The epigenetic mark indicative of DNA UV damage or double-strand breaks is achieved via a common pathway regardless of the cause of damage.
Abstract
Chromatin modifications are an important component of the of DNA damage response (DDR) network that safeguard genomic integrity. Recently, we demonstrated nucleotide excision repair (NER)–dependent histone H2A ubiquitination at sites of ultraviolet (UV)-induced DNA damage. In this study, we show a sustained H2A ubiquitination at damaged DNA, which requires dynamic ubiquitination by Ubc13 and RNF8. Depletion of these enzymes causes UV hypersensitivity without affecting NER, which is indicative of a function for Ubc13 and RNF8 in the downstream UV–DDR. RNF8 is targeted to damaged DNA through an interaction with the double-strand break (DSB)–DDR scaffold protein MDC1, establishing a novel function for MDC1. RNF8 is recruited to sites of UV damage in a cell cycle–independent fashion that requires NER-generated, single-stranded repair intermediates and ataxia telangiectasia–mutated and Rad3-related protein. Our results reveal a conserved pathway of DNA damage–induced H2A ubiquitination for both DSBs and UV lesions, including the recruitment of 53BP1 and Brca1. Although both lesions are processed by independent repair pathways and trigger signaling responses by distinct kinases, they eventually generate the same epigenetic mark, possibly functioning in DNA damage signal amplification.
Introduction
Endogenous and environmental agents continuously damage DNA, compromise its normal functioning, and are associated with accelerated ageing and malignant transformation. DNA damage response (DDR) mechanisms, including diverse repair and cell cycle control pathways, protect organisms against the adverse effects of genomic insults (Hoeijmakers, 2001). DDR-associated chromatin modifications play an important role in regulating both DNA repair and checkpoints (Bennett and Harper, 2008), as illustrated by the involvement of the ataxia telangiectasia–mutated (ATM) kinase in DNA double-strand break (DSB)–induced DDR. ATM is the upstream kinase responsible for the phosphorylation of H2AX on serine 193 (γH2AX) in response to DSBs (Rogakou et al., 1998). This early damage marker subsequently recruits MDC1 (mediator of DNA damage checkpoint protein 1), which is an important step for the subsequent recruitment of 53BP1 and BRCA1 at the damaged chromatin (Stucki et al., 2005), thereby mediating the checkpoint signaling toward the effector kinases CHK1 and CHK2 (Kim and Chen, 2008). Additional ATM recruitment results in enhanced accumulation of DNA repair factors. The collective association of a large number of diverse DDR factors at the damaged chromatin results in microscopically detectable structures referred to as ionizing radiation (IR)–induced foci (IRIF; Bekker-Jensen et al., 2006).
UV-induced DNA damage results in helix-distorting DNA lesions predominantly consisting of cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts (6-4PPs). In mammals, these DNA damages are removed by nucleotide excision repair (NER) that eliminates a wide spectrum of helix-distorting lesions in a multistep “cut and patch”–type reaction. The damage is excised as a 25–30 oligonucleotide DNA fragment followed by gap filling through DNA repair synthesis and restoration of an intact DNA duplex by a final ligation step (Hoeijmakers, 2001; Gillet and Schärer, 2006). Although the function of the various proteins essential for the core DNA repair process is well understood, its connection with the UV-induced DNA damage signaling is less well characterized. In contrast to DSB repair, NER does not take place in subnuclear structures like IRIF. These IRIF are linked with DNA damage–induced large-scale chromatin modifications. Although a variety of chromatin modifications have been associated with NER (Dinant et al., 2008), the biological significance of these changes is poorly understood. However, the best-described UV-induced damage signaling, involving RAD17, the 9-1-1 complex, ATM and Rad3-related (ATR), and Chk1, is linked to replication stress rather than to the repair process itself (Niida and Nakanishi, 2006). However, Giannattasio et al. (2004) identified a clear NER-dependent signaling pathway in yeast and mammalian cells (Giannattasio et al., 2004). One of the currently best-known players in the UV-induced DDR is the phosphatidylinositol 3-kinase ATR, which is activated upon UV-induced replication stress (Zou and Elledge, 2003; Falck et al., 2005). This activation is caused through recruitment of ATR by ATR-interacting protein to replication protein A (RPA)–coated single-stranded DNA, which occurs at stalled replication forks (Cortez et al., 2001). It has recently become clear that UV damage also induces ATR activation and γH2AX in a cell cycle–independent fashion (O'Driscoll et al., 2003; Hanasoge and Ljungman, 2007; Matsumoto et al., 2007; Stiff et al., 2008), which may trigger similar large-scale chromatin modification as observed after DSB. Similar to the observed NER-dependent Chk1 activation, this activation is also dependent on active NER. During NER, single-stranded DNA repair intermediates are responsible for RPA recruitment, resulting in ATR activation (Hanasoge and Ljungman, 2007).
Recently, we have identified a novel UV-induced, NER-dependent chromatin modification, ubiquitination of H2A (Bergink et al., 2006), although the molecular mechanism and its function in UV– remain elusive. These findings were recently confirmed in another study by Zhu et al. (2009). In addition, other core histones are also ubiquitinated upon UV damage like H2B (Robzyk et al., 2000; Giannattasio et al., 2005), H3, and H4 (Wang et al., 2006), although to a much lower extent in absolute number of histone molecules being modified. A similar chromatin mark on H2A (Mailand et al., 2007; Nicassio et al., 2007) and H2AX (Huen et al., 2007; Ikura et al., 2007) was later identified in response to DSB. In contrast to DSB–DDR, within UV-induced DDR, the connection between NER, chromatin modifications, and signaling is less well established. Next to the molecular mechanism and function of the observed UV-induced H2A ubiquitination, an appealing question is to which extent this epigenetic DDR is mechanistically conserved between these two types of DNA damage that are repaired by entirely different repair machineries.
Results
UV-induced H2A ubiquitination is a continuous and dynamic process and depends on Ubc13 and RNF8
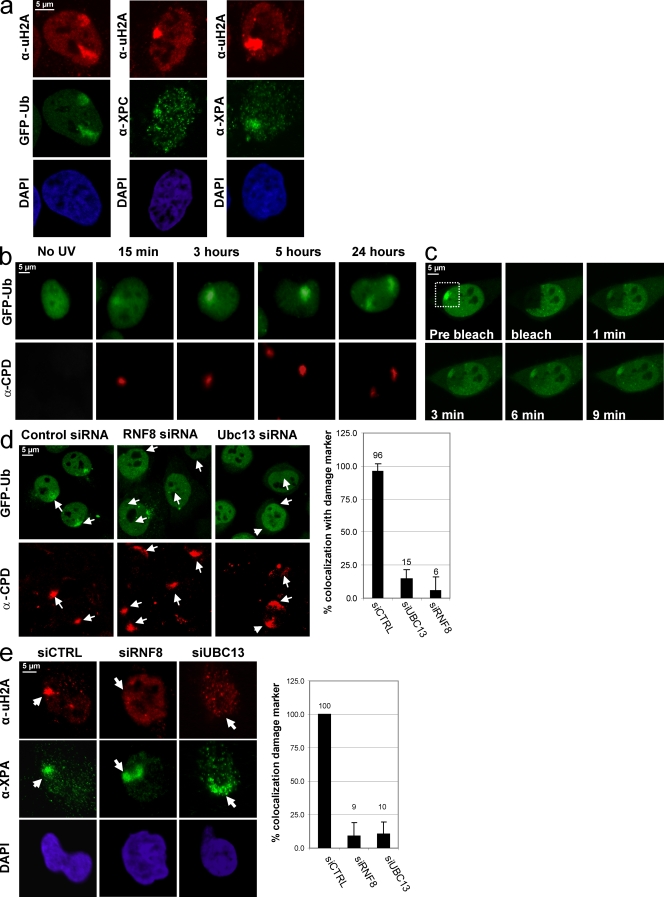
H2A ubiquitination can be visualized using antibodies recognizing specifically ubiquitinated H2A on Western blot or immunofluorescence (IF; Baarends et al., 2005) or indirect in living cells studying GFP-ubiquitin (Ub; Dantuma et al., 2006). Previous experiments in our laboratory (Bergink et al., 2006) have identified a UV-induced H2A ubiquitination as shown by IF (Fig. 1 a) and Western blot analysis (Fig. S1 a). GFP-Ub was found to be accumulated after local UV damage (LUD) infliction at subnuclear regions that were enriched for Ub-H2A and colocalize with the core NER factors XPC (Xeroderma pigmentosum group C) and XPA (Fig. 1 a) and with the UV damage marker anti-CPD (antibody that specifically recognizes the major UV-induced DNA lesion, CPD; Fig. 1 b; Bergink et al., 2006). These experiments indicate that GFP-Ub can be used as a sensitive live cell marker for DNA damage–induced H2A ubiquitination. In this study, we analyzed the kinetics of this UV-induced H2A ubiquitination using cells stably expressing GFP-Ub (Fig. 1 b; Bergink et al., 2006). GFP-Ub accumulation at LUD slowly increases after irradiation and reaches a maximum after ∼3 h. Surprisingly, local recruitment of GFP-Ub remains detectable up to at least 24 h after UV when most NER proteins do not accumulate anymore at the site of damage (see Fig. 4 f; Hoogstraten et al., 2008), suggesting that this histone modification represents a persisting chromatin mark. We tested the binding dynamics of Ub at LUD 3.5 h after irradiation. A swift recovery of GFP-Ub was observed upon photobleaching both at the damaged and nondamaged site (Fig. 1 c). To test whether this is caused by exchange of histone H2A at the nucleosome or by a continuous cycle of ubiquitination and deubiquitination of H2A while residing in the nucleosome, we compared the mobility of GFP-tagged H2A with the mobility of GFP-Ub (Fig. S1 b). Although GFP-Ub is fully recovered within 12 min after photobleaching (both within and outside of the LUD), only a minor part of GFP-H2A is recovered within 12 min, suggesting that H2A is constantly ubiquitinated and deubiquitinated within the context of an intact nucleosome. These data indicate that local UV-induced H2A ubiquitination is a dynamic process and further suggest that Ub ligases are constantly targeted to chromatin containing damage to maintain the UV-induced H2A ubiquitination up to 24 h after the initial damage.
