT or the mutant (Fig. 3A). The yield of vK1L−C7L−/GFP+ was increasingly reduced by prolonging the time of hIFN-β treatment from 2 h to 8 h before infection but was not reduced further beyond 8 h of treatment (Fig. 3B). In contrast, the yield of WT-GFP+ was reduced only slightly by pretreating the cells with hIFN-β for 2 h and was not further reduced by increasing the time of hIFN-β treatment.
FIG. 3.
The replication of vK1L−C7L−/GFP+ is inhibited by treating the cells with hIFN-β before but not during the infection. (A) hIFN-β was effective at inhibiting vK1L−C7L−/GFP+ replication only when it was added to the cells before the infection. Huh7 cells were left untreated (−) or were treated (+) with 200 U/ml of hIFN-β before and/or during the infection, as indicated. The cells were infected at an MOI of 5, and viral yields at 24 hpi were determined by plaque assay. (B) The yield of vK1L−C7L−/GFP+ is inversely related to the length of time the cells were pretreated with hIFN-β. Huh7 cells were treated with 200 U/ml of hIFN-β for 0, 2, 4, 6, 8, or 24 h before they were infected with vK1L−C7L−/GFP+ or WT-GFP+ at an MOI of 5. Viral yields at 24 hpi were determined by plaque assay.
To confirm that the lack of K1L and C7L is responsible for the phenotype of vK1L−C7L−/GFP+, we tested the IFN sensitivities of vK1L−C7L− (19), another K1L and C7L deletion mutant, and the revertants of vK1L−C7L−. vK1L−C7L− differs from vK1L−C7L−/GFP+ in that it does not encode GFP, and it also replicates productively in Huh7 and MCF-7 cells (data not shown). Two revertants of vK1L−C7L−, K1L-rev and C7L-rev, were derived from vK1L−C7L− by putting back the K1L or C7L gene, respectively, along with a GFP cassette (18, 19). Similarly to vK1L−C7L−/GFP+, vK1L−C7L− was sensitive to hIFN-β (Fig. 4). In contrast, both K1L-rev and C7L-rev were resistant to hIFN-β, suggesting that K1L and C7L function equivalently at antagonizing IFN activities. This experiment was performed with both Huh7 and MCF-7 cells with similar results (Fig. 4), indicating that K1L and C7L can antagonize IFN activities in a variety of human cells.
FIG. 4.
Restoring either K1L or C7L to vK1L−C7L− restores the IFN resistance phenotype. Huh7 (A) or MCF-7 (B) cells were pretreated with 0 or 200 U/ml of hIFN-β for 24 h and infected by the indicated viruses at an MOI of 5. The viral yields at 24 hpi were determined. vK1L−C7L− is a VACV mutant specifically deleted of K1L and C7L genes, as previously reported (19). C7L-rev and K1L-rev were derived from vK1L−C7L− by restoring C7L and K1L, respectively (18, 19).
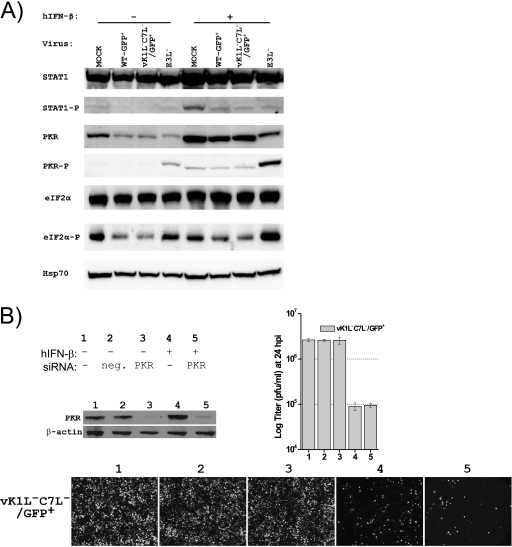
IFN treatment blocks the replication of vK1L−C7L−/GFP+ at the step of translating viral intermediate mRNAs.
