Brain-specific Drp1 knockout mice demonstrate that Drp1-mediated organelle division is important for development, mitochondrial morphogenesis, and apoptosis.
Abstract
The dynamin-related guanosine triphosphatase Drp1 mediates the division of mitochondria and peroxisomes. To understand the in vivo function of Drp1, complete and tissue-specific mouse knockouts of Drp1 were generated. Drp1-null mice die by embryonic day 11.5. This embryonic lethality is not likely caused by gross energy deprivation, as Drp1-null cells showed normal intracellular adenosine triphosphate levels. In support of the role of Drp1 in organelle division, mitochondria formed extensive networks, and peroxisomes were elongated in Drp1-null embryonic fibroblasts. Brain-specific Drp1 ablation caused developmental defects of the cerebellum in which Purkinje cells contained few giant mitochondria instead of the many short tubular mitochondria observed in control cells. In addition, Drp1-null embryos failed to undergo developmentally regulated apoptosis during neural tube formation in vivo. However, Drp1-null embryonic fibroblasts have normal responses to apoptotic stimuli in vitro, suggesting that the apoptotic function of Drp1 depends on physiological cues. These findings clearly demonstrate the physiological importance of Drp1-mediated organelle division in mice.
Introduction
Mitochondria are highly dynamic organelles that continuously fuse and divide within cells (Karbowski and Youle, 2003; Osteryoung and Nunnari, 2003; Rube and van der Bliek, 2004; Okamoto and Shaw, 2005; Anesti and Scorrano, 2006; Chan, 2006; McBride et al., 2006; Cerveny et al., 2007a; Hoppins et al., 2007). A balance between mitochondrial fusion and division is critical for the maintenance of normal mitochondrial structures and cellular physiology. For example, defects in mitochondrial fusion typically result in mitochondrial fragmentation as a result of excessive division. However, defects in mitochondrial division produce highly connected mitochondrial tubules as a result of excessive mitochondrial fusion. The importance of a balance between fusion and division in the regulation of organelle morphogenesis is highlighted by the observations that mitochondria lacking both fusion and division functions have wild-type morphology (Sesaki and Jensen, 1999).
Normal mitochondrial dynamics are critical for human health and disease (Knott and Bossy-Wetzel, 2008; Mattson et al., 2008). Mitochondrial fusion defects cause human neuropathies, including autosomal dominant optic atrophy and Charcot-Marie-Tooth disease type 2A (Alexander et al., 2000; Delettre et al., 2000; Züchner et al., 2004). Abnormalities in division have also been implicated in Charcot-Marie-Tooth type 4A, Parkinson's disease, Alzheimer's disease, and abnormal brain development (Cuesta et al., 2002; Waterham et al., 2007; Deng et al., 2008; Poole et al., 2008; Yang et al., 2008; Cho et al., 2009). The neurodegeneration associated with these diseases suggests that neuronal function and survival depend on mitochondrial fusion and division, possibly due to a high demand for ATP and specialized cellular morphologies (Knott and Bossy-Wetzel, 2008).
A dynamin-related GTPase, Dnm1p (yeast)/Drp1 (mammals), functions in mitochondrial division (Bleazard et al., 1999; Labrousse et al., 1999; Sesaki and Jensen, 1999; Smirnova et al., 1999) and distribution (Otsuga et al., 1998; Cerveny et al., 2007b). Studies have suggested that Dnm1p/Drp1 self-assembles into spirals that wrap around mitochondria and pinches off their membranes as a mechanochemical enzyme (Yoon et al., 2001; Ingerman et al., 2005). Alternatively, Dnm1p/Drp1 may function as a classical regulatory GTPase (Fukushima et al., 2001). Most Drp1 is cytosolic and is recruited to the surface of mitochondria by proteins such as Fis1p/Fis1, Mdv1p, Caf4p, and Num1p (Mozdy et al., 2000; Tieu and Nunnari, 2000; Cerveny et al., 2001; James et al., 2003; Yoon et al., 2003: Griffin et al., 2005; Cerveny et al., 2007b). The association of Drp1 with mitochondria and its activity are regulated by phosphorylation, ubiquitination, and sumoylation (Santel and Frank, 2008). Drp1 is also involved in peroxisome division, indicating that a common mechanism exists for mitochondrial and peroxisomal division (Li and Gould, 2003; Koch et al., 2004; Kobayashi et al., 2007). In addition, previous studies have shown that Drp1 regulates apoptosis; however, its exact role in apoptosis remains to be determined (Frank et al., 2001; Germain et al., 2005; Jagasia et al., 2005; Parone et al., 2006; Abdelwahid et al., 2007; Goyal et al., 2007; Breckenridge et al., 2008; Cassidy-Stone et al., 2008; James and Martinou, 2008; Suen et al., 2008). The RNAi knockdown of Drp1 was examined in Caenorhabditis elegans and revealed an important role in embryonic development (Labrousse et al., 1999). However, the physiological role of Drp1 in mitochondrial division has not been studied in mammals. In this study, we describe for the first time the characterization of complete and brain-specific Drp1 knockout mice. Our data demonstrate the physiological importance of Drp1-mediated organelle division for embryonic development, brain development, mitochondrial morphogenesis, and developmentally regulated apoptosis.
Results and discussion
Drp1 deletion causes embryonic lethality in mice
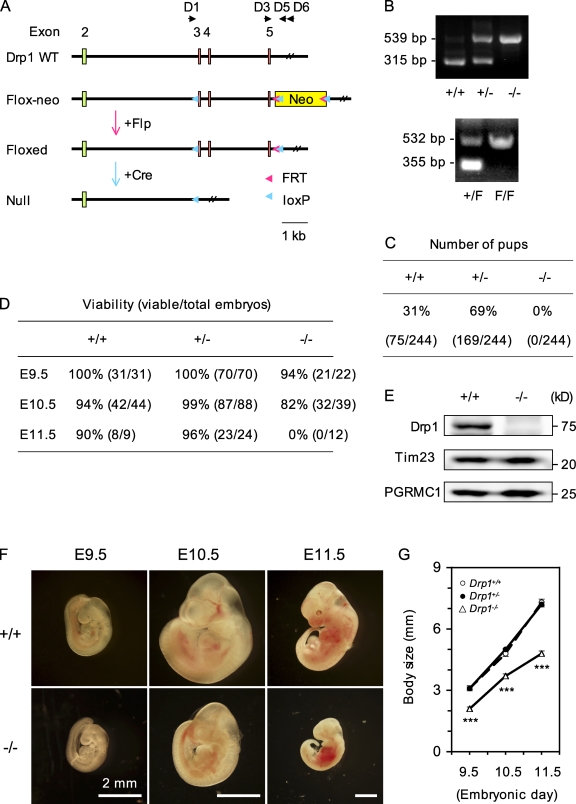
To investigate the in vivo function of Drp1, we generated complete and conditional mouse knockouts as described in Materials and methods (Fig. 1). To examine the effect of complete loss of Drp1, we analyzed the genotype of 244 progeny from crosses of heterozygous mice. Drp1+/− heterozygous mice express a partially reduced level of Drp1 (∼75% of wild-type level; Fig. S1) and are viable, fertile, normal in size, and do not display any gross physical or behavioral abnormalities. None of the progeny were homozygous for deletion of Drp1 (Fig. 1 C). These results suggest that homozygous deletion of Drp1 leads to embryonic lethality. To determine how loss of Drp1 affects embryonic development, we dissected embryos of various stages from pregnant female mice after timed breeding. We found Drp1−/− embryos at approximately expected frequencies between embryonic days (E) 9.5 and 11.5 (Fig. 1 D). However, the viability of embryos, as determined by the presence of a beating heart, was compromised in Drp1−/− embryos. At E11.5, we found no viable Drp1−/− embryos, and most showed internal hemorrhage (Fig. 1 F). In addition, Drp1−/− embryos appeared smaller than littermate Drp1+/+ and Drp1+/− embryos, suggesting a developmental delay (Fig. 1, F and G). Loss of Drp1 expression was confirmed in whole embryos by immunoblotting (Fig. 1 E). Thus, Drp1 is essential for embryonic development in mice.
Figure 1.
Drp1 is required for embryonic mouse development. (A) Design of Drp1 knockout alleles. WT, wild type. (B) Confirmation of knockout by PCR. Genomic DNAs were prepared from embryos, yolk sack, or tails of adult mice for PCR. To distinguish Drp1+/+, Drp1+/−, and Drp1−/−, D1, D3, and D5 were used. Wild-type and null alleles produced 315-bp and 539-bp PCR products, respectively. For floxed alleles, D3 and D6 were used. Wild-type (+) and floxed (F) alleles generated 355-bp and 532-bp PCR products, respectively. (C) Genotypes of offspring from breeding of Drp1+/− mice. (D) Number of viable embryos. (E) Immunoblotting of embryos. (F) Images of embryos. (G) Crown rump length of embryos. ***, P < 0.001 (n ≥ 14).
