Nonspecific forces, in addition to biophysical properties, lead to the careful arrangement of chromosomes in the nucleus.
Abstract
Chromosomes are not distributed randomly in nuclei. Appropriate positioning can activate (or repress) genes by bringing them closer to active (or inactive) compartments like euchromatin (or heterochromatin), and this is usually assumed to be driven by specific local forces (e.g., involving H bonds between nucleosomes or between nucleosomes and the lamina). Using Monte Carlo simulations, we demonstrate that nonspecific (entropic) forces acting alone are sufficient to position and shape self-avoiding polymers within a confining sphere in the ways seen in nuclei. We suggest that they can drive long flexible polymers (representing gene-rich chromosomes) to the interior, compact/thick ones (and heterochromatin) to the periphery, looped (but not linear) ones into appropriately shaped (ellipsoidal) territories, and polymers with large terminal beads (representing centromeric heterochromatin) into peripheral chromocenters. Flexible polymers tend to intermingle less than others, which is in accord with observations that gene-dense (and so flexible) chromosomes make poor translocation partners. Thus, entropic forces probably participate in the self-organization of chromosomes within nuclei.
Introduction
The paths followed by diffusing molecules, bacteria, foraging animals, share prices, and long polymers have all been described by random walks, and this has led to a rich theory (Chandrasekhar, 1943; Berg, 1993). Random walks are trajectories consisting of uncorrelated steps; in the case of polymers like chromosomes, steps are biochemical links between successive monomers. Notwithstanding the success of this theory, there remain few theoretical analyses of individual polymers in confined spaces (van Vliet et al., 1992; Jun and Mulder, 2006; Jun, 2008), where excluded volume effects become important (which are not easily captured by theory) and simulations require high performance computing. There are also few attempts to validate results experimentally (for important studies, see Haber et al., 2000; Tegenfeldt et al., 2004; Cohen and Moerner, 2007; Bonthuis et al., 2008); this is largely because it is so difficult to detect single molecules. As there is such a wealth of experimental data on individual human chromosomes confined in nuclei, we compared this with new data obtained using Monte Carlo simulations of model polymers subject solely to entropic forces and found remarkable agreement.
Human chromosomes are not distributed randomly in nuclei (Gilbert et al., 2005; Cremer and Cremer, 2006; Branco and Pombo, 2007; Takizawa et al., 2008). For example, gene-poor chromosomes in lymphocytes tend to be peripheral and gene-rich ones internal, inactive heterochromatin often aggregates at the periphery, and centromeres may cluster into chromocenters. Such positioning has important consequences, for example, in repressing genes by bringing them closer to inactive heterochromatin. It also underpins the production of cancer-promoting translocations. These arise early during tumorigenesis when chromosomes broken accidentally are repaired by rejoining the wrong fragments, and this can reposition the relevant territories. As a result, there is interest in detecting such repositioning as it may allow early diagnosis, especially in the major solid tumors that are refractory to the karyotyping used routinely (Takizawa et al., 2008).
It is usually assumed that chromosomal shape and positioning result from the action of specific forces acting locally, for example, through hydrogen bonds, van der Waals forces, charge interactions, or hydrophobic forces acting between one nucleosome and another or between nucleosomes and the lamina. However, it is instructive to estimate whether nonspecific (entropic) forces might have similar effects, especially when (a) theory suggests that they should become significant when acting on polymers as long as individual human chromosomes, which are fibers with contour lengths of several millimeters, confined in micrometer-sized nuclei, (b) simulations indicate that such forces can segregate whole daughter chromosomes to the opposite ends of rod-shaped bacteria (Jun and Mulder, 2006), and (c) experiments confirm that those forces can position plastic beads larger than chromosomes at the periphery of lipid vesicles roughly the size of nuclei (Dinsmore et al., 1998).