To find out the stage at which the replication of vK1L−C7L−/GFP+ was blocked by IFN treatment, we assessed the transcription and translation of several representative VACV genes in infected cells (Fig. 5). In cells that were not treated with hIFN-β, viral proteins of all three temporal classes can be detected by Western blotting at 8 hpi, and there was no difference in viral protein levels between WT-GFP+- and vK1L−C7L−/GFP+-infected cells. hIFN-β treatment did not affect viral protein levels in WT-GFP+-infected cells except for slightly reducing the level of the WR148 (late) protein. In contrast, hIFN-β treatment, while having very little effect on the level of the D12 (early) and E3 (early) proteins, greatly reduced the level of the G8 (intermediate), GFP (late), and WR148 proteins in vK1L−C7L−/GFP+-infected cells. The levels of D12L and G8R mRNAs, as measured by RT-PCR, were similar in cells infected by either virus (Fig. 5B). Altogether, these results indicate that hIFN-β blocks the replication of vK1L−C7L−/GFP+ at the stage of translating intermediate viral mRNAs.
FIG. 5.
hIFN-β blocks the replication of vK1L−C7L−/GFP+ at the step of translating viral intermediate mRNAs. (A) hIFN-β treatment greatly reduced the level of VACV intermediate and late proteins expressed by vK1L−C7L−/GFP+. Huh7 cells were left untreated (−) or were treated (+) with 200 U/ml of hIFN-β for 24 h and infected at an MOI of 5 for 8 h. Representative VACV early, intermediate, and late protein levels were then determined by Western blotting. (B) hIFN-β treatment did not inhibit the transcription of VACV intermediate gene G8R. The cells were treated and infected as described for panel A. Total RNA was extracted, and the levels of the D12L (early), G8R (intermediate), and human GAPDH (control) transcripts were assessed by RT-PCR.
K1L and C7L are not required for VACV to antagonize IFN signaling.
That the replication of vK1L−C7L−/GFP+ was blocked by pretreating the cells with hIFN-β but not by adding hIFN-β during the infection suggests that K1L and C7L are required for VACV to antagonize IFN effectors but are not required for inhibition of IFN signaling. To further test this idea, we examined how IFN-induced transcription of ISGs was affected by WT-GFP+ and vK1L−C7L−/GFP+. hIFN-β significantly induced the transcription of both MxA and ISG15 in Huh7 cells, as measured by RT-PCR (Fig. 6). This induction was not affected when VACV was added to the cells after hIFN-β treatment but was inhibited when VACV was added to the cells along with hIFN-β. WT-GFP+ and vK1L−C7L−/GFP+ were equally capable of inhibiting hIFN-β-induced MxA and ISG15 transcription (Fig. 6). In addition, infection of hIFN-β-treated cells by either WT-GFP+ or vK1L−C7L−/GFP+ greatly reduced the level of phospho-STAT1 in the cells (Fig. 7A). Altogether, these results indicate that K1L and C7L are not required for VACV to inhibit IFN signaling.
FIG. 6.
K1L and C7L are not required for VACV to antagonize IFN signaling. (A) Huh7 cells were left untreated (−) or were treated (+) with 200 U/ml of hIFN-β for 24 h and then infected by the indicated viruses at an MOI of 5 for 8 h. vE3L− was constructed by deleting the E3L gene from WT WR VACV. (B) Huh7 cells were infected by the indicated viruses at an MOI of 5 for 8 h in the presence (+) or absence (−) of 200 U/ml of hIFN-β. For both panel A and panel B, total RNA was extracted from the cells, and the levels of the MxA, ISG15, and GAPDH (control) transcripts were assessed by RT-PCR.