Drp1−/− embryos are defective in placental development and show decreased cardiomyocyte beat rates
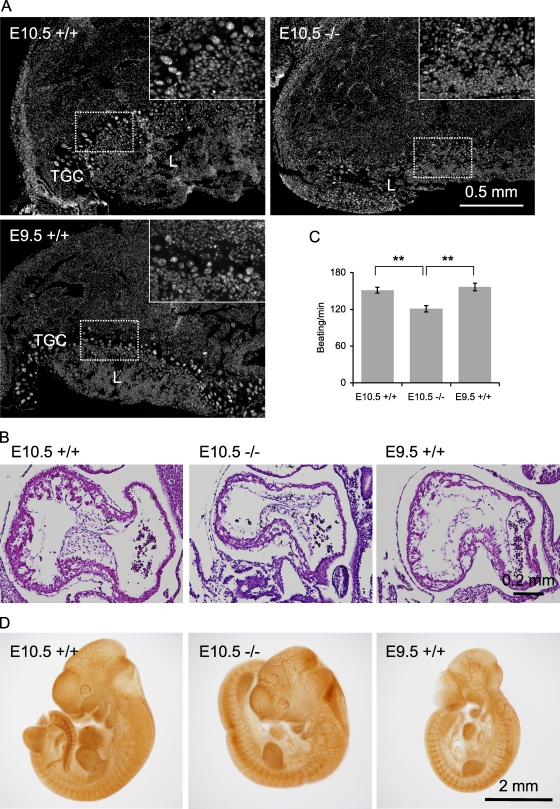
To determine the causes of embryonic lethality at E11.5, we examined the placenta, heart, and angiogenesis, which are important for embryo survival at this developmental stage. Strikingly, E10.5 Drp1−/− embryos lacked the trophoblast giant cell layer in the placenta as shown by DAPI staining (Fig. 2 A). To confirm that this was not simply caused by developmental delays, we also examined E9.5 Drp1+/+ embryos, which were size matched to the E10.5 Drp1−/− embryos, and observed trophoblast giant cell layers. These results clearly show that Drp1 is required for placental development and suggest that a defective exchange of nutrients and oxygen between the mother and fetus is likely involved in the embryonic lethality of Drp1−/− mice. Also, the heart was clearly beating in E9.5–10.5 Drp1−/− embryos, and in histological analysis of serial sagittal sections, the architecture of the heart appeared normal (Fig. 2 B). However, cardiomyocytes isolated from E10.5 Drp1−/− embryos had decreased beating rates relative to those isolated from E9.5 and 10.5 Drp1+/+ embryos (Fig. 2 C). This suggests that heart defects also contribute to the lethality of Drp1−/− embryos. Finally, we examined angiogenesis in whole embryos by immunohistochemistry using antibodies to PECAM and found indistinguishable patterns of blood vessels in Drp1+/+ and Drp1−/− embryos (Fig. 2 D).
Figure 2.
Characterization of Drp1−/− embryos. (A) DAPI-stained sections of placenta of age-matched E10.5 Drp1+/+ and Drp1−/− embryos and an E9.5 Drp1+/+ embryo, which was size matched to the E10.5 Drp1−/− embryos. Insets show enlarged images of boxed regions. Trophoblast giant cell (TGC) and labyrinth (L) layers are indicated. (B) H&E stains of sagittal sections of embryonic heart. (C) Beat rates in isolated embryonic cardiomyocytes were measured using differential interference contrast microscopy. (D) Whole-mount immunohistochemistry of embryos with anti-PECAM antibodies. **, P < 0.01 (n ≥ 39). Error bars indicate mean ± SEM.
The timing of Drp1−/− embryonic lethality is similar to the timing of lethality in mice defective in Mfn1, Mfn2, and Opa1, which are all necessary for mitochondrial fusion (Chen et al., 2003; Alavi et al., 2007; Davies et al., 2007). This suggests that proper mitochondrial dynamics are critical for embryonic development in mice. For example, like Drp1-null mice, Mfn2-null mice lack trophoblast giant cell layers (Chen et al., 2003). Although Drp1 controls the division of mitochondria and peroxisomes, it is likely that the embryonic lethality of Drp1−/− mice results from defects in mitochondrial division rather than peroxisomal defects because mice lacking Pex11 β, a peroxisomal protein specifically required for peroxisomal division, die after birth (Li et al., 2002).
Defects in mitochondrial division and peroxisomal shape in Drp1−/− mouse embryonic fibroblasts (MEFs)
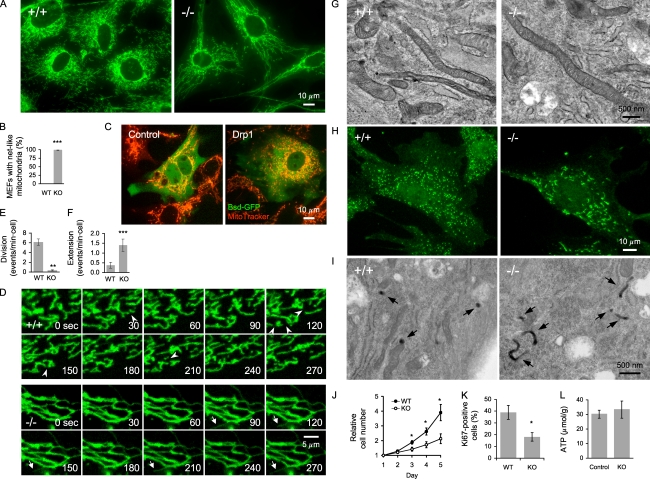
To examine mitochondrial shape, we isolated MEFs from E10.5 embryos, immunostained them using anti-Tom20 antibodies, and performed a quantitative analysis (Fig. 3, A and B). To exclude the possibility that transformation affects the characteristics of MEFs, we used nonimmortalized MEFs in all experiments. In Drp1+/+ MEFs, mitochondria formed short tubules with occasional branches. In contrast, most Drp1−/− MEFs contained highly connected, elongated mitochondrial tubules that could be rescued by reexpression of Drp1 (Fig. 3, A and C). This morphology confirms that Drp1 is required for mitochondrial division. To directly examine mitochondrial division, we used time-lapse microscopy of MEFs expressing matrix-targeted Su9-GFP (Fig. 3 D and Videos 1 and 2; Eura et al., 2003). The frequency of mitochondrial division was decreased by 17-fold in Drp1−/− MEFs compared with Drp1+/+ MEFs (Fig. 3 D). However, there appeared to be residual mitochondrial division in Drp1−/− MEFs (Fig. 3 E). Time-lapse microscopy also showed that the frequency of extension and retraction of mitochondria tubules with free ends at the edge of mitochondrial networks was increased by approximately threefold in Drp1−/− MEFs (Fig. 3, D and F). To analyze the ultrastructure of mitochondria, we used EM and showed that mitochondria were elongated, whereas the inner membrane cristae appeared normal in Drp1−/− MEFs (Fig. 3 G). This is in sharp contrast to the dramatic alterations in the cristae membrane structures observed in mitochondria defective in membrane fusion (Olichon et al., 2003; Griparic et al., 2004; Chen et al., 2007).
Figure 3.
Characterization of Drp1−/− MEFs. (A) MEFs were immunostained with anti-Tom20 antibodies. (B) Quantitation of MEFs with netlike mitochondria (n ≥ 3; >100 cells/experiment). (C) Drp1−/− MEFs carrying pTracer-EF/Bsd (control; Invitrogen) and pTracer-EF/Bsd-Drp1 were stained with MitoTracker (Invitrogen). (D) Frames from time-lapse confocal microscopy (Videos 1 and 2). Arrowheads indicate mitochondrial division, and arrows show extension and retraction of mitochondrial tubules. (E and F) Quantitation of mitochondrial division (E) and extension (F; n ≥ 8). (G) EM of mitochondria in MEFs. (H) Peroxisomal morphology. MEFs were immunostained using anti-Pex14 antibodies. (I) EM of peroxisomes in MEFs. Peroxisomes were identified using cytochemical staining for catalase with DAB (arrows). (J) Cell proliferation (n ≥ 9). (K) Quantitation of Ki67-positive MEFs (n = 6; >100 cells/experiment). (L) Intracellular ATP levels in control (Drp1+/+ and Drp1+/−) and Drp1−/− MEFs (n ≥ 6). WT, wild type; KO, knockout. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars indicate mean ± SEM.
