Abstract
Autophagy or “self-eating” is a highly conserved pathway that enables cells to degrade pieces of themselves in autolysosomes to enable their survival in times of stress, including nutrient deprivation. The formation of these degradative compartments requires cytosolic proteins, some of which are autophagy specific, as well as intracellular organelles, such as the ER and Golgi, and the endosome–lysosome system. Here we discuss the cross talk between autophagy and intracellular compartments, highlighting recent exciting data about the role and regulation of the Vps34 class III phosphatidylinositol (PI) 3-kinase in autophagy.
Introduction
Macroautophagy (hereafter called autophagy) is an intracellular membrane trafficking pathway involved in delivery of cytoplasmic material to the lysosomes for degradation. During induction of autophagy, cytoplasmic components are sequestered by a membrane called the phagophore or isolation membrane (IM), which expands to form double-membrane vesicles called autophagosomes. The autophagosomes can then fuse with vesicles of the endocytic pathway to form amphisomes, which eventually will fuse with lysosomes where the sequestered material becomes degraded (Fig. 1). The resulting macromolecules are released from the lysosome and can then be used for the maintenance of vital cellular functions during conditions of cellular stress, for example starvation, hypoxia, and infection. Autophagy was originally characterized as a nonspecific pathway involving random sequestration of cytoplasmic components, but it is now evident that cargo, such as aberrant protein aggregates, organelles, and bacteria can also be selectively and exclusively incorporated into autophagosomes. Autophagy is known to be important to meet amino acid–poor growth conditions during developmental processes and has been implicated in several human diseases, including neurodegeneration and cancer (for review see Mizushima et al., 2008).
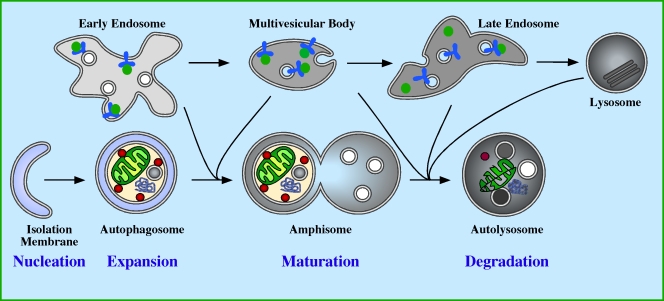
Figure 1.
Autophagy in mammalian cells. The autophagic pathway comprises four stages: nucleation, expansion, maturation, and degradation. Nucleation occurs in response to a signal transmitted downstream of a cellular stress, causing the isolation membrane (IM) to start to grow. The IM is also known as the phagophore (Seglen and Bohley, 1992). Expansion of the IM occurs, enabling it to grow in all dimensions sequestering cytosolic proteins, organelles, and aggregated proteins. Expansion is complete when the membrane closes to form the double membrane autophagosome vesicle. After closure of the autophagosome, maturation begins by fusion with endocytic compartments, including early endosomes, multivesicular bodies, late endosomes, and lysosomes. The step-wise, or individual fusion events of the autophagosome with the endocytic compartment create an amphisome that has both autophagosomal and endosomal content. During maturation the lumen of the amphisome acidifies, and the membrane acquires LAMPs (lysosmomal membrane proteins), hydrolytic enzymes, and lipases. This leads to the formation of an autolysosome, within which the sequestered content is degraded and recycled back to the cytosol.
The molecular mechanisms underlying the process of autophagy have been extensively studied in yeast using genetic screens to identify autophagy-related (Atg) mutants (Klionsky et al., 2003). Inactivation of Atg orthologues in higher eukaryotes has revealed that the autophagic machinery is highly conserved. 31 Atg genes have been identified in yeast, 18 of which encode core components of the machinery (Atg1-10, Atg12-14, Atg16-18, Atg29, and Atg31) involved in starvation-induced biogenesis of autophagosomes. These core Atg proteins can be divided into four subgroups: (1) Atg1/unc-51–like kinase (ULK) and their regulators; (2) Vps34/class III phosphatidylinositol 3-phosphate (PI3P) kinase complex I (PI3K complex I); (3) the Atg9/mAtg9 cycling complex; and (4) the ubiquitin-like proteins Atg12 and Atg8/LC3 and their conjugation systems. Most of these proteins are recruited to the yeast preautophagosomal structure (PAS), the autophagosome assembly site in yeast, as well as to the expanding IM in mammalian cells. In addition to these core components, some other Atg proteins are required for cargo-specific autophagy pathways, such as the cytoplasm-to-vacuole targeting (Cvt) pathway and pexophagy, which sequester aminopeptidase I and peroxisomes, respectively, in yeast (for review see Klionsky, 2005).
Autophagic activity has been found to be regulated by PI 3-kinases (PI3K), which phosphorylate phosphatidylinositol (PtdIns) or phosphoinositides (PIs, phosphorylated derivatives of PtdIns) at the 3-position of the inositol ring. The mammalian class I PI3K and its product PI(3,4,5)P3 have been found to inhibit autophagy through activation of Akt/PKB, which activates mTOR (mammalian target of rapamycin) kinase—the major negative regulator of autophagy. However, the class III PI3K (PI3KC3) and its product PI3P are required for autophagy (for review see Lindmo and Stenmark, 2006). For a comprehensive overview of the regulation and function of Vps34 in the mTOR pathway, see Backer (2008). PIK3C3 is the orthologue of yeast Vps34 protein, the only PI3K found in yeast. Vps34 was identified in a yeast vacuolar protein sorting (Vps) screen, providing the first evidence that PI3P is involved in membrane trafficking (Odorizzi et al., 2000). Later, it was found that Vps34, its regulatory subunit Vps15, as well as the two accessory proteins Atg14 and Vps30/Atg6, form a complex and are all required for autophagy in yeast (Kihara et al., 2001). This complex is referred to as the PI3K complex I, as Vps34–Vps15–Vps30/Atg6 also can engage in a complex with Vps38 (PI3K complex II), known to play a role in the endosomal Vps pathway (for review see Klionsky, 2005). The nomenclature PI3K complex I and II is also used to describe the mammalian Vps34 class III complexes that contain Atg14L, and Vps38, respectively, and should not be confused with the mammalian class I and class II PI3Ks. Most recently there has been an explosion of new information about the mammalian Vps34 complexes, and as the lipid kinase activity of Vps34 appears to be one of the most essential requirements for autophagy, these recent results provide important insight into autophagy.
In this review, we will discuss the present knowledge about the mechanisms involved in formation and maturation of autophagosomes, with particular emphasis on the role of the Vps34 complexes and how this pathway communicates with other cellular vesicular trafficking pathways.
Origin of the autophagosome membrane
Despite the incredible advances in understanding the functions of autophagy and the identification of many Atg proteins over the last decade, neither the source of the IM nor the mechanism of its formation is known. The origin of the IM has been a question of intense debate ever since the first morphological description of autophagosomes in the early 1960s and two models for autophagosome formation have prevailed. The first model suggests that the autophagosomal membrane originates from a preexisting organelle, whereas the second model implies de novo formation from localized lipid synthesis. Although in yeast, all autophagosomes apparently arise from a single PAS localized close to the vacuole, it appears in mammalian cells that they can be generated anywhere in the cytoplasm. There is no evidence for a PAS in mammalian cells and the IM could either be derived from a structure equivalent to the yeast PAS, or in fact be the PAS itself.
Various organelles, such as the ER, the Golgi complex, and the plasma membrane, have been suggested as IM sources. However, whereas compartment-specific markers have been detected in early autophagosomes, others have failed to detect them (for review see Reggiori, 2006). Although these discrepancies could be explained by different experimental approaches and techniques used in the various laboratories, currently it cannot be excluded that autophagosomes arise from and/or contain membranes derived from more than one organelle. However, the fact that the autophagosome membrane seems to be devoid of transmembrane proteins (Fengsrud et al., 2000) indicates that these membranes differ structurally from other subcellular organelles, supporting the de novo formation model. Thus, if the IMs arise from preexisting sources, integral membrane components must be actively segregated away from the lipid bilayers used to form autophagosomes or be rapidly retrieved from the expanding IM.
