The soluble and membrane-anchored isoforms of Mgm1 are only active when they work together (in trans).
Abstract
Two dynamin-related protein (DRP) families are essential for fusion of the outer and inner mitochondrial membranes, Fzo1 (yeast)/Mfn1/Mfn2 (mammals) and Mgm1 (yeast)/Opa1 (mammals), respectively. Fzo1/Mfns possess two medial transmembrane domains, which place their critical GTPase and coiled-coil domains in the cytosol. In contrast, Mgm1/Opa1 are present in cells as long (l) isoforms that are anchored via the N terminus to the inner membrane, and short (s) isoforms were predicted to be soluble in the intermembrane space. We addressed the roles of Mgm1 isoforms and how DRPs function in membrane fusion. Our analysis indicates that in the absence of a membrane, l- and s-Mgm1 both exist as inactive GTPase monomers, but that together in trans they form a functional dimer in a cardiolipin-dependent manner that is the building block for higher-order assemblies.
Introduction
Mitochondrial fusion is a conserved process whose fundamental function is likely to create a more connected compartment that facilitates content exchange and access to mitochondrial DNA (Hoppins et al., 2007). Two dynamin-related protein (DRP) families are essential for fusion: Fzo1 (yeast)/Mfn1/2 (mammals) and Mgm1 (yeast)/Opa1 (mammals), which drive outer and inner mitochondrial membrane fusion, respectively (Meeusen et al., 2004, 2006). Outer and inner membrane tethering is mediated by the self assembly of mitochondrial fusion DRPs via intermolecular interactions (Ishihara et al., 2004; Koshiba et al., 2004; Meeusen et al., 2004, 2006; Griffin and Chan, 2006). Analysis of mutant alleles of the fusion DRPs indicates that membrane tethering is separable from subsequent lipid content mixing and that fusion DRPs are essential at each stage (Meeusen et al., 2006).
The localization and topologies of the mitochondrial outer and inner membrane DRPs are distinct. Fzo1/Mfn1/2 possess two medial transmembrane domains that target and anchor them in the mitochondrial outer membrane and place the critical GTPase and coiled-coil regions in the cytosol, with a short loop in the intermembrane space (Hermann et al., 1998; Rapaport et al., 1998). Mgm1/Opa1 are targeted to the mitochondrial inner membrane via an N-terminal stop-transfer signal, placing the GTPase domain proximal to the membrane (Herlan et al., 2003).
Two isoforms of Mgm1/Opa1 are generated during their biosynthesis by divergent proteolytic mechanisms: long (l) isoforms are anchored via the N terminus to the inner membrane, and short (s) isoforms are predicted to be soluble in the intermembrane space. (Esser et al., 2002; Herlan et al., 2003, 2004; McQuibban et al., 2003; Sesaki et al., 2003, Cipolat et al., 2006; Duvezin-Caubet et al., 2006; Ishihara et al., 2006; Griparic et al., 2007; Song et al., 2007). Functional studies have demonstrated that both long and short isoforms are critical for efficient fusion (Herlan et al., 2003, 2004; McQuibban et al., 2003; Griparic et al., 2007; Song et al., 2007). In mammalian cells, dissipation of membrane potential is associated with increased proteolysis of l-Opa1 isoforms, which leads to an attenuation of mitochondrial fusion and thus the linking of mitochondrial function and fusion to facilitate the separation of dysfunctional mitochondria (Duvezin-Caubet et al., 2006; Griparic et al., 2007; Song et al., 2007). Through an analysis of the simpler yeast s- and l-Mgm1 isoforms, we provide insight into their respective roles in fusion and the mechanism of Mgm1's inner membrane specificity.
Results and discussion
l- and s-Mgm1 isoforms exist as inactive monomers
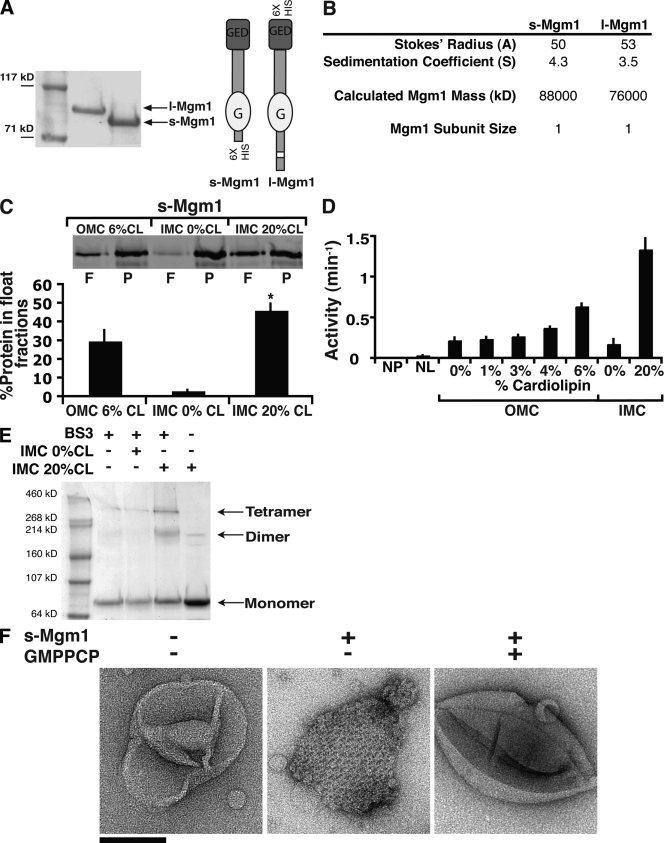
To understand their roles in fusion, we expressed and purified s- and l-Mgm1 and characterized their kinetic and structural properties (Fig. 1 A). l-Mgm1 uniquely required detergent and glycerol to maintain its solubility (1.5% vs. critical micelle concentration of 1.5–2.0%). We examined the ability of s- and l-Mgm1 to hydrolyze GTP, which for DRPs depends on self-assembly (Danino and Hinshaw, 2001). We observed no detectable GTPase activity for s- or l-Mgm1 over a range of protein and GTP concentrations (unpublished data; s-Mgm1 0.03 min−1; l-Mgm1 0.00 min−1). Hydrodynamic analysis of s- or l-Mgm1 by sucrose gradient centrifugation and gel filtration chromatography revealed that both exist as monomers (Fig. 1 B). In contrast to Mgm1, the membrane division DRPs—Dnm1 and dynamin—exist as stable dimers, which in their unassembled form possess a basal rate of GTP hydrolysis (Ingerman et al., 2005; Ramachandran et al., 2007).
Figure 1.
s-Mgm1 assembly is regulated by CL. (A) Purified l- and s-Mgm1 (40 pmol of each) analyzed by SDS-PAGE. Schematic representations of Mgm1 isoforms are shown (right). (B) Hydrodynamic analysis of l- and s-Mgm1. (C) s-Mgm1 preferentially associates with IMC liposomes. 0.5 µM s-Mgm1 was incubated with OMC 6% CL, IMC 0% CL, or IMC 20% CL liposomes and analyzed by floatation in sucrose gradients. A representative SDS-PAGE and Western analysis of float (F) and pellet (P) fractions is shown. Quantification from three experiments is shown as the mean + SEM (error bars). (D) 1 µM s-Mgm1 was analyzed alone (NL, no liposomes; NP, no protein) or preincubated with liposomes of OMC or IMC composition with CL present at the indicated amounts. Data from three experiments are shown as the mean + SEM (error bars). (E) s-Mgm1 self-assembles as a dimer. A representative SDS-PAGE Coomassie-stained gel of chemically cross-linked s-Mgm1 under indicated conditions is shown. (F) s-Mgm1 assembles into lattices in a GTP-regulated manner. Negative-stain EM analysis of IMC 20% CL liposomes with or without 1 µM s-Mgm1 as indicated. Bar, 200 nm.
s-Mgm1 preferentially associates with liposomes containing the inner membrane–enriched anionic lipid cardiolipin (CL), which stimulates self assembly–driven GTP hydrolysis
The significance of monomeric species of Mgm1 was revealed when we examined how s-Mgm1 interacts with liposomes, as assessed by floatation using equilibrium sucrose gradient centrifugation. This analysis indicated that s-Mgm1 preferentially associates with liposomes whose composition mimics the inner mitochondrial membrane composition (IMC) as compared with liposomes with an outer membrane composition (Fig. 1 C, OMC 6% CL vs. IMC 20% CL, composition described in Materials and methods). These data indicate that the critical lipid species required for s-Mgm1 association with IMC liposomes was the inner membrane–enriched dianionic phospholipid, CL (Fig. 1 C, IMC 20% CL vs. IMC 0% CL; and not depicted).