FIG. 7.
hIFN-β inhibits the replication of vK1L−C7L−/GFP+ through a PKR-independent pathway. (A) K1L and C7L are not required for VACV to inhibit the phosphorylation of PKR or eIF2α. Huh7 cells were left untreated (−) or were treated (+) with 200 U/ml of hIFN-β for 24 h and infected with the indicated viruses at an MOI of 5 for 8 h. The levels of total and phosphorylated STAT1, PKR, and eIF2α were determined by Western blotting. The Hsp70 level was determined as the gel loading control. (B) Knocking down PKR expression with siRNA did not rescue the replication of vK1L−C7L−/GFP+ in hIFN-β-treated Huh7 cells. Huh7 cells were not transfected (−) or were transfected with negative-control siRNA (neg.) or PKR siRNA for 48 h and treated with no (−) or 200 U/ml of hIFN-β (+) for 24 h. One set of cells was used to confirm the knockdown of PKR protein expression by Western blot analysis of PKR and β-actin as a control. Another set of cells was infected by vK1L−C7L−/GFP+ at an MOI of 5 for 24 h. The GFP-expressing cells were photographed, and viral yields were determined by plaque assay.
K1L and C7L are not required for VACV to inhibit the phosphorylation of PKR or eIF2α.
Among IFN effectors, PKR is known to affect protein translation upon its activation through autophosphorylation. Since IFN blocked the replication of vK1L−C7L−/GFP+ at the step of translating intermediate viral mRNAs, we examined whether K1L and C7L were required for VACV to inhibit the phosphorylation and activation of PKR. Consistent with previous reports (1, 32), we found that the infection of Huh7 cells by vE3L−, a VACV mutant with a deletion in E3L, resulted in PKR phosphorylation. The phosphorylated PKR migrated slightly more slowly than unphosphorylated PKR in sodium dodecyl sulfate-polyacrylamide gel electrophoresis and was readily detected by an antibody against phospho-PKR by Western blotting (Fig. 7A). In contrast to infection by vE3L−, infection by WT-GFP+ and vK1L−C7L−/GFP+ did not result in any PKR phosphorylation in Huh7 cells. Treating the cells with hIFN-β resulted in a higher level of PKR and the detection of a weak band with phospho-PKR antibody. This weak band most likely represents nonspecific recognition of a high level of unphosphorylated PKR by the phospho-PKR antibody, since it migrated at the same location in a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel as unphosphorylated PKR. A similar weak band was also present in hIFN-β-treated cells that were subsequently infected by either WT-GFP+ or vK1L−C7L−/GFP+, indicating that neither virus activated PKR and that K1L and C7L are not required for VACV to inhibit PKR phosphorylation. In contrast, vE3L− induced a much higher level of phospho-PKR and phospho-eIF2α in hIFN-β-treated cells, consistent with E3 being the VACV protein that antagonizes PKR. The levels of phospho-eIF2α and total PKR in WT-GFP+- and vK1L−C7L−/GFP+-infected cells were even lower than in mock-infected cells, suggesting that VACV inhibited the basal levels of eIF2α phosphorylation and PKR production in Huh7 cells.
Previously, it was shown that the replication of an E3L deletion mutant in IFN-treated Huh7 cells can be rescued by silencing PKR expression (1). To further prove that K1L and C7L function differently than E3L at antagonizing IFN activities, we tested whether knocking down PKR could rescue the replication of vK1L−C7L−/GFP+ in hIFN-β-treated Huh7 cells. The level of PKR protein in Huh7 cells was greatly reduced by transfection of siRNA against PKR (Fig. 7B). However, hIFN-β remained effective at inhibiting the replication of vK1L−C7L−/GFP+, as indicated by viral yields at 24 hpi and by GFP expression as an indication of VACV late protein expression (Fig. 7B).
The ability of K1L to antagonize IFN in Huh7 cells correlates with its host range regulative function.