In this study, we used Monte Carlo simulations to generate and analyze different sets of polymers (representing interphase chromosomes) confined within a sphere (representing a nucleus). Each polymer was a string of beads that adopted an ideal random walk. To model a chromosome more realistically, each fiber/bead had a finite thickness (and so was self-avoiding, so that each fiber/bead could not occupy the same space as another). The fiber was also endowed with elastic properties, and, in some cases, an appropriate set of interaction or self-interaction potentials. Motivated by the fact that gene-rich and -poor segments have different physical properties, we explicitly distinguished between eu- and heterochromatin. We used relatively fine graining, basing our analysis on 30-nm monomers. (For various other polymer models of chromosomes, see Marko and Siggia, 1997; Odenheimer et al., 2005; Langowski, 2006; Shopland et al., 2006; Jhunjhunwala et al., 2008; Nicodemi et al., 2008; Rosa and Everaers, 2008; de Nooijer et al., 2009; Mateos-Langerak et al., 2009.) As knowledge of the biophysical properties of chromatin remains sketchy, our results suggest qualitative trends rather than quantitative outcomes. We interpret them in terms of an entropic bias that resolves sometimes conflicting forces to position stiff, compact, and thick fibers at the periphery, create asymmetric territories, and drive chromocenters to the periphery, which is all in agreement with experimental data. The idea that the nucleus can functionally self-organize (Misteli, 2001) is growing (Cook, 2002; Kaiser et al., 2008; Rajapakse et al., 2009), and we suggest that nonspecific entropic forces act in concert with specific ones to position chromosomes within nuclei. Our results were obtained assuming equilibrium, but the entropic forces uncovered will inevitably bias out-of-equilibrium systems like nuclei.
Results
The radial positioning of chromosome territories
Gene-poor chromosomes tend to lie peripherally within the spherical nuclei of human lymphocytes and gene-rich ones internally; this organization is evolutionarily conserved among primates. G/C content proves to be one of the best predictors of such radial position (Gilbert et al., 2005; Cremer and Cremer, 2006; Branco and Pombo, 2007; Küpper et al., 2007; Neusser et al., 2007; Takizawa et al., 2008); a high value correlates with an interior position, a high gene content, transcriptional activity, and an increased flexibility and decompaction of the chromatin fiber (Versteeg et al., 2003; Gilbert et al., 2005; Solovei et al., 2009). Therefore, we investigated how these properties affect positioning of model polymers. (Chromosome length plays a more important role in fibroblast nuclei, which are more like flattened ellipsoids than spheres [Bolzer et al., 2005; Neusser et al., 2007].)
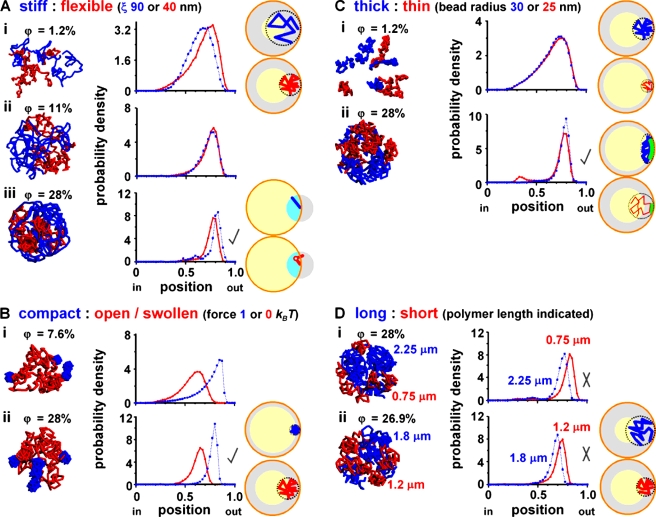
We first considered five stiff and five flexible self-avoiding fibers, representing hetero- and euchromatic fragments of ∼150 kbp, within a confining sphere, the nuclear envelope; each fiber was a string of closely packed 30-nm beads, with each bead representing ∼3 kbp of DNA in chromatin. Although the persistence length (ξ), which is a measure of flexibility, has not been determined precisely in vivo, experiments in vitro yielded values between 40 and 200 nm (Langowski, 2006), and so we selected ones in this range. We then determined (using Monte Carlo simulations) the radial probability density of monomers within each fiber and found a crossover between different behaviors as the fraction of nuclear volume occupied by chromatin (φ) increased. At low packing fractions, stiffer fibers were found, on average, closer to the center of the sphere (which is at position 0; Fig. 1 A, i). Above a fraction of ∼10%, this trend reversed, and stiffer polymers became statistically more likely to lie near the periphery (which is at position 1; Fig. 1 A, ii and iii). As chromatin occupies ∼10% nuclear volume and as many other molecules are also packed into nuclei (e.g., nuclei contain an equal weight of ribonucleoprotein), the pattern found in Fig. 1 A (iii) should be the closest to that found in vivo.
Figure 1.