Figure 1.
RNF8 and Ubc13 knockdown inhibits the continuous UV-induced H2A ubiquitination. (a) GPF-Ub–expressing HeLa cells (left) or nontransfected HeLa cells were locally UV irradiated (60 J/m2) and stained with an antibody-recognizing Ub conjugated to H2A (uH2A). uH2A colocalizes with GFP-Ub (left) and with the damage markers XPC (middle) or XPA (right). (b) HeLa cells stably expressing GFP-Ub were locally UV irradiated (60 J/m2) and fixed after the indicated times. The local UV-irradiated area was visualized using CPD counterstaining. The GFP-Ub accumulation at LUD is visible up to 24 h. (c) The mobility of GFP-Ub in HeLa cells was determined by FRAP. 3.5 h after local UV exposure (60 J/m2), GFP-Ub–expressing HeLa cells were subject to live cell imaging. A nucleus containing a local Ub accumulation was photobleached inside the indicated white box for three iterations at 100% of laser. Pictures were acquired at the indicated times after photobleaching. 9 min after photobleaching, the GFP-Ub was almost completely redistributed. (d) GFP-Ub–expressing HeLa cells were transfected with the indicated siRNA oligonucleotides. 48 h after transfection, the cells were locally UV exposed with 60 J/m2 and 3 h later were stained for CPD. Knockdown of RNF8 or Ubc13 results in an almost absence of GFP-Ub accumulation at LUD. The percentage of colocalization of GFP-Ub with CPD is plotted for the different siRNA transfections. (e) HeLa cells were similarly treated as in d and stained for ubiquitinated H2A together with XPA as a damage marker. siRNA-mediated depletion of RNF8 or Ubc13 caused a severely reduced accumulation of uH2A at LUD. The percentage of colocalization of uH2A with XPA after LUD is plotted in the graph for the different siRNA transfections. Arrows and arrowheads indicate local damage sites. Error bars indicate SEM.
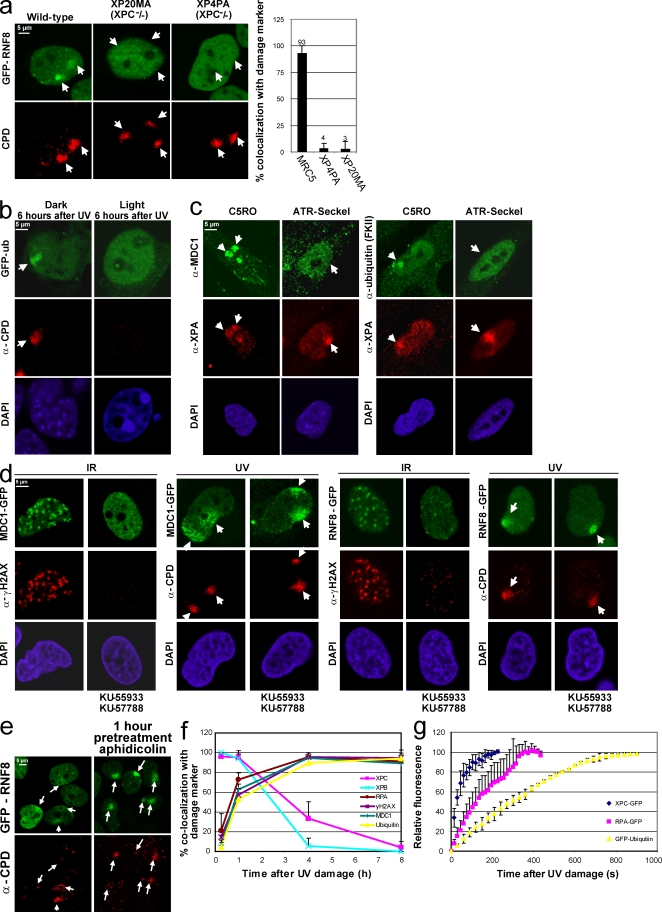
Figure 4.
NER intermediates trigger RNF8 accumulation in a DNA damage– and ATR-dependent manner. (a) RNF8-GFP stably expressing NER-proficient (MRC5) or NER-deficient (XPC-negative cell lines XP20MA and XP4PA) were locally UV exposed (60 J/m2). 2 h later, damaged cells were immunostained for CPDs. The percentage of cells in which RNF8-GFP colocalizes with CPD after LUD is plotted in the graph. RNF8 does not accumulate in NER-deficient cells. (b) Mouse embryonic fibroblasts expressing both CPD and 6-4PP photolysases were stably transfected with GFP-Ub. 1.5 h after local UV exposure (60 J/m2), cells were cultured either in the dark (left) or were photoreactivated with visible light for 2 h (right). 6 h after initial UV damage, cells were fixed and immunostained for CPDs. After UV lesion removal, GFP-Ub does not accumulate at LUD. (c) ATR hypomorphic human cells (Seckel) and C5RO cells with a wild-type ATR status were locally UV irradiated (25 J/m2) and immunostained after 2 h with antibodies recognizing MDC1 or conjugated Ub (FKII). XPA was used as a damage marker, indicating that MDC1 and Ub accumulate in an ATR-dependent manner. (d) Stably expressing MDC1-GFP (left) or RNF8-GFP (right) cells were incubated for 90 min with ATM and DNA-PK inhibitors (KU-55933 and KU-57788) or with an equal volume of DMSO. Cells were exposed to IR (10 Gy) or UV (60 J/m2), and after 2 h, they were immunostained for γH2AX or CPD. Although the addition of these inhibitors clearly inhibits the IR-induced foci formation of MDC1, MDC1 still accumulates after LUD. (e) Stable MRC5 RNF8-GFP–expressing cells were locally exposed to 20 J/m2 UV with or without a pretreatment for 1 h with 1 µg/ml aphidicolin. 1 h after UV damage, cells were fixed and immunostained for CPDs. A strong increase of RNF8 accumulation at the damaged DNA after aphidicolin pretreatment was observed. (f) The percentage of GFP-MDC1, XPB, RPA, γH2AX, GFP-Ub, and XPC-GFP colocalization with a damage marker (either CPD or XPA) at LUD (irradiated with 45 J/m2) is plotted for the different time points at 15 min, 1 h, 4 h, and 8 h after LUD. Although XPC and XPB are colocalizing in almost all cells 15 min after UV damage, the other factor colocalizes significantly with the used damage markers 1 h after LUD. (g) Cells stably expressing the XPC-GFP, GFP-Ub, and GFP-RPAp70 proteins were UV damaged using UV-C (266 nm) laser irradiation. GFP fluorescence intensities at the site of UV damage were measured by real time imaging until they reached a maximum. Assembly kinetic curves were derived from at least six cells for each protein. Relative fluorescence was normalized on 0 (before damage) and 100% (maximum level of accumulation). Arrows and arrowheads indicate local damage sites. Error bars indicate SEM.
Recently, it was found that DSB-induced H2A and H2AX ubiquitination is mediated by the E3 Ub ligase RNF8 and the E2-conjugating enzyme Ubc13 (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Nicassio et al., 2007). We tested whether these enzymes were also responsible for the GFP-Ub accumulation at LUD by depleting RNF8 or Ubc13 in GFP-Ub–expressing HeLa cells. siRNA-mediated knockdown of RNF8 and Ubc13, as confirmed by immunoblotting (Fig. S1 c), resulted in a strongly decreased accumulation of GFP-Ub (Fig. 1 d) and ubiquitinated H2A (Fig. 1 e) at LUD. These results were confirmed by immunostaining for endogenous conjugated Ub (Fig. S1 d). In mammalian nuclei, 5–15% of H2A is monoubiquitinated, explaining the relative abundant nuclear localization of GFP-Ub (Dantuma et al., 2006). Depletion of RNF8 does not affect the total pool of nuclear conjugated Ub (mainly H2A-Ub), suggesting that RNF8 is a DNA damage–specific E3 ligase.