Recently, it was found that PI3P-enriched structures (named omegasomes due to their shape) form in close proximity to ER membranes in response to amino acid starvation (Axe et al., 2008). These structures also contain the autophagy-specific proteins LC3/Atg8 and Atg5, and newly formed autophagosomes seem to emanate from the omegasome (Fig. 2). The fact that ER-targeted PI3P-binding proteins, even transmembrane proteins, translocate to these structures upon starvation strongly suggests that the omegasomes are being formed from ER membranes and might indicate that the ER could form a platform for PI3P synthesis and autophagosome formation. The ER contains little PI3P under normal conditions, and thus the Vps34 complex must be recruited specifically to the omegasomes in response to starvation to initiate autophagosome formation. This is an intriguing mechanism, but several questions remain to be answered: what is the mechanism involved in activation and specific recruitment of the Vps34 complex to the ER/omegasome? Which PI3K complex is responsible for PI3P formation in the ER? Which PI3P effector protein(s) are involved? How can the double-membrane autophagosome vesicles bud off from the ER?
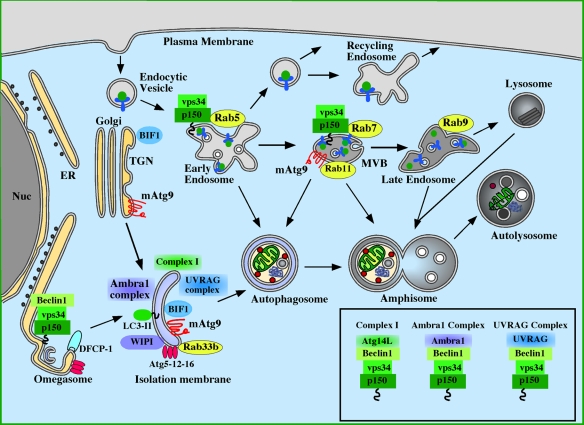
Figure 2.
Molecules involved in the induction of autophagy: Atg proteins, Rab proteins, and the class III PI3 kinase. The source of the isolation membrane is unknown, but has been proposed to be derived from the endoplasmic reticulum (ER) or trans-Golgi network (TGN). The omegasome is continuous with the ER and is also proposed to contribute to the IM formation. Class III PI3K complex I (p150–Vps34–Beclin1–Atg14L) is localized to the IM and is required for autophagy (Itakura et al., 2008; Sun et al., 2008; Matsunaga et al., 2009; Zhong et al., 2009). A subset of this complex (p150-Vps34) is required for endocytosis and is found on early endosomes and multivesicular bodies (MVBs), and interacts with Rab5 and Rab7. A Beclin1-containing subcomplex is required for function of the omegasome and the recruitment of DFCP-1 (Axe et al., 2008). Recent data also point to two additional p150–Vps34–Beclin1 complexes, Ambra1 and UVRAG complexes, being required for autophagy. Ambra1 associates directly with Beclin1 and is required for induction of autophagy (Fimia et al., 2007). UVRAG likewise associates directly with Beclin1, and is required for autophagy (Liang et al., 2008). BIF-1, previously shown to be required for fission of Golgi carriers (Yang et al., 2006), associates with UVRAG and has been shown to be required for autophagy (Takahashi et al., 2007). The activity of the PI3K Vps34 is enhanced by interaction with Rab5, Rab7, and Beclin1-associated UVRAG together with BIF-1, but it is not known how Ambra1 association with Beclin1 affects the activity of the PI3K. The composition of these complexes is shown in the box.
Although clues to some of these questions exist, the exact mechanisms remain to be elucidated. The small GTPase Rab5, known to form a complex with Vps34 and Beclin1, was shown to be necessary for autophagosome formation (Ravikumar et al., 2008) and to be required for formation of peripheral ER tubules in Caenorhabditis elegans (Audhya et al., 2007). Thus, Rab5 might play a role in formation of ER-associated omegasomes and recruitment of the Vps34–Beclin1 complex to initiate PI3P production and autophagosome formation. Recently, it was found that the Salmonella typhimurium phosphoinositide phosphatase SopB is responsible for recruitment of Rab5 and Vps34 to the bacteria-containing vacuole (Mallo et al., 2008) and that Atg14L, a member of the PI3K complex I, is required for autophagy-mediated elimination of these bacteria (Sun et al., 2008). Thus, one might speculate that a similar Rab5-dependent mechanism is involved in formation of autophagic membranes from the omegasome as in sequestration of the bacteria. As Rab5 interacts with the Vps15 orthologue p150, the regulatory protein kinase component of the Vps34 complex (Christoforidis et al., 1999), and PI3P is found at early endocytic membranes (Gillooly et al., 2000), it cannot be excluded that this machinery is delivered to the forming autophagosomes by fusion of early endosomes with the IM. However, the fact that Atg14L is not found on early endosomes (Matsunaga et al., 2009) suggests an autophagy-specific role of Rab5. It will be interesting to see whether Atg14L, as well as other Beclin1-interacting proteins, are recruited to the omegasomes.
Molecular machinery involved in autophagosome formation
Regardless of the origin of the IM membrane and the many questions that still remain unanswered, our knowledge of the basic molecular machinery involved in autophagosome formation has increased dramatically during the last decade. The process of autophagosome formation can be divided in three steps; nucleation, expansion, and closure of the IM membrane (see Fig. 1).
Nucleation
Although the exact order of recruitment of the Atg proteins and other factors required for the nucleation step of autophagy still is not completely understood, the Atg1/ULK kinase and the PI3K complex I could be considered the two most important for nucleation.
Atg1/ ULK and their regulators.
The yeast serine/threonine kinase Atg1, which exists in a complex with other Atg proteins, including Atg13 and Atg17, is known to function downstream of TOR kinase. The mammalian homologue of Atg1, ULK1, is required for induction of autophagy. ULK1 and the closely related ULK2 are found in a complex with Atg13, and FIP200, the mammalian homologue of Atg17 (Chan et al., 2009; Chang and Neufeld, 2009; Hara and Mizushima, 2009; Hosokawa et al., 2009; Jung et al., 2009). Recently a new mammalian Atg protein, Atg101, was also found to interact with the ULK1–mAtg13–FIP200 complex in an Atg13-dependent manner (Mercer et al., 2009). These recent data demonstrate the regulation of the mammalian complex is significantly different from yeast and has been recently reviewed (Chan and Tooze, 2009). Yeast Atg1 complex (Reggiori et al., 2004a) and ULK1 and Atg13 are also required for the cycling of Atg9 (Young et al., 2006; Chan et al., 2009), the only multi-spanning membrane protein in the core autophagy machinery.
Vps34/class III PI3K complex I.
The yeast PI3K complex I is required for induction of autophagy. Mammalian homologues of Vps34, Vps15, and Vps30/Atg6 are called Vps34, p150, and Beclin1, respectively (see Fig. 2). The mammalian orthologue of yeast Atg14 was recently identified by several groups and was called Atg14-like protein (Atg14L) (Itakura et al., 2008) or Barkor (Sun et al., 2008), hereafter referred to as Atg14L. Atg14L is a 55-kD protein with 15% overall identity to yeast Atg14. Both proteins have coiled-coil regions that are important for their interaction with Beclin1 and Vps34. As in yeast, Atg14L was found to exist in complex with Vps34, p150, and Beclin1 and to be excluded from the mammalian equivalent of complex II, which contains UVRAG (ultraviolet irradiation resistant-associated gene). Depletion of Atg14L inhibited recruitment of Atg16L and LC3 to IMs, and autophagosome formation (Itakura et al., 2008; Matsunaga et al., 2009; Zhong et al., 2009). Under nutrient-rich conditions, ectopically expressed Atg14L was observed in a diffuse or reticular pattern, which partly colocalized with ER markers, but was recruited to Atg16L (IM) (described below) and LC3-positive (autophagosomes) structures during starvation. Interestingly, Atg14L can localize to Atg16-positive puncta independently of Vps34 and Beclin1, as well as when the Vps34 activity is inhibited (Itakura et al., 2008; Sun et al., 2008). Moreover, overexpressed Atg14L was found to stimulate the kinase activity of Vps34 (Zhong et al., 2009), whereas Atg14L-depleted cells showed reduced PI3P production (Sun et al., 2008). In addition, the levels of Atg14L influence the stability of both Beclin1 and Vps34 (Itakura et al., 2008). Collectively, these data suggest that Atg14L functions to mediate recruitment of the PI3K complex I to the IM where it can stimulate its activity, which is essential for the initial step of autophagosome formation. Thus, understanding how Atg14L is localized to the IM may be the key to understanding the dynamics of PI3P production at the IM in response to various cellular autophagy-inducing signals.