Monte Carlo simulations of two sets of five polymers confined within a sphere. Polymers were allowed to diffuse in the computer until they reached equilibrium. In each case, typical configurations (left), normalized radial probabilities (middle), and cartoons illustrating major determinants of position (right) are shown for the volume density (φ) indicated. All polymers are self-avoiding (i.e., no bead occupies the same space as another), and, unless stated otherwise, there are 50 beads (diameter of 30 nm) in a polymer with a contour length of 1.5 µm (representing 150 kbp) and a persistence length of 40 nm. (In each case, increasing length 10-fold yields essentially similar patterns [see Monte Carlo simulations of confined chromatin segments].) (A) Five stiff (blue) and five flexible (red) polymers with persistence lengths (ξ) of 90 and 40 nm. (i) At low volume fractions, stiff polymers tend to lie more internally than flexible ones (difference significant, P < 0.0001; unpaired Student’s t test). The cartoon shows that stiff polymers statistically occupy more volume and so lose more entropy when positioned in the (larger) gray volume close to the wall; as a consequence, their centers of mass tend to be concentrated in the yellow volume at the center. (ii and iii) At volume fractions >11%, this trend reverses, and stiff polymers tend to be peripheral (for iii, difference significant, P < 0.0001; unpaired Student’s t test). This is what is seen experimentally (check), where stiff, heterochromatic regions are often peripheral. Data with different length and/or persistence lengths give qualitatively similar trends (see Materials and methods). For a description of the cartoon, see The radial positioning of chromosome territories and Monte Carlo simulations of confined chromatin segments. (B) Five compact (blue) and five open/swollen (red) polymers. Compaction is achieved by allowing monomers in one set to interact with other monomers in the same polymer with an attractive potential of 1 kBT in the range between 30 and 50 nm (center to center distances). (i and ii) Compact polymers are more peripheral at both low and high volume fractions, which is in accord with what is seen experimentally (as heterochromatic regions are more compact; check). The cartoon shows that this entropic bias is the result of compact fibers being able to approach closer to the confining wall (i.e., the inaccessible gray volume is less, and the accessible yellow volume is more). (C) Five thick (30-nm beads; blue) and five thin (25-nm beads; red) polymers. (i) At low volume fractions, thicker polymers tend to be slightly more internal (difference significant, P < 0.005; Student’s t test). The cartoon shows that thicker polymers tend to be excluded from a larger gray volume. (ii) At high volume fractions, thicker polymers tend to be peripheral (difference significant, P < 0.0001; Student’s t test); this is what is seen experimentally (check). For a description of the cartoon, see The radial positioning of chromosome territories and Monte Carlo simulations of confined chromatin segments. (D) Five long (blue) and five short (red) polymers. (i and ii) Longer polymers tend to be more internal, which is not found experimentally (X). The cartoon shows that the long polymers occupy more volume and so tend to be excluded from the larger gray volume (to occupy the smaller yellow volume toward the center).
We describe the combination of entropic forces acting here as an “entropic centrifuge” that works as follows. At low packing fractions, a stiff polymer statistically occupies more volume than a flexible one (it has a larger radius of gyration), and, when it approaches the confining wall, it “feels” the wall sooner to lose more entropy; therefore, it tends to be found more toward the interior. Thus, in the cartoon on the right of Fig. 1 A (i; see also Materials and methods), the stiffer blue polymer is larger and more surface-phobic and so tends to be excluded from the (larger) gray volume at the periphery; as a result, it is more frequently found toward the center in the smaller yellow volume. But at high packing fractions, the entropic effect illustrated in the cartoon in Fig. 1 A (iii) becomes significant. Here, one end of the first persistence length in a stiff polymer (represented by the blue rod) abuts the confining wall. If we imagine this blue rod is tethered to the wall, it can access all conformations in the light blue volume but not the gray volume outside of the confining wall. If the rod is now divided into two (shown in red) to increase flexibility, the light blue volume still remains accessible, but the particular conformation shown is not permissible (as the second half of the red rod penetrates the wall). This qualitatively suggests that flexible (red) polymers lose more configurations (and so entropy) when squashed against the wall. (This effect is quantified by additional simulations in Materials and methods.) Then, they have become the most surface phobic and so tend to be found internally where they lose less entropy. This is what is seen experimentally (Fig. 1 A, iii, check), where heterochromatic (stiffer) regions are often (albeit not always) peripheral (Solovei et al., 2009).