We examined whether the loss of Drp1 affected the levels of other proteins involved in mitochondrial fusion (Mfn1, Mfn2, and Opa1) and division (Fis1). Levels of Mfn1 and 2, but not Opa1 or Fis1, were decreased by ∼50% in Drp1−/− MEFs (Fig. S1). It would be interesting to test whether these changes in protein levels alter mitochondrial fusion to compensate for the changes in mitochondrial morphology.
We also examined the shapes of peroxisomes by immunofluorescence microscopy using antibodies to Pex14, a peroxisomal membrane protein (Will et al., 1999), and EM of peroxisomal catalase staining (Angermüller and Fahimi, 1981). The peroxisomes were relatively round in Drp1+/+ MEFs, whereas Drp1−/− MEFs contained elongated peroxisomes (Fig. 3, H and I; and Fig. S2), indicating that defects occurred in peroxisomal division, which supports previous observations (Koch et al., 2004; Schrader, 2006; Kobayashi et al., 2007). Therefore, our data show that Drp1 controls both mitochondrial and peroxisomal division.
Normal intracellular ATP levels in Drp1−/− MEFs
We found that Drp1−/− MEFs grew slower than Drp1+/+ MEFs and showed reduced levels of Ki67, a marker for cell proliferation (Fig. 3, J and K). Consistent with these results, Drp1−/− embryonic stem (ES) cells have been shown to be defective in normal cell proliferation (Ishihara et al., 2009). To determine whether the loss of Drp1 affected cellular energy levels, we measured ATP levels in control (Drp1+/+ and Drp1+/−) and Drp1−/− MEFs using a luciferase-based assay. The ATP levels were comparable in these cells (Fig. 3 L). Therefore, Drp1 was not required for the maintenance of intracellular ATP levels, and the reduced cell proliferation rates of Drp1−/− MEFs and embryonic lethality in Drp1−/− mice were not caused by reduced ATP levels. Interestingly, Drp1+/+ and Drp1−/− MEFs grew similarly when they were transformed and immortalized using the SV40 large T antigen (Ishihara et al., 2009). These results suggest that Drp1−/− MEFs, which are compromised in cell proliferation, can be compensated by alternations in gene expression induced by transformation (Ahuja et al., 2005).
Roles of Drp1 in apoptosis in vivo and in vitro
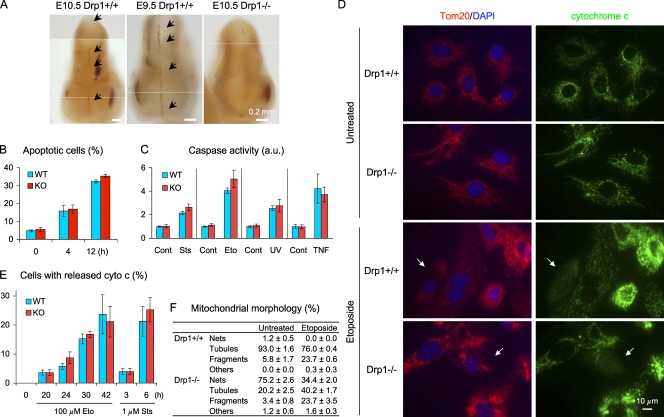
Different roles of Drp1 in apoptosis have been suggested. Some studies showed that Drp1 regulates the mitochondrial outer membrane permeability to release cytochrome c independent of mitochondrial division (Frank et al., 2001; Germain et al., 2005; Cassidy-Stone et al., 2008; James and Martinou, 2008; Suen et al., 2008). Other studies showed that Drp1 only has a minor role in apoptosis (Parone et al., 2006; Breckenridge et al., 2008; Sheridan et al., 2008). To determine whether Drp1 regulates apoptosis in vivo, we examined apoptosis in neural tube formation during development (Fig. 4 A). It has been shown that cells located at the site of closure of neural tubes undergo apoptosis (Kuan et al., 1999; Papaioannou and Behringer, 2005). We isolated embryos at E9.5 and 10.5 and immunostained them for activated caspase-3 (Fig. 4 A). As expected, in Drp1+/+ embryos, cells located at the site of neural tube closure were positive for activated caspase-3. Conversely, Drp1−/− embryos showed considerably weaker signals and decreased numbers of caspase-3–positive cells, demonstrating that Drp1 facilitates developmentally regulated apoptosis during neural tube formation.
Figure 4.
Differential requirements for Drp1 in apoptosis, in vivo, and in vitro. (A) Whole-mount immunohistochemistry of embryos using antibodies to activated caspase-3. Arrows indicate apoptotic cells at the site of neural tube closure. (B) MEFs were incubated with 1 µM staurosporine for the indicated times, stained with Alexa Fluor 488–annexin V and propidium iodine, and analyzed by flow cytometry (n ≥ 3). (C) Quantitation of caspase activity (caspase-3/7) using the fluorogenic substrate DEVD-AFC. MEFs were left untreated (Cont) or treated with the following death stimuli and harvested at the indicated time points: 1 µM staurosporin (Sts; 6 h), 100 µM etoposide (Eto; 24 h), 600 J/m2 UV (24 h), and 10 ng/ml TNF-α plus 10 µg/ml cycloheximide (16 h; n ≥ 6). (D) Cytochrome c release during apoptosis. MEFs were incubated with 100 µM etoposide for 30 h in the presence of Z-VAD-FMK and immunostained using antibodies to cytochrome c and Tom20. Arrows indicate cells with released cytochrome c. (E) Quantitation of cytochrome c release (n = 3; >100 cells/experiment). (F) Quantitation of mitochondrial shape (n = 3; >100 cells/experiment). WT, wild type; KO, knockout. Error bars indicate mean ± SEM.
In contrast to the in vivo results, we found that Drp1 is not required for apoptosis in nonimmortalized MEFs in vitro. We incubated MEFs with 1 µM staurosporine for 0, 4, or 12 h and stained them using Alexa Fluor 488–labeled annexin V and propidium iodide. Alexa Fluor 488–annexin V–positive and propidium iodide–negative cells were scored as apoptotic using flow cytometry. The number of apoptotic cells gradually increased and were indistinguishable in Drp1+/+ and Drp1−/− MEFs (Fig. 4 B). Furthermore, we treated MEFs using intrinsic (staurosporine, etoposide, and UV) and extrinsic apoptotic stimuli (TNF-α plus cycloheximide) and examined caspase activity using fluorescent substrate (Fig. 4 C). The levels of caspase activation were indistinguishable in Drp1+/+ and Drp1−/− cells for all stimuli examined. Similarly, during apoptosis, Drp1+/+ and Drp1−/− MEF mitochondria divided and fragmented (Fig. 4, D and F). Comparable numbers of Drp1+/+ and Drp1−/− MEFs released cytochrome c (Fig. 4, D and E). The majority of MEFs that released cytochrome c contained completely fragmented mitochondria (Drp1+/+, 89.2 ± 3.9%; Drp1−/−, 79.6 ± 5.5%; mean ± SEM, n = 3). Thus, nonimmortalized MEFs can initiate mitochondrial division and release cytochrome c during apoptosis independently of Drp1. Collectively, these results suggest that the requirement of Drp1 in apoptosis likely depends on cell type and physiological apoptotic cue, which accounts for the discrepancies of previous studies (Frank et al., 2001; Germain et al., 2005; Jagasia et al., 2005; Parone et al., 2006; Abdelwahid et al., 2007; Goyal et al., 2007; Breckenridge et al., 2008; Cassidy-Stone et al., 2008; James and Martinou, 2008; Suen et al., 2008). Supporting our conclusion, a recent study using ES cells and transformed MEFs showed that the effects of Drp1 deficiency on cytochrome c release vary depending on the cell type and apoptotic stimulus (Ishihara et al., 2009).