Association of s-Mgm1 with CL-containing IMC liposomes stimulated GTP hydrolysis in a CL-dependent manner, to a maximal velocity of 1.3 min−1 (Fig. 1 D, compare IMC 20% CL vs. IMC 0% CL). When compared with other DRPs this is relatively slow, but it is similar to recently published kinetic data for assembled s-Mgm1 (Meglei and McQuibban, 2009). Although CL is enriched in the inner membrane, biochemical analysis of mitochondria indicates that it may also comprise up to 6% of the outer membrane (Sperka-Gottlieb et al., 1988; Daum and Vance, 1997). In the presence of OMC liposomes containing up to 6% CL, s-Mgm1 was significantly less active as compared with s-Mgm1 in the presence of IMC 20% CL liposomes, which is consistent with its role in inner membrane fusion (Fig. 1 D; Meeusen et al., 2006).
These observations suggest that CL-containing liposomes stimulate self-assembly of s-Mgm1. To test this possibility, we chemically cross-linked s-Mgm1 in the presence and absence of CL-containing IMC membranes with bis(sulfosuccinimidyl)suberate (Fig. 1 E, BS3). Analysis of cross-linked products revealed that s-Mgm1 dimers and tetramers form in a CL-specific manner (Fig. 1 E). These data indicate that dimeric s-Mgm1 is the building block for the assembly of larger structures, which is consistent with previous cross-linking analysis of Mgm1 and Opa1, and further suggests that the formation of a DRP dimer interface is required for GTP hydrolysis and is a general property of the DRP family (Frezza et al., 2006; Gasper et al., 2009; Meglei and McQuibban, 2009).
Thus, in contrast to membrane division DRPs, our data indicate that the s-Mgm1 monomer-to-dimer assembly step is regulated by CL. CL-saturated detergent micelles did not activate s-Mgm1 GTP hydrolysis, indicating that CL stimulates s-Mgm1 self assembly only in the context of a lipid bilayer, which may serve as a 2D platform for the stimulation of s-Mgm1 assembly (unpublished data). This CL membrane regulatory step has likely been harnessed in vivo to couple inner membrane targeting of Mgm1 to its assembly and activation, which is critical given that s-Mgm1 has access to both mitochondrial membranes.
Short Mgm1 assembles into a parallel dimer that further assembles into a novel DRP structure
To gain insight into organization of assembled s-Mgm1 structures, we examined IMC liposomes containing s-Mgm1 by negative-stain EM. This analysis revealed that in the absence of nucleotide, s-Mgm1 associates with CL-liposomes and self-assembles into an extended, organized lattice (Fig. 1 F). The structural features of this lattice are striking in their novelty for a DRP. Division DRPs form curved filaments or helical structures in the absence or presence of GTP, respectively (Ingerman et al., 2005). In the presence of GTP or nonhydrolyzable GTP analogues, s-Mgm1 remained associated with IMC liposomes, but s-Mgm1 lattices were not observed, indicating that GTP binding and hydrolysis facilitates conformational changes (Fig. 1 F and not depicted).
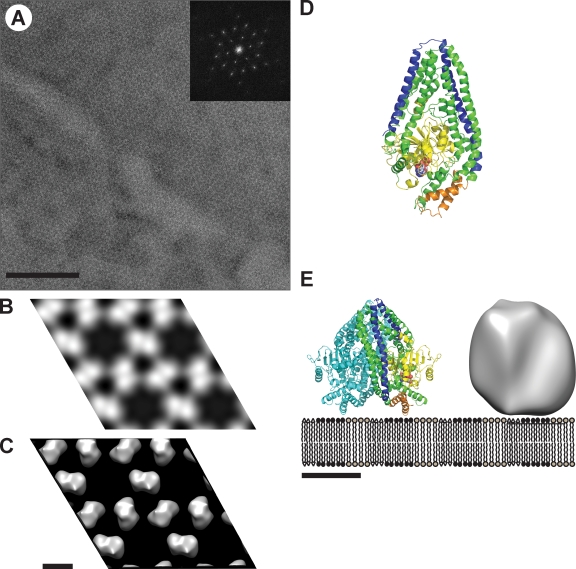
The uniform organization of the s-Mgm1 lattice prompted us to attempt 2D crystallization of s-Mgm1 on CL-containing monolayers (Fig. 2). Crystals of ∼1 µm were obtained and imaged as negative-stained preparations in the transmission EM (Fig. 2 A). The calculated power spectrum of recorded 2D crystal images showed diffraction spots with up to 3 nm resolution before image unbending. Comparison of phase residuals with the ALLSPACE program revealed a P3 symmetry. The unit cell parameters were a = b = 200 Å, γ = 120°. A total of eight images from the nontilted images were merged in 2dx_merge, yielding a projection map at 3.1 nm resolution (overall phase residual: 29.4° [IQ weighted]; Fig. 2 B). Six images of 30° tilted and nine images of 45° tilted negatively stained s-Mgm1 2D crystals were collected and merged with the nontilted data, yielding a 3D reconstruction at 3.1 nm resolution, applying P3 symmetry (Fig. 2 C). This 3D reconstruction showed a trimer of densities that clearly separated into dimers at higher contouring thresholds that were assembled in a hexameric ringlike structure. This structural model is consistent with our cross-linking data, which indicates that a dimer is the building block for self assembly of higher-order structures (Fig. 1 E). A contour level was chosen to include the volume of the s-Mgm1 dimer of 2 × 84 kD, assuming a density of 0.82 D/Å3 (Fig. 2 C).
Figure 2.
Structural analysis of s-Mgm1. (A) EM analysis of a negatively stained 2D crystal of s-Mgm1. The power spectrum calculated from the raw image is shown in the inset and shows strong diffraction up to 3 nm resolution. (B) P3 symmetrized projection map of s-Mgm1, calculated from merged data from nine processed images of negatively stained s-Mgm1 2D crystals. 2 × 2 unit cells are shown. One unit cell has dimensions of a = b = 200 Å, γ = 120°. (C) 3D reconstruction from images of tilted negatively stained 2D crystals of s- Mgm1 as seen from the direction perpendicular to the membrane plane. 2 × 2 unit cells are shown. (D) Homology model for s-Mgm1. This model was created using Modeller (Martí-Renom et al., 2000) and is based on the structures of dynamin A, dynamin-1, and a bacterial dynamin-like protein (PDB accession nos. 1JWY, 2AKA, and 2J69). The monomer is depicted as seen from the dimer interface. The model is color coded as follows: the GTPase domains is yellow, the GED is blue, and the pair of helices that putatively bind the membrane are orange. GDP–Mg complexes are depicted as spheres. (E) Schematic representation of the proposed parallel s-Mgm1 dimer bound to a lipid bilayer. The homology-modeled dimer and the corresponding 3D reconstruction based on the 2D crystallographic data are shown at the same scale. One monomer is colored as in D. Bars: (A) 200 nm; (C) 10 nm; (E) 5 nm.
The GTPase domain of Mgm1 has a high sequence homology to both dynamin A and dynamin 1 (from Dictyostelium discoideum and Rattus norvegicus, respectively) and the remainder of Mgm1 exhibits weak homology to Nostoc punctiforme bacterial dynamin-like protein, whose structure has been determined by x-ray crystallography (Low and Löwe, 2006). To obtain an atomic model for s-Mgm1, we used a combination of homology and threading modeling. The resulting model contains 682 residues spanning the GTPase domain and part of the C-terminal coiled-coil domain (from residue 145 to 829) in which the N and C termini are colocalized in an antiparallel helix bundle (Fig. 2 D). The homology model was docked into the 3D reconstruction, as described in Materials and methods. The available data are most consistent with a parallel dimeric arrangement of two s-Mgm1 models (Fig. 2 E).