Other Beclin1-associated proteins.
In recent studies UVRAG was found to be excluded from the Atg14L-containing complex I (Itakura et al., 2008; Sun et al., 2008; Matsunaga et al., 2009; Zhong et al., 2009) and to compete with Atg14L for binding to the coiled-coil region of Beclin1 (Sun et al., 2008; Matsunaga et al., 2009) and the N-terminal C2 domain of Vps34 (Itakura et al., 2008). In fact, UVRAG is likely to function as the putative Vps38 homologue in the PI3K complex II (discussed below). However, UVRAG has also been reported to positively regulate autophagosome formation; ectopic expression of UVRAG in a colon cancer cell line with a monoallelic UVRAG mutation increased the levels of LC3-II (described below) and GFP-LC3 puncta (Liang et al., 2006). Interestingly, UVRAG has been found to interact with BIF-1, also known as endophilin B1 (Takahashi et al., 2007). BIF-1 was shown to be required for autophagy and to colocalize with GFP-Atg5 and GFP-LC3 on the surface of autophagosomes. This exciting result suggests that recruitment of BIF-1 via UVRAG may provide the machinery to deform membranes, as BIF-1 has an N-BAR domain, known to bind membranes and cause them to undergo curvature (Itoh and De Camilli, 2006). Interestingly, BIF-1 is also required for COPI (coat protein complex I) vesicle budding via interaction with ARF-GAP, the GTPase-activating protein (GAP) for the small GTPase ARF, thus providing additional support for its role in membrane deformation (Yang et al., 2005). Finally, BIF-1 was shown to colocalize with mammalian Atg9 (Takahashi et al., 2008). However, further work is required to clarify the role of UVRAG because Itakura et al. (2008) did not detect a role for UVRAG in autophagy. As Atg14L and UVRAG clearly exist in different PI3K complexes, it is possible that the Atg14L-containing complex I regulates nucleation, whereas the UVRAG-BIF-1 complex is involved in membrane expansion and curvature.
Beclin1 could be considered to be the master regulator of the Vps34 complex specificity. In addition to binding to Vps34, Atg14L, and UVRAG, as described above (and see Fig. 2), Beclin1 also interacts with Ambra1 (activating molecule in Beclin1-regulated autophagy) (Fimia et al., 2007). Ambra1, which contains WD repeats and binds directly to Beclin1, is a positive regulator of Beclin1 function and autophagy, but its exact functional mechanism is still not clear. The activity of Beclin1 is also negatively regulated by interaction with the anti-apoptotic protein Bcl-2 in normal conditions (Pattingre et al., 2005). Recently, it was found that starvation induces phosphorylation of Bcl-2 by the stress-activated signaling molecule, Jun N-terminal protein kinase 1 (JNK1), leading to dissociation of Bcl-2 from Beclin1 and induction of autophagy (Wei et al., 2008).
IM expansion and closure
Atg12 and Atg8/LC3 and their conjugation systems.
Studies in yeast and mammalian cells have shown that elongation of the IM requires the ubiquitin-like proteins Atg12 and Atg8/LC3 and their respective conjugation machineries. Atg12 is conjugated to Atg5 by the combined action of the E1- and E2-like enzymes Atg7 and Atg10, respectively. The Atg5-12 conjugate then associates with Atg16L, followed by homodimerization of the trimeric complex to form a large multimeric complex called the Atg16L complex. Atg8, as well as its mammalian homologue LC3, is conjugated to phosphatidylethanolamine (PE), a major component of cellular membranes, through the action of Atg7 and the E2-like protein Atg3. Before LC3 lipidation, the cytosolic form of LC3 (LC3-I) is cleaved by the cysteine protease Atg4. The lipidated form of LC3 (LC3-II) is the most widely used marker to study autophagy, as it remains membrane bound throughout the pathway. However, Atg4 can also mediate removal of LC3-II from the autophagosome membrane, and this has been found to be critical for fusion of autophagosomes with endosomes/lysosomes (for review see Klionsky, 2005).
Recent data indicate that there is a complex interrelationship between the Atg16L complex and the LC3 conjugation machinery. The Atg16L complex was found to be required for specific recruitment of LC3 to the sites of autophagosome formation (Fujita et al., 2008). In fact, ectopic expression of Atg16L containing a plasma membrane targeting motif (C-terminal prenylation motif, CAAX) caused the redistribution and lipidation of LC3 on the plasma membrane. The Atg5–Atg12 conjugate has thus been proposed to act as an E3 ligase for the Atg8–PE conjugation and thereby directly regulate LC3 lipidation (Hanada et al., 2007). Vice versa, the LC3 conjugation machinery seems to be important for formation of the Atg16L complex: in Atg3−/− mouse embryonic fibroblasts (MEFs) which have no LC3-II, less conjugation of Atg12 to Atg5 was observed (Sou et al., 2008). Interestingly, autophagosomes, but no autolysosomes, were formed in response to starvation; however, the autophagosomes were generally much smaller and often open and multilamellar, indicating that LC3 lipidation is critical for IM closure. This is supported by in vitro data showing that Atg8–PE mediates membrane tethering and fusion (Nakatogawa et al., 2007), suggesting that it can catalyze the IM elongation process and maybe also its final closure.
Finally, localization of the Atg12 and Atg8/LC3 proteins and conjugation machinery to the PAS and the IM, as well as IM elongation, has also been found to depend on Atg9 and the PI3K complex I (for review see Longatti and Tooze, 2009). The formation of aberrant IMs in Atg3−/− MEFs was sensitive to the PI3K inhibitor wortmannin (Sou et al., 2008), indicating that PI3P formation is a prerequisite for recruitment of the Atg16L complex to sites of autophagosome formation.
PI3P effector proteins.
Why is Vps34 activity, and by extension PI3P production, required for recruitment of the Atg12 and LC3 conjugation proteins to the PAS/IM and for IM expansion? Several PI3P-binding proteins (called PI3P effectors) are known, and most of them have been localized to early endosomes where PI3P is highly enriched; however, autophagic PI3P effectors have also been described. Most PI3P effectors contain one of the two known PI3P-binding domains; the FYVE (for conserved in Fab1, YOTB, Vac1, and EEA1) zinc finger domain and the conserved Phox-homology (PX) domain (for review see Lindmo and Stenmark, 2006), although also PI3P effector proteins lacking either of these domains are know to bind PI3P.
The PI3P-binding protein DFCP1 (double FYVE domain containing protein 1), which normally localizes to ER and Golgi membranes, was found to translocate to the omegasomes in a PI3P-dependent manner upon starvation (see Fig. 2). DFCP1 overexpression was found to inhibit autophagy, probably by sequestering PI3P and thereby preventing recruitment of autophagic PI3P effector proteins. However, DFCP1 itself seemed not required for autophagosome formation (Axe et al., 2008).