Heterochromatin is more compact than euchromatin as the result of specific interactions between hypoacetylated and hypermethylated residues (for example, at H3K9, H3K27, and H4K20; Weidemann et al., 2003; Dehghani et al., 2005). Therefore, we considered two sets of polymers that self-interact to different degrees. Monomers in one set were as before (i.e., in physicist’s terms, they interact solely via a hard core repulsion), whereas those in the second attracted other monomers in the same fiber with an energy of 1 kBT if their centers were within 30–50 nm (the hard core diameter being 30 nm; kB is the Boltzmann constant, and T is the temperature). As a result (Fig. 1 B, left), half of the polymers had “open” or “swollen” conformations (like euchromatin), and the other half had compact ones (like heterochromatin). (Open and swollen are terms used by biologists and physicists, respectively, to describe roughly the same property.) Compact fibers were found, on average, closer to the edge; this entropic bias is found at low and high packing densities and is the result of compact fibers being able to approach closer to the confining wall (Fig. 1 B). Mild interactions (range of 0.5–2.0 kBT) led to similarly well-separated profiles. These results are also in accord with experimental observations: compact heterochromatin is often peripheral (Solovei et al., 2009).
We next considered two populations of fibers with 25- and 30-nm beads, representing different compactions of the 11-nm nucleosomal string. All fibers were equally flexible (i.e, ξ = 40 nm). Increasing confinement again triggered an exchange. At low volume fractions, thinner segments were slightly more likely to be peripheral because of their smaller size (Fig. 1 C, i). But at high packing fractions, they were more likely to be internal (Fig. 1 C, ii). This effect was smaller than those seen in Fig. 1 (A and B; but the differences are nevertheless significant). We imagine that an entropic “depletion attraction” (Asakura and Oosawa, 1958; Marenduzzo et al., 2006a) drives this switch. Here, the center of mass of each bead of radius r cannot enter the volume that extends a distance r away from any other bead or the confining wall. As a bead approaches the wall, this depleted volume around the bead overlaps that around the wall to increase the volume available to other beads (increasing their entropy). This overlap volume (green in the cartoon in Fig. 1 C, ii) is larger for larger beads, so positioning thicker polymers near the wall increases the entropy of the system. In other words, thicker polymers are more surface philic. This result is again in accord with experimental ones; thicker heterochromatic regions are often peripheral (Solovei et al., 2009).
We then considered two sets of fibers with different lengths; longer fragments were found internally at both low (not depicted) and high packing fractions (Fig. 1 D). This is because long polymers occupy more volume and tend to be found in a smaller volume in the interior (as in the cartoon). This result is not found experimentally because longer chromosomes in spherical nuclei have a slight tendency to be peripheral (Cremer et al., 2001; Habermann et al., 2001). However, this length effect is now known to be small (Mayer et al., 2005), so we suggest that other drivers like flexibility and compaction play more important roles. We confirmed this by running simulations with 10 long but compact fibers (with 100 beads, a persistence length of 40 nm, and compacted as before by a 1 kBT attraction) and 10 short but flexible fibers (with 20 beads and the same persistence length); then, the long fibers were peripheral (unpublished data). This is exactly the organization found in chicken nuclei (Habermann et al., 2001), which contain long macrochromosomes (which are gene poor and so have a higher compaction) and short microchromosomes (which are gene rich and so more flexible).
Our results thus far concern short chromosome fragments and highlight entropic biases caused by heterogeneity in chromatin. However, such biases should persist at all scales, and our simulations can represent longer chromosomes (with the sole exception of those relating to stiff/flexible fibers, as persistence length has little meaning when each bead represents hundreds to thousands of kilobase pairs). For instance, one bead in Fig. 1 (B–D) may represent 2 Mbp (packed into a diameter of 260 nm) in a 100-Mbp human chromosome or 15 kbp (packed into a diameter 51 nm) in a 0.75-Mbp yeast chromosome. (We have also repeated the simulations in Fig. 1 with 2- and 10-fold longer chromosomes and found the same trends [Fig. 1 legend].)