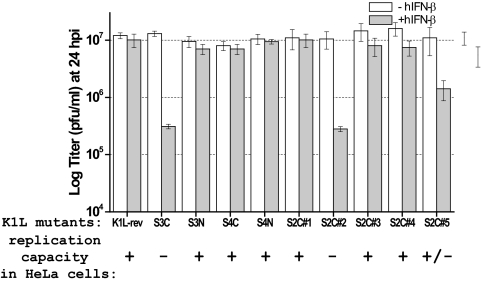
Similar to our finding that VACV requires either K1L or C7L to antagonize IFN activities in Huh7 cells, VACV requires either K1L or C7L to replicate in many mammalian cells, including HeLa cells. To explore whether the function of K1L in antagonizing IFN activities in Huh7 cells is related to its host range regulative function, we tested the IFN sensitivity of a panel of VACV K1L mutants that were derived from vK1L−C7L− and encode a K1 protein with specific amino acid substitutions at its ankyrin repeats. These K1L mutants were previously shown to have different replication capacities in HeLa cells (19), which are summarized in Fig. 8. We found that all of the K1L mutants replicated equally well in Huh7 cells but displayed various degrees of IFN sensitivity (Fig. 8). The mutants S3C and S3C#2, which failed to replicate in HeLa cells, also failed to replicate in hIFN-β-treated Huh7 cells, as measured by viral yield at 24 hpi. The S3N, S4C, S4N, S2C#1, S2C#3, and S2C#4 mutants, which replicated at the WT rate in HeLa cells, replicated in hIFN-β-treated Huh7 cells with efficiencies similar to that of K1L-rev. The S2C#5 mutant, which was partially defective at replication in HeLa cells, was also partially sensitive to hIFN-β in Huh7 cells. Altogether, these results show a very consistent correlation between the replication capacities of the mutants in HeLa cells and their sensitivities to hIFN-β in Huh7 cells.
FIG. 8.
K1L's ability to antagonize hIFN-β in Huh7 cells correlates with its host range function in HeLa cells. Huh7 cells were left untreated (−) or were treated (+) with 200 U/ml of hIFN-β for 24 h and infected with a panel of K1L mutant VACV strains at an MOI of 5. Viral yields at 24 hpi were determined by plaque assay. The mutants were all derived from vK1L−C7L− and encode a K1 protein with specific amino acid substitutions at its ankyrin repeats (19). The previously reported replication capacities of the mutants in HeLa cells are summarized below the name of the mutant virus. +, WT level of replication; −, no viral replication; ±, partial defect in viral replication.
DISCUSSION
In this study, we revealed an essential role for VACV host range genes K1L and C7L in antagonizing antiviral activities induced by type I IFN. Type I IFNs play a critical role in innate immune defense against viruses and in modulating the adaptive immune response to infection and tumor formation (4). Viruses have evolved diverse strategies to down-modulate the production or the activities of IFNs. Uncovering and understanding these strategies have provided great insight into the mechanisms of viral pathogenesis and IFN functions. Among viruses, poxviruses are particularly adept at modulating IFN activities and encode multiple antagonists that block IFN action at different steps. Extracellularly, VACV type I IFN binding protein B18 prevents IFN from binding and activating the IFN-α/β receptor (26). Intracellularly, VACV H1, E3, and K3 block IFN signaling or specific IFN effectors (6, 10, 22). Our finding that K1L and C7L play an essential and nonredundant role in blocking antiviral effects of IFNs suggests that K1L and C7L are novel IFN antagonists, functioning differently than the IFN antagonists previously identified in VACV.
We discovered this novel function of K1L and C7L by studying K1L and C7L deletion mutants (vK1L−C7L−/GFP+ and vK1L−C7L−) in human Huh7 and MCF-7 cells. vK1L−C7L− was previously known to replicate in only a few selected cell lines, none of which was of human origin. Therefore, deducing the function of K1L and C7L from analyzing vK1L−C7L− in nonpermissive human cells was hampered by the fact that many VACV genes, in addition to K1L and C7L, were not expressed or were expressed at lower levels in nonpermissive cells. This problem, however, was eliminated in this study by the use of Huh7 and MCF-7 cells, which we found to be fully permissive for the replication of vK1L−C7L−. The growth curve and Western blot analyses showed that WT and vK1L−C7L− replicated with equal efficiencies and expressed viral proteins of all temporal classes at the same level in these permissive human cells. The identification of the permissive human cell lines for vK1L−C7L− is important for this and future studies, as any phenotypic difference between vK1L−C7L− and WT VACV in these human cell lines can now be attributed to K1L and C7L.