A second PI3P effector candidate protein is a mammalian homologue of yeast Atg18, WD-repeat protein interacting with phosphoinositides (WIPI)-49, also called WIPI-1, which is a member of a family of four proteins called WIPI-1–4 (Jeffries et al., 2004; Proikas-Cezanne et al., 2004). WIPI-1 is recruited to autophagic membranes in a PI3P-dependent manner (Proikas-Cezanne et al., 2004). However, WIPI-1 is also localized to both endosomal and Golgi membranes, where it regulates the trafficking of proteins involved in the mannose-6-phosphate receptor recycling pathway (Jeffries et al., 2004). Yeast Atg18p, together with its homologues Atg21p and Ygr223c, form a novel family of PI3P-binding proteins. Atg21p is required for the yeast-specific Cvt pathway (Strømhaug et al., 2004), whereas Ygr223c recently was found to play a role in the autophagic process micronucleophagy (also called piecemeal microautophagy of the nucleus, PMN, during which portions of the nucleus are sequestered and degraded) (Krick et al., 2008a,b). These proteins were all found to localize also to endosomes in a Vps34 complex II–dependent manner. Interestingly, Atg18 is known to regulate the activity of the PI3P 5-kinase Fab1/PIKfyve, generating PI(3,5)P2 on late endosomes (Efe et al., 2007), and Fab1 was found to be required for maturation of autolysosomes (Rusten et al., 2007). Thus, Atg18 and its mammalian homologues might work as PI3P sensors, tightly regulating cellular PI3P levels and thereby autophagic activity.
Clearly, further work is required to understand the role of DFCP1 and the Atg18/WIPI family in the crucial PI3P-dependent steps in autophagosome formation. Other autophagic PI3P effector proteins probably exist, due to the complexity of autophagy and the essential role of Vps34 in this process. Such proteins are likely to be revealed in the near future, as specific PI3P-binding domains (FYVE and PX) allow us to predict with a certain degree of confidence the most likely PI3P-binding proteins encoded by the human genome.
Very recently the PI3P phosphatase Jumpy (also known as MTMR14) was found to negatively regulate autophagosome formation (Vergne et al., 2009). Jumpy localizes to Atg16L-positive membranes and its depletion was found to increase both basal and starvation-induced autophagy, as well as accumulation of WIPI-1 on autophagic membranes. In contrast, ectopic expression of Jumpy inhibited autophagy and increased the association of mAtg9 with autophagic membranes. Interestingly, expression of a Jumpy mutant with a defect phosphatase activity, corresponding to a mutation found in patients with centronuclear myopathy, did not down-regulate autophagy. Thus, negative regulation of autophagic PI3P levels clearly has important physiological functions.
In conclusion, formation of autophagosomes is a tightly regulated process that involves a complex hierarchical interrelationship of the Atg and Vps proteins in which PI3K complex I plays a key role, but the full molecular details of how the IM expands and closes remain to be understood. Another open question is whether similar mechanisms are involved in IM expansion during selective autophagy, for example protein aggregate–mediated autophagy, as the autophagy-inducing signals during selective autophagy are likely to be different than signals inducing autophagy downstream of amino acid starvation.
Molecular regulation of autophagosome maturation
The process of autophagy culminates at the maturation step when the nascent autophagosomes fuse with the endosomal/lysosomal system to create a fully functional degradative compartment, the autolysosome (Fig. 3). Maturation into an autolysosome may occur by a single- or multi-step fusion process. Single-step fusion of autophagosomes with lysosomes would generate a fully degradative autolysosome, which seems to be the normal process in yeast, but evidence that this occurs in mammalian cells is lacking. Thus, it has been proposed that maturation occurs by sequential fusions with different endosomal populations (Dunn, 1990).
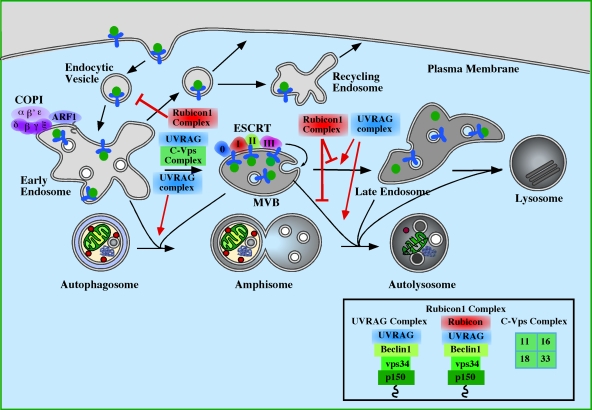
Figure 3.
The role of endosomes and Class III PI3-kinase complexes in autophagosome maturation. Maturation of autophagosomes has been shown to require functional early endosomes, multivesicular bodies (MVBs) and late endosomes. COPI is important for early steps in endocytosis and for autophagosome maturation (Razi et al., 2009). Similarly, the ESCRT complexes participate in autophagosome maturation and later steps of endocytosis (Raiborg and Stenmark, 2009). Loss of either COPI or ESCRT results in the accumulation of autophagosomes and amphisomes. Loss of the Class III PI3K subcomplex p150-Vps34 (see Fig. 2) would also be predicted to have a similar inhibition of maturation as it is required to maintain endosomal function on Rab5- and Rab7-positive endosomes. Of the Class III PI3K complexes, the UVRAG complex, described in Fig. 2 and the Rubicon complex (p150–Vps34–Beclin1–UVRAG–Rubicon) regulate autophagosome maturation (Matsunaga et al., 2009; Zhong et al., 2009). UVRAG, required for endosome function, positively regulates maturation and autophagy, while the association of Rubicon to UVRAG causes an inhibition of autophagic maturation. Note: Rubicon complex also reduces the activity of the Class III PI3K. The composition of the complexes is shown in the box. UVRAG can also associate with the core class C–Vps complex, containing Vps11, 16, 18, and 33, independently of Beclin1, and this interaction accelerates autophagy (Liang et al., 2008), perhaps though stimulation of Rab7 activity.
Compared with the formation process, more is known about maturation of autophagosomes. However, among the many questions about maturation, two are relevant here: first, when is the autophagosome ready to undergo fusion with endosomes; and second, how does fusion with the endosomal/lysosomal system occur? As mentioned above, loss of the Atg16L complex and LC3-II from the surface of the autophagosome appears to be one requirement for fusion of autophagosomes with endosomes, but the composition of the fusion-ready autophagosome is not known. In particular, is the PI3K still active on the autophagosome? And is PI3P present on the surface of the autophagosome? Recent data in yeast would suggest that there is no PI3P on the outer surface of the closed autophagosomes, and in fact PI3P is accumulated on the inner surface (Obara and Ohsumi, 2008), but this remains to be examined in mammalian cells. Thus, whereas PI3P and its effector(s) clearly are directly involved in autophagosome formation, their role in autophagosome maturation is likely to be indirect, through recruitment of the endocytic machinery required to obtain functional endocytic compartments that can fuse with autophagosomes to form amphisomes. Significant progress in understanding this process has been made recently by exploiting alterations of the machinery involved in the endocytic pathway, either through siRNA-mediated protein depletion or from genetic models. In addition, recent knowledge about the class III PI3K complex II (Vps34, p150, Beclin1, and UVRAG) has also advanced our understanding of the maturation process, and highlights the close intimacy between the autophagic and endocytic pathways.
The endosomal pathway functions to internalize into cells nutrients and growth factors bound to their cognate receptors, thereby mediating cell survival, as well as transmitting signals to control growth and differentiation. Briefly, the endocytic pathway is comprised of early compartments having a neutral pH, and that can be identified by the association of the small GTPase Rab5. Whereas the nutrient receptors partition into vesicles that recycle to the plasma membrane, growth factor receptors become internalized into the intraluminal vesicles of multivesicular bodies (MVBs). These mature into or later fuse with more acidic compartments that are positive for Rab7, and finally the receptors are degraded in the lysosome (see Figs. 2 and 3). This entire process requires the action of the class III PI3K, as well as the COPI, ESCRT, and HOPS complexes. Other information about the molecules involved in the fusion event (SNARE proteins and Rab proteins, for example) will not be discussed, as they have been recently reviewed elsewhere (Longatti and Tooze, 2009).