Drp1 is required for cerebellar development
Mitochondrial division is implicated in many neurodegenerative diseases (Knott and Bossy-Wetzel, 2008), and heterozygous dominant-negative somatic mutation of Drp1 leads to postnatal death with developmental defects in the brain and retina (Waterham et al., 2007). To investigate the role of Drp1 in the brain, we used a conditional allele of Drp1 (Fig. 1 A) and an engrailed 1 (En1)–Cre (En1-Cre) transgenic line, which starts expressing Cre recombinase in the cerebellum and other tissues, including the midbrain and limb buds during development (Kimmel et al., 2000). By breeding Drp1flox/flox mice to Drp1flox/+::En1-Cre (or Drp1−/+::En1-Cre) mice, we generated mice that carry Drp1flox/−::En1-Cre and Drp1flox/flox::En1-Cre, which were phenotypically indistinguishable and are collectively termed En1-Drp1KO hereafter. As controls, we used Drp1flox/+::En1-Cre mice, which are phenotypically wild type. En1-Drp1KO mice were born at normal frequencies (23 control and 23 En1-Drp1KO mice in 11 litters) and had body sizes indistinguishable from littermate controls. However, En1-Drp1KO mice died within 36 h of birth. By visual inspection, En1-Drp1KO newborn mice had no milk in their stomachs. When we manually fed control and En1-Drp1KO mice, both mice could drink milk, ruling out the possibility that En1-Drp1KO mice were defective in swallowing.
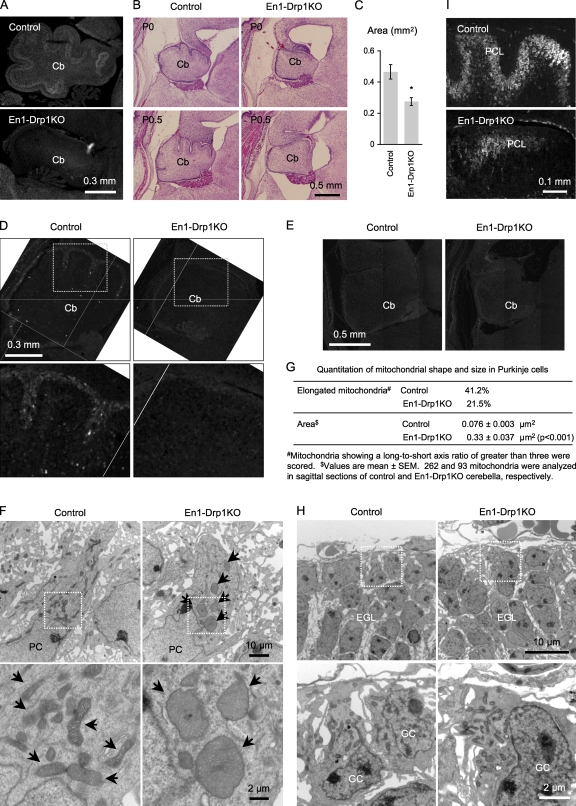
We found cerebellar development to be defective in En1-Drp1KO mice. We confirmed the expression of En1-Cre in the cerebellum using a reporter strain (Fig. S3 A; Soriano, 1999) and the loss of Drp1 by immunostaining (Fig. 5 A). The cerebellum largely develops postnatally and undergoes morphological changes after birth, including the formation of lobule and migration of Purkinje cells (PCs; Armstrong and Hawkes, 2000; Chizhikov and Millen, 2003). In control mice, within 1 h of birth (postnatal day [P] 0), the cerebellum was round, but the lobule formations were relatively shallow fissures (Fig. 5 B). In contrast, the cerebella of En1-Drp1KO mice had completely smooth surfaces, and their size was decreased to ∼60% of control cerebella (Fig. 5, B and C). At the P0.5 stage, control cerebellum had developed further, showing multiple lobules distinguished by deeper fissures. However, in the cerebella of En1-Drp1KO mice, the fissures were barely detectable. To confirm that the cerebellum defect was not caused by starvation, we separated control and En1-Drp1KO mice from their mothers, manually fed them with milk, and obtained similar results.
Figure 5.
Defects in cerebellar development in En1-Drp1KO mice. (A) Immunofluorescence of cerebellum (Cb) at P1 using anti-Drp1 antibodies. (B) H&E stains of sagittal sections of cerebella. (C) Area of cerebella in sagittal sections around the median line was determined (n = 3). (D and E) Sagittal cerebellar sections were immunostained using antibodies to Ki67 (D) or activated caspase-3 (E). (F) EM of PCs at P0. Arrows indicate mitochondria. Sagittal sections of cerebella are shown. (G) Quantitation of mitochondrial shape and size in PCs. (H) EM of granule cells (GC) in the external granular layer (EGL). Sagittal sections of cerebella are shown. (I) Immunofluorescence of sagittal cerebellar sections around the median line using anti-Car8 antibodies. PCL, PC layer. (D, F, and H) Boxed areas are enlarged in the corresponding panels below. *, P < 0.05. Error bars indicate mean ± SEM.
The cerebellar developmental defects in En1-Drp1KO mice likely resulted from decreased cell proliferation but not from increased apoptosis. When we immunostained the cerebellum using antibodies against Ki67 for cell proliferation and activated caspase-3 for apoptosis, Ki67 signals were lost in the En1-Drp1KO cerebella (Fig. 5 D). In contrast, signals for activated caspase-3 were barely detectable in both control and En1-Drp1KO mice (Fig. 5 E).
Enlarged mitochondria of PCs in En1-Drp1KO cerebella
When we examined the cerebellum using EM, we found that mitochondrial shape was strikingly altered in PCs but not in granule cells in the En1-Drp1KO mice. In control cerebella, mitochondria had short tubular and small round structures in PCs (Fig. 5, F and G). In contrast, mitochondria in En1-Drp1KO PCs had large round structures. The number of mitochondria was also considerably decreased in En1-Drp1KO mice. The inner membrane maintained relatively normal cristae morphology. Unlike PCs, granule cells in the external granular layer showed normal mitochondrial morphology in En1-Drp1KO mice (Fig. 5 H). Our results are consistent with the previous observation that Drp1 is more highly expressed in PCs than granule cells in the cerebellum (Lein et al., 2007) and suggest that mitochondrial shape and dynamics may be regulated by a Drp1-independent mechanism in granule cells.
We then examined PCs in En1-Drp1KO mice using immunofluorescence microscopy with antibodies against Car8, a marker for PCs (Patrizi et al., 2008). PCs gradually form a monolayer and establish a long axon and highly branched dendrites with a large number of synapses within 2–3 wk of birth (Armstrong and Hawkes, 2000; Chizhikov and Millen, 2003). In P0 control mice, PCs were slightly elongated, formed thick layers, and lacked dendritic processes (Fig. 5 I). In P0 En1-Drp1KO mice, the number of PCs was dramatically decreased and formed a discontinuous cell layer (Fig. 5 I). These results suggest that Drp1 is required for normal proliferation and development of PCs in the cerebellum.
Importantly, the loss of Drp1 led to distinct alterations in mitochondrial morphology in MEFs, PCs, and granule cells. Although it remains to be determined how defects in mitochondrial division resulted in distinct organelle shapes in different cell types, organelle–cytoskeleton interactions may contribute to such differences. Alternatively, lipid compositions may be different in these cell types. Also, specific cell types may use different modes of mitochondrial fusion, which likely create these highly connected or enlarged mitochondria. In support of this idea, Mfn2 is required for cell survival of PCs but not granule cells (Chen et al., 2007). Therefore, we suggest that mitochondrial dynamics is essential for cell type–specific mitochondrial morphogenesis and function.
In this study, using a null allele of Drp1, we generated a mammalian model defective in mitochondrial division for the first time and demonstrated that mitochondrial division is required for embryonic development in mice. During development, Drp1−/− mice showed defects in trophoblast giant cells and cardiomyocytes. Because loss of Drp1 does not severely reduce total intracellular ATP, we suggest that mitochondrial division is important for efficient local ATP delivery. Alternatively, other cellular functions that have been shown to be regulated by mitochondrial division, such as calcium signaling, the control of reactive oxygen species production, apoptosis, and senescence, may be affected. In addition, using a conditional allele of Drp1, we showed that Drp1 is required for the development of the cerebellum and mitochondrial morphology in PCs.