Role of RNF8 and Ubc13 in UV-induced DDR
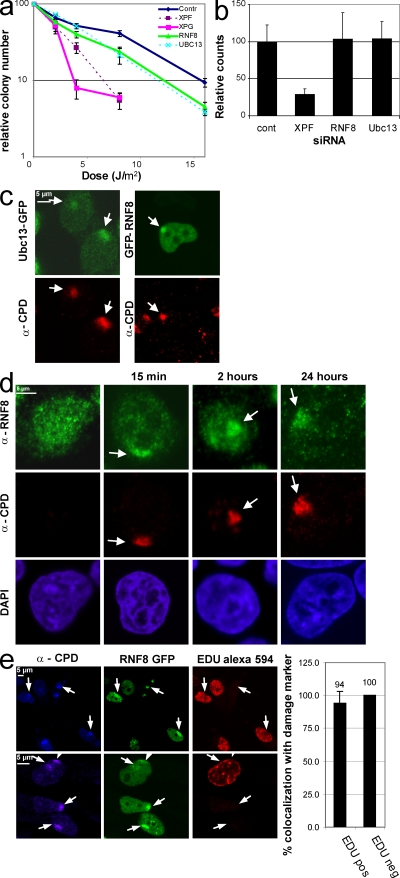
We tested the involvement of RNF8 and Ubc13 in UV-induced DDR by determining the UV sensitivity of HeLa cells depleted for these proteins. RNF8- and Ubc13-depleted cells were more UV sensitive than control siRNA–transfected cells (Fig. 2 a) but not as sensitive as cells depleted for the essential NER endonucleases XPF or XPG. These results suggest that UV-induced H2A ubiquitination plays an important role in UV survival but is not as essential as depletion of the NER factors XPF or XPG. To further investigate the role of RNF8 and Ubc13 in NER, we measured the activity of the final step of the NER process (Hoeijmakers, 2001), the gap-filling DNA synthesis. This unscheduled DNA synthesis (UDS) was analyzed in cells treated with siRNA against XPF, RNF8, and Ubc13 (Fig. 2 b). In contrast to the severe UDS reduction by XPF depletion, knockdown of RNF8 or Ubc13 does not affect UDS despite their function in UV survival, as revealed by UV hypersensitivity in RNF8- or Ubc13-depleted cells. These results indicate that RNF8 and Ubc13 play a role in UV–DDR but do not directly affect the core NER repair process.
Figure 2.
RNF8 and Ubc13 depletion sensitizes cells to UV. (a) UV survival using HeLa cells transiently transfected with siRNA targeting RNF8, Ubc13, XPF, XPG, or control siRNA. The percentage of surviving cells is plotted against the applied UV-C dose (J/m2). RNF8 and Ubc13 depletion sensitizes cells to UV. (b) Gap-filling DNA repair synthesis (UDS) was measured by autoradiography. Wild-type primary fibroblasts (C5RO) were transfected with the indicated siRNA. 48 h after transfection, cells were pulse labeled with [3H]thymidine after exposure to 16 J/m2 UV-C. UDS was quantified by counting autoradiographic grains in 50 nuclei, and the number of counts in control-transfected cells was set at 100%. RNF8 and Ubc13 have no effect on UDS. (c) HeLa cells stably expressing Ubc13-GFP or Mrc5 cells expressing RNF8-GFP were locally UV irradiated (60 J/m2) and immunostained for CPD. Both Ubc13 and RNF8 accumulate at LUD. (d) Localization of endogenous RNF8 before and after local UV exposure (60 J/m2). Immunostainings were performed using RNF8 and CPD antibodies. RNF8 accumulation was found up to 24 h after UV damage. (e) RNF8-GFP–expressing cells were exposed to LUD and directly after damage, were incubated for 2 h in medium containing EdU (BrdU analogue). Cells that were in S phase after the UV exposure stain positive for EdU visualized using Alexa Fluor 594. CPD was used as a damage marker. The graph adjacent to the images shows a similar (∼100%) colocalization of RNF8 with CPDs in S phase and non–S phase cells. Arrows indicate local damage sites. Error bars indicate SEM.
RNF8 is recruited to LUD by MDC1
The observed dynamic damage-induced H2A ubiquitination argued for a continuous presence of the responsible E3 Ub ligase and E2 Ub–conjugating enzyme at the damaged DNA. The recruitment of these enzymes to LUD was tested in cells expressing full-length GFP-tagged RNF8 or Ubc13 (Fig. S2 a). We found a clear accumulation of both GFP-RNF8 and GFP-Ubc13 at LUD, colocalizing with CPD (Fig. 2 c) and Ub (Fig. S2 b). Endogenous RNF8 was also found to accumulate at UV lesions (Fig. 2 d) and remains, like Ub, 24 h after UV enriched at LUD. This supports a model of continuous H2A ubiquitination at DNA damage. Importantly, RNF8 recruitment to UV lesions occurs in a cell cycle–independent manner, as shown by EdU incorporation (Fig. 2 e; Salic and Mitchison, 2008) and cyclin A costaining (Fig. S2 c).
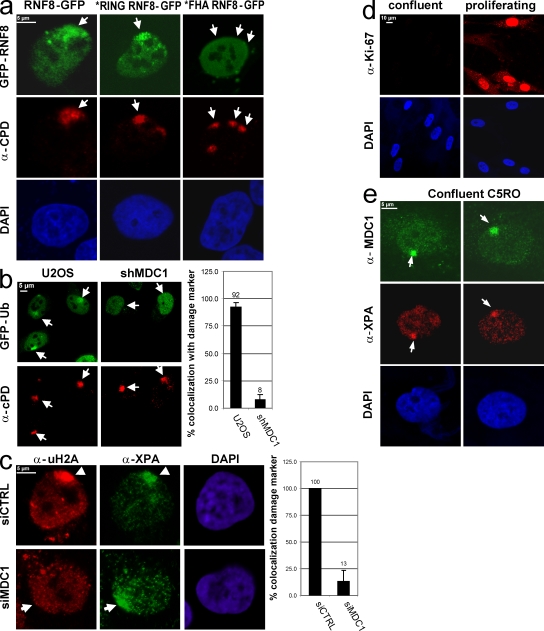
RNF8 harbors an N-terminal forkhead-associated (FHA) domain (Ito et al., 2001; Plans et al., 2006), which is commonly implicated in phosphorylation-dependent protein–protein interactions (Hammet et al., 2003), and a C-terminal RING finger (Ito et al., 2001; Plans et al., 2006), which is known to interact with E2-conjugating enzymes and is essential for its Ub ligase activity (Fang and Weissman, 2004). We observed that the RING finger point mutant (C403S; Mailand et al., 2007) localizes to LUD similar as wild-type RNF8, whereas the FHA domain point mutant (R42A; Mailand et al., 2007) is unable to accumulate at lesions (Fig. 3 a). These data suggest that RNF8 is recruited to UV-damaged DNA via a phospho-specific protein–protein interaction involving its FHA domain. After IR, RNF8 is recruited to DSBs through binding of its FHA domain to phosphorylated MDC1 (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007). Therefore, we tested whether the scaffold protein MDC1 is also involved in loading RNF8 and Ub to LUD using cells that stably express MDC1 short hairpin RNA (shRNA) (Fig. S3 a). In the absence of MDC1, both GFP-Ub (Fig. 3 b) and GFP-RNF8 (Fig. S3 b) failed to accumulate at LUD. These results were confirmed by immunostaining with an antibody specific for ubiquitinated H2A in cells transfected with siRNA targeting MDC1 (Fig. 3 c). Together, these data argue that MDC1 is required for UV-induced H2A ubiquitination. Surprisingly, thus far, MDC1 was only found to be implicated in DSB–DDR, and its implication in UV–DDR is unprecedented. To rule out the possibility that its recruitment during UV–DDR was caused by replication stress–induced DSBs after collapsed replication forks, we showed that MDC is recruited to LUD independent of replication using MDC1-GFP–expressing cells in combination with staining for EdU incorporation (Fig. S3 c; Salic and Mitchison, 2008). Finally, in noncycling confluent primary fibroblasts confirmed by Ki-67 staining (Fig. 3 d), endogenous MDC1 accumulated at LUD (Fig. 3 e). This uncovers a novel function for this DSB–DDR protein in UV–DDR.
Figure 3.
MDC1 is essential for Ub accumulation at local damage. (a) U20S cells stably expressing wild-type, *RING (C403S), or *FHA (R42A) forms of GFP-tagged RNF8 were locally UV irradiated followed by CPD immunostaining. The FHA domain of RNF8 is essential for the observed RNF8 accumulation at the damaged area. (b) Wild-type or stably expressing shRNA targeting MDC1 U2OS cells were transiently transfected with GFP-Ub. 36 h after transfection, cells were locally UV exposed and were immunostained for CPDs 2 h later. The percentage of cells in which GFP-Ub colocalizes with CPD after LUD is plotted in the graph. MDC1 is essential for the Ub accumulation at the damaged DNA. (c) HeLa cells were transfected with control siRNA or siRNA targeting MDC1. 36 h after siRNA transfection, cells were UV irradiated (60J/m2) and stained for ubiquitinated H2A and XPA. The percentage of cells in which uH2A colocalizes with XPA after LUD is plotted in the graph. (d) Proliferation status of primary human C5RO fibroblasts was checked using the Ki-67 proliferation marker. Proliferating cells are positive for Ki-67 (right), whereas C5RO cells grown confluent for 7 d are negative for Ki-67 (left), indicating that all cells are in G0. (e) These nonproliferating human primary C5RO fibroblasts were locally UV exposed (60 J/m2), and after 2 h, they were immunostained for endogenous MDC1 and the essential NER protein XPA. MDC1 accumulates at the local damage in nonproliferating cells. Arrows and arrowheads indicate local damage sites. Error bars indicate SEM.