COPI.
Coat protein complex I (COPI) is required for ER–Golgi transport (Lee et al., 2004), and maintenance of endosomal/lysosomal function, which has been shown to depend on three of the seven coatomer subunits, α-, β-, and ϵ-COP, of the COP coat (Whitney et al., 1995; Guo et al., 1996; Daro et al., 1997; Gu et al., 1997; Styers et al., 2008). More recently, in yeast COPI has been shown to be required for endosomal transport to the vacuole, similar to the class E yeast vacuolar protein sorting (Vps) mutants involved in MVB formation, and indeed ϵ-COP has been shown to interact with Vps27/Hrs, suggesting COPI recruitment may also be sensitive to PI3P levels (Gabriely et al., 2007). siRNA-mediated depletion of coatomer subunits α, β, and β′ results in the accumulation of GFP-LC3–positive autophagosomes, which are not degradative (Razi et al., 2009). These autophagosomes are not acidic, but are Golgi (TGN46) and LAMP-2 (lysosomal membrane protein 2) positive, and a small percent also contain early endocytic marker EEA1 (early endosomal autoantigen 1) and an internalized fluid phase marker, Rhodamine dextran, demonstrating they are amphisomes that contain early endosomal markers. Furthermore, loss of COPI results in the accumulation of p62/SQSTM1 (an LC3-binding protein sequestered into autophagosomes) and ubiquitin-positive vesicular structures, and p62 and ubiquitin-positive amphisomes. As expected from its role in ER–Golgi transport, COPI depletion disrupts the Golgi complex, but the effect on autophagy is due solely to the disruption of its function on the early endosome. Although the accumulation of p62- and Ub-labeled structures is similar to the phenotype observed in mouse models defective in autophagy, it remains to be determined if alterations in autophagy due to loss of COPI have physiological consequences, similar to that observed with loss of ESCRTs (see below) in neurodegenerative diseases. Finally, as mentioned above, the UVRAG-associated protein BIF-1 is also required for COPI vesicle budding via interaction with ARF-GAP (Yang et al., 2005). Thus, the role of COPI on the endosome, and in autophagosome maturation, might be related to the interaction of UVRAG/BIF-1 with ARF-GAP.
ESCRTs.
The endosomal sorting complex required for transport (ESCRT), first characterized as vacuolar protein sorting (Vps) mutants in yeast (Babst et al., 1998), consists of four multi-protein subcomplexes (ESCRT-0, -I, -II, and -III) that are required for MVB formation and sorting of ubiquitinated integral membrane receptors into the intraluminal vesicles of the MVB (see Fig. 3). PI3P production in the endosomal membrane is essential for membrane recruitment of the ESCRTs; the ESCRT-0 complex is recruited to endosomal membranes through the binding of the FYVE domain–containing protein Vps27p/Hrs to PI3P in the endosomal membrane. Moreover, PI3P binding of the ESCRT-II subunit Vps36p/EAP45 is important for its function in vacuolar sorting, suggesting that PI3P also contributes to the endosomal recruitment of ESCRT-II. The ESCRT complexes dissociate from the endosomal membrane by the action of the AAA ATPase Vps4/SKD-1 upon formation of an intraluminal vesicle (for review see Raiborg and Stenmark, 2009). Loss of any of the ESCRT components or Vps4/SKD-1 results in an inhibition of MVB formation and retention of the internalized receptor on the surface of the endosome, which has severe consequences on the duration of the growth factor signaling pathway which might cause cancer (Haglund et al., 2007).
Studies in nematodes, flies, and mammals have revealed that several ESCRT subunits are essential for autophagy (Rusten and Stenmark, 2009). Deletion of ESCRT-0 (Hrs), ESCRT-I (Vps28/Tsg101), ESCRT-II (Vps22, Vps25), ESCRT-III (Vps2B, Vps24, Vps32), or Vps4 (AAA-ATPase) was generally found to result in accumulation of nondegradative autophagosomes and sometimes accumulation of cytoplasmic protein aggregates containing ubiquitinated proteins and p62. The fact that overexpression of an ESCRT-III subunit mutant, Vps2B/CHMP2BIntron5, corresponding to a mutation found in patients with frontotemporal dementia (FTD) in cortical neurons of mice (Lee et al., 2007) or in HeLa cells (Filimonenko et al., 2007) caused a similar phenotype, implies that ESCRT-mediated control of autophagy is physiologically important. Thus, a functional pool of MVBs seems to be essential for autophagosome maturation by providing target organelles for fusion. However, it cannot be entirely excluded that loss of ESCRT function resulted in a closure defect, and that expanded but not closed IMs accumulated. In fact, the membrane topology of a closing autophagosome is the same as that involving the abscission of an inward-budding vesicle on the endosome, abscission of a virus from the plasma membrane, and of two cells during the final stage of cytokinesis, all known to require the ESCRT machinery. Thus, one might speculate that components of the ESCRT machinery, as well as Vps4, may be responsible for final closure of autophagosomes (for review see Longatti and Tooze, 2009; Raiborg and Stenmark, 2009). However, the fact that deletion of ESCRT proteins in yeast does not seem to affect autophagy (Epple et al., 2003; Reggiori et al., 2004b) argues against this hypothesis and in favor of the endosome fusion defect hypothesis, as yeast autophagosomes fuse directly with the yeast vacuole. One can also not rule out the possibility that the increased number of autophagosomes found in ESCRT-depleted cells in part is due to elevated autophagosome formation, as JNK, which induces autophagy, was found to become activated in fly tissue lacking functional ESCRT subunits (Herz et al., 2006; Rodahl et al., 2009).
The evidence demonstrating that COPI and ESCRT are both required for autophagy supports the stepwise fusion model. Fusion of autophagosomes with early endosomes may allow delivery of proton pumps and LAMPs, whereas subsequent fusion with MVBs may provide mannose-6-phosphate receptors, which bind and deliver lysosomal enzymes to acidic late endosomes. In addition, fusion of autophagosomes with the early endosome would provide the fused amphisome with proteins of the early endosomes, including, but not only SNARE proteins, thus facilitating recognition of the autophagosome by the late endosome, and likewise further fusion with late endosomes would facilitate recognition with lysosomes. One could hypothesize that this mechanism provides an increase in specificity and efficiency of fusion during autophagosome maturation.
PI3K complex II and C-Vps complexes.
The Vps34 PI3K and its adaptor p150 have previously been found to interact both with the early endosome GTPase Rab5 (Christoforidis et al., 1999) and with late endosomal Rab7 (Stein et al., 2005). It is known that PI3P production on early endosomes is essential for recruitment of PI3P-binding proteins to early endosomes (e.g., EEA1 and ESCRT subunits) and thus for endosome fusion and MVB formation. What is not known is whether Rab5- and Rab7-associated Vps34 exist in a complex containing Beclin1 and any of its interacting partners, but it is likely that the PI3K complex II, known to regulate endocytic trafficking, also can engage in a complex with Rab5 or Rab7.
UVRAG, proposed to be the putative Vps38 orthologue, was found in a complex with Vps34, p150, and Beclin1 (Itakura et al., 2008; Sun et al., 2008; Matsunaga et al., 2009; Zhong et al., 2009). A role of UVRAG as a complex II subunit is supported by recent data showing that UVRAG colocalizes with Rab9-positive endosomes (Itakura et al., 2008) and interacts with the core class C Vps complex (C-Vps; see Fig. 3), containing the proteins Vps11, Vps16, Vps18, and Vps33 (Liang et al., 2008). This complex, together with the subunits Vps39/Vam6 and Vps41, make up the HOPS (homotypic vacuole fusion and protein sorting) complex, known to regulate tethering and fusion of endosomes to the yeast vacuole, as well as lysosomal delivery and autophagosome maturation in Drosophila (Sevrioukov et al., 1999; Pulipparacharuvil et al., 2005; Lindmo et al., 2006). HOPS is an established GEF for Rab 7 (Wurmser et al., 2000), but also interacts with Rab 5 and has been found to be required for Rab 5-to-Rab 7 conversion (Rink et al., 2005). However, Vps38 also functions in the retromer-dependent transport pathway in yeast (Burda et al., 2002), a role so far not attributed to UVRAG.