Materials and methods
Generation of knockout mice
All animal work was performed according to guidelines established by the Johns Hopkins University Committee on Animal Care. We inserted a neomycin-resistant marker flanked by flipase (Flp) recombinase target and loxP sites next to exons 3 and 5, which are located in an essential GTPase domain (Fig. 1 A, Flox-neo). The target region was ∼3.3 kb and included exons 3–5. The targeting vector was transfected into C57BL/6-129/SvEv ES cells by electroporation. G418-resistant colonies were screened by PCR. Targeted ES cells were injected into C57BL/6 blastocysts to create chimeric mice (Wakabayashi et al., 2003). To create the null Drp1 allele, we crossed Flox-neo mice to a transgenic strain that expresses Cre recombinase from the EIIa promoter (Lakso et al., 1996) and confirmed loxP recombination by PCR (Fig. 1, A [Null] and B). In addition, upon loxP recombination, a stop codon was generated immediately after exon 2 as a result of a frame shift. To generate the conditional allele, Flox-neo mice were crossed to a transgenic strain that ubiquitously expresses Flp recombinase (Dymecki, 1996) and confirmed Flp recombinase target recombination by PCR (Fig. 1, A [Floxed] and B). We bred these strains to a wild-type strain and isolated mice heterozygous for the null or conditional allele but not Cre or Flp recombinase. To distinguish Drp1+/+, Drp1+/−, and Drp1−/− by PCR, the PCR primers D1 (5′-CACTGAGAGCTCTATATGTAGGC-3′), D3 (5′-ACCAAAGTAAGGAATAGCTGTTG-3′), and D5 (5′-GAGTACCTAAAGTGGACAAGAGGTCC-3′) were used. For floxed alleles, D3 and D6 (5′-ATGCGCTGATAATACTATCAACC-3′) were used.
Histology
After timed breeding and confirmation of vaginal plugs, embryos were isolated at E9.5–11.5. For histological analysis of the heart, embryos were fixed in PBS containing 4% paraformaldehyde overnight and embedded in paraffin at the Johns Hopkins University reference histology facility. Paraffin sections were stained using hematoxylin and eosin (H&E). H&E-stained sections were viewed on an upright microscope (BX51; Olympus) equipped with 4× 0.16 NA and 10× 0.3 NA objectives (UIS2; Olympus), and a color camera (DP-70; Olympus) was used.
Placenta were dissected from embryos, fixed, and embedded in paraffin. Paraffin sections were stained with 1 µg/ml DAPI. Sections were viewed on a motorized upright microscope (BX61; Olympus) equipped with a charge-coupled device (CCD) camera (CoolSnap HQ; Roper Scientific).
For whole-mount immunohistochemistry, embryos were fixed in 4% paraformaldehyde in PBS overnight at 4°C (Nagy et al., 2003). After washes in PBS, samples were incubated in 80% methanol containing 6% H2O2 for 1 h at room temperature. Samples were immersed twice in 100% methanol for 10 min and in 3 ml 1:1 methanol/DMSO. 0.75 ml 10% Triton X-100 was added to the methanol/DMSO solution. After 15–30-min incubation, samples were washed three times in TST (100 mM Tris-HCl, pH 7.8, 0.1% Triton X-100, and 154 mM NaCl) and blocked in 5% skim milk in TST overnight at 4°C. Embryos were stained with antibodies against cleaved caspase-3 (1:200; clone 5A1; Cell Signaling Technology) and PECAM/CD31 (1:10; clone MEC13.3; BD) for 24 h at 4°C. After several washes, samples were incubated with appropriate secondary antibodies conjugated with horseradish peroxidase (1:100; GE Healthcare) overnight. Samples were incubated in TST containing 250 µg/ml 3,3′-diaminobenzidine for ∼30 min on ice and 250 µg/ml 3,3′-diaminobenzidine containing 0.003% H2O2 for 15–30 min on ice. After extensive washes in TST, samples were postfixed in 4% paraformaldehyde in PBS at room temperature for 20 min. Samples were viewed using a stereo microscope (SZX12; Olympus) equipped with a color camera (DP-70; Olympus).
For immunofluorescence of the cerebellum, newborn pups were fixed by perfusing 4% paraformaldehyde in PBS. After removing the skin, the head was further fixed for 2–3 h. The brain was dissected and fixed overnight at 4°C. After embedding in paraffin, sections were cut, deparaffinized, washed in PBS, and blocked in MOM-blocking reagent according to the manufacturer's protocol (Vector Laboratories). Samples were incubated with anti-Drp1 antibodies (1:100; clone 8; BD) at room temperature for 2 h and then with Alexa Fluor 488 anti–mouse IgG (1:400). For immunofluorescence of PCs, paraffin sections were stained with anti-Car8 antibodies (1:400; Patrizi et al., 2008) followed by Alexa Fluor 488–labeled secondary antibodies (1:400; Invitrogen). Samples were observed on a motorized upright microscope (BX61; Olympus).
Immunoblotting of embryos
Proteins were extracted from embryos in RIPA buffer and analyzed by immunoblotting using antibodies against Drp1 (clone 8; BD), the mitochondrial protein Tim23 (clone 32; BD), and the ER protein PGRMC1 (Hughes et al., 2007).
Isolation of MEFs and cardiomyocytes
After removing brain and heart, E10.5 embryos were digested with 200 µl 0.05% trypsin 0.02% EDTA at 37°C for 10 min. After addition of 800 µl Iscove's modified Dulbecco's medium (Invitrogen) containing 10% fetal bovine serum (Invitrogen) and 100 µg/ml primocin (Invitrogen), MEFs were dispersed by pipetting (Wakabayashi et al., 2004). After washes, MEFs were cultured in Iscove's modified Dulbecco's medium containing 10% fetal bovine serum and 100 µg/ml primocin at 37°C in a humidified atmosphere of 5% CO2 (Wakabayashi et al., 2004). In all of the experiments, MEFs of at least three independent preparations per genotype were used without transformation or immortalization.
For isolation of cardiomyocytes, hearts were digested with 200 µl 0.05% trypsin 0.02% EDTA at 37°C for 20 min (Nakamura et al., 1993). After washes, cells were incubated in Iscove's modified Dulbecco's medium containing 10% fetal bovine serum and 100 µg/ml primocin at 37°C in a humidified atmosphere of 5% CO2 for 2 h. Culture media containing unattached cardiomyocytes were collected. 6 d after isolation, cardiomyocytes were observed at 37°C using a microscope (Axio Observer Z1; Carl Zeiss, Inc.) equipped with an environmentally controlled enclosure to monitor CO2 levels, temperature, and humidity (Precision Plastic).
Observation of organelles in MEFs
For visualization of mitochondria, MEFs were fixed in prewarmed PBS containing 4% paraformaldehyde for 30 min and permeabilized with 0.1% Triton X-100 in PBS for 10 min. After blocking in 0.5% BSA, cells were incubated with anti-Tom20 antibodies (1:100; Santa Cruz Biotechnology, Inc.) followed by Alexa Fluor 488–labeled secondary antibodies (1:2,000; Invitrogen). For time-lapse observations, MEFs were grown in 8-well chambered coverglass (Laboratory-Tek II; Thermo Fisher Scientific), transfected with a plasmid carrying Su9-GFP (Eura et al., 2003) using GeneJuice (EMD), and observed using a custom-assembled microscope comprised of a fully automated microscope (Axio Observer Z1), a spinning-disc confocal (CSU22; Yokogawa), and a camera (Cascade II 512b; Roper Scientific) with an environmentally controlled enclosure to monitor CO2 levels, temperature, and humidity.
For observation of peroxisomes, MEFs were grown in 8-well chamber slides (Laboratory-Tek II), fixed in prewarmed PBS containing 4% paraformaldehyde for 30 min, and permeabilized with 0.1% Triton X-100 in PBS for 10 min. After blocking in 0.5% BSA, cells were incubated for 3 h with anti-Pex14 antibodies (1:200) followed by Alexa Fluor 488–labeled secondary antibodies (1:2,000; Invitrogen). Cells were viewed on a motorized upright microscope (BX61) equipped with a 40× 0.75 NA objective (UIS2) and a CCD camera (CoolSnap HQ). SlideBook (3i), QuickTime 7 (Apple, Inc.), Photoshop (Adobe), and ImageJ (National Institutes of Health) software were used to analyze images.
MEF proliferation
Drp1+/+ and Drp1−/− MEFs were isolated, cultured for 5 d, and plated at a density of 2–4 × 104 cells/well in 24-well dishes on day 0. Cell numbers were counted at the indicated time points and normalized relative to day 1.
ATP measurements
MEFs were collected by centrifugation, incubated with 10 µl 5% TCA, and lysed in 90 µl 1% Triton X-100 and 100 mM Tris-actate, pH 8.0, according to the manufacturer's instructions (Enliten ATP assay system; Promega). Luminescence was measured using a Fluostar Optima (BMG Labtech).
Apoptosis in MEFs
MEFs were used in all of the apoptosis assays within three weeks after isolation. MEFs were incubated with 1 µM staurosporine for 0, 4, or 12 h, collected, and stained with Alexa Fluor 488–annexin V and propidium iodine according to the manufacturer's instructions (Invitrogen). Cells were analyzed using FACScan (BD). To measure caspase activity, MEFs were incubated with various apoptotic stimuli, including 1 µM staurosporine for 6 h, 100 µM etoposide for 24 h, 600 J/m2 UV for 24 h, and 10 ng/ml TNF-α plus 10 µg/ml cycloheximide for 16 h, lysed, and incubated with a fluorogenic substrate according to the manufacturer's instructions (Apo-ONE homogeneous caspase-3/7 assay kit; Promega). Fluorescence was measured using Fluostar Optima.