The phenotypic difference between vK1L−C7L− and WT VACV that we found in this study is their distinct difference in type I IFN sensitivity. While hIFN-β had no or very limited effect on the growth of WT VACV, it inhibited the replication of vK1L−C7L− in a dose-dependent manner. In hIFN-β-treated cells, WT VACV increased its titer more than 100-fold over a 48-h period, but the vK1L−C7L− titer did not increase. At the molecular level, hIFN-β blocked the translation of intermediate and late proteins of vK1L−C7L−. The IFN sensitivity of vK1L−C7L− is independent of a specific type I IFN or a specific cell type, as at least seven different IFN-α subtypes and IFN-β blocked the replication of vK1L−C7L− in MCF-7 as well as Huh7 cells. There are some variations in the degrees of inhibition imposed by the different hIFN-α subtypes. This may be due to different activities of the commercial IFNs. It is also possible that various hIFN-α subtypes, although they bind to the same receptor, may interact with the receptor components in different ways and induce different genes (13). The IFN-sensitive phenotype of vK1L−C7L− is not due to any unintended mutation introduced during the construction of the mutant, as revertant viruses were resistant to IFN treatment. Based on the fact that the revertant viruses with either K1L or C7L were equally resistant to IFN, we concluded that K1L and C7L function equivalently at antagonizing IFN.
How do K1L and C7L antagonize IFN activities? Our data suggest that they do so not by inhibiting IFN signaling but by antagonizing antiviral factors induced by IFN. We found that the deletion of K1L and C7L from VACV did not affect the ability of the virus to inhibit IFN signaling. vK1L−C7L− was fully capable of blocking IFN-induced STAT1 phosphorylation and ISG transcription. This is not very surprising, since vK1L−C7L− still encodes B18, which binds type I IFNs, and H1, which dephosphorylates STAT1. Although vK1L−C7L− was able to inhibit IFN signaling and replicate in the presence of IFN, it failed to replicate in IFN-treated cells, where the transcription and translation of hundreds of ISGs had already been induced. In these cells, blocking IFN signaling would not have benefited viral replication. Instead, the antagonizing of preexisting antiviral factors is necessary for VACV to replicate successfully. Since WT VACV but not vK1L−C7L− was able to replicate in IFN-treated cells, K1L and C7L must antagonize an antiviral factor(s) that is induced by IFN.
IFN induces the expression of more than 300 ISGs (11), only a few of which have been characterized for their antiviral activities. In this study, we have eliminated the possibility that some of the well-characterized ISGs are the targets for K1L and C7L. We found that IFN-β inhibited only the translation of intermediate mRNAs and did not affect the level of intermediate gene transcripts, so it is unlikely that the 2′-5′ oligoadenylate synthetase (OAS)/RNase L pathway might have inhibited the replication of vK1L−C7L− by degrading viral mRNA. Previously, we also showed that IFN-treated Huh7 cells did not express RNase L (1). Among ISGs, PKR is well known for its effect on protein translation through eIF2α phosphorylation. We found that both WT and vK1L−C7L− were able to block the phosphorylation of PKR and eIF2α. Furthermore, knocking down PKR expression with siRNA failed to rescue the replication of vK1L−C7L− in IFN-treated cells, indicating that PKR was not the antiviral factor that restricted the replication of vK1L−C7L−. Overall, our data suggest that type I IFN can induce a PKR-independent pathway to block VACV protein translation and that this pathway can be inhibited by K1L and C7L. Interestingly, some recent studies with norovirus and picornavirus also suggest that a PKR-independent pathway is induced by type I IFN to inhibit protein translation in cells infected by these RNA viruses (8, 20). Identification of the target for K1L and C7L may lead to the identification of a novel antiviral host factor/pathway that has broad antiviral effects. In an effort to identify the target of K1L and C7L, we performed an initial screening for an ISG that is able to specifically inhibit vK1L−C7L− replication in Huh7 cells (data not shown). We selected 13 ISGs (BST2, CASP1, CXCL10, GBP1, HLA-F, IFI-16, IFIT1, IFITM1, ISG15, OAS1, OAS2, IFI-6, and IFI-27) that are highly induced by IFN-β in Huh7 cells and expressed them individually with a mammalian expression vector in Huh7 cells. The expression of these ISGs in transfected Huh7 cells was confirmed by RT-PCR, but none of these ISGs showed any specific inhibitory effect on vK1L−C7L− replication, suggesting that they are not the target of K1L and C7L (data not shown). Large-scale screenings of hundreds of uncharacterized ISGs will have to be performed to identify the target.