Another C-Vps endosomal tethering complex, CORVET (class C core vacuole/endosome tethering), containing Vps3 and Vps8 in addition to the core subunits, was found to interact with the yeast Rab GTPase Vps21 (Rab5) (Peplowska et al., 2007). Interestingly, the CORVET and HOPS complexes can also interconvert, and two additional intermediate complexes were found. The C-Vps complexes are known to couple Rab activation and SNARE assembly during endosome fusion (Seals et al., 2000). Thus, it is likely that modular assembled tethering complexes, regulated by Vps34–Beclin1 complexes and Rab proteins, define organelle biogenesis in the endocytic as well as in the autophagic pathway.
In addition, a separate stable Vps34–Vps15–Beclin1–UVRAG complex also containing the protein Rubicon (RUN domain and cysteine-rich domain containing, Beclin1-interacting protein) was identified (Matsunaga et al., 2009; Zhong et al., 2009) (see Fig. 3). In contrast to Atg14L, Rubicon was found to reduce Vps34 activity and inhibit autophagy; although depletion of Rubicon caused increased autophagy and endocytosis, ectopic expression of Rubicon resulted in aberrant late endosomal/lysosomal structures and impaired autophagosome maturation. It seems likely that the PI3K complex II previously found at early and late endosomes is also regulated by Rubicon, as depletion of Rubicon accelerates EGFR degradation (Matsunaga et al., 2009). Rubicon appears to colocalize both with early (EEA1-positive), MVB (LBPA-positive), and late endosomes (Lamp1) (Matsunaga et al., 2009; Zhong et al., 2009). Thus, its expression level might regulate autophagy at multiple steps, starting from fusion with early endosomes. This is supported by the findings that the loss of Rubicon drove the formation of GFP-LC3-LAMP1–positive autolysosomes (Matsunaga et al., 2009). The fact that Rubicon contains a RUN domain, a domain found in several proteins involved in regulation of Ras-like GTPase activity (e.g., Rab proteins), makes it tempting to speculate that Rubicon could be a Rab effector protein which might regulate the activity of endocytic Rab proteins.
In conclusion, a burst of recent papers has provided evidence that several mammalian Vps34–Beclin1 complexes exist that all seem to closely regulate the autophagic pathway, either at the early stage of autophagosome formation and/or at later stages when the autophagosomes mature by fusion with endocytic compartments. The localization and activity of the Vps34 complexes seems to be tightly interconnected with other vesicular trafficking components, in particular Rab proteins, the ESCRT machinery, and C-Vps complexes. Future studies will likely elucidate the intricate network of intracellular compartments whose function is modulated and manipulated by key regulators, such as PI3K, to respond to amplification of stress–response pathways such as autophagy.
Footnotes
Abbreviations used in this paper:
- Atg
- autophagy related
- COPI
- coat protein complex I
- ESCRT
- endosomal sorting complex required for transport
- IM
- isolation membrane
- MVB
- multivesicular body
- PAS
- preautophagosomal structure
- PI3P
- phosphatidylinositol 3-phosphate
- ULK
- unc-51–like kinase
- UVRAG
- ultraviolet irradiation resistant–associated gene
References
- Audhya A., Desai A., Oegema K. 2007. A role for Rab5 in structuring the endoplasmic reticulum.J. Cell Biol. 178:43–56 doi:10.1083/jcb.200701139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., Ktistakis N.T. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum.J. Cell Biol. 182:685–701 doi:10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E.J., Emr S.D. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function.EMBO J. 17:2982–2993 doi:10.1093/emboj/17.11.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer J.M. 2008. The regulation and function of Class III PI3Ks: novel roles for Vps34.Biochem. J. 410:1–17 doi:10.1042/BJ20071427 [DOI] [PubMed] [Google Scholar]
- Burda P., Padilla S.M., Sarkar S., Emr S.D. 2002. Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase.J. Cell Sci. 115:3889–3900 doi:10.1242/jcs.00090 [DOI] [PubMed] [Google Scholar]
- Chan E.Y., Tooze S.A. 2009. Evolution of Atg1 function and regulation.Autophagy. 5:758–765 [DOI] [PubMed] [Google Scholar]
- Chan E.Y., Longatti A., McKnight N.C., Tooze S.A. 2009. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism.Mol. Cell. Biol. 29:157–171 doi:10.1128/MCB.01082-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.-Y., Neufeld T.P. 2009. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation.Mol. Biol. Cell. 20:2004–2014 doi:10.1091/mbc.E08-12-1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis S., Miaczynska M., Ashman K., Wilm M., Zhao L., Yip S.-C., Waterfield M.D., Backer J.M., Zerial M. 1999. Phosphatidylinositol-3-OH kinases are Rab5 effectors.Nat. Cell Biol. 1:249–252 doi:10.1038/12075 [DOI] [PubMed] [Google Scholar]
- Daro E., Sheff D., Gomez M., Kreis T., Mellman I. 1997. Inhibition of endosome function in CHO cells bearing a temperature-sensitive defect in the coatomer (COPI) component epsilon-COP.J. Cell Biol. 139:1747–1759 doi:10.1083/jcb.139.7.1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W.A., Jr. 1990. Studies on the mechanisms of autophagy: maturation of the autophagic vacuole.J. Cell Biol. 110:1935–1945 doi:10.1083/jcb.110.6.1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efe J.A., Botelho R.J., Emr S.D. 2007. Atg18 regulates organelle morphology and Fab1 kinase activity independent of its membrane recruitment by phosphatidylinositol 3,5-bisphosphate.Mol. Biol. Cell. 18:4232–4244 doi:10.1091/mbc.E07-04-0301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple U.D., Eskelinen E.L., Thumm M. 2003. Intravacuolar membrane lysis in Saccharomyces cerevisiae. Does vacuolar targeting of Cvt17/Aut5p affect its function? J. Biol. Chem. 278:7810–7821 doi:10.1074/jbc.M209309200 [DOI] [PubMed] [Google Scholar]
- Fengsrud M., Erichsen E.S., Berg T.O., Raiborg C., Seglen P.O. 2000. Ultrastructural characterization of the delimiting membranes of isolated autophagosomes and amphisomes by freeze-fracture electron microscopy.Eur. J. Cell Biol. 79:871–882 doi:10.1078/0171-9335-00125 [DOI] [PubMed] [Google Scholar]
- Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerød L., Fisher E.M., Isaacs A., Brech A., Stenmark H., Simonsen A. 2007. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease.J. Cell Biol. 179:485–500 doi:10.1083/jcb.200702115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia G.M., Stoykova A., Romagnoli A., Giunta L., Di Bartolomeo S., Nardacci R., Corazzari M., Fuoco C., Ucar A., Schwartz P., et al. 2007. Ambra1 regulates autophagy and development of the nervous system.Nature. 447:1121–1125 [DOI] [PubMed] [Google Scholar]