H2A ubiquitination induced by UV requires ATR and NER intermediates
Within DSB–DDR, recruitment of RNF8 and H2A ubiquitination are early events and occur with almost similar kinetics as the loading of the primary DSB-recognizing proteins (Mailand et al., 2007). In contrast, H2A ubiquitination within UV–DDR is a relatively slow process, reaching a maximum only ∼3 h after UV (Fig. 1 b), whereas lesion binding of UV damage–recognizing NER proteins is very quick (Luijsterburg et al., 2007; Hoogstraten et al., 2008). Moreover, depletion of the responsible E2-conjugating enzyme or E3 ligase does not directly affect NER efficiency (Fig. 2 b). Together, these data suggest that UV-induced H2A ubiquitination occurs late in UV–DDR. To investigate at which point during UV–DDR RNF8 becomes implicated, we monitored GFP-RNF8 binding to LUD in NER-deficient cells. We found that in two independent cell lines derived from Xeroderma pigmentosum patients mutated in the damage-recognizing and NER-initiating protein XPC, RNF8-GFP fails to bind to LUD (Fig. 4 a). This indicates that the RNF8 recruitment, similar to the subsequent GFP-Ub accumulation (Bergink et al., 2006), is dependent on active NER. Surprisingly, both RNF8 and Ub accumulation to LUD was still visible 24 h after UV (Fig. 1 b and Fig. 2 d), whereas presence of NER factors at LUD faded within 4–6 h after UV (Fig. 4 f; Hoogstraten et al., 2008). Within this time frame, the bulk of the UV-induced 6-4PPs are removed by the NER machinery (Fig. S4 a; Hoogstraten et al., 2008), indicating that detectable accumulation of NER factors at LUD follows the repair kinetics of 6-4PP. This triggers the question of why this epigenetic mark remains far beyond removal of 6-4PP but still requires functional NER. However, it should be noted that in the majority of UV-induced lesions, CPDs (∼60%) are slowly repaired by NER, and a significant fraction remains unrepaired even 24 h after UV (Mitchell et al., 1985). To test whether the repair of photolesions in general causes this chromatin response after UV, we removed these lesions by photoreactivation. We expressed GFP-Ub in mouse embryonic fibroblasts derived from a transgenic mouse model expressing both 6-4PP and CPD photolyases (Jans et al., 2005). These photolyases directly reverse photolesions after exposure to visible light (Fig. S4 b) independent of NER (Jans et al., 2005). In photoreactivated cells, no accumulation of GFP-Ub at LUD could be detected 6 h after UV irradiation (Fig. 4 b, right), whereas in nonphotoreactivated control cells, GFP-Ub clearly accumulated at LUD (Fig. 4 b, left). These data indicate that H2A is continuously ubiquitinated as long as DNA lesions, including the poorly repaired CPDs, are present.
Having established that active NER is required to trigger H2A ubiquitination, the molecular mechanism remains unknown. Within DSB–DDR, H2A ubiquitination depends on the DNA damage signaling kinase ATM (Mailand et al., 2007). However, ATM does not become directly activated by UV irradiation in nonreplicating cells (Stiff et al., 2006). Thus far, the best-characterized damage signaling kinase in UV–DDR is ATR (Abraham, 2004; Hanasoge and Ljungman, 2007). We found that in fibroblasts carrying a hypomorphic ATR mutation that causes severely reduced ATR expression (O'Driscoll et al., 2003), both endogenous MDC1 and conjugated Ub accumulation at LUD were significantly attenuated as compared with wild-type cells (Fig. 4 c). In addition, we found that when UV-irradiated cells were treated with the ATM- and DNA-PK–specific inhibitors (KU-55933 and KU-57788, respectively), MDC1 and RNF8 loading to LUD was not affected (Fig. 4 d). Addition of both inhibitors to cells exposed to IR completely abolished H2AX phosphorylation and formation of MDC1 or RNF8 foci, indicating that the inhibitors efficiently down-regulated the activity of both kinases. These results indicate that the UV-induced MDC1 and RNF8 recruitment depends on ATR.
Besides the involvement of different kinases, a clear difference also exists in the time of RNF8 pathway activation between DSB– and UV–DDR. The RNF8 pathway is activated before DSB repair (Huen et al., 2007; Mailand et al., 2007); however, after UV damage, it is activated as a consequence of NER. This implies that ATR becomes activated in an NER-dependent fashion. It is established that ATR activation after UV damage is caused by replication stress that creates single-stranded DNA, which is a substrate for RPA/ATR-interacting protein, resulting in the subsequent ATR activation (Shechter et al., 2004). Nonetheless, we found that MDC1 and RNF8 binding also occurs outside S phase. It was recently described that in non–S phase cells, single-stranded repair intermediates generated during NER by the excision of the damaged strand activate ATR (Hanasoge and Ljungman, 2007; Stiff et al., 2008). To study whether these NER-mediated repair intermediates are responsible for the observed RNF8 pathway activation, we increased the amount of single-stranded NER intermediates (and consequently increased ATR activation) by blocking the DNA repair synthesis using the DNA polymerase inhibitor aphidicolin. To visualize a possible increase of RNF8 activation, we exposed the cells to a relative low dose of local UV damage (20 J/m2) and shortly after UV (1.5 h) as compared with the previously used conditions, i.e., suboptimal conditions for visualizing H2A-Ub and GFP-RNF8 accumulation at LUD. As expected, without repair replication inhibition, only a faint accumulation could be detected (Fig. 4 e, left). However, a very clear LUD accumulation of GFP-RNF8 was observed (Fig. 4 e, right) in cells treated with aphidicolin for 1 h.
This indicates that MDC1, RNF8, and the subsequent GFP-Ub accumulation are relatively late events triggered by the DNA repair intermediates, which subsequently activate ATR. This would suggest that the MDC1–RNF8 pathway is activated at a hierarchical step in the UV–DDR, occurring after the initial incision step mediated by the core NER machinery. To test this hypothesis in more detail, we have analyzed the time-dependent accumulation of several proteins involved in different steps during UV–DDR by IF at different time points after LUD (Fig. 4 f and Fig. S4 e). Core NER factors like the damage-recognizing protein XPC or the DNA helicase of the TFIIH complex XPB completely colocalize with the used damage markers 15 min after UV damage. This colocalization remains for a few hours and gradually decreases to background levels at 4 h after UV damage, basically after the repair kinetics of 6-4PPs. In contrast, RPA, which is known to interact with single-stranded DNA repair intermediates (Zou and Elledge, 2003), followed an entirely different kinetic profile. Shortly after LUD, only ∼20% colocalized with a damage marker and reached a maximum of almost-full colocalization ∼4 h after LUD that persisted until 8 h after UV. Similar kinetic profiles were found for phosphorylated H2AX, MDC1, and Ub, indicating that these events represent late, postincision events of the UV–DDR pathway. To further analyze the order of accumulation, we have measured the real-time live cell accumulation of XCP-GFP, RPAp70-GFP, and Ub-GFP by confocal imaging. After applying LUD using a UV-C laser (Dinant et al., 2007), we measured the assembly kinetics of GFP-tagged proteins at LUD (Fig. 4 g). A striking difference in the assembly kinetics was observed between these proteins in which XPC reaches equilibrium the fastest, representative for an early event (damage detecting), followed by the recruitment of RPA, which represents the formation of single-stranded DNA repair intermediates, and finally GFP-Ub, which reaches its plateau much later. Together, these data provide insight into the hierarchy of the UV–DDR response: (1) initiation of the NER machinery results in (2) repair intermediates that finally activate the (3) MDC1–RNF8 pathway, resulting in the recruitment of Ub at the LUD.