For analysis of cytochrome c release, MEFs were preincubated with 40 µM Z-VAD-FMK for 1 h, treated with 1 µM staurosporine for 3 and 6 h or100 µM etoposide for 20, 24, 30, and 42 h in the presence of 40 µM Z-VAD-FMK, fixed in prewarmed PBS containing 4% paraformaldehyde for 30 min, and permeabilized with 0.1% Triton X-100 in PBS for 10 min. After blocking in 0.5% BSA, cells were incubated with antibodies against cytochrome c (1:200; clone 6H2.B4; BD) and Tom20 (1:100; Santa Cruz Biotechnology, Inc.) overnight at 4°C followed by appropriate secondary antibodies.
Immunofluorescence for Ki67 and activated caspase-3
MEFs were fixed in prewarmed PBS containing 4% paraformaldehyde for 30 min and permeabilized with 0.1% Triton X-100 in PBS for 10 min. After blocking in 0.5% BSA, cells were incubated with antibodies against Ki67 (1:100; AB15580; Abcam) followed by Alexa Fluor 594–labeled secondary antibodies.
Sagittal sections of control and En1-Drp1KO cerebella were immunostained using antibodies to Ki67 (1:250; Abcam) or activated caspase-3 (1:200; clone 5A1; Cell Signaling Technology) as a marker for apoptosis and Alexa Fluor 488–labeled secondary antibodies. Samples were viewed on a microscope (BX61) equipped with a 40× 0.75 NA objective (UIS2) and a CCD camera (CoolSnap HQ) for Ki67 and on a confocal microscope (510 Meta; Carl Zeiss, Inc.) for activated caspase-3.
EM
The heads of pups were dissected and fixed in 3% paraformaldehyde, 1.5% glutaraldehyde, 2.5% sucrose, and 100 mM cacodylate, pH 7.4, overnight. Brains were dissected and further fixed for 2 d. After washes, samples were postfixed in 2.7% OsO4 and 167 mM cacodylate, pH 7.4, for 1 h on ice. After washes in water, samples were incubated in 2% uranyl acetate for 30 min. After dehydration using 50, 70, 90, and 100% ethanol and 100% propylene oxide, samples were embedded in epon resin (Ted Pella, Inc.). Ultrathin sections were obtained using a Reichert-Jung ultracut E, stained with 2% uranyl acetate and lead citrate, and viewed on a transmission electron microscope (H-7600; Hitachi) equipped with a dual CCD camera (Advanced Microscopy Techniques). ImageJ software was used to measure the length and size of mitochondria.
MEFs were fixed in prewarmed 2% glutaraldehyde, 2.5% sucrose, 3 mM CaCl2, and 100 mM Hepes, pH 7.4, for 1 h. After washes, MEFs were postfixed using reduced OsO4 (1% OsO4, 10 mg/ml potassium ferrocyanide, 1.25% sucrose, and 100 mM cacodylate, pH 7.4) for 1 h on ice. After washes in water, cells were incubated in 2% uranyl acetate for 30 min. After dehydration using 50, 70, 90, and 100% ethanol, samples were embedded in epon resin. For cytochemical staining of catalase, MEFs were fixed (4% paraformaldehyde, 0.05% glutaraldehyde, 2.5% sucrose, 3 mM CaCl2, and 100 mM Hepes, pH 7.4) for 1 h. After washes, MEFs were incubated in 2 mg/ml DAB, 0.15% H2O2, and 100 mM glycine NaOH, pH 10.5, at 37°C for 1 h (Angermüller and Fahimi, 1981). Samples were postfixed, dehydrated, and embedded as described in the previous paragraph.
Statistical analysis
All values are mean ± SEM. Results were statistically analyzed using t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Online supplemental material
Fig. S1 shows the characterization of Drp1+/+ and Drp1+/− mice and Drp1+/+ and Drp1−/− MEFs using immunoblot analysis. Fig. S2 shows Drp1+/+ and Drp1−/− MEFs immunostained with anti-Pex14 antibodies. Fig. S3 shows that En1-Cre is expressed in the whole cerebellum and H&E stains of the sections adjacent to those shown in Fig. 5 D. Video 1 shows the characterization of Drp1+/+ MEFs expressing matrix-targeted Su9-GFP using time-lapse confocal microscopy. Video 2 shows the characterization of Drp1−/− MEFs expressing matrix-targeted Su9-GFP using time-lapse confocal microscopy. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200903065/DC1.
Acknowledgments
We thank S. Gould, P. Watkinson, P. Espenshade, and M. Watanabe for antibodies, N. Ishihara and K. Mihara for Su9-GFP plasmids, A. Joyner, S. Dymecki, H. Westphal, J. Nathans, X. Ye, T. Badea, and T. Rotolo for transgenic mice, and C. Mendis-Handagama for help with EM for peroxisomes.
This work was supported by the American Heart Association (M. Iijima and H. Sesaki), the National Institutes of Health, and the Muscular Dystrophy Association (H. Sesaki).
Footnotes
Abbreviations used in this paper:
- CCD
- charge-coupled device
- En1
- engrailed 1
- ES
- embryonic stem
- Flp
- flipase
- MEF
- mouse embryonic fibroblast
- PC
- Purkinje cell
References
- Abdelwahid E., Yokokura T., Krieser R.J., Balasundaram S., Fowle W.H., White K. 2007. Mitochondrial disruption in Drosophila apoptosis.Dev. Cell. 12:793–806 doi:10.1016/j.devcel.2007.04.004 [DOI] [PubMed] [Google Scholar]
- Ahuja D., Sáenz-Robles M.T., Pipas J.M. 2005. SV40 large T antigen targets multiple cellular pathways to elicit cellular transformation.Oncogene. 24:7729–7745 doi:10.1038/sj.onc.1209046 [DOI] [PubMed] [Google Scholar]
- Alavi M.V., Bette S., Schimpf S., Schuettauf F., Schraermeyer U., Wehrl H.F., Ruttiger L., Beck S.C., Tonagel F., Pichler B.J., et al. 2007. A splice site mutation in the murine Opa1 gene features pathology of autosomal dominant optic atrophy.Brain. 130:1029–1042 doi:10.1093/brain/awm005 [DOI] [PubMed] [Google Scholar]
- Alexander C., Votruba M., Pesch U.E., Thiselton D.L., Mayer S., Moore A., Rodriguez M., Kellner U., Leo-Kottler B., Auburger G., et al. 2000. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28.Nat. Genet. 26:211–215 doi:10.1038/79944 [DOI] [PubMed] [Google Scholar]
- Anesti V., Scorrano L. 2006. The relationship between mitochondrial shape and function and the cytoskeleton.Biochim. Biophys. Acta. 1757:692–699 doi:10.1016/j.bbabio.2006.04.013 [DOI] [PubMed] [Google Scholar]
- Angermüller S., Fahimi H.D. 1981. Selective cytochemical localization of peroxidase, cytochrome oxidase and catalase in rat liver with 3,3′-diaminobenzidine.Histochemistry. 71:33–44 doi:10.1007/BF00592568 [DOI] [PubMed] [Google Scholar]
- Armstrong C.L., Hawkes R. 2000. Pattern formation in the cerebellar cortex.Biochem. Cell Biol. 78:551–562 doi:10.1139/bcb-78-5-551 [PubMed] [Google Scholar]
- Bleazard W., McCaffery J.M., King E.J., Bale S., Mozdy A., Tieu Q., Nunnari J., Shaw J.M. 1999. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast.Nat. Cell Biol. 1:298–304 doi:10.1038/13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckenridge D.G., Kang B.H., Kokel D., Mitani S., Staehelin L.A., Xue D. 2008. Caenorhabditis elegans drp-1 and fis-2 regulate distinct cell-death execution pathways downstream of ced-3 and independent of ced-9.Mol. Cell. 31:586–597 doi:10.1016/j.molcel.2008.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Stone A., Chipuk J.E., Ingerman E., Song C., Yoo C., Kuwana T., Kurth M.J., Shaw J.T., Hinshaw J.E., Green D.R., Nunnari J. 2008. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization.Dev. Cell. 14:193–204 doi:10.1016/j.devcel.2007.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny K.L., McCaffery J.M., Jensen R.E. 2001. Division of mitochondria requires a novel DMN1-interacting protein, Net2p.Mol. Biol. Cell. 12:309–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveny K.L., Tamura Y., Zhang Z., Jensen R.E., Sesaki H. 2007a. Regulation of mitochondrial fusion and division.Trends Cell Biol. 17:563–569 doi:10.1016/j.tcb.2007.08.006 [DOI] [PubMed] [Google Scholar]