- Fujita N., Hayashi-Nishino M., Fukumoto H., Omori H., Yamamoto A., Noda T., Yoshimori T. 2008. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure.Mol. Biol. Cell. 19:4651–4659 doi:10.1091/mbc.E08-03-0312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriely G., Kama R., Gerst J.E. 2007. Involvement of specific COPI subunits in protein sorting from the late endosome to the vacuole in yeast.Mol. Cell. Biol. 27:526–540 doi:10.1128/MCB.00577-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly D.J., Morrow I.C., Lindsay M., Gould R., Bryant N.J., Gaullier J.M., Parton R.G., Stenmark H. 2000. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells.EMBO J. 19:4577–4588 doi:10.1093/emboj/19.17.4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu F., Aniento F., Parton R.G., Gruenberg J. 1997. Functional dissection of COP-I subunits in the biogenesis of multivesicular endosomes.J. Cell Biol. 139:1183–1195 doi:10.1083/jcb.139.5.1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q., Penman M., Trigatti B.L., Krieger M. 1996. A single point mutation in epsilon-COP results in temperature-sensitive, lethal defects in membrane transport in a Chinese hamster ovary cell mutant.J. Biol. Chem. 271:11191–11196 doi:10.1074/jbc.271.19.11191 [DOI] [PubMed] [Google Scholar]
- Haglund K., Rusten T.E., Stenmark H. 2007. Aberrant receptor signaling and trafficking as mechanisms in oncogenesis.Crit. Rev. Oncog. 13:39–74 [DOI] [PubMed] [Google Scholar]
- Hanada T., Noda N.N., Satomi Y., Ichimura Y., Fujioka Y., Takao T., Inagaki F., Ohsumi Y. 2007. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy.J. Biol. Chem. 282:37298–37302 doi:10.1074/jbc.C700195200 [DOI] [PubMed] [Google Scholar]
- Hara T., Mizushima N. 2009. Role of ULK-FIP200 complex in mammalian autophagy: FIP200, a counterpart of yeast Atg17? Autophagy. 5:85–87 [DOI] [PubMed] [Google Scholar]
- Herz H.M., Chen Z., Scherr H., Lackey M., Bolduc C., Bergmann A. 2006. vps25 mosaics display non-autonomous cell survival and overgrowth, and autonomous apoptosis.Development. 133:1871–1880 doi:10.1242/dev.02356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S.-i., Natsume T., Takehana K., Yamada N., et al. 2009. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy.Mol. Biol. Cell. 20:1981–1991 doi:10.1091/mbc.E08-12-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi C., Inoue K., Mizushima N. 2008. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG.Mol. Biol. Cell. 19:5360–5372 doi:10.1091/mbc.E08-01-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., De Camilli P. 2006. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature.Biochim. Biophys. Acta. 1761:897–912 [DOI] [PubMed] [Google Scholar]
- Jeffries T.R., Dove S.K., Michell R.H., Parker P.J. 2004. PtdIns-specific MPR pathway association of a novel WD40 repeat protein, WIPI49.Mol. Biol. Cell. 15:2652–2663 doi:10.1091/mbc.E03-10-0732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C.H., Jun C.B., Ro S.-H., Kim Y.-M., Otto N.M., Cao J., Kundu M., Kim D.-H. 2009. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery.Mol. Biol. Cell. 20:1992–2003 doi:10.1091/mbc.E08-12-1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Noda T., Ishihara N., Ohsumi Y. 2001. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae.J. Cell Biol. 152:519–530 doi:10.1083/jcb.152.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J. 2005. The molecular machinery of autophagy: unanswered questions.J. Cell Sci. 118:7–18 doi:10.1242/jcs.01620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Cregg J.M., Dunn W.A., Jr., Emr S.D., Sakai Y., Sandoval I.V., Sibirny A., Subramani S., Thumm M., Veenhuis M., Ohsumi Y. 2003. A unified nomenclature for yeast autophagy-related genes.Dev. Cell. 5:539–545 doi:10.1016/S1534-5807(03)00296-X [DOI] [PubMed] [Google Scholar]
- Krick R., Henke S., Tolstrup J., Thumm M. 2008a. Dissecting the localization and function of Atg18, Atg21 and Ygr223c.Autophagy. 4:896–910 [DOI] [PubMed] [Google Scholar]
- Krick R., Muehe Y., Prick T., Bremer S., Schlotterhose P., Eskelinen E.L., Millen J., Goldfarb D.S., Thumm M. 2008b. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes.Mol. Biol. Cell. 19:4492–4505 doi:10.1091/mbc.E08-04-0363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.A., Beigneux A., Ahmad S.T., Young S.G., Gao F.B. 2007. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration.Curr. Biol. 17:1561–1567 doi:10.1016/j.cub.2007.07.029 [DOI] [PubMed] [Google Scholar]
- Lee M.C., Miller E.A., Goldberg J., Orci L., Schekman R. 2004. Bi-directional protein transport between the ER and Golgi.Annu. Rev. Cell Dev. Biol. 20:87–123 doi:10.1146/annurev.cellbio.20.010403.105307 [DOI] [PubMed] [Google Scholar]
- Liang C., Feng P., Ku B., Dotan I., Canaani D., Oh B.-H., Jung J.U. 2006. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG.Nat. Cell Biol. 8:688–699 doi:10.1038/ncb1426 [DOI] [PubMed] [Google Scholar]
- Liang C., Lee J.S., Inn K.-S., Gack M.U., Li Q., Roberts E.A., Vergne I., Deretic V., Feng P., Akazawa C., Jung J.U. 2008. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking.Nat. Cell Biol. 10:776–787 doi:10.1038/ncb1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindmo K., Stenmark H. 2006. Regulation of membrane traffic by phosphoinositide 3-kinases.J. Cell Sci. 119:605–614 doi:10.1242/jcs.02855 [DOI] [PubMed] [Google Scholar]
- Lindmo K., Simonsen A., Brech A., Finley K., Rusten T.E., Stenmark H. 2006. A dual function for Deep orange in programmed autophagy in the Drosophila melanogaster fat body.Exp. Cell Res. 312:2018–2027 doi:10.1016/j.yexcr.2006.03.002 [DOI] [PubMed] [Google Scholar]
- Longatti A., Tooze S.A. 2009. Vesicular trafficking and autophagosome formation.Cell Death Differ. 16:956–965 doi:10.1038/cdd.2009.39 [DOI] [PubMed] [Google Scholar]
- Mallo G.V., Espina M., Smith A.C., Terebiznik M.R., Alemán A., Finlay B.B., Rameh L.E., Grinstein S., Brumell J.H. 2008. SopB promotes phosphatidylinositol 3-phosphate formation on Salmonella vacuoles by recruiting Rab5 and Vps34.J. Cell Biol. 182:741–752 doi:10.1083/jcb.200804131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K., Saitoh T., Tabata K., Omori H., Satoh T., Kurotori N., Maejima I., Shirahama-Noda K., Ichimura T., Isobe T., et al. 2009. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages.Nat. Cell Biol. 11:385–396 doi:10.1038/ncb1846 [DOI] [PubMed] [Google Scholar]
- Mercer C.A., Kaliappan A., Dennis P.B. 2009. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy.Autophagy. 5:649–662 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A.M., Klionsky D.J. 2008. Autophagy fights disease through cellular self-digestion.Nature. 451:1069–1075 doi:10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H., Ichimura Y., Ohsumi Y. 2007. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion.Cell. 130:165–178 doi:10.1016/j.cell.2007.05.021 [DOI] [PubMed] [Google Scholar]
- Obara K., Ohsumi Y. 2008. Dynamics and function of PtdIns(3)P in autophagy.Autophagy. 4:952–954 [DOI] [PubMed] [Google Scholar]
- Odorizzi G., Babst M., Emr S.D. 2000. Phosphoinositide signaling and the regulation of membrane trafficking in yeast.Trends Biochem. Sci. 25:229–235 doi:10.1016/S0968-0004(00)01543-7 [DOI] [PubMed] [Google Scholar]
- Pattingre S., Tassa A., Qu X., Garuti R., Liang X.H., Mizushima N., Packer M., Schneider M.D., Levine B. 2005. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy.Cell. 122:927–939 doi:10.1016/j.cell.2005.07.002 [DOI] [PubMed] [Google Scholar]