The findings in this study also provide a much-needed new clue to the mechanism by which K1L and C7L modulate the cellular tropism of VACV. How the host range genes such as K1L and C7L control cellular tropism of poxviruses is one of the most enigmatic questions in the poxvirus field. Previously, it was shown that K1L inhibits host NF-κB activation and that C7L inhibits cellular apoptosis (23, 25). However, it was also shown that inhibition of NF-κB activation or cellular apoptosis was not critical for regulating cellular tropism of VACV (7, 9). It was also unclear what difference between permissive and nonpermissive cells makes them either support or inhibit the replication of vK1L−C7L−. One possibility is that the permissive cells have some additional host factors that could substitute for K1L and C7L for an essential function in VACV replication. Another possibility is that the nonpermissive cells have some additional host factors that restrict the replication of vK1L−C7L− and are antagonized by K1L and C7L. Our finding that the permissive cells for vK1L−C7L− could be turned into nonpermissive cells simply by inducing ISG expression with IFN indicates that the latter possibility is the case. Furthermore, we suggest that the above-mentioned IFN-inducible antiviral factors in Huh7 cells are also the host factors that restrict the replication of vK1L−C7L− in many nonpermissive cells. In untreated HeLa cells and IFN-treated Huh7 cells, the replication of vK1L−C7L− is similarly blocked at the step of translating viral intermediate mRNAs. In addition, we found a perfect correlation between the abilities of a panel of K1L mutant viruses to antagonize IFN activities in Huh7 cells and their abilities to replicate in HeLa cells. These mutants are derived from the same parental virus, and they express K1 proteins that differ from each other at only a few amino acids. The fact that K1L's functions in regulating host range and antagonizing IFN are genetically nonsegregable suggests that these are the same function.
Type I IFNs are known to regulate cellular tropism of both DNA and RNA viruses. The cellular tropism of poliovirus is partly determined by whether the cells can rapidly produce type I IFNs in response to the virus (31). Myxoma virus, a rabbit poxvirus, replicates abortively in primary mouse cells because of the restriction posed by virus-induced type I IFNs (29). However, in the case of host range restriction of vK1L−C7L−, it is very unlikely that virus-induced type I IFN, acting in an autocrine fashion, restricts the replication of vK1L−C7L− in nonpermissive cells. vK1L−C7L− is able to block IFN signaling and replicate in the presence of IFN in permissive Huh7 cells. In addition, the replication of vK1L−C7L− in nonpermissive HeLa cells cannot be rescued by adding neutralizing antibodies against type I IFNs during infection (data not shown). The putative host factors that restrict the replication of vK1L−C7L− in nonpermissive cells are most likely present at constitutively high levels in these cells or could be rapidly induced by a signal other than IFN.
Acknowledgments
We thank members of the Xiang lab for critical readings of the manuscript.
This work was supported by National Institutes of Health grants AI065731 and AI079217.
Footnotes
Published ahead of print on 5 August 2009.