- Cerveny K.L., Studer S.L., Jensen R.E., Sesaki H. 2007b. Yeast mitochondrial division and distribution require the cortical num1 protein.Dev. Cell. 12:363–375 doi:10.1016/j.devcel.2007.01.017 [DOI] [PubMed] [Google Scholar]
- Chan D.C. 2006. Mitochondria: dynamic organelles in disease, aging, and development.Cell. 125:1241–1252 doi:10.1016/j.cell.2006.06.010 [DOI] [PubMed] [Google Scholar]
- Chen H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E., Chan D.C. 2003. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development.J. Cell Biol. 160:189–200 doi:10.1083/jcb.200211046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., McCaffery J.M., Chan D.C. 2007. Mitochondrial fusion protects against neurodegeneration in the cerebellum.Cell. 130:548–562 doi:10.1016/j.cell.2007.06.026 [DOI] [PubMed] [Google Scholar]
- Chizhikov V., Millen K.J. 2003. Development and malformations of the cerebellum in mice.Mol. Genet. Metab. 80:54–65 doi:10.1016/j.ymgme.2003.08.019 [DOI] [PubMed] [Google Scholar]
- Cho D.H., Nakamura T., Fang J., Cieplak P., Godzik A., Gu Z., Lipton S.A. 2009. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury.Science. 324:102–105 doi:10.1126/science.1171091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuesta A., Pedrola L., Sevilla T., García-Planells J., Chumillas M.J., Mayordomo F., LeGuern E., Marín I., Vílchez J.J., Palau F. 2002. The gene encoding ganglioside-induced differentiation-associated protein 1 is mutated in axonal Charcot-Marie-Tooth type 4A disease.Nat. Genet. 30:22–25 doi:10.1038/ng798 [DOI] [PubMed] [Google Scholar]
- Davies V.J., Hollins A.J., Piechota M.J., Yip W., Davies J.R., White K.E., Nicols P.P., Boulton M.E., Votruba M. 2007. Opa1 deficiency in a mouse model of autosomal dominant optic atrophy impairs mitochondrial morphology, optic nerve structure and visual function.Hum. Mol. Genet. 16:1307–1318 doi:10.1093/hmg/ddm079 [DOI] [PubMed] [Google Scholar]
- Delettre C., Lenaers G., Griffoin J.M., Gigarel N., Lorenzo C., Belenguer P., Pelloquin L., Grosgeorge J., Turc-Carel C., Perret E., et al. 2000. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy.Nat. Genet. 26:207–210 doi:10.1038/79936 [DOI] [PubMed] [Google Scholar]
- Deng H., Dodson M.W., Huang H., Guo M. 2008. The Parkinson's disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila.Proc. Natl. Acad. Sci. USA. 105:14503–14508 doi:10.1073/pnas.0803998105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymecki S.M. 1996. Flp recombinase promotes site-specific DNA recombination in embryonic stem cells and transgenic mice.Proc. Natl. Acad. Sci. USA. 93:6191–6196 doi:10.1073/pnas.93.12.6191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eura Y., Ishihara N., Yokota S., Mihara K. 2003. Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion.J. Biochem. 134:333–344 doi:10.1093/jb/mvg150 [DOI] [PubMed] [Google Scholar]
- Frank S., Gaume B., Bergmann-Leitner E.S., Leitner W.W., Robert E.G., Catez F., Smith C.L., Youle R.J. 2001. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis.Dev. Cell. 1:515–525 doi:10.1016/S1534-5807(01)00055-7 [DOI] [PubMed] [Google Scholar]
- Fukushima N.H., Brisch E., Keegan B.R., Bleazard W., Shaw J.M. 2001. The GTPase effector domain sequence of the Dnm1p GTPase regulates self-assembly and controls a rate-limiting step in mitochondrial fission.Mol. Biol. Cell. 12:2756–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain M., Mathai J.P., McBride H.M., Shore G.C. 2005. Endoplasmic reticulum BIK initiates DRP1-regulated remodelling of mitochondrial cristae during apoptosis.EMBO J. 24:1546–1556 doi:10.1038/sj.emboj.7600592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal G., Fell B., Sarin A., Youle R.J., Sriram V. 2007. Role of mitochondrial remodeling in programmed cell death in Drosophila melanogaster.Dev. Cell. 12:807–816 doi:10.1016/j.devcel.2007.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin E.E., Graumann J., Chan D.C. 2005. The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria.J. Cell Biol. 170:237–248 doi:10.1083/jcb.200503148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griparic L., van der Wel N.N., Orozco I.J., Peters P.J., van der Bliek A.M. 2004. Loss of the intermembrane space protein Mgm1/OPA1 induces swelling and localized constrictions along the lengths of mitochondria.J. Biol. Chem. 279:18792–18798 doi:10.1074/jbc.M400920200 [DOI] [PubMed] [Google Scholar]
- Hoppins S., Lackner L., Nunnari J. 2007. The machines that divide and fuse mitochondria.Annu. Rev. Biochem. 76:751–780 doi:10.1146/annurev.biochem.76.071905.090048 [DOI] [PubMed] [Google Scholar]
- Hughes A.L., Powell D.W., Bard M., Eckstein J., Barbuch R., Link A.J., Espenshade P.J. 2007. Dap1/PGRMC1 binds and regulates cytochrome P450 enzymes.Cell Metab. 5:143–149 doi:10.1016/j.cmet.2006.12.009 [DOI] [PubMed] [Google Scholar]
- Ingerman E., Perkins E.M., Marino M., Mears J.A., McCaffery J.M., Hinshaw J.E., Nunnari J. 2005. Dnm1 forms spirals that are structurally tailored to fit mitochondria.J. Cell Biol. 170:1021–1027 doi:10.1083/jcb.200506078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Nomura M., Jofuku A., Kato H., Suzuki S.O., Masuda K., Otera H., Nakanishi Y., Nonaka I., Goto Y., et al. 2009. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice.Nat. Cell Biol. 11:958–966 doi:10.1038/ncb1907 [DOI] [PubMed] [Google Scholar]
- Jagasia R., Grote P., Westermann B., Conradt B. 2005. DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans.Nature. 433:754–760 doi:10.1038/nature03316 [DOI] [PubMed] [Google Scholar]
- James D.I., Martinou J.C. 2008. Mitochondrial dynamics and apoptosis: a painful separation.Dev. Cell. 15:341–343 doi:10.1016/j.devcel.2008.08.011 [DOI] [PubMed] [Google Scholar]
- James D.I., Parone P.A., Mattenberger Y., Martinou J.C. 2003. hFis1, a novel component of the mammalian mitochondrial fission machinery.J. Biol. Chem. 278:36373–36379 doi:10.1074/jbc.M303758200 [DOI] [PubMed] [Google Scholar]
- Karbowski M., Youle R.J. 2003. Dynamics of mitochondrial morphology in healthy cells and during apoptosis.Cell Death Differ. 10:870–880 doi:10.1038/sj.cdd.4401260 [DOI] [PubMed] [Google Scholar]
- Kimmel R.A., Turnbull D.H., Blanquet V., Wurst W., Loomis C.A., Joyner A.L. 2000. Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation.Genes Dev. 14:1377–1389 [PMC free article] [PubMed] [Google Scholar]
- Knott A.B., Bossy-Wetzel E. 2008. Impairing the mitochondrial fission and fusion balance: a new mechanism of neurodegeneration.Ann. N. Y. Acad. Sci. 1147:283–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S., Tanaka A., Fujiki Y. 2007. Fis1, DLP1, and Pex11p coordinately regulate peroxisome morphogenesis.Exp. Cell Res. 313:1675–1686 doi:10.1016/j.yexcr.2007.02.028 [DOI] [PubMed] [Google Scholar]
- Koch A., Schneider G., Lüers G.H., Schrader M. 2004. Peroxisome elongation and constriction but not fission can occur independently of dynamin-like protein 1.J. Cell Sci. 117:3995–4006 doi:10.1242/jcs.01268 [DOI] [PubMed] [Google Scholar]
- Kuan C.Y., Yang D.D., Samanta Roy D.R., Davis R.J., Rakic P., Flavell R.A. 1999. The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development.Neuron. 22:667–676 doi:10.1016/S0896-6273(00)80727-8 [DOI] [PubMed] [Google Scholar]
- Labrousse A.M., Zappaterra M.D., Rube D.A., van der Bliek A.M. 1999. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane.Mol. Cell. 4:815–826 doi:10.1016/S1097-2765(00)80391-3 [DOI] [PubMed] [Google Scholar]