The propensity of s-Mgm1 to readily self-assemble into an ordered 2D lattice indicates that it possesses distinct interfaces that mediate higher-order self-assembly interactions: a monomer–monomer interface for dimer formation and a dimer–dimer interface for the assembly of a higher-order hexameric s-Mgm1 ring. Our structural model of assembled s-Mgm1 also predicts that an open interface exists, which contains the helical region that is likely orthologous to HR2 in Mfn1, proposed to mediate trans interactions responsible for membrane tethering (Koshiba et al., 2004).
l-Mgm1 preferentially associates with and reconstitutes into liposomes containing CL, but cannot hydrolyze GTP
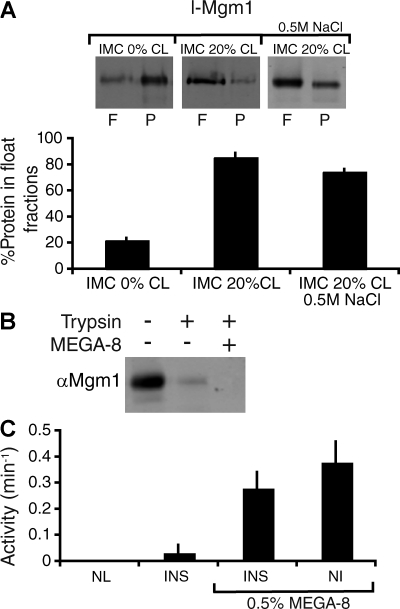
Similar to s-Mgm1, l-Mgm1 was preferentially inserted into the bilayer of IMC liposomes containing CL (Fig. 3 A, IMC 20% CL vs. IMC 0% CL). Stable insertion of l-Mgm1 into IMC liposomes was assessed by treatment with high salt followed by OptiPrep gradient centrifugation, which indicated that the efficiency of l-Mgm1 membrane insertion was ∼74 ± 1.9%, n = 3 (Fig. 3 A, IMC 20% CL, 0.5 M NaCl). Protease protection analysis of reconstituted l-Mgm1 indicated that the membrane-inserted form of l-Mgm1 is in its native topology, with its GTPase and coiled-coil regions facing outwards (Fig. 3 B).
Figure 3.
l-Mgm1 GTPase activity is inhibited when inserted into a membrane bilayer. (A) l-Mgm1 inserts into IMC liposomes. l-Mgm1 was reconstituted into IMC 0% (left) and IMC 20% CL (right) liposomes as described in Materials and methods and fractionated by floatation on sucrose gradients. A 0.5 M NaCl treatment was performed to remove uninserted l-Mgm1 before floatation (right). A representative SDS-PAGE and Western blot of equivalent amounts of the float (F) and pellet (P) fractions is shown. Quantification from three experiments is shown as the mean + SEM (error bars). (B) l-Mgm1 inserts in the correct orientation in IMC liposomes. Reconstituted l-Mgm1 liposomes were treated with trypsin in the presence and absence of MEGA-8 as described (see Materials and methods). (C) GTPase activity of l-Mgm1 was determined as described alone (left; NL, no lipids), after reconstitution into IMC 20% CL liposomes (left; INS), after reconstitution into IMC 20% CL liposomes and subsequent addition of 0.5% MEGA-8 (right; INS), and upon addition of detergent-solubilized l-Mgm1 to IMC 20% CL liposomes (right; NI). Data from three experiments are shown as the mean + SEM (error bars).
In contrast to s-Mgm1, the GTPase activity of inserted l-Mgm1 in IMC liposomes was undetectable (Fig. 3 C, INS). However, l-Mgm1 GTPase activity was observed upon treatment of inserted l-Mgm1 IMC liposomes with concentrations of the detergent MEGA-8 that saturated the IMC membranes, and under noninsertion conditions produced by the addition of MEGA-8–solubilized l-Mgm1 to IMC liposomes (Fig. 3 C, INS and NI, 0.5% MEGA-8, respectively). These observations indicate that insertion of l-Mgm1, placing its GTPase domain proximal to the membrane, likely constrains and attenuates its ability to hydrolyze GTP (Fig. 1 A). The topology of l-Mgm1 and its effects on GTPase activity are also likely responsible for the coevolution of divergent proteolytic pathways in yeast and mammalian cells that function to create the active GTPase, s-Mgm1. These data further suggest that a heterotypic s-Mgm1/l-Mgm1 dimer is the functional unit for fusion, where l-Mgm1 uniquely contributes a transmembrane region required for accurate inner membrane targeting and other fusion activities, whereas s-Mgm1 contributes an active GTPase domain.
Short and long Mgm1 act together in trans to create a heterodimeric functional unit that mediates mitochondrial fusion
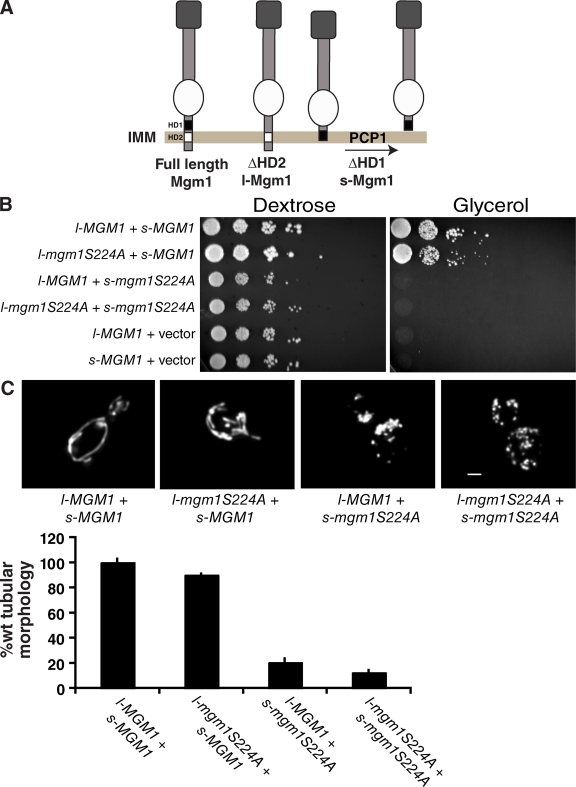
We tested this hypothesis in vivo by engineering separate versions of s- and l-Mgm1, and used these constructs to individually create s-Mgm1S224A and l-Mgm1S224A mutants (Fig. 4 A; Herlan et al., 2004). S224A is a recessive mutation in the G1 motif of the GTPase domain that is predicted to abolish GTPase activity. This mutation does not interfere with the expression and targeting of the protein, but is unable to support mitochondrial fusion in cells (Wong et al., 2003). Our biochemical analyses indicate that neither s-Mgm1S224A nor l-Mgm1S224A possess significant, detectable GTPase activity. We observed that mitochondrial fusion is restored in Δmgm1 cells when expressing a combination of s-Mgm1 and l-Mgm1S224A, but not when expressing the reciprocal combination of s-Mgm1S224A and l-Mgm1, as assessed by the ability of cells to grow on glycerol and by the presence of tubular mitochondrial structures in cells (Fig. 4, B and C). These observations are in agreement with recently published data indicating that the GTPase domain of l-Mgm1 is not required for fusion in vivo (Zick et al., 2009).
Figure 4.
The GTPase activity of l-Mgm1 is not essential for fusion in vivo. (A) Schematic of in vivo l- and s-Mgm1 constructs. Isoforms were constructed by deletion of the first hydrophobic domain (HD1) to produce s-Mgm1 and the second hydrophobic domain (HD2) to produce l-Mgm1. l-Mgm1S224A is functional in vivo as assessed by growth of yeast strains on glycerol media (B) and by mitochondrial morphology (C). Bar, 1 µm. A quantification of mitochondrial morphology in Δmgm1 cells expressing the indicated l- and s-Mgm1 combinations is shown. Data were normalized to Δmgm1 + l-Mgm1 + s-Mgm1 and are shown as the mean + SEM (error bars; three experiments, >50 cells/experiment).