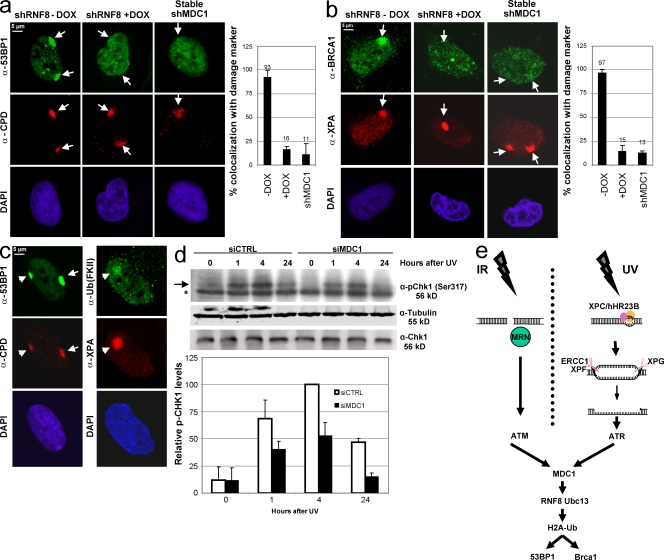
RNF8 is essential for 53BP1 and BRCA1 accumulation after UV damage
Despite important mechanistic and kinetic differences, our data suggest that both DSB and UV lesions trigger a similar epigenetic mark, H2A ubiquitination. During DSB–DDR, the RNF8-dependent H2A or H2AX ubiquitination eventually promotes the recruitment of 53BP1 and BRCA1 to chromatin near breaks (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007). It is suggested that ubiquitinated H2A/H2AX is recognized by the Ub-interacting motifs of RAP80, which through its interaction with CCDC98 recruits BRCA1/BARD1 to chromatin (Kim et al., 2007; Sobhian et al., 2007; Wang et al., 2007). It is currently unknown how 53BP1 is recruited in an RNF8- and MDC1-dependent way. To study whether the observed UV-induced H2A ubiquitination also triggers the recruitment of 53BP1 and BRCA1, we tested whether these thus-far DSB-specific factors also accumulate at LUD. Both endogenous 53BP1 and BRCA1 (Fig. 5, a and b) accumulate at LUD in an RNF8- and MDC1-dependent fashion. As in the DSB response, the UV damage–induced accumulation of BRCA1 at LUD most likely occurs via its interaction with RAP80, which also accumulates at UV-induced LUD, colocalizing with CPD or H2AX (Fig. S5 a). Both the accumulation of 53BP1 and conjugated Ub also take place in nonproliferating cells (Fig. 5 c), ruling out the possibility that these proteins accumulate at DSBs originating from stalled replication forks. The 53BP1 accumulation in primary nonproliferating cells was further studied using different UV doses. Even at a relative low dose of 5 J/m2, the 53BP1 colocalization with CPD was still clearly visible (Fig. S5 b). Together, these results show that like DSBs, UV-induced DNA damage results in a strong increase in local concentration of checkpoint mediators like MDC1, RNF8, and ubiquitinated H2A, suggesting that this robust epigenetic mark is a conserved DNA damage signal–amplifying process between DSB processing and NER.
Figure 5.
53BP1 and BRCA1 accumulate at LUD in an RNF8- and MDC1-dependent manner. (a and b) U2OS cells conditionally expressing doxycycline (Dox)-inducible shRNA targeting RNF8 were induced or not with doxycycline for 48 h, and cells stably expressing shRNA targeting MDC1 were locally UV damaged and stained for 53BP1 and CPD (a) or for BRCA1 and XPA (b). Both 53BP1 and Brca1 are recruited to the site of UV damage in an RNF8- and MDC1-dependent manner. The percentage of cells in which 53BP1 or BRCA1 colocalize with the used damage marker after LUD is plotted in the graphs. (c) Nonproliferating human primary fibroblasts (C5RO), as determined by a negative Ki-67 staining (not depicted), were stained for endogenous 53BP1 and conjugated Ub accumulation after local UV exposure (60 J/m2). CPD and XPA were used as a damage marker. (d) HeLa cells were transfected with siRNA targeting MDC1 or control siRNA. 36 h after transfection, cells were UV exposed (10 J/m2) and lysed at the indicated time points after UV damage. Chk1 phosphorylation status (Ser317) was analyzed using a phospho-specific antibody, and tubulin staining was used as a loading control. Arrow, phosphorylated Ser317 Chk1; asterisk, α-specific band. The relative amount of phosphorylated Chk1 is plotted in the graph (two blots were quantified, relative phospho-Chk1 levels were normalized, and the highest phospho-Chk1 level was set at 100%), indicating that MDC1 knockdown result is a reduction of the UV-induced Chk1 phosphorylation. (e) Model for the UV-induced DDR. UV-induced lesions are repaired by the core NER machinery, thereby generating single-stranded DNA repair intermediates, which subsequently activate ATR. This results in recruitment of MDC1 at the chromatin, which is essential for the RNF8 accumulation at the DNA damage. In concert with Ubc13, RNF8 ubiquitinates H2A, which triggers the recruitment of 53BP1 and BRCA1. From MDC1 recruitment onwards, the IR- and UV-induced DDR are similar. DSBs are recognized and bound by the Mre11–Rad50–Nbs1 (MRN) complex. Arrows and arrowheads indicate local damage sites. Error bars indicate SEM.
MDC1 is involved in UV-dependent checkpoint control
One of the proposed functions of this MDC1–RNF8 pathway after, for example, IR-induced DSBs, is to induce and maintain cell cycle checkpoint control, thereby protecting cells from mutagenesis (Huen et al., 2007). To test whether the described MDC1–RNF8 pathway might also play a role in the amplification of the checkpoint after UV damage, we have studied the phosphorylation status of Chk1 in control or MDC1 siRNA–transfected cells after UV damage (Fig. 5 d). siRNA-mediated depletion of MDC1 results in a decrease of Chk1 phosphorylation after UV damage. These results were confirmed by IF staining of global UV-irradiated cells (Fig. S5 c). Together, these data indicate that one of the main functions of the UV-induced activation of the MDC1–RNF8 pathway at the damaged DNA is to potentiate the cell cycle regulation via Chk1.
Discussion
In this study, we present novel insight into the molecular mechanism of UV-induced H2A ubiquitination. Our data show that after UV damage, the Ub E3 ligase RNF8, in concert with the E2-conjugating enzyme Ubc13, is essential for the NER-dependent Ub modification of histone H2A. In a previous study, we reported that UV-induced ubiquitination of histone H2A required Ring2 (Bergink et al., 2006). Notably, Ring2 knockdown also results in a severe reduction of basal H2A ubiquitination as well as the nuclear Ub pool. Thus, Ring2 knockdown is likely to disturb the cellular Ub equilibrium (Groothuis et al., 2006) and may affect downstream-specific ubiquitination reactions such as the RNF8-dependent H2A ubiquitination. In contrast, RNF8 knockdown inhibits only UV-induced and not basal H2A ubiquitination (Fig. 1 c; Huen et al., 2007; Mailand et al., 2007). Furthermore, we show that RNF8 is recruited to the damaged DNA, suggesting that RNF8 is the DNA damage–specific H2A E3 ligase.
Intriguingly, a significant proportion of Ubc13 was found to accumulate at the site of DNA damage, indicating that a large part of the pool of Ubc13 proteins plays a role during UV-induced DNA repair. Recently, it was also observed that a large part of the total amount of Ubc13 is involved during the DSB-induced DDR, as was shown by the accumulation of Ubc13 at laser-induced DSB damage (Ikura et al., 2007). This indicates that Ubc13 plays an important role during DNA repair in addition to its involvement in many other biological processes (Pickart, 2000). Because RNF8 recruitment to damaged DNA is independent on Ubc13 (unpublished data), we assume that Ubc13 depends on RNF8 recruitment, which is in line with the described interaction of Ubc13 with the RING finger domain of RNF8 (Plans et al., 2006). The strong accumulation of Ubc13 at the site of damage, which is surprising for such a usually less-specific functioning, E2-conjugating enzyme (in contrast to the more substrate-specific E3 ligases), indicates its important role during UV-induced DDR. Ubc13 is, for the majority, not involved in the core NER machinery because Ubc13 depletion has no detectable effect on UDS (Fig. 2 b) or XPA accumulation (Fig. S1 d). Most likely, Ubc13 plays a role in the downstream UV-induced DDR together with RNF8.
Our data (Fig. 5 e) show that the molecular mechanism that induces ubiquitination of H2A after genotoxic stress, including the recruitment of downstream factors 53BP1 and BRCA1, is highly conserved between DSB- and UV-induced DDR. This is remarkable considering the important mechanistic differences between DSB and UV damage repair, which may explain the variation in observed timing. MDC1 and RNF8 recruitment are early events in the DSB-induced DDR (Mailand et al., 2007), although it is a relatively late event after UV-induced DNA damage. It is possible that this difference is caused by a much faster processing of DSB breaks, resulting in a fast ATM activation compared with a slower ATR activation by UV lesion processing.
Importantly, we found factors (e.g., MDC1, 53BP1, and Brca1) to be recruited to UV-damaged DNA that were previously known only to play a role in the DSB response. Until now, their involvement in UV–DDR was unanticipated, although recently, some were found to be phosphorylated by ATR after UV damage, such as MDC1 (Stewart et al., 2003), RAP80 (Yan et al., 2008), and 53BP1 (Jowsey et al., 2007). Although these proteins are maximally phosphorylated within minutes upon IR via ATM, after UV, their phosphorylation becomes apparent after 60 min and keeps increasing in time, showing that the UV–DDR response via this RNF8 pathway has indeed slower kinetics.