REFERENCES
- 1.Arsenio, J., Y. Deschambault, and J. Cao. 2008. Antagonizing activity of vaccinia virus E3L against human interferons in Huh7 cells. Virology 377:124-132. [DOI] [PubMed] [Google Scholar]
- 2.Bart, P. A., R. Goodall, T. Barber, A. Harari, A. Guimaraes-Walker, M. Khonkarly, N. C. Sheppard, Y. Bangala, M. J. Frachette, R. Wagner, P. Liljestrom, J. P. Kraehenbuhl, M. Girard, J. Goudsmit, M. Esteban, J. Heeney, Q. Sattentau, S. McCormack, A. Babiker, G. Pantaleo, and J. Weber. 2008. EV01: a phase I trial in healthy HIV negative volunteers to evaluate a clade C HIV vaccine, NYVAC-C undertaken by the EuroVacc Consortium. Vaccine 26:3153-3161. [DOI] [PubMed] [Google Scholar]
- 3.Beattie, E., E. B. Kauffman, H. Martinez, M. E. Perkus, B. L. Jacobs, E. Paoletti, and J. Tartaglia. 1996. Host-range restriction of vaccinia virus E3L-specific deletion mutants. Virus Genes 12:89-94. [DOI] [PubMed] [Google Scholar]
- 4.Borden, E. C., G. C. Sen, G. Uze, R. H. Silverman, R. M. Ransohoff, G. R. Foster, and G. R. Stark. 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 6:975-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, H. W., and B. L. Jacobs. 1993. Identification of a conserved motif that is necessary for binding of the vaccinia virus E3L gene products to double-stranded RNA. Virology 194:537-547. [DOI] [PubMed] [Google Scholar]
- 6.Chang, H. W., J. C. Watson, and B. L. Jacobs. 1992. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. USA 89:4825-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, S.-J., J.-C. Hsiao, S. Sonnberg, C.-T. Chiang, M.-H. Yang, D.-L. Tzou, A. A. Mercer, and W. Chang. 2009. Poxvirus host range protein CP77 contains an F-box-like domain that is necessary to suppress NF-κB activation by tumor necrosis factor alpha but is independent of its host range function. J. Virol. 83:4140-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Changotra, H., Y. Jia, T. N. Moore, G. Liu, S. M. Kahan, S. V. Sosnovtsev, and S. M. Karst. 2009. Type I and type II interferons inhibit the translation of murine norovirus proteins. J. Virol. 83:5683-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung, C. S., I. A. Vasilevskaya, S. C. Wang, C. H. Bair, and W. Chang. 1997. Apoptosis and host restriction of vaccinia virus in RK13 cells. Virus Res. 52:121-132. [DOI] [PubMed] [Google Scholar]
- 10.Davies, M. V., H.-W. Chang, B. L. Jacobs, and R. J. Kaufman. 1993. The E3L and K3L vaccinia virus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J. Virol. 67:1688-1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Veer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69:912-920. [PubMed] [Google Scholar]
- 12.Falkner, F. G., and B. Moss. 1988. Escherichia coli gpt gene provides dominant selection for vaccinia virus open reading frame expression vectors. J. Virol. 62:1849-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster, G. R, and N. B. Finter. 1998. Are all Type I human interferons equivalent? J. Viral Hepat. 5:143-152. [DOI] [PubMed] [Google Scholar]
- 14.Guerra, S., A. Caceres, K. P. Knobeloch, I. Horak, and M. Esteban. 2008. Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog. 4:e1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, Y. G., M. Muralinath, T. Brandt, M. Pearcy, K. Hauns, K. Lowenhaupt, B. L. Jacobs, and A. Rich. 2003. A role for Z-DNA binding in vaccinia virus pathogenesis. Proc. Natl. Acad. Sci. USA 100:6974-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langland, J. O., and B. L. Jacobs. 2002. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology 299:133-141. [DOI] [PubMed] [Google Scholar]