Looping promotes territory formation
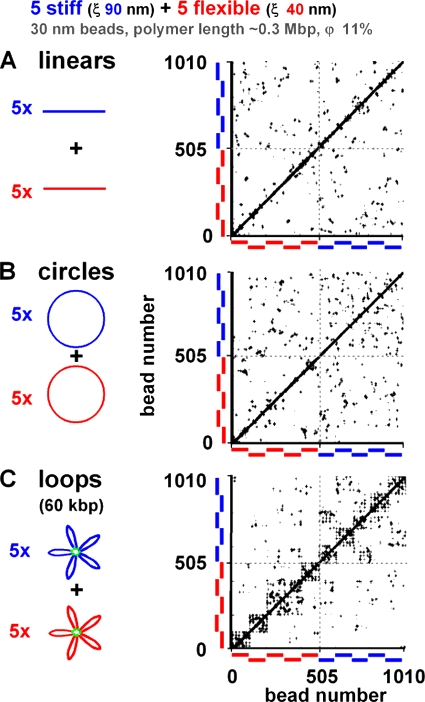
The fibers illustrated in Fig. 1 often interpenetrate without forming discrete territories. (The compact fibers in Fig. 1 B are the sole exceptions.) They provide a simple case that is easy to analyze, analogous to the chromosomes of budding yeast which do not form obvious territories, although heterochromatic telomeres are often peripheral (Berger et al., 2008). Therefore, we next investigated how global fiber conformation affects the formation of territories like those in higher eukaryotes. We began with a control. Five stiff (ξ = 90 nm) and five flexible (ξ = 40 nm) fibers were allowed to equilibrate, and contacts between each 30-nm bead and all others were determined. In the contact map in Fig. 2 A, crosses mark positions of beads lying within 60 nm of each other (30 nm being the center to center distance of two touching beads). Beads were numbered from the first to last bead on the first (flexible) fiber (i.e., from number 1 to 101), through other flexible fibers, then to the first bead on the first stiff fiber (i.e., number 506), and on to the last bead on the last (stiff) fiber (i.e., number 1,010). Beads rarely contacted others on the same fiber but often touched those on other fibers; this confirms that territories did not form. However, there were slightly more contacts in the top right quadrant than the bottom left one; contacts between stiff fibers were slightly more numerous, indicating there was a partial phase separation. We interpret this in terms of a greater reduction in the number of possible conformations when two flexible polymers are positioned next to each other (compared with that obtained with two stiff polymers) and, to a lesser extent, a consequence of the differing radial concentrations of the two types of polymer (Fig. 1 A). Here, the centrifuge drove stiffer polymers both together (Fig. 2 A) and to the wall (Fig. 1 A). Roughly the same pattern was found with circles (Fig. 2 B), although there were more stiff–stiff contacts (top right quadrant) and fewer flexible–flexible ones (bottom left quadrant). These results indicate that linear and circular polymers intermingle without forming discrete territories. (The theoretical reason underlying this behavior is that intermingling requires only a few kilocalories/mole per loop/fiber [Jun, 2008].)
Figure 2.
Contacts within and between polymers with different conformations determined using Monte Carlo simulations. Sets of 10 linear, circular, or looped polymers (each a string of 101 30-nm beads representing 303 kbp) were confined in a sphere (radius of 0.3 µm; φ = 11%) and allowed to diffuse in the computer until they reached equilibrium; contacts (defined as ≤60 nm center to center distance) that each bead made with others are marked by a cross (+; which appears as a dot at low magnification) on the plot. In each, five polymers were flexible (ξ = 40 nm; beads 1–101, 102–202,…, 405–505), and five were stiff (ξ = 90 nm; beads 506–606, 607–707,…, 910–1,010). (A) Beads are not so likely to contact other beads in the same fiber, indicating that fibers intermingle without forming discrete territories (inter- and intrachain contacts, 30% and 70%, respectively). There are also more contacts in the top right quadrant than the bottom left one, showing that contacts between stiff fibers are more numerous (flexible–flexible contacts, 3,480 [16%]; stiff–flexible contacts, 850 [39%]; stiff–stiff contacts, 978 [45%]). (B) The pattern is largely the same (inter- and intrachain contacts, 30% and 70%, respectively). There are more stiff–stiff contacts (top right quadrant) and fewer flexible–flexible ones (bottom left quadrant; flexible–flexible contacts, 390 [17%]; stiff–flexible contacts, 796 [35%]; stiff–stiff contacts, 1,100 [48%]). (C) Rosettes, each with five 60-kbp loops, generated by tethering together beads (green) at positions 1, 21, 41, 61, 81, and 101 in each fiber. The 10 6 × 6 grids along the diagonal mark these forced contacts. There are many other contacts within each grid (and many fewer interfiber contacts), indicating that a bead in one fiber is more likely to contact others in the same fiber; this reflects the formation of more discrete territories (inter- and intrachain contacts, 20% and 80%, respectively). Stiff fibers are again more likely to contact other stiff fibers (flexible–flexible contacts, 730 [34.6%]; stiff–flexible contacts, 311 [14.7%]; stiff–stiff contacts, 848 [50.7%]).