The question of why these DSB–DDR factors (including MDC1, 53BP1, and Brca1) are also recruited to chromatin after UV damage is more difficult to answer. Genetic insults may ring a “general alarm bell,” resulting in the assembly of a toolbox of diverse DDR proteins near the lesion irrespective of the type of DNA damage. This enables the cell to facilitate different response pathways, but only a specific subset will be used if needed (Harper and Elledge, 2007). This might also explain why RNF8 is observed to accumulate at stalled replication forks (Sakasai and Tibbetts, 2008). The recruitment of the DDR proteins such as MDC1, 53BP1, and BRCA1 may represent an extra line of defense against the UV-induced damage. Normally, only the NER-specialized DNA repair enzymes will be used. However, in the rare event that a DSB originates from a UV lesion during replication, the DSB-involved proteins that were present “just in case” at the damaged chromatin can swiftly be used.
Alternatively, the same pathways, and thus the same proteins, are used as cells use similar crisis management strategies triggered by different repair machineries. H2A ubiquitination increases the local concentration of 53BP1 and BRCA1 that enhances activation of the cell cycle checkpoint kinases Chk1 and Chk2 (Stucki and Jackson, 2006). siRNA-mediated down-regulation of MDC1 results in a lower UV-induced Chk1 phosphorylation, indicating that this pathway is involved in the checkpoint induction and maintenance (Fig. 5 d). Chk1 and Chk2 play a crucial role in DDR by, for example, regulating the S to G2 and G2/M checkpoints (Niida and Nakanishi, 2006). Both checkpoints are implicated in the cellular survival of IR and UV, which may explain the use of a common DDR pathway.
Materials and methods
Cell culture and transfection
HeLa, U2OS, MRC5, and the human primary fibroblasts wild-type control (C5RO), XPC-deficient patient cell lines XP4PA and XP20MA, and ATR hypomorphic GM18366 fibroblasts (obtained from The Coriell Institute, Camden, NJ) were cultured in Ham's F10 (Invitrogen) supplemented with antibiotics and 10% fetal calf serum at 37°C with 5% CO2. Cell lines stably expressing fluorescent-tagged proteins (GFP-RNF8, Ubc13-GFP, and RFP-Ub) were isolated by FACS sorting and neomycin selection. Other cell lines were used as described previously: GFP-Ub (Dantuma et al., 2006), MDC1-GFP (Bekker-Jensen et al., 2006), RNF8-GFP (wild type, ΔRING, and ΔFHA; Mailand et al., 2007), shRNF8 (Mailand et al., 2007), and shMDC1 (Bekker-Jensen et al., 2006). Aphidicolin was used at 5 µg/ml, and ATM inhibitor (KU-55933) and DNA-PK inhibitor (KU-57788) were used at a concentration of 10 µM. For the local UV irradiation, cells were treated with a UV-C germicidal lamp (254 nm; Phillips) through a 5-µm microporous filter at the indicated dose (Moné et al., 2001). Cells were exposed to IR using a 137Cs source at the specified dose. siRNA oligonucleotides were synthesized (Thermo Fisher Scientific) to the following sequences: MDC1, 5′-GUCUCCCAGAAGACAGUGA-3′; RNF8, 5′-GGACAAUUAUGGACAACAA-3′; Ubc13, 5′-GGGACUUUUAAACUUGAAC-3′; XPF SMARTpool, 5′-UGACAAGGGUACUACAUGA-3′, 5′-GUAGGAUACUUGUGGUUGA-3′, 5′-ACAAGACAAUCCHCCAUUA-3′, and 5′-AAGAXGAGCUCACGAGUAU-3′; and XPG SMARTpool, 5′-CAUGAAAUCUUGACUGAUA-3′, 5′-GAACGCACCUGCUGCUGUA-3′, 5′-GAAAGAAGAUGCUAAACGU-3′, and 5′-GAACGAACUUUGCCCAUAU-3′. siRNA transfections were performed using Lipofectamine2000 (Invitrogen) according to the manufacturer's protocol. DNA transfections were performed using Fugene (Roche).
IF microscopy and Western blotting
Cells were fixed using 2% paraformaldehyde in the presence of 0.1% Triton X-100. Samples were processed as described previously (Rademakers et al., 2003). For uH2A staining, cells were permeabilized with 0.5% Triton X-100 for 5 min before fixation with 4% paraformaldehyde for 15 min at 4°C. Immunofluorescent images were obtained using the Aristoplan Flu (134795; Leitz) or confocal microscope (LSM 510 META; Carl Zeiss, Inc.) equipped with a 63× 1.4 NA Plan Apochromat oil immersion lens (Carl Zeiss, Inc.). LSM image browser acquisition software (version 4.0; Carl Zeiss, Inc.) was used. The following antibodies were used: anti-MDC1 (Abcam), anti-RNF8 (Abcam), anti-53BP1 (H-300; Santa Cruz Biotechnology, Inc.), anti-BRCA1 (D9; Santa Cruz Biotechnology, Inc.), anti-Ub (FK2; BIOMOL International L.P.), anti–cyclin A (GNS; Santa Cruz Biotechnology, Inc.), anti-γH2AX (07-164; Millipore), anti–Ki-67 (Abcam), anti-XPB (clone IB3; Giglia-Mari et al., 2006), anti-RPA32 (clone 9H8; Abcam), anti-ubiquitinated H2A (uH2A; Abcam), RAP80 (rabbit polyclonal; Bethyl Laboratories, Inc.), phospho–S317-Chk1 (Western blotting, Bethyl Laboratories, Inc.; IF, Cell Signaling Technology), and anti-GFP (rabbit polyclonal; Abcam) in combination with the corresponding secondary antibodies labeled with Alexa Fluor 350, 488, or 594 as indicated (Invitrogen; The Jackson Laboratory). DNA was stained using DAPI Vectashield (Vector Laboratories). As marker for detecting LUD, anti-hXPA (rabbit polyclonal anti–human XPA) or mouse anti-CPD (TDM-2; MBL International) were used, depending on the species in which the other used antibody was raised in. Colocalization was quantified by counting at least 50 cells per experiment in different fields. Colocalization was defined as a more than twofold increase in intensity at the LUD defined by the presence of the used damage marker (XPA or CPD). EdU (5-ethynyl-2'-deoxyuridine) incorporation was visualized using Click-iT Alexa Fluor 594 according to the manufacturer's protocol (Invitrogen). Western blotting was performed with the indicated primary antibody according to the manufacturer's protocol. Alexa Fluor 680 goat anti–mouse or –rabbit (Invitrogen) were used to visualize the Western blotting using an infrared imaging system (Odyssey; LI-COR Biosciences). H2A ubiquitination after UV damage was studied using HeLa cells stably expressing His-tagged Ub (Choudhury et al., 2004). Ubiquitinated proteins were isolated as described previously (Marteijn et al., 2007) and subject for immunoblotting using uH2A and FKII antibodies.
Live cell confocal laser-scanning microscopy
Confocal laser-scanning microscopy images were obtained using a confocal microscope (LSM 510 META) with a 63× oil Plan Apochromat 1.4 NA oil immersion lens (Carl Zeiss, Inc.) equipped with a cell culture microscopy stage. GFP fluorescence imaging was recorded after excitation with a 488-nm argon laser and a 515–540-nm band-pass filter. FRAP was performed as described previously (Houtsmuller and Vermeulen, 2001). The indicated areas in Fig. 1 c and Fig. S1 b were photobleached by three iterations using 100% 488-nm laser power. The cytoplasm of the GFP-Ub cell line was photobleached (three iterations; 100% 488-nm laser power) preceding the photobleaching of the indicated areas (Fig. 1 c and Fig. S1 b). The recovery of fluorescence in the photobleached box was followed by imaging every minute. Kinetics of GFP-tagged RPA, Ub, and XPC accumulation were performed using a UV-C (266 nm) laser irradiation for 1.5 s as described previously (Dinant et al., 2007).
UDS and UV survival
3 d before the UDS assay, C5RO cells were siRNA transfected. UDS was performed as described previously (Vermeulen et al., 1986). Cells were UV irradiated with 16 J/m2 and incubated for 2 h in culture medium containing 10 µCi (methyl[3H]) thymidine (110 Ci/mmol/ml; GE Healthcare). Repair capacity was quantified by counting autoradiographic grains. Cellular survival of HeLa cells was determined using a colony assay. Cells were plated in 6-cm dishes at various dilutions. After 16 h, cells were exposed to different doses of UV-C (254 nm; TUV lamp; Phillips) and left to grow for 7 d, fixed, and stained, and colonies were counted to assess the colony-forming ability.
Online supplemental material
Fig. S1 shows the UV-induced H2A ubiquitination, the mobility of H2A-GFP, and the siRNA-mediated knockdown efficiency of RNF8 and Ubc13. Fig. S2 shows the characterization of the stable RNF8-GFP and Ubc13-GFP cell line, the colocalization of RNF8-GFP with RFP-Ub, and the cell cycle–independent accumulation of RNF8 at the site of UV damage. Fig. S3 shows the MDC1 dependency of RNF8 accumulation and the accumulation of MDC1 in both replicating and nonreplicating cells. Fig. S4 shows the effect of the 6-4PP and CPD photolyases, the effect of aphidicolin, and the kinetics of different DDR-involved proteins using IF. Fig. S5 shows the RAP80 accumulation at the site of damage, the 53BP1 accumulation in nonproliferating C5RO cells at different UV doses, and the role of MDC1 on the UV-induced Chk1 phosphorylation using IF. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200902150/DC1.