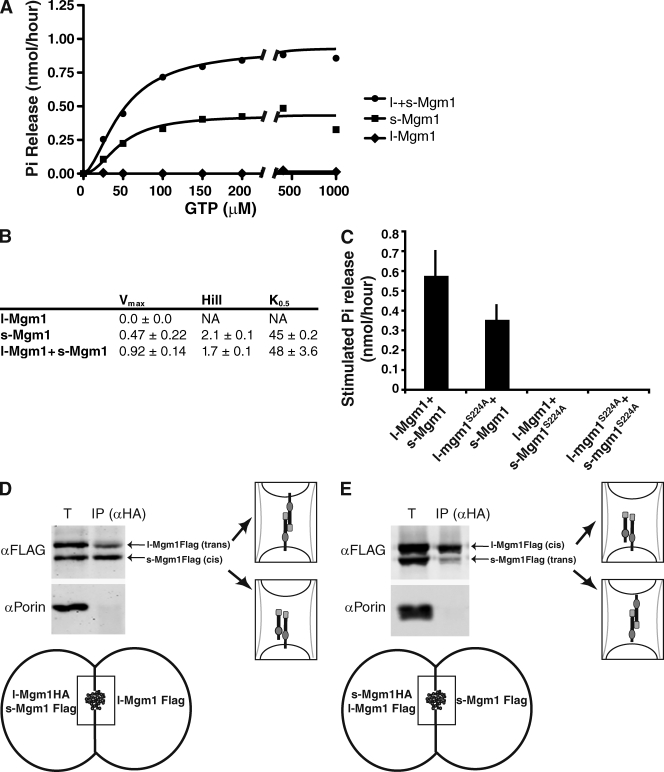
To further test whether s- and l-Mgm1 interact, we examined the ability of a mixture of wild-type recombinant s- and l-Mgm1 proteins to hydrolyze GTP in the presence of IMC liposomes. In reactions with both s-Mgm1 and inserted l-Mgm1, we observed a synergistically stimulated maximal rate of GTP hydrolysis, as compared with maximal GTP hydrolysis rates observed in independent l-Mgm1 or s-Mgm1 reactions (Fig. 5, A–C). Full kinetic analysis of GTP hydrolysis revealed that either s-Mgm1 with liposomes or s-Mgm1 together with inserted l-Mgm1 are positively cooperative with respect to GTP, which is consistent with their coassembly (Fig. 5 B).
Figure 5.
l-Mgm1 stimulates s-Mgm1 GTPase activity. (A) Kinetics of GTP hydrolysis of l-Mgm1, s-Mgm1, and l- + s-Mgm1 (s-Mgm1, 0.5 µM; l-Mgm1, 0.3 µM). A representative kinetic plot fit to the Hill equation is shown. (B) Kinetic parameters for l-Mgm1, s-Mgm1, and l- + s-Mgm1. (C) GTP hydrolysis of s-Mgm1 is stimulated by reconstituted l-Mgm1. The indicated combinations of s- and l-Mgm1/IMC liposomes were analyzed as described in Materials and methods. l- and s-Mgm1–stimulated phosphate release was calculated by subtracting phosphate released for each individual protein from phosphate released when combined. Data from three experiments are shown as the mean + SEM (error bars). (D and E) l- and s-Mgm1 interact in mitochondrial outer membrane fused intermediates in vitro. In outer membrane fused intermediates in vitro, l-Mgm1HA (D) or s-Mgm1HA (E) was immunoprecipitated using α-HA antibodies, and coimmunoprecipitated proteins were analyzed by SDS-PAGE and Western analysis with the indicated antibodies as described (see Materials and methods). l-Mgm1HA/s-Mgm1Flag (cis), l-Mgm1Flag/l-Mgm1Flag (trans), l-Mgm1HA/l-Mgm1Flag (trans), and s-Mgm1HA/s-Mgm1Flag (trans) complexes were detected and are depicted schematically.
To assess which isoform was stimulated under these conditions, we determined maximal rates of GTP hydrolysis in reactions containing combinations of wild-type and S224A mutant l- and s-Mgm1 proteins. Consistent with our in vivo analysis, synergistic stimulation of GTP hydrolysis was observed in reactions containing s-Mgm1 and l-Mgm1S224A, but not in those containing the reciprocal combination of s-Mgm1S224A and l-Mgm1 (Fig. 5 C). These data are consistent with our model that l- and s-Mgm1 assemble together to form a functional unit required for mitochondrial fusion. Our data further demonstrate that within assembled l-Mgm1/s-Mgm1 structures, the s-Mgm1 GTPase domain is activated. Thus, although l-Mgm1 is not able to hydrolyze GTP, it can assemble with and activate the GTPase domain of s-Mgm1.
We also examined how l- and s-Mgm1 interact within mitochondria during fusion in vitro. Previously, under outer membrane fusion conditions in vitro, we observed a physical Mgm1–Mgm1 interaction from opposing inner membranes (Wong et al., 2003; Meeusen et al., 2004, 2006). Using our MGM1 constructs engineered to separately express s- and l-Mgm1 (Fig. 2 A), we resolved whether l- and s-Mgm1 interactions occur on the same membrane and/or on opposing inner membranes in outer membrane fused mitochondria. We mixed mitochondria isolated from cells expressing l-Mgm1-HA and s-Mgm1-FLAG with mitochondria expressing l-Mgm1-FLAG, and after chemical cross-linking under stage 1 conditions, we immunoprecipitated Mgm1 using anti-HA antibodies (Fig. 5 D, see schematic). Western analysis indicated that in outer membrane fused intermediates, l-Mgm1 interacts with l-Mgm1 on opposing inner membranes, and s-Mgm1 interacts with l-Mgm1 on the same membrane (Fig. 5 D). In analogous immunoprecipitation experiments, s-Mgm1 interacted with s-Mgm1 on opposing inner membranes (Fig. 5 E). These data confirm that s- and l-Mgm1 interact with each other across adjacent inner membranes, as well as within the same membrane in mitochondrial fusion intermediates that are arrested at and poised to undergo inner membrane fusion.
Together, our genetic, biochemical, and structural data suggest a model in which the assembly of heterotypic l-/s-Mgm1 structures facilitates the fusion of the mitochondrial inner membrane. EM analysis, however, indicated that in contrast to s-Mgm1, higher-ordered structures are not observed on IMC liposomes containing inserted l-Mgm1 alone or inserted l-Mgm1 in combination with s-Mgm1 in the absence or presence of GTP or nonhydrolyzable GTP analogues (unpublished data). Thus, based on our structural analysis, we postulate that l- and s-Mgm1 self assemble together into shorter range structures, not detectable by negative stain EM analysis, that function to mediate mitochondrial membrane fusion by locally deforming the inner membrane. In this context, we think it is likely that l-Mgm1, as the integral membrane protein, functions to both tether inner membranes together and harness GTP-dependent conformational changes of s-Mgm1 that are needed to destabilize lipid bilayers for fusion.
We can also speculate on the potential roles of homotypic l- and s-Mgm1 structures in mitochondria. One obvious function for homotypic membrane-anchored l-Mgm1 dimers is in the maintenance of cristae structures that have previously been shown to require Mgm1/Opa1 function (Frezza et al., 2006; Meeusen et al., 2006). Our observation that the GTPase domain of l-Mgm1 is inactive and that fusion requires a heterodimeric l-Mgm1/s-Mgm1 structure predicts that l-Mgm1 dimers that form from adjacent membranes could function as membrane tethers that would not promote membrane fusion events. Such l-Mgm1 dimers could provide stability to cristae structures and proximity to adjacent inner membranes in the process of fusion. Although s-Mgm1 dimers possess intrinsic affinity for inner membranes, a role as an inner membrane tether seems unlikely given that they do not possess a transmembrane region. However, s-Mgm1 dimers could play a role in inner membrane organization and cristae structure by forming lattices to create a more lamellar inner membrane structure.