Can looping promote territory formation? Each of the 10 linear strings was then forced to form loops by tethering together beads 1, 21, 41, 61, 81, and 101 in each string (creating a rosette with five loops of 60 kbp in each string). The resulting forced contacts are seen as 10 6 × 6 grids along the diagonal in Fig. 2 C. There were then many other contacts within each 6 × 6 grid; beads in one fiber had become more likely to contact others in the same fiber. The number of interfiber contacts provides a good measure of territory formation, which looping clearly promotes. To quantify intermingling more precisely, we determined the percentage of contacts between beads on different fibers; for linear fibers and circles in Fig. 2 (A and B), this percentage was between 30 and 40%, whereas it dropped to ∼20% for rosettes with 60-kbp loops (Fig. 2 C) and to <10% for rosettes with 30-kbp loops (not depicted). These values compare with the ∼20% seen in human lymphocytes (Branco and Pombo, 2007). Flexible–flexible contacts were again less likely (Fig. 2 C, bottom left quadrants); this is in accord with observations that gene-dense (and so flexible; Gilbert et al., 2005) chromosomes make poor translocation partners (Bickmore and Teague, 2002). An obvious next step is to model fibers that more closely resemble real chromosomes (e.g., with varying flexibility along their length) and compare interpenetration of loci like MYC and IGH, which are frequently involved in the translocations seen in Burkitt’s lymphoma (Roix et al., 2003).
Positioning territories
Many fibers in Fig. 1 did not form discrete territories; therefore, we checked the radial positions of the two types of fibers that did generate them. 10 territories formed in different ways, 5 by compaction and 5 by rosetting, were examined. Each set was generated as before either by an attraction of 1 kBT between beads in each linear string or by tethering together every 10th bead. (We illustrated this example using rosettes with 10 loops of 30 kbp, but those with 5 loops of 60 kbp gave similar patterns. For example, rosettes with 60 kbp and 30 kbp loops gave 80% and 94% intrachain contacts, respectively, and only 20% and 6% interchain contacts [unpublished data].) Then, compact territories were peripheral and rosettes internal (Fig. 3, A and B). The contact map confirms that both formed territories with few interchromosomal contacts (Fig. 3 C). As the two types of territory have roughly the same volumes (unpublished data), the centrifuge works here mainly through differences in flexibility and compactness. Again, our results confirm those seen experimentally; active regions of the genome are probably organized into shorter loops (Faro-Trindade and Cook, 2006) and tend to be more internal than inactive (heterochromatic) regions, which are often more compact and peripheral (Solovei et al., 2009).
Figure 3.
Positioning rosettes and compact (linear) fibers. Sets of five rosettes (each with 10 30-kbp loops; red) and five compact (linear) fibers (blue) were confined in a sphere (radius of 0.3 µm; φ = 12.6%) and allowed to diffuse in the computer until they reached equilibrium. Each fiber was a string of 101 30-nm beads representing 303 kbp with 40-nm persistence length. Fibers 1–5 were rosettes and made of beads 1–101, 102–202,…, 405–505; each rosette (with 10 30-kbp loops) was formed by tethering together beads at positions 1, 11, 21, etc., in the fiber. Fibers 6–10 were compact and made of beads 506–606, 607–707,…, 910–1,010; compaction was achieved using an interaction of 1 kBT in the range of 30–50 nm between each monomer in any one fiber. (A) Snapshot of one simulation. Compact fibers (blue) are expected to be more heterochromatic and are more peripheral, as seen experimentally. (B) Normalized radial probability distributions of beads in rosettes (red) and compact fibers (blue); the latter tend to be peripheral. (C) Contact map (determined as in Fig. 2; crosses [+; which appear as dots at low magnification] mark contacts). The five 11 × 11 grids along the diagonal nearest the origin mark forced contacts in rosettes; additional contacts within each grid reflect many other interfiber contacts and territory formation. The five clusters on the diagonal distant from the origin mark interfiber contacts within each of the five compact fibers; they also form territories (inter- and intrachain contacts, 4% and 96%, respectively). There were many contacts in the quadrants at top left and bottom right, showing that rosettes preferred to pack against compact fibers (rosette–rosette contacts, 136 [3.8%]; rosette–compact contacts, 266 [46.5%]; compact–compact contacts, 170 [29.7%]).