- Peplowska K., Markgraf D.F., Ostrowicz C.W., Bange G., Ungermann C. 2007. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis.Dev. Cell. 12:739–750 doi:10.1016/j.devcel.2007.03.006 [DOI] [PubMed] [Google Scholar]
- Proikas-Cezanne T., Waddell S., Gaugel A., Frickey T., Lupas A., Nordheim A. 2004. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy.Oncogene. 23:9314–9325 doi:10.1038/sj.onc.1208331 [DOI] [PubMed] [Google Scholar]
- Pulipparacharuvil S., Akbar M.A., Ray S., Sevrioukov E.A., Haberman A.S., Rohrer J., Krämer H. 2005. Drosophila Vps16A is required for trafficking to lysosomes and biogenesis of pigment granules.J. Cell Sci. 118:3663–3673 doi:10.1242/jcs.02502 [DOI] [PubMed] [Google Scholar]
- Raiborg C., Stenmark H. 2009. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins.Nature. 458:445–452 doi:10.1038/nature07961 [DOI] [PubMed] [Google Scholar]
- Ravikumar B., Imarisio S., Sarkar S., O'Kane C.J., Rubinsztein D.C. 2008. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease.J. Cell Sci. 121:1649–1660 doi:10.1242/jcs.025726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razi M., Chan E.Y., Tooze S.A. 2009. Early endosomes and endosomal coatomer are required for autophagy.J. Cell Biol. 185:305–321 doi:10.1083/jcb.200810098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F. 2006. 1. Membrane origin for autophagy.Curr. Top. Dev. Biol. 74:1–30 doi:10.1016/S0070-2153(06)74001-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Tucker K.A., Stromhaug P.E., Klionsky D.J. 2004a. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure.Dev. Cell. 6:79–90 doi:10.1016/S1534-5807(03)00402-7 [DOI] [PubMed] [Google Scholar]
- Reggiori F., Wang C.W., Nair U., Shintani T., Abeliovich H., Klionsky D.J. 2004b. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae.Mol. Biol. Cell. 15:2189–2204 doi:10.1091/mbc.E03-07-0479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J., Ghigo E., Kalaidzidis Y., Zerial M. 2005. Rab conversion as a mechanism of progression from early to late endosomes.Cell. 122:735–749 doi:10.1016/j.cell.2005.06.043 [DOI] [PubMed] [Google Scholar]
- Rodahl L.M., Haglund K., Sem-Jacobsen C., Wendler F., Vincent J.P., Lindmo K., Rusten T.E., Stenmark H. 2009. Disruption of Vps4 and JNK function in Drosophila causes tumour growth.PLoS One. 4:e4354 doi:10.1371/journal.pone.0004354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten T.E., Stenmark H. 2009. How do ESCRT proteins control autophagy? J. Cell Sci. 122:2179–2183 doi:10.1242/jcs.050021 [DOI] [PubMed] [Google Scholar]
- Rusten T.E., Vaccari T., Lindmo K., Rodahl L.M., Nezis I.P., Sem-Jacobsen C., Wendler F., Vincent J.P., Brech A., Bilder D., Stenmark H. 2007. ESCRTs and Fab1 regulate distinct steps of autophagy.Curr. Biol. 17:1817–1825 doi:10.1016/j.cub.2007.09.032 [DOI] [PubMed] [Google Scholar]
- Seals D.F., Eitzen G., Margolis N., Wickner W.T., Price A. 2000. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion.Proc. Natl. Acad. Sci. USA. 97:9402–9407 doi:10.1073/pnas.97.17.9402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen P.O., Bohley P. 1992. Autophagy and other vacuolar protein degradation mechanisms.Experientia. 48:158–172 doi:10.1007/BF01923509 [DOI] [PubMed] [Google Scholar]
- Sevrioukov E.A., He J.P., Moghrabi N., Sunio A., Krämer H. 1999. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila.Mol. Cell. 4:479–486 doi:10.1016/S1097-2765(00)80199-9 [DOI] [PubMed] [Google Scholar]
- Sou Y.-S., Waguri S., Iwata J.-I., Ueno T., Fujimura T., Hara T., Sawada N., Yamada A., Mizushima N., Uchiyama Y., et al. 2008. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice.Mol. Biol. Cell. 19:4762–4775 doi:10.1091/mbc.E08-03-0309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M.P., Cao C., Tessema M., Feng Y., Romero E., Welford A., Wandinger-Ness A. 2005. Interaction and functional analyses of human VPS34/p150 phosphatidylinositol 3-kinase complex with Rab7.Methods Enzymol. 403:628–649 doi:10.1016/S0076-6879(05)03055-7 [DOI] [PubMed] [Google Scholar]
- Strømhaug P.E., Reggiori F., Guan J., Wang C.W., Klionsky D.J. 2004. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy.Mol. Biol. Cell. 15:3553–3566 doi:10.1091/mbc.E04-02-0147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styers M.L., O'Connor A.K., Grabski R., Cormet-Boyaka E., Sztul E. 2008. Depletion of beta-COP reveals a role for COP-I in compartmentalization of secretory compartments and in biosynthetic transport of caveolin-1.Am. J. Physiol. Cell Physiol. 294:C1485–C1498 doi:10.1152/ajpcell.00010.2008 [DOI] [PubMed] [Google Scholar]
- Sun Q., Fan W., Chen K., Ding X., Chen S., Zhong Q. 2008. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase.Proc. Natl. Acad. Sci. USA. 105:19211–19216 doi:10.1073/pnas.0810452105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Coppola D., Matsushita N., Cualing H.D., Sun M., Sato Y., Liang C., Jung J.U., Cheng J.Q., Mulé J.J., et al. 2007. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis.Nat. Cell Biol. 9:1142–1151 doi:10.1038/ncb1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Meyerkord C.L., Wang H.G. 2008. BARgaining membranes for autophagosome formation: Regulation of autophagy and tumorigenesis by Bif-1/Endophilin B1.Autophagy. 4:121–124 [DOI] [PubMed] [Google Scholar]
- Vergne I., Roberts E., Elmaoued R.A., Tosch V., Delgado M.A., Proikas-Cezanne T., Laporte J., Deretic V. 2009. Control of autophagy initiation by phosphoinositide 3-phosphatase jumpy.EMBO J. 28:2244–2258 doi:10.1038/emboj.2009.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Sinha S., Levine B. 2008. Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation.Autophagy. 4:949–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney J.A., Gomez M., Sheff D., Kreis T.E., Mellman I. 1995. Cytoplasmic coat proteins involved in endosome function.Cell. 83:703–713 doi:10.1016/0092-8674(95)90183-3 [DOI] [PubMed] [Google Scholar]
- Wurmser A.E., Sato T.K., Emr S.D. 2000. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion.J. Cell Biol. 151:551–562 doi:10.1083/jcb.151.3.551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.S., Lee S.Y., Spanò S., Gad H., Zhang L., Nie Z., Bonazzi M., Corda D., Luini A., Hsu V.W. 2005. A role for BARS at the fission step of COPI vesicle formation from Golgi membrane.EMBO J. 24:4133–4143 doi:10.1038/sj.emboj.7600873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.S., Zhang L., Lee S.Y., Gad H., Luini A., Hsu V.W. 2006. Key components of the fission machinery are interchangeable.Nat. Cell Biol. 8:1376–1382 doi:10.1038/ncb1503 [DOI] [PubMed] [Google Scholar]
- Young A.R.J., Chan E.Y.W., Hu X.W., Köchl R., Crawshaw S.G., High S., Hailey D.W., Lippincott-Schwartz J., Tooze S.A. 2006. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes.J. Cell Sci. 119:3888–3900 doi:10.1242/jcs.03172 [DOI] [PubMed] [Google Scholar]
- Zhong Y., Wang Q.J., Li X., Yan Y., Backer J.M., Chait B.T., Heintz N., Yue Z. 2009. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex.Nat. Cell Biol. 11:468–476 doi:10.1038/ncb1854 [DOI] [PMC free article] [PubMed] [Google Scholar]