- Lakso M., Pichel J.G., Gorman J.R., Sauer B., Okamoto Y., Lee E., Alt F.W., Westphal H. 1996. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage.Proc. Natl. Acad. Sci. USA. 93:5860–5865 doi:10.1073/pnas.93.12.5860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A.F., Boguski M.S., Brockway K.S., Byrnes E.J., et al. 2007. Genome-wide atlas of gene expression in the adult mouse brain.Nature. 445:168–176 doi:10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Li X., Gould S.J. 2003. The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11.J. Biol. Chem. 278:17012–17020 doi:10.1074/jbc.M212031200 [DOI] [PubMed] [Google Scholar]
- Li X., Baumgart E., Morrell J.C., Jimenez-Sanchez G., Valle D., Gould S.J. 2002. PEX11 beta deficiency is lethal and impairs neuronal migration but does not abrogate peroxisome function.Mol. Cell. Biol. 22:4358–4365 doi:10.1128/MCB.22.12.4358-4365.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson M.P., Gleichmann M., Cheng A. 2008. Mitochondria in neuroplasticity and neurological disorders.Neuron. 60:748–766 doi:10.1016/j.neuron.2008.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride H.M., Neuspiel M., Wasiak S. 2006. Mitochondria: more than just a powerhouse.Curr. Biol. 16:R551–R560 doi:10.1016/j.cub.2006.06.054 [DOI] [PubMed] [Google Scholar]
- Mozdy A.D., McCaffery J.M., Shaw J.M. 2000. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p.J. Cell Biol. 151:367–380 doi:10.1083/jcb.151.2.367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A., Gertsenstein M., Vintersten K., Behringer R. 2003. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 764 pp [Google Scholar]
- Nakamura T.Y., Goda K., Okamoto T., Kishi T., Nakamura T., Goshima K. 1993. Contractile and morphological impairment of cultured fetal mouse myocytes induced by oxygen radicals and oxidants. Correlation with intracellular Ca2+ concentration.Circ. Res. 73:758–770 [DOI] [PubMed] [Google Scholar]
- Okamoto K., Shaw J.M. 2005. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes.Annu. Rev. Genet. 39:503–536 doi:10.1146/annurev.genet.38.072902.093019 [DOI] [PubMed] [Google Scholar]
- Olichon A., Baricault L., Gas N., Guillou E., Valette A., Belenguer P., Lenaers G. 2003. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis.J. Biol. Chem. 278:7743–7746 doi:10.1074/jbc.C200677200 [DOI] [PubMed] [Google Scholar]
- Osteryoung K.W., Nunnari J. 2003. The division of endosymbiotic organelles.Science. 302:1698–1704 doi:10.1126/science.1082192 [DOI] [PubMed] [Google Scholar]
- Otsuga D., Keegan B.R., Brisch E., Thatcher J.W., Hermann G.J., Bleazard W., Shaw J.M. 1998. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast.J. Cell Biol. 143:333–349 doi:10.1083/jcb.143.2.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaioannou V., Behringer R. 2005. Mouse Phenotypes: A Handbook of Mutation Analysis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 235 pp [Google Scholar]
- Parone P.A., James D.I., Da Cruz S., Mattenberger Y., Donzé O., Barja F., Martinou J.C. 2006. Inhibiting the mitochondrial fission machinery does not prevent Bax/Bak-dependent apoptosis.Mol. Cell. Biol. 26:7397–7408 doi:10.1128/MCB.02282-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrizi A., Scelfo B., Viltono L., Briatore F., Fukaya M., Watanabe M., Strata P., Varoqueaux F., Brose N., Fritschy J.M., Sassoè-Pognetto M. 2008. Synapse formation and clustering of neuroligin-2 in the absence of GABAA receptors.Proc. Natl. Acad. Sci. USA. 105:13151–13156 doi:10.1073/pnas.0802390105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole A.C., Thomas R.E., Andrews L.A., McBride H.M., Whitworth A.J., Pallanck L.J. 2008. The PINK1/Parkin pathway regulates mitochondrial morphology.Proc. Natl. Acad. Sci. USA. 105:1638–1643 doi:10.1073/pnas.0709336105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rube D.A., van der Bliek A.M. 2004. Mitochondrial morphology is dynamic and varied.Mol. Cell. Biochem. 256–257:331–339 doi:10.1023/B:MCBI.0000009879.01256.f6 [DOI] [PubMed] [Google Scholar]
- Santel A., Frank S. 2008. Shaping mitochondria: The complex posttranslational regulation of the mitochondrial fission protein DRP1.IUBMB Life. 60:448–455 doi:10.1002/iub.71 [DOI] [PubMed] [Google Scholar]
- Schrader M. 2006. Shared components of mitochondrial and peroxisomal division.Biochim. Biophys. Acta. 1763:531–541 doi:10.1016/j.bbamcr.2006.01.004 [DOI] [PubMed] [Google Scholar]
- Sesaki H., Jensen R.E. 1999. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape.J. Cell Biol. 147:699–706 doi:10.1083/jcb.147.4.699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C., Delivani P., Cullen S.P., Martin S.J. 2008. Bax- or Bak-induced mitochondrial fission can be uncoupled from cytochrome C release.Mol. Cell. 31:570–585 doi:10.1016/j.molcel.2008.08.002 [DOI] [PubMed] [Google Scholar]
- Smirnova E., Shurland D.L., Newman-Smith E.D., Pishvaee B., van der Bliek A.M. 1999. A model for dynamin self-assembly based on binding between three different protein domains.J. Biol. Chem. 274:14942–14947 doi:10.1074/jbc.274.21.14942 [DOI] [PubMed] [Google Scholar]
- Soriano P. 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain.Nat. Genet. 21:70–71 doi:10.1038/5007 [DOI] [PubMed] [Google Scholar]
- Suen D.F., Norris K.L., Youle R.J. 2008. Mitochondrial dynamics and apoptosis.Genes Dev. 22:1577–1590 doi:10.1101/gad.1658508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu Q., Nunnari J. 2000. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division.J. Cell Biol. 151:353–366 doi:10.1083/jcb.151.2.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi N., Itoh K., Wakabayashi J., Motohashi H., Noda S., Takahashi S., Imakado S., Kotsuji T., Otsuka F., Roop D.R., et al. 2003. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation.Nat. Genet. 35:238–245 doi:10.1038/ng1248 [DOI] [PubMed] [Google Scholar]
- Wakabayashi N., Dinkova-Kostova A.T., Holtzclaw W.D., Kang M.I., Kobayashi A., Yamamoto M., Kensler T.W., Talalay P. 2004. Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers.Proc. Natl. Acad. Sci. USA. 101:2040–2045 doi:10.1073/pnas.0307301101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham H.R., Koster J., van Roermund C.W., Mooyer P.A., Wanders R.J., Leonard J.V. 2007. A lethal defect of mitochondrial and peroxisomal fission.N. Engl. J. Med. 356:1736–1741 doi:10.1056/NEJMoa064436 [DOI] [PubMed] [Google Scholar]
- Will G.K., Soukupova M., Hong X., Erdmann K.S., Kiel J.A., Dodt G., Kunau W.H., Erdmann R. 1999. Identification and characterization of the human orthologue of yeast Pex14p.Mol. Cell. Biol. 19:2265–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Ouyang Y., Yang L., Beal M.F., McQuibban A., Vogel H., Lu B. 2008. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery.Proc. Natl. Acad. Sci. USA. 105:7070–7075 doi:10.1073/pnas.0711845105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y., Pitts K.R., McNiven M.A. 2001. Mammalian dynamin-like protein DLP1 tubulates membranes.Mol. Biol. Cell. 12:2894–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Y., Krueger E.W., Oswald B.J., McNiven M.A. 2003. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1.Mol. Cell. Biol. 23:5409–5420 doi:10.1128/MCB.23.15.5409-5420.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züchner S., Mersiyanova I.V., Muglia M., Bissar-Tadmouri N., Rochelle J., Dadali E.L., Zappia M., Nelis E., Patitucci A., Senderek J., et al. 2004. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A.Nat. Genet. 36:449–451 doi:10.1038/ng1341 [DOI] [PubMed] [Google Scholar]