Aspherical chromosome territories
Intuition suggests that isolated random coils like DNA molecules will be spherically symmetric, and they are, at least when large populations are averaged over time. However, both theory and experiment show that they are instantaneously more likely to be prolate ellipsoids (i.e., like American footballs). (In the language of polymer physics, this is because the resulting increase in volume increases the number of constituent blobs, and so degrees of freedom, and the entropic gains outweigh the losses [Rudnick and Gaspari, 1987; Haber et al., 2000].) Chromosome territories within spherical mouse pro-B nuclei are also ellipsoidal, with a ratio of principal axes (a:b:c) of 1:2.9:4.5 (Khalil et al., 2007). Various properties of randomly packed ellipsoids are of interest here (Donev et al., 2004; Man et al., 2005). First, they can be packed even more tightly than spheres of equivalent volume, so we might expect nature to exploit this when packing chromosomes into crowded nuclei. We might also guess that squashing a spherical territory against a spherical wall would yield an oblate ellipsoid (like an M&M or lentil). Second, tightly packed ellipsoids touch more neighbors than do spheres, which may go some way to explain why so many interterritory contacts are seen experimentally by chromosome conformation capture (Simonis and de Laat, 2008). Third, they are less likely than spheres to become locally jammed, as they have one thinner axis and so can escape through thinner gaps in the surrounding cage formed by nearest neighbors, and this should have entropic consequences. Therefore, we investigated the shapes of various fibers (Table I). In this case, we analyzed 46 fibers. For flexible fibers, our approach could be scaled. For example, each bead could then be 208 nm in diameter (a size chosen to maintain the packing fraction) and contain 1 Mbp, whereas each fiber would represent a 100-Mbp chromosome and all 46 fibers the complete chromosomal set (see Materials and methods). At low packing fractions, linear fibers (whether flexible or stiff) were ellipsoidal (as expected), and they became more spherical as the packing fraction increased (i.e., the longest axis came closer to unity). In contrast, rosettes (containing 30 or 60 kbp loops) were close to spherical at low packing fractions and became more aspherical as the packing fraction increased to give structures more like those seen experimentally. Stiff rosettes tended to be even more aspherical. Once again, nonspecific entropic forces acting alone could generate the required organization.
Table I.
The shapes of polymers within confining spheres
| Packing fraction | Axial ratios |
| % | |
| Linear (no territories) | |
| Flexiblea | |
| 1.2 | 1:1.88:4.34 |
| 9.4 | 1:1.77:3.77 |
| 21.7 | 1:1.69:3.45 |
| Stiffb | |
| 1.2 | 1:1.99:4.46 |
| 9.4 | 1:2.01:3.49 |
| 21.7 | 1:1.71:2.58 |
| Rosettes (30-kbp loops; territories) | |
| Flexiblea | |
| 1.2 | 1:1.16:1.34 |
| 9.4 | 1:1.17:1.36 |
| 21.7 | 1:1.33:1.71 |
| Stiffb | |
| 1.2 | 1:1.19:1.37 |
| 9.4 | 1:1.20:1.38 |
| 21.7 | 1:1.64:2.85c |
| Rosettes (60-kbp loops; territories) | |
| Flexiblea | |
| 1.2 | 1:1.35:1.68 |
| 9.4 | 1:1.34:1.71 |
| 21.7 | 1:1.46:2.37c |
| Stiffb | |
| 1.2 | 1:1.44:1.92 |
| 9.4 | 1:1.52:2.12 |
| 21.7 | 1:1.72:2.89c |
Sets of 46 linear fibers or 46 rosettes (with 10 loops of 30 kbp, as in Fig. 3, or 5 loops of 60 kbp, as in Fig. 2 C) were confined in a sphere and allowed to diffuse in the computer until they reached equilibrium; the shapes of the resulting fibers were then examined. Values are given for the packing fraction and axial ratios (for each of the three principal axes of the ellipsoid of inertia, a, b, and c). Territories in mouse lymphocytes have axial ratios of 1:2.9:4.5 (Khalil et al., 2007).