Acknowledgments
We thank Dr. T. Thomsom for sharing the GFP-RNF8 plasmid and Dr. Ikura for the GFP-Ubc13 construct. We thank Professor J. Hoeijmakers for critically reading this manuscript, Mr. D. Warmerdam for helpful discussions, Mrs. A. Fazalalikhan, Mr. N. Wijgers, and Mr. A.F. Theil for technical assistance, and Drs. A.B. Houtsmuller and W.A. van Cappellen for providing the imaging facility. We also thank Dr. G. Smith of KuDOS Pharmaceuticals for providing ATM and DNA-PK inhibitors and Dr. Baer for providing us with the stably expressing His-Ub cell line.
This study was financed by the Dutch Organization for Scientific Research (grants ZonMW 917-46-364 to W. Vermeulen and ZonMW Veni 917-96-120 to J.A. Marteijn), the Human Frontiers in Science Program (grant RGP0007/2004-C to H. Lans), the American Institute for Cancer Research (grant 09-0084 to H. Lans), the European Union Integrated Project DNA repair FP6 program (grant LSHG-CT-2005-512113 to J.A. Marteijn), the Swedish Cancer Society, and the Swedish Research Council (N.P. Dantuma).
Footnotes
Abbreviations used in this paper:
- 6-4PP
- 6-4 photoproduct
- ATM
- ataxia telangiectasia mutated
- ATR
- ATM and Rad3-related
- CPD
- cyclobutane pyrimidine dimer
- DDR
- DNA damage response
- DSB
- double-strand break
- FHA
- forkhead associated
- IF
- immunofluorescence
- IR
- ionizing radiation
- IRIF
- IR-induced foci
- LUD
- local UV damage
- NER
- nucleotide excision repair
- RPA
- replication protein A
- shRNA
- short hairpin RNA
- Ub
- ubiquitin
- UDS
- unscheduled DNA synthesis
References
- Abraham R.T. 2004. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways.DNA Repair (Amst.). 3:883–887 doi:10.1016/j.dnarep.2004.04.002 [DOI] [PubMed] [Google Scholar]
- Baarends W.M., Wassenaar E., van der Laan R., Hoogerbrugge J., Sleddens-Linkels E., Hoeijmakers J.H., de Boer P., Grootegoed J.A. 2005. Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis.Mol. Cell. Biol. 25:1041–1053 doi:10.1128/MCB.25.3.1041-1053.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M.B., Bartek J., Lukas J. 2006. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks.J. Cell Biol. 173:195–206 doi:10.1083/jcb.200510130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett E.J., Harper J.W. 2008. DNA damage: ubiquitin marks the spot.Nat. Struct. Mol. Biol. 15:20–22 doi:10.1038/nsmb0108-20 [DOI] [PubMed] [Google Scholar]
- Bergink S., Salomons F.A., Hoogstraten D., Groothuis T.A., de Waard H., Wu J., Yuan L., Citterio E., Houtsmuller A.B., Neefjes J., et al. 2006. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A.Genes Dev. 20:1343–1352 doi:10.1101/gad.373706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury A.D., Xu H., Baer R. 2004. Ubiquitination and proteasomal degradation of the BRCA1 tumor suppressor is regulated during cell cycle progression.J. Biol. Chem. 279:33909–33918 doi:10.1074/jbc.M403646200 [DOI] [PubMed] [Google Scholar]
- Cortez D., Guntuku S., Qin J., Elledge S.J. 2001. ATR and ATRIP: partners in checkpoint signaling.Science. 294:1713–1716 doi:10.1126/science.1065521 [DOI] [PubMed] [Google Scholar]
- Dantuma N.P., Groothuis T.A., Salomons F.A., Neefjes J. 2006. A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling.J. Cell Biol. 173:19–26 doi:10.1083/jcb.200510071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinant C., de Jager M., Essers J., van Cappellen W.A., Kanaar R., Houtsmuller A.B., Vermeulen W. 2007. Activation of multiple DNA repair pathways by sub-nuclear damage induction methods.J. Cell Sci. 120:2731–2740 doi:10.1242/jcs.004523 [DOI] [PubMed] [Google Scholar]
- Dinant C., Houtsmuller A.B., Vermeulen W. 2008. Chromatin structure and DNA damage repair.Epigenetics Chromatin. 1:9 doi:10.1186/1756-8935-1-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J., Coates J., Jackson S.P. 2005. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage.Nature. 434:605–611 doi:10.1038/nature03442 [DOI] [PubMed] [Google Scholar]
- Fang S., Weissman A.M. 2004. A field guide to ubiquitylation.Cell. Mol. Life Sci. 61:1546–1561 doi:10.1007/s00018-004-4129-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M., Lazzaro F., Longhese M.P., Plevani P., Muzi-Falconi M. 2004. Physical and functional interactions between nucleotide excision repair and DNA damage checkpoint.EMBO J. 23:429–438 doi:10.1038/sj.emboj.7600051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M., Lazzaro F., Plevani P., Muzi-Falconi M. 2005. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1.J. Biol. Chem. 280:9879–9886 doi:10.1074/jbc.M414453200 [DOI] [PubMed] [Google Scholar]
- Giglia-Mari G., Miquel C., Theil A.F., Mari P.O., Hoogstraten D., Ng J.M., Dinant C., Hoeijmakers J.H., Vermeulen W. 2006. Dynamic interaction of TTDA with TFIIH is stabilized by nucleotide excision repair in living cells.PLoS Biol. 4:e156 doi:10.1371/journal.pbio.0040156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet L.C., Schärer O.D. 2006. Molecular mechanisms of mammalian global genome nucleotide excision repair.Chem. Rev. 106:253–276 doi:10.1021/cr040483f [DOI] [PubMed] [Google Scholar]
- Groothuis T.A., Dantuma N.P., Neefjes J., Salomons F.A. 2006. Ubiquitin crosstalk connecting cellular processes.Cell Div. 1:21 doi:10.1186/1747-1028-1-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammet A., Pike B.L., McNees C.J., Conlan L.A., Tenis N., Heierhorst J. 2003. FHA domains as phospho-threonine binding modules in cell signaling.IUBMB Life. 55:23–27 doi:10.1080/1521654031000070636 [DOI] [PubMed] [Google Scholar]
- Hanasoge S., Ljungman M. 2007. H2AX phosphorylation after UV irradiation is triggered by DNA repair intermediates and is mediated by the ATR kinase.Carcinogenesis. 28:2298–2304 doi:10.1093/carcin/bgm157 [DOI] [PubMed] [Google Scholar]
- Harper J.W., Elledge S.J. 2007. The DNA damage response: ten years after.Mol. Cell. 28:739–745 doi:10.1016/j.molcel.2007.11.015 [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J.H. 2001. Genome maintenance mechanisms for preventing cancer.Nature. 411:366–374 doi:10.1038/35077232 [DOI] [PubMed] [Google Scholar]
- Hoogstraten D., Bergink S., Ng J.M., Verbiest V.H., Luijsterburg M.S., Geverts B., Raams A., Dinant C., Hoeijmakers J.H., Vermeulen W., Houtsmuller A.B. 2008. Versatile DNA damage detection by the global genome nucleotide excision repair protein XPC.J. Cell Sci. 121:2850–2859 doi:10.1242/jcs.031708 [DOI] [PubMed] [Google Scholar]
- Houtsmuller A.B., Vermeulen W. 2001. Macromolecular dynamics in living cell nuclei revealed by fluorescence redistribution after photobleaching.Histochem. Cell Biol. 115:13–21 [DOI] [PubMed] [Google Scholar]
- Huen M.S., Grant R., Manke I., Minn K., Yu X., Yaffe M.B., Chen J. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly.Cell. 131:901–914 doi:10.1016/j.cell.2007.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T., Tashiro S., Kakino A., Shima H., Jacob N., Amunugama R., Yoder K., Izumi S., Kuraoka I., Tanaka K., et al. 2007. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics.Mol. Cell. Biol. 27:7028–7040 doi:10.1128/MCB.00579-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Adachi S., Iwakami R., Yasuda H., Muto Y., Seki N., Okano Y. 2001. N-Terminally extended human ubiquitin-conjugating enzymes (E2s) mediate the ubiquitination of RING-finger proteins, ARA54 and RNF8.Eur. J. Biochem. 268:2725–2732 doi:10.1046/j.1432-1327.2001.02169.x [DOI] [PubMed] [Google Scholar]
- Jans J., Schul W., Sert Y.G., Rijksen Y., Rebel H., Eker A.P., Nakajima S., van Steeg H., de Gruijl F.R., Yasui A., et al. 2005. Powerful skin cancer protection by a CPD-photolyase transgene.Curr. Biol. 15:105–115 doi:10.1016/j.cub.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Jowsey P., Morrice N.A., Hastie C.J., McLauchlan H., Toth R., Rouse J. 2007. Characterisation of the sites of DNA damage-induced 53BP1 phosphorylation catalysed by ATM and ATR.DNA Repair (Amst.). 6:1536–1544 doi:10.1016/j.dnarep.2007.04.011 [DOI] [PubMed] [Google Scholar]