Materials and methods
Strains and plasmids
Strains used were W303 (ade-1; leu2-3; his3-11, 15; trp1-1, ura3-1; can1-100 MatA) and W303 Δmgm1 (ade-1; leu2-3; his3-11, 15; trp1-1, ura3-1; can1-100 MatA; Δmgm1:HIS3). pRS425 dnm1G385D, as described previously (Naylor et al., 2006), was subcloned into pRS316 using the XhoI and NotI sites. To create epitope-tagged alleles of Mgm1, we engineered 3′ restriction sites with an amino acid linker (GCGCGC) by PCR (Meeusen et al., 2006), and cloned the resulting fragment into pRS425. HA and FLAG tags were then subcloned from pFA6a-3HA-TRP1 and pFA6a-3FLAG- into pRS425-Mgm1GCGCGC at the 3′ end (Longtine et al., 1998; Hoppins et al., 2009). s-Mgm1 FLAG was made by deleting the first hydrophobic domain (residues L73-Y90) by site-directed mutagenesis of pRS424-MGM1HA using a PCR-based method with complementary primers of 20–30 nucleotides on either side of the altered nucleotides. Whole plasmid amplification was performed and PCR products were digested with DpnI for 2 h at 37°C to remove template DNA. The amplified plasmid DNA was transformed into DH5α cells. Plasmid DNA was isolated from selected colonies and sequenced to confirm mutations. l-MGM1HA and l-MGM1FLAG were created by deleting the second hydrophobic domain (residues G135-L148) by site-directed mutagenesis of pRS425-MGM1HA and pRS425-MGM1FLAG, respectively, and confirmed by sequencing. l-mgm1S224A and s-mgm1S224A were created by site-directed mutagenesis of pRS425–l-Mgm1Flag and pRS424–s-MGM1HA and confirmed by sequencing. All Δmgm1 strains expressing wild-type, epitope-tagged, or mutant mgm1 alleles were generated by a plasmid shuffle with Dnm1G385D on a URA plasmid.
For expression in baculovirus-infected insect cells, Mgm1 was PCR amplified beginning at residue A151 and cloned into pFastBacHT using the NcoI and XbaI sites, which adds an N-terminal 6xHis tag to yield pFastBacHT–6xHis–s-Mgm1 for purification of s-Mgm1. For expression and purification of l-Mgm1, full-length Mgm1 was first cloned into pFastBac1 using XhoI and XbaI sites with a GCGCGC linker before the C-terminal 6xHis tag. Subsequently, the second hydrophobic domain of Mgm1 containing the processing site to generate s-Mgm1 was deleted by site-directed mutagenesis resulting in pFastBac1–l-Mgm1His.
Growth assay and mitochondrial morphology
For serial dilutions, Δmgm1 cells containing the indicated plasmids were grown to log phase in minimal raffinose. 0.5 OD of each strain was spotted on minimal dextrose and yeast extract, peptone, ethanol, glycerol plates with sequential 1:5 dilutions. To observe mitochondrial morphology, strains containing mtGFP were grown to log phase in minimal raffinose and assessed by fluorescence microscopy at room temperature with a microscope (IX70 Deltavision; Olympus) using a 60× 1.4 N.A. oil objective lens (Olympus), and a 100-watt mercury lamp (Applied Precision, LLC). 3D light microscopy data were collected using an integrated, cooled charge-coupled device (CCD)-based camera (Micromax; Princeton Instruments) equipped with an interline chip (Sony). 3D datasets were processed using DeltaVision's iterative, constrained 3D deconvolution method to remove out-of-focus light. Deconvolved images were analyzed in SoftWorx (Applied Precision, LLC).
Immunoprecipitation
Immunoprecipitations were performed to detect cis and trans Mgm1 interactions during outer membrane fusion (Meeusen et al., 2006). Mitochondria were isolated from cells expressing s-Mgm1Flag/l-Mgm1HA or l-Mgm1Flag/s-Mgm1 (for l-Mgm1 trans interactions), or s-Mgm1HA/l-Mgm1FLAG or l-Mgm1/s-Mgm1FLAG (for s-Mgm1 trans interactions), and then mixed and subjected to outer mitochondrial membrane fusion (Meeusen et al., 2004). After fusion, mitochondria were subjected to chemical cross-linking with 1 mM dithiobis succinimidyl propionate (Thermo Fischer Scientific) for 1 h on ice. Reactions were quenched with 100 mM glycine and proteins were subjected to TCA precipitation. Proteins were denatured and solubilized in 100 ml MURB (100 mM MES, pH 7, 1% SDS, and 3 M urea) followed by the addition of 900 µl TWIP (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 0.5% Tween 20, and 0.1% EDTA). Insoluble proteins were removed by centrifugation at 16,000 g at 4°C for 15 min. Lysate was preadsorbed with 75 µl TWIP-equilibrated protein A agarose beads (Santa Cruz Biotechnology, Inc.) for 30 min at 4°C, then incubated with 8 µl polyclonal HA antibody (Covance) for 1.5 h at 4°C followed by incubation with 75 µl TWIP-equilibrated protein A agarose beads for 45 min at 4°C. Agarose beads were washed three times with 500 µl TWIP and protein was eluted with MURB + 10% β-mercaptoethanol to reduce cross links. Analysis by SDS-PAGE and Western blotting was performed on the total fraction and the eluate fractions with monoclonal α-Flag antibody (Sigma-Aldrich) and monoclonal α-Porin (Covance).
Liposome preparation
Lipids were obtained from Avanti Polar Lipids in chloroform. The following lipids were used in biochemical assays: tetraoleoyl-CL, palmitoyl-oleoyl phosphatidylcholine, palmitoyl-oleoyl phosphatidylethanolamine, soybean phosphatidylinositol, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphate (POPA), and 1-palmitoyl-2-oleoyl-sn-glycero-3-(phosphor-l-serine). Liposomes were mixed in the appropriate ratios and dried under nitrogen gas. The dried lipids were placed under vacuum for at least 1 h, and hydrated in 100 mM NaCl for several hours to overnight. Hydrated liposomes were extruded through a 1-mm nanopore membrane (GE Healthcare) a minimum of 11 times.
Compositions of liposomes used in this study were as follows. IM composition: 39% phosphatidylcholine (PC), 29% phosphatidylethanolamine (PE), 20% CL, 8% phosphatidylinositol (PI), 2% phosphatidic acid (PA), and 2% phosphatidylserine (PS). OM composition: 43% PC, 22% PE, 20% PI, 3% PA, 2% PS, and 6% CL, as described in Zinser and Daum (1995). When the percentage of CL was varied, compensatory changes were made in the percentage of PC present in the lipid mixture. Headgroup-labeled lissamine rhodamine B phosphatidylethanolamine was added to all reactions in tracer amounts.
Purification of s- and l-Mgm1
To express s- and l-Mgm1 in insect cells for expression, we first made bacmid by transforming the baculovirus expression vectors described in “Strains and plasmids” into DH10Bac cells. Bacmid was then transfected into Sf9 insect cells using Transfection Buffer Set A and B (BD). Primary virus was harvested after 5 d and amplified twice in Sf9 cells to make high-titer 3° virus. The high-titer virus was used to infect Hi5 suspension cells (a gift from B. Hammock, University of California, Davis, Davis, CA) for protein expression. Hi5 cells were harvested after 48 h and stored at −80°C.