- 17.McFadden, G. 2005. Poxvirus tropism. Nat. Rev. Microbiol. 3:201-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meng, X., J. Chao, and Y. Xiang. 2008. Identification from diverse mammalian poxviruses of host-range regulatory genes functioning equivalently to vaccinia virus C7L. Virology 372:372-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng, X., and Y. Xiang. 2006. Vaccinia virus K1L protein supports viral replication in human and rabbit cells through a cell-type-specific set of its ankyrin repeat residues that are distinct from its binding site for ACAP2. Virology 353:220-233. [DOI] [PubMed] [Google Scholar]
- 20.Morrison, J. M., and V. R. Racaniello. 2009. Proteinase 2Apro is essential for enterovirus replication in type I interferon-treated cells. J. Virol. 83:4412-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moss, B. 2007. Poxviridae: the viruses and their replication, p. 2905-2946. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 22.Najarro, P., P. Traktman, and J. A. Lewis. 2001. Vaccinia virus blocks gamma interferon signal transduction: viral VH1 phosphatase reverses Stat1 activation. J. Virol. 75:3185-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nájera, J. L., C. E. Gómez, E. Domingo-Gil, M. M. Gherardi, and M. Esteban. 2006. Cellular and biochemical differences between two attenuated poxvirus vaccine candidates (MVA and NYVAC) and role of the C7L gene. J. Virol. 80:6033-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perkus, M. E., S. J. Goebel, S. W. Davis, G. P. Johnson, K. Limbach, E. K. Norton, and E. Paoletti. 1990. Vaccinia virus host range genes. Virology 179:276-286. [DOI] [PubMed] [Google Scholar]
- 25.Shisler, J. L., and X.-L. Jin. 2004. The vaccinia virus K1L gene product inhibits host NF-κB activation by preventing IκBα degradation. J. Virol. 78:3553-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Symons, J. A., A. Alcami, and G. L. Smith. 1995. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell 81:551-560. [DOI] [PubMed] [Google Scholar]
- 27.Tartaglia, J., M. E. Perkus, J. Taylor, E. K. Norton, J. C. Audonnet, W. I. Cox, S. W. Davis, J. van der Hoeven, B. Meignier, M. Riviere, et al. 1992. NYVAC: a highly attenuated strain of vaccinia virus. Virology 188:217-232. [DOI] [PubMed] [Google Scholar]
- 28.Vijaysri, S., G. Jentarra, M. C. Heck, A. A. Mercer, C. J. McInnes, and B. L. Jacobs. 2008. Vaccinia viruses with mutations in the E3L gene as potential replication-competent, attenuated vaccines: intranasal vaccination. Vaccine 26:664-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, F., Y. Ma, J. W. Barrett, X. Gao, J. Loh, E. Barton, H. W. Virgin, and G. McFadden. 2004. Disruption of Erk-dependent type I interferon induction breaks the myxoma virus species barrier. Nat. Immunol. 5:1266-1274. [DOI] [PubMed] [Google Scholar]
- 30.Weaver, J. R., M. Shamim, E. Alexander, D. H. Davies, P. L. Felgner, and S. N. Isaacs. 2007. The identification and characterization of a monoclonal antibody to the vaccinia virus E3 protein. Virus Res. 130:269-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshikawa, T., T. Iwasaki, M. Ida-Hosonuma, M. Yoneyama, T. Fujita, H. Horie, M. Miyazawa, S. Abe, B. Simizu, and S. Koike. 2006. Role of the alpha/beta interferon response in the acquisition of susceptibility to poliovirus by kidney cells in culture. J. Virol. 80:4313-4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, P., B. L. Jacobs, and C. E. Samuel. 2008. Loss of protein kinase PKR expression in human HeLa cells complements the vaccinia virus E3L deletion mutant phenotype by restoration of viral protein synthesis. J. Virol. 82:840-848. [DOI] [PMC free article] [PubMed] [Google Scholar]