- Kim H., Chen J. 2008. New players in the BRCA1-mediated DNA damage responsive pathway.Mol. Cells. 25:457–461. [PMC free article] [PubMed] [Google Scholar]
- Kim H., Chen J., Yu X. 2007. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response.Science. 316:1202–1205 doi:10.1126/science.1139621 [DOI] [PubMed] [Google Scholar]
- Kolas N.K., Chapman J.R., Nakada S., Ylanko J., Chahwan R., Sweeney F.D., Panier S., Mendez M., Wildenhain J., Thomson T.M., et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase.Science. 318:1637–1640 doi:10.1126/science.1150034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijsterburg M.S., Goedhart J., Moser J., Kool H., Geverts B., Houtsmuller A.B., Mullenders L.H., Vermeulen W., van Driel R. 2007. Dynamic in vivo interaction of DDB2 E3 ubiquitin ligase with UV-damaged DNA is independent of damage-recognition protein XPC.J. Cell Sci. 120:2706–2716 doi:10.1242/jcs.008367 [DOI] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins.Cell. 131:887–900 doi:10.1016/j.cell.2007.09.040 [DOI] [PubMed] [Google Scholar]
- Marteijn J.A., van der Meer L.T., van Emst L., van Reijmersdal S., Wissink W., de Witte T., Jansen J.H., Van der Reijden B.A. 2007. Gfi1 ubiquitination and proteasomal degradation is inhibited by the ubiquitin ligase Triad1.Blood. 110:3128–3135 doi:10.1182/blood-2006-11-058602 [DOI] [PubMed] [Google Scholar]
- Matsumoto M., Yaginuma K., Igarashi A., Imura M., Hasegawa M., Iwabuchi K., Date T., Mori T., Ishizaki K., Yamashita K., et al. 2007. Perturbed gap-filling synthesis in nucleotide excision repair causes histone H2AX phosphorylation in human quiescent cells.J. Cell Sci. 120:1104–1112 doi:10.1242/jcs.03391 [DOI] [PubMed] [Google Scholar]
- Mitchell D.L., Haipek C.A., Clarkson J.M. 1985. (6-4)Photoproducts are removed from the DNA of UV-irradiated mammalian cells more efficiently than cyclobutane pyrimidine dimers.Mutat. Res. 143:109–112 [DOI] [PubMed] [Google Scholar]
- Moné M.J., Volker M., Nikaido O., Mullenders L.H., van Zeeland A.A., Verschure P.J., Manders E.M., van Driel R. 2001. Local UV-induced DNA damage in cell nuclei results in local transcription inhibition.EMBO Rep. 2:1013–1017 doi:10.1093/embo-reports/kve224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicassio F., Corrado N., Vissers J.H., Areces L.B., Bergink S., Marteijn J.A., Geverts B., Houtsmuller A.B., Vermeulen W., Di Fiore P.P., Citterio E. 2007. Human USP3 is a chromatin modifier required for S phase progression and genome stability.Curr. Biol. 17:1972–1977 doi:10.1016/j.cub.2007.10.034 [DOI] [PubMed] [Google Scholar]
- Niida H., Nakanishi M. 2006. DNA damage checkpoints in mammals.Mutagenesis. 21:3–9 doi:10.1093/mutage/gei063 [DOI] [PubMed] [Google Scholar]
- O'Driscoll M., Ruiz-Perez V.L., Woods C.G., Jeggo P.A., Goodship J.A. 2003. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome.Nat. Genet. 33:497–501 doi:10.1038/ng1129 [DOI] [PubMed] [Google Scholar]
- Pickart C.M. 2000. Ubiquitin in chains.Trends Biochem. Sci. 25:544–548 doi:10.1016/S0968-0004(00)01681-9 [DOI] [PubMed] [Google Scholar]
- Plans V., Scheper J., Soler M., Loukili N., Okano Y., Thomson T.M. 2006. The RING finger protein RNF8 recruits UBC13 for lysine 63-based self polyubiquitylation.J. Cell. Biochem. 97:572–582 doi:10.1002/jcb.20587 [DOI] [PubMed] [Google Scholar]
- Rademakers S., Volker M., Hoogstraten D., Nigg A.L., Moné M.J., Van Zeeland A.A., Hoeijmakers J.H., Houtsmuller A.B., Vermeulen W. 2003. Xeroderma pigmentosum group A protein loads as a separate factor onto DNA lesions.Mol. Cell. Biol. 23:5755–5767 doi:10.1128/MCB.23.16.5755-5767.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robzyk K., Recht J., Osley M.A. 2000. Rad6-dependent ubiquitination of histone H2B in yeast.Science. 287:501–504 doi:10.1126/science.287.5452.501 [DOI] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139.J. Biol. Chem. 273:5858–5868 doi:10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- Sakasai R., Tibbetts R. 2008. RNF8-dependent and RNF8-independent regulation of 53BP1 in response to DNA damage.J. Biol. Chem. 283:13549–13555 doi:10.1074/jbc.M710197200 [DOI] [PubMed] [Google Scholar]
- Salic A., Mitchison T.J. 2008. A chemical method for fast and sensitive detection of DNA synthesis in vivo.Proc. Natl. Acad. Sci. USA. 105:2415–2420 doi:10.1073/pnas.0712168105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter D., Costanzo V., Gautier J. 2004. Regulation of DNA replication by ATR: signaling in response to DNA intermediates.DNA Repair (Amst.). 3:901–908 doi:10.1016/j.dnarep.2004.03.020 [DOI] [PubMed] [Google Scholar]
- Sobhian B., Shao G., Lilli D.R., Culhane A.C., Moreau L.A., Xia B., Livingston D.M., Greenberg R.A. 2007. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites.Science. 316:1198–1202 doi:10.1126/science.1139516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G.S., Wang B., Bignell C.R., Taylor A.M., Elledge S.J. 2003. MDC1 is a mediator of the mammalian DNA damage checkpoint.Nature. 421:961–966 doi:10.1038/nature01446 [DOI] [PubMed] [Google Scholar]
- Stiff T., Walker S.A., Cerosaletti K., Goodarzi A.A., Petermann E., Concannon P., O'Driscoll M., Jeggo P.A. 2006. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling.EMBO J. 25:5775–5782 doi:10.1038/sj.emboj.7601446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiff T., Cerosaletti K., Concannon P., O'Driscoll M., Jeggo P.A. 2008. Replication independent ATR signalling leads to G2/M arrest requiring Nbs1, 53BP1 and MDC1.Hum. Mol. Genet. 17:3247–3253 doi:10.1093/hmg/ddn220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M., Jackson S.P. 2006. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes.DNA Repair (Amst.). 5:534–543 doi:10.1016/j.dnarep.2006.01.012 [DOI] [PubMed] [Google Scholar]
- Stucki M., Clapperton J.A., Mohammad D., Yaffe M.B., Smerdon S.J., Jackson S.P. 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks.Cell. 123:1213–1226 doi:10.1016/j.cell.2005.09.038 [DOI] [PubMed] [Google Scholar]
- Vermeulen W., Osseweijer P., de Jonge A.J., Hoeijmakers J.H. 1986. Transient correction of excision repair defects in fibroblasts of 9 xeroderma pigmentosum complementation groups by microinjection of crude human cell extracts.Mutat. Res. 165:199–206 [DOI] [PubMed] [Google Scholar]
- Wang H., Zhai L., Xu J., Joo H.Y., Jackson S., Erdjument-Bromage H., Tempst P., Xiong Y., Zhang Y. 2006. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage.Mol. Cell. 22:383–394 doi:10.1016/j.molcel.2006.03.035 [DOI] [PubMed] [Google Scholar]
- Wang B., Matsuoka S., Ballif B.A., Zhang D., Smogorzewska A., Gygi S.P., Elledge S.J. 2007. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response.Science. 316:1194–1198 doi:10.1126/science.1139476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Yang X.P., Kim Y.S., Jetten A.M. 2008. RAP80 responds to DNA damage induced by both ionizing radiation and UV irradiation and is phosphorylated at Ser 205.Cancer Res. 68:4269–4276 doi:10.1158/0008-5472.CAN-07-5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Wani G., Arab H.H., El-Mahdy M.A., Ray A., Wani A.A. 2009. Chromatin restoration following nucleotide excision repair involves the incorporation of ubiquitinated H2A at damaged genomic sites.DNA Repair (Amst.). 8:262–273 doi:10.1016/j.dnarep.2008.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Elledge S.J. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes.Science. 300:1542–1548 doi:10.1126/science.1083430 [DOI] [PubMed] [Google Scholar]