For s-Mgm1, cells were thawed and resuspended in wash buffer (25 mM Hepes, pH 7.0, 25 mM Pipes, pH 7.0, 500 mM NaCl, 80 mM imidazole, pH 7.4, and 0.1% MEGA-8 [Dojindo]) with protease inhibitor cocktail I (EMD) and 2 mM PMSF. Cells were then lysed by sonication and passage through a 27-gauge/0.5-inch needle. The lysate was centrifuged at 60,000 g for 40 min. The cleared lysate was loaded onto a HiTrap metal chelating column (GE Healthcare) attached to an AKTA prime system (GE Healthcare). s-Mgm1 was eluted from the column by using a linear gradient of 25 mM Hepes, pH 7.0, 25 mM Pipes, pH 7.0, 500 mM NaCl, 500 mM imidazole, pH 7.4, and 0.1% MEGA-8. s-Mgm1 was further purified over a HiLoad 16/60 Superdex 200 preparative gel filtration column (GE Healthcare) in 20 mM Tris-Cl, pH 8.0, 2 mM EDTA, 375 mM NaCl, and 0.01% (vol/vol) β-mercaptoethanol. Glycerol was added to a final concentration of 20%, rendering the following short freezing buffer (S-FB): 16 mM Tris-Cl, pH 8.0, 2 mM EDTA, 300 mM NaCl, and 20% glycerol. The protein was aliquoted, flash frozen in liquid N2, and stored at −80°C.
l-Mgm1 was prepared identically with the following changes. Lysis of l-Mgm1 cells was done in buffer containing 25 mM Hepes, pH 7.0, 25 mM Pipes, pH 7.0, 200 mM NaCl, 80 mM imidazole, pH 7.4, and 10% glycerol. After the centrifugation step at 60,000 g, 0.5% Triton X-100 (Sigma-Aldrich) was added to the cleared lysate and loaded onto a HiTrap metal chelating column (GE Healthcare). Triton X-100 was exchanged for 1.5% MEGA-8 during a wash step. l-Mgm1 was eluted in 25 mM Hepes, pH 7.0, 25 mM Pipes, pH 7.0, 200 mM NaCl, 80 mM imidazole, pH 7.4, 10% glycerol, and 1.5% MEGA-8. Glycerol was added to 20%, rendering the following long freezing buffer (L-FB): 23 mM Hepes/Pipes, 450 mM imidazole, 1.4% MEGA-8, and 20% glycerol. The protein was aliquoted, flash frozen in liquid N2, and stored at −80°C. The protein was stable at 4°C for at least 1 mo. Concentrations of s- and l-Mgm1 were determined by BCA Protein assay (Thermo Fisher Scientific).
Reconstitution of l-Mgm1
l-Mgm1 was reconstituted in 100-µl reactions containing liposomes by saturating 0.2 mg liposomes with 0.5% MEGA-8 detergent. 7.0 µM l-Mgm1 was added to the detergent liposomes so the final MEGA-8 concentration was 0.9%. The lipid, protein, and detergent mixture was incubated for 1 h and then rapidly diluted 10-fold with 20 mM Tris, pH 8.0, and 100 mM NaCl. After reconstitution, l-Mgm1 was at a final concentration of 0.7 µM. Salt extraction of inserted l-Mgm1 was performed to remove associated protein by adding NaCl to l-Mgm1 to a final concentration of 0.5M. To remove uninserted protein, we performed equilibrium centrifugation using an OptiPrep gradient. Specifically, 860 µl of a 53% OptiPrep solution (0.5 M NaCl and 20 mM Tris; Sigma-Aldrich) was added to 500 µl of salt-washed l-Mgm1 to make a 34% OptiPrep solution. This solution was layered with 2 ml 23% OptiPrep floatation buffer (OFB; 20 mM Tris, and 100 mM NaCl), 1 ml 6% OFB, and 750 µl 0% OFB. Liposomes were floated at 200,000 g for 2.5 h (SW-55 rotor; Sorvall) and then extracted from the gradient in a 500-µl fraction.
GTPase assays
All assays were performed in 5 mM MgCl2, 100 mM NaCl, and 20 mM Tris, pH 8.0. The malachite green assay was used to detect free phosphate as described previously (Quan and Robinson, 2005). s-Mgm1 GTPase reactions were performed by first incubating 1 µM s-Mgm1 with 0.4 mg/ml liposomes for 15 min at RT. Buffer was added to give a final concentration of 0.2 mg/ml liposomes in 5 mM MgCl2, 20 mM Tris, pH 8.0, 100 mM NaCl, and 500 µM GTP (unless otherwise stated). The reaction was stopped after 1 h by addition of 125 mM EDTA. 200 µl of malachite green was added to 25 µl of reaction and the absorbance at 650 nm was determined using a Spectra Max Plus plate reader (MDS Analytical Technologies). GTP hydrolysis was determined by subtracting the amount of phosphate release in a no-protein control from that of the protein reactions.
l-Mgm1 and l-Mgm1 + s-Mgm1 GTPase assays were done after reconstitution by adding 0.3 µM of inserted l-Mgm1 to 0.1 mg/ml liposomes and 0.5 µM s-Mgm1 or buffer. The proteins were incubated for 30 min at RT before adding reaction buffer (25 mM MgCl2 and 20 mM Tris-Cl, pH 8.0) with the nucleotide to yield 0.3 mg/ml liposomes, 0.2% MEGA-8, 5 mM MgCl2, 20 mM Tris, 100 mM NaCl, and 500 µM GTP (unless otherwise stated). GTP hydrolysis activity was determined using malachite green as described above.
Equilibrium sucrose gradient centrifugation of liposomes
Purified s-Mgm1 or freezing buffer was preincubated with 0.4 mg/ml liposomes for 20 min in a volume of 50 µl. The protein/lipid mix was diluted to 100 µL in floatation buffer (FlB) so that the final liposome concentration was 0.2 mg/ml in 20 mM Tris, pH 8.0, 100 mM NaCl, 5 mM MgCl2, and 300 µM nucleotide, then incubated for 1 h. 400 µl of FlB + 50% sucrose (FlB50) was added and mixed thoroughly, and the resulting 40% sucrose reaction mix was transferred to a centrifuge tube. The 40% sucrose layer was overlayed with 1 mL FlB30, 500 µl FlB10, and 250 µl FlB0.
Sucrose step gradients were centrifuged in a SW-55 rotor at 200,000 g for 2.5 h at 4°C. Five 450-µl fractions were pipetted from the top and the pellet was resuspended in 450 µl FB, resulting in a total of six fractions. The top three fractions were pooled as the float fractions (F) because the majority of liposomes were found in these fractions, as determined by measuring the rhodamine fluorescence of each fraction using a Spectra Max Plus plate reader with the excitation and emission monochromators set at 550 nm and 590 nm, respectively. The bottom two fractions were combined with the pellet and were pooled as a single fraction (P). For quantification of the fraction of protein that floated with the liposomes, equal volumes of F and P fractions were analyzed by SDS-PAGE and Western analysis using the primary antibody, α-Mgm1, and an IRDye 800 CW secondary antibody (LI-COR Biosciences) for detection. The immunoreactive bands were detected with the Odyssey Infrared Imaging System (LI-COR Biosciences) and quantified using the accompanying software.
Hydrodynamic analysis
The Svedberg coefficients of s- and l- Mgm1 were determined as described previously (Ingerman et al., 2005). 100 µl of 0.5 µM Mgm1 in 5% sucrose was loaded on top of a 2 ml 5–20% sucrose gradient and subjected to a 4-h ultracentrifugation spin at 200,000 g in an SW-55 rotor. Molecular mass markers with known sedimentation coefficients (17–0445-01; GE Healthcare) were simultaneously run over a separate gradient. 10 × 0.2 ml fractions were collected manually from the top of the gradients for analysis. Svedberg coefficients were estimated from a standard curve from fraction number versus known sedimentation coefficients of the protein standards.
To determine the Stokes’ radii of s- and l-Mgm1, purified protein at 0.5 µg/µl or gel filtration standards (Bio-Rad Laboratories) were run over a HiLoad16/60 Superset 200 preparative gel filtration column using an AKTA prime system (GE Healthcare). For each protein component of gel filtration standards, values of Kd1/3 were plotted versus Stokes’ radii to construct a Porath correlation standard curve. Molecular mass was calculated from combined sucrose gradient and gel filtration data using the method developed by Siegel and Monty (1966). l-Mgm1 was analyzed identically to s-Mgm1, except the 5–20% sucrose gradient contained 0.75% MEGA-8.
s-Mgm1 cross-linking
s-Mgm1 was dialyzed into 25 mM Hepes/Pipes, 300 mM NaCl, and 20% glycerol for 1 h at 4°C. A 10-fold molar excess of the nonreversible cross-linker bis(sulfosuccinimidyl)suberate (BS3; Thermo Fisher Scientific) was added to s-Mgm1 preincubated with 0.2 mg/ml liposomes or liposome buffer for 30 min. The reactions were quenched with 80 mM Tris-Cl, pH 8.0, for 15 min at RT. Cross-linked products were resolved on a 3–8% Tris-acetate gel (NuPage Novel; Invitrogen) and visualized by Coomassie blue staining. HiMark molecular weight markers (Invitrogen) were used as standards.
Mgm1 structural analysis
2D crystals were grown under a lipid monolayer covering the air–water interface, as described by Lévy et al., (1999). In brief, ∼60-µl wells in a Teflon block were filled with Tris 20 mM, pH 8.0, such that a flat air–water interface was formed. A 0.5-ml drop of IMC lipid mixture at 0.1 mg/ml concentration dissolved in a 9:1 chloroform/methanol mixture was spread on the water surface. After 24 h of incubation time at RT, 5 µl of 0.1 mg/ml s-Mgm1 suspension was added through a side well. After an additional 24 h of incubation time, the monolayers were recovered by applying nonglow discharged carbon-coated grids to the surface of the wells. Grids were removed, blotted, stained with 2% uranyl acetate, blotted again, air dried, and transferred into a transmission electron microscope (JEM-1230; JEOL, Ltd.), operated at an acceleration voltage of 120 kV. Images were recorded at 30,000× nominal magnification with a F214 CCD camera (2,048 × 2,048 pixels; TVIPS GmbH), which corresponds to a pixel size of 3.89Å at the specimen level.
Images were processed using the 2dx program system (Gipson et al., 2007a,b), which is based on the MRC software (Crowther et al., 1996). A total of nine images of nontilted negatively stained s-Mgm1 2D crystals were processed and merged, yielding a projection map at 31 Å resolution. Images from six tilted negatively stained 2D crystals at a 30° sample tilt and eight tilted crystals at a 45° sample tilt were processed and merged into a 3D reconstruction, and the same 31-Å resolution cutoff was applied. The final map was created from 1,033 observed amplitudes and phases, and had an overall phase residual of 26.8°. The overall weighted phase residual was 9.6°.
Homology modeling of s-Mgm1
Homology modeling was performed with the program Modeller (Martí-Renom et al., 2000) using the structures of dynamin A, dynamin 1, and dynamin-like bacterial protein (PDB accession nos. 1JWY, 2AKA, and 2J69) as references. Sequence alignment for Mgm1, dynamin A, and dynamin 1 was performed within Modeller. Because of the low homology between Mgm1 and the dynamin-like bacterial protein, the sequence alignment for the latter was obtained from the mGenThreader server (McGuffin and Jones, 2003). The full alignment was checked with the mumsa program (Lassmann and Sonnhammer, 2002), and 1,000 models were generated using Modeller (Martí-Renom et al., 2000). The five models with the lowest DOPE score were manually inspected, and the best model that did not show knots in coil regions was selected. This model had a DOPE score of −59,372.
The dimeric form of s-Mgm1 was obtained by 100 rounds of blind docking with the GRAMM-X server (Tovchigrechko and Vakser, 2005), followed by a local search with Rosetta (Gray et al., 2003). From the 1,000 docked models, the best 10 dimeric arrangements were selected by manual verification of the presence of a twofold symmetry and a head-to-head arrangement of the monomers. The model that showed the best fit with the experimental 3D reconstruction and the lowest number of atoms outside of the volume was manually selected (Fig. 4 D).
Figures of the 3D reconstruction were produced with Chimera (Pettersen et al., 2004) and the homology model was rendered using POV-Ray (http://www.povray.org).
Negative stain EM
Reactions containing l-Mgm1, s-Mgm1, and l- + s-Mgm1 or corresponding storage buffers were prepared as described for the GTPase assays, and nucleotide was added as indicated. When the reactions were complete, a glow-discharged carbon-coated grid was placed on a 10-µl drop of the reaction mix for 1 min. The grid was then stained with 2% uranyl acetate for 1.5 min, blotted, and air dried. The grids were imaged at 37,000× using a transmission EM (CM120 [Phillips] with a biotwin lens [FEI]) operating at 80 kV, and images were captured digitally using a MegaScan CCD camera (Gatan) and the Digital Micrograph software package (Gatan).
Acknowledgments
We would like to thank Dr. Marijn Ford for advice on protein purification and members of the Nunnari laboratory for discussion and comments on the manuscript. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institutes of Health P41 RR-01081) and The PyMOL Molecular Graphics System (DeLano Scientific, San Carlos, CA). We are thankful to V. Vu for assistance in computer image processing.
This work was supported by the National Institutes of Health (NIH) grants R01GM062942 to J. Nunnari and U54GM074929 to H. Stahlberg and L. Dominguez-Ramirez. R. DeVay was supported by a Howard Hughes Medical Institute translation Fellowship to the University of California, Davis (S-HHMI01-GSRRD) and by an American Heart Preodoctoral Award (0715023Y), L. Dominguez-Ramirez was supported by the Pew Latin American Fellow Program (no. MCA4934SC), and L.L. Lackner is supported by NIH postdoctoral fellowship 1F32GM078749.
Footnotes
Abbreviations used in this paper:
- CCD
- charge-coupled device
- CL
- cardiolipin
- DRP
- dynamin-related protein
- IMC
- inner mitochondrial membrane composition
- OMC
- outer membrane composition
References
- Cipolat S., Rudka T., Hartmann D., Costa V., Serneels L., Craessaerts K., Metzger K., Frezza C., Annaert W., D'Adamio L., et al. 2006. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling.Cell. 126:163–175 doi:10.1016/j.cell.2006.06.021 [DOI] [PubMed] [Google Scholar]
- Crowther R.A., Henderson R., Smith J.M. 1996. MRC image processing programs.J. Struct. Biol. 116:9–16 doi:10.1006/jsbi.1996.0003 [DOI] [PubMed] [Google Scholar]
- Danino D., Hinshaw J.E. 2001. Dynamin family of mechanoenzymes.Curr. Opin. Cell Biol. 13:454–460 doi:10.1016/S0955-0674(00)00236-2 [DOI] [PubMed] [Google Scholar]
- Daum G., Vance J.E. 1997. Import of lipids into mitochondria.Prog. Lipid Res. 36:103–130 doi:10.1016/S0163-7827(97)00006-4 [DOI] [PubMed] [Google Scholar]
- Duvezin-Caubet S., Jagasia R., Wagener J., Hofmann S., Trifunovic A., Hansson A., Chomyn A., Bauer M.F., Attardi G., Larsson N., et al. 2006. Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J. Biol Chem.J. Biol. Chem. 281:37972–37979 [DOI] [PubMed] [Google Scholar]
- Esser K., Tursun B., Ingenhoven M., Michaelis G., Pratje E. 2002. A novel two-step mechanism for removal of a mitochondrial signal sequence involves the mAAA complex and the putative rhomboid protease Pcp1.J. Mol. Biol. 323:835–843 doi:10.1016/S0022-2836(02)01000-8 [DOI] [PubMed] [Google Scholar]
- Frezza C., Cipolat S., Martins de Brito O., Micaroni M., Beznoussenko G.V., Rudka T., Bartoli D., Polishuck R.S., Danial N.N., De Strooper B., Scorrano L. 2006. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion.Cell. 126:177–189 doi:10.1016/j.cell.2006.06.025 [DOI] [PubMed] [Google Scholar]
- Gasper R., Meyer S., Gotthardt K., Sirajuddin M., Wittinghofer A. 2009. It takes two to tango: regulation of G proteins by dimerization.Nat. Rev. Mol. Cell Biol. 10:423–429 doi:10.1038/nrm2689 [DOI] [PubMed] [Google Scholar]
- Gipson B., Zeng X., Stahlberg H. 2007a. 2dx_merge: data management and merging for 2D crystal images.J. Struct. Biol. 160:375–384 doi:10.1016/j.jsb.2007.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gipson B., Zeng X., Zhang Z.Y., Stahlberg H. 2007b. 2dx—user-friendly image processing for 2D crystals.J. Struct. Biol. 157:64–72 doi:10.1016/j.jsb.2006.07.020 [DOI] [PubMed] [Google Scholar]
- Gray J.J., Moughon S., Wang C., Schueler-Furman O., Kuhlman B., Rohl C.A., Baker D. 2003. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations.J. Mol. Biol. 331:281–299 doi:10.1016/S0022-2836(03)00670-3 [DOI] [PubMed] [Google Scholar]
- Griffin E.E., Chan D.C. 2006. Domain interactions within Fzo1 oligomers are essential for mitochondrial fusion.J. Biol. Chem. 281:16599–16606 doi:10.1074/jbc.M601847200 [DOI] [PubMed] [Google Scholar]
- Griparic L., Kanazawa T., van der Bliek A.M. 2007. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage.J. Cell Biol. 178:757–764 doi:10.1083/jcb.200704112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlan M., Vogel F., Bornhovd C., Neupert W., Reichert A.S. 2003. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA.J. Biol. Chem. 278:27781–27788 doi:10.1074/jbc.M211311200 [DOI] [PubMed] [Google Scholar]
- Herlan M., Bornhövd C., Hell K., Neupert W., Reichert A.S. 2004. Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor.J. Cell Biol. 165:167–173 doi:10.1083/jcb.200403022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann G.J., Thatcher J.W., Mills J.P., Hales K.G., Fuller M.T., Nunnari J., Shaw J.M. 1998. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p.J. Cell Biol. 143:359–373 doi:10.1083/jcb.143.2.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S., Lackner L., Nunnari J. 2007. The machines that divide and fuse mitochondria.Annu. Rev. Biochem. 76:751–780 doi:10.1146/annurev.biochem.76.071905.090048 [DOI] [PubMed] [Google Scholar]
- Hoppins S., Horner J., Song C., McCaffery J.M., Nunnari J. 2009. Mitochondrial outer and inner membrane fusion requires a modified carrier protein.J. Cell Biol. 184:569–581 doi:10.1083/jcb.200809099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerman E., Perkins E.M., Marino M., Mears J.A., McCaffery J.M., Hinshaw J.E., Nunnari J. 2005. Dnm1 forms spirals that are structurally tailored to fit mitochondria.J. Cell Biol. 170:1021–1027 doi:10.1083/jcb.200506078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Eura Y., Mihara K. 2004. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity.J. Cell Sci. 117:6535–6546 doi:10.1242/jcs.01565 [DOI] [PubMed] [Google Scholar]
- Ishihara N., Fujita Y., Oka T., Mihara K. 2006. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1.EMBO J. 25:2966–2977 doi:10.1038/sj.emboj.7601184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba T., Detmer S.A., Kaiser J.T., Chen H., McCaffery J.M., Chan D.C. 2004. Structural basis of mitochondrial tethering by mitofusin complexes.Science. 305:858–862 doi:10.1126/science.1099793 [DOI] [PubMed] [Google Scholar]
- Lassmann T., Sonnhammer E.L. 2002. Quality assessment of multiple alignment programs.FEBS Lett. 529:126–130 doi:10.1016/S0014-5793(02)03189-7 [DOI] [PubMed] [Google Scholar]
- Lévy D., Mosser G., Lambert O., Moeck G.S., Bald D., Rigaud J.L. 1999. Two-dimensional crystallization on lipid layer: A successful approach for membrane proteins.J. Struct. Biol. 127:44–52 doi:10.1006/jsbi.1999.4155 [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae.Yeast. 14:953–961 doi:10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U [DOI] [PubMed] [Google Scholar]
- Low H.H., Löwe J. 2006. A bacterial dynamin-like protein.Nature. 444:766–769 doi:10.1038/nature05312 [DOI] [PubMed] [Google Scholar]
- Martí-Renom M.A., Stuart A.C., Fiser A., Sánchez R., Melo F., Sali A. 2000. Comparative protein structure modeling of genes and genomes.Annu. Rev. Biophys. Biomol. Struct. 29:291–325 doi:10.1146/annurev.biophys.29.1.291 [DOI] [PubMed] [Google Scholar]
- McGuffin L.J., Jones D.T. 2003. Improvement of the GenTHREADER method for genomic fold recognition.Bioinformatics. 19:874–881 doi:10.1093/bioinformatics/btg097 [DOI] [PubMed] [Google Scholar]
- McQuibban G.A., Saurya S., Freeman M. 2003. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease.Nature. 423:537–541 doi:10.1038/nature01633 [DOI] [PubMed] [Google Scholar]
- Meeusen S., McCaffery J.M., Nunnari J. 2004. Mitochondrial fusion intermediates revealed in vitro.Science. 305:1747–1752 doi:10.1126/science.1100612 [DOI] [PubMed] [Google Scholar]
- Meeusen S., DeVay R., Block J., Cassidy-Stone A., Wayson S., McCaffery J.M., Nunnari J. 2006. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1.Cell. 127:383–395 doi:10.1016/j.cell.2006.09.021 [DOI] [PubMed] [Google Scholar]
- Meglei G., McQuibban G.A. 2009. The dynamin-related protein Mgm1p assembles into oligomers and hydrolyzes GTP to function in mitochondrial membrane fusion (dagger).Biochemistry. 48:1774–1784 doi:10.1021/bi801723d [DOI] [PubMed] [Google Scholar]
- Naylor K., Ingerman E., Okreglak V., Marino M., Hinshaw J.E., Nunnari J. 2006. Mdv1 interacts with assembled dnm1 to promote mitochondrial division.J. Biol. Chem. 281:2177–2183 [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. 2004. UCSF Chimera—a visualization system for exploratory research and analysis.J. Comput. Chem. 25:1605–1612 doi:10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Quan A., Robinson P.J. 2005. Rapid purification of native dynamin I and colorimetric GTPase assay.Methods Enzymol. 404:556–569 doi:10.1016/S0076-6879(05)04049-8 [DOI] [PubMed] [Google Scholar]
- Ramachandran R., Surka M., Chappie J.S., Fowler D.M., Foss T.R., Song B.D., Schmid S.L. 2007. The dynamin middle domain is critical for tetramerization and higher-order self-assembly.EMBO J. 26:559–566 doi:10.1038/sj.emboj.7601491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapaport D., Brunner M., Neupert W., Westermann B. 1998. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae.J. Biol. Chem. 273:20150–20155 doi:10.1074/jbc.273.32.20150 [DOI] [PubMed] [Google Scholar]
- Siegel L.M., Monty K.J. 1966. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases.Biochim. Biophys. Acta. 112:346–362 [DOI] [PubMed] [Google Scholar]
- Sesaki H., Southard S.M., Hobbs A.E., Jensen R.E. 2003. Cells lacking Pcp1p/Ugo2p, a rhomboid-like protease required for Mgm1p processing, lose mtDNA and mitochondrial structure in a Dnm1p-dependent manner, but remain competent for mitochondrial fusion.Biochem. Biophys. Res. Commun. 308:276–283 doi:10.1016/S0006-291X(03)01348-2 [DOI] [PubMed] [Google Scholar]
- Song Z., Chen H., Fiket M., Alexander C., Chan D.C. 2007. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L.J. Cell Biol. 178:749–755 doi:10.1083/jcb.200704110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperka-Gottlieb C.D., Hermetter A., Paltauf F., Daum G. 1988. Lipid topology and physical properties of the outer mitochondrial membrane of the yeast, Saccharomyces cerevisiae.Biochim. Biophys. Acta. 946:227–234 doi:10.1016/0005-2736(88)90397-5 [DOI] [PubMed] [Google Scholar]
- Tovchigrechko A., Vakser I.A. 2005. Development and testing of an automated approach to protein docking.Proteins. 60:296–301 doi:10.1002/prot.20573 [DOI] [PubMed] [Google Scholar]
- Wong E.D., Wagner J.A., Scott S.V., Okreglak V., Holewinske T.J., Cassidy-Stone A., Nunnari J. 2003. The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion.J. Cell Biol. 160:303–311 doi:10.1083/jcb.200209015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick M., Duvezin-Caubet S., Schäfer A., Vogel F., Neupert W., Reichert A.S. 2009. Distinct roles of the two isoforms of the dynamin-like GTPase Mgm1 in mitochondrial fusion.FEBS Lett. 583:2237–2243 doi:10.1016/j.febslet.2009.05.053 [DOI] [PubMed] [Google Scholar]
- Zinser E., Daum G. 1995. Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae.Yeast. 11:493–536 doi:10.1002/yea.320110602 [DOI] [PubMed] [Google Scholar]