ξ = 40 nm.
ξ = 150 nm.
Only rosettes at the high packing fraction give both territories and ratios that approach those seen in cells.
Driving chromocenters to the periphery
All mouse chromosomes bear centromeres at one end (unlike some human chromosomes, which are metacentric), and in lymphocytes, these centromeres are often found at the edge of individual territories, whereas those in different territories cluster together at the nuclear periphery into chromocenters (Weierich et al., 2003; Mayer et al., 2005; de Nooijer et al., 2009). We previously suggested that the depletion attraction might act through the large clumps of heterochromatin in the centromeres to drive such positioning (Marenduzzo et al., 2006a). Therefore, we investigated various types of fiber, linear, (linear) compact, and rosettes carrying a large terminal bead of 120 nm representing a heterochromatic centromere (Fig. 4). With all types, the large terminal bead was often found at the periphery of its territory (Fig. 4, left; and not depicted). Moreover, beads in different territories often clustered together (reflected by the peaks in zone q) at the periphery of the nucleus (reflected by the peaks in zone u). (Similar patterns were given by terminal beads of 90–150 nm [unpublished data].) We suggest that the depletion attraction acts through the large terminal bead to drive this organization, and again, entropic forces are sufficient to position the terminal bead in the appropriate way.
Figure 4.
Positioning of centromeres in nuclei. Sets of 10 linear, compact, and rosetted fibers (red; ξ = 40 nm) bearing one large terminal bead of 120 nm representing a telocentric centromere (blue) were confined in a sphere (radius of 0.3 µm; φ = 12.5%) and allowed to diffuse in the computer until they reached equilibrium. Typical snapshots are shown on the left, distances (in simulation units, where 1 unit corresponds to 30 nm) between terminal beads in the middle, and normalized probabilities that territories are found at a given radial position on the right. In all cases, terminal beads are often peripheral and clustered. In zones p and v, probabilities are zero, as the center of mass of a terminal bead cannot approach within 120 nm of that of another terminal bead or within 60 nm of the confining wall. The peaks in zone q reflect clustering of terminal beads, and those in zone r reflect the increased probability that two terminal beads will be found randomly in the larger volume found at these separations. The small peak in zone s arises because probabilities seen with a random distribution rise progressively toward the periphery, but here, terminal beads tend to be concentrated in zone u to leave fewer in zone t. (A) 10 linear fibers of 300 kbp, each with one additional large terminal bead. (B) 10 (linear) compact fibers of 300 kbp plus a large terminal bead; compaction is driven by 1 kBT interaction as in Fig. 1 B. (C) Nine 30-kbp loops formed as in Fig. 2 C, each with one linear 30-kbp extension bearing the large terminal bead.
Discussion
In this study, we used Monte Carlo simulations to demonstrate that nonspecific (entropic) forces acting alone can position and shape self-avoiding polymers within crowded nuclei in the ways seen experimentally. Polymers composed of strings of beads are allowed to diffuse in a confining sphere until they reach equilibrium. Flexible polymers (like gene-rich chromosomes) tend to be found more toward the center (Fig. 1 A, iii) and compact/thick ones (like heterochromatin) toward the periphery (Fig. 1, B [ii] and C [ii]). Looped polymers but not linear ones also form discrete territories (Fig. 2; de Nooijer et al., 2009) with the appropriate aspherical shapes (Table I). In addition, flexible territories tend to intermingle less with others (Fig. 2 C), which is in accord with observations that gene-dense (and so flexible) chromosomes make poor translocation partners (Bickmore and Teague, 2002). If the polymers carry a large terminal bead (to represent centromeric heterochromatin at one end of a telocentric chromosome), beads are found both at the edge of their own territories and cluster at the nuclear periphery (Fig. 4), again as found in vivo (de Nooijer et al., 2009).
We do not wish to suggest that these entropic forces act alone, without contribution from additional specific interactions; rather, we imagine that the ultimate outcome is determined by resolution of the combined forces, which sometimes may be conflicting. For example, the centrifuge may drive heterochromatin to the periphery in higher animals and plants, with specific nucleosome–lamin interactions being involved in animals (Polioudaki et al., 2001) but not in plants, as they lack lamin proteins. And although heterochromatin is often found at the periphery, the rod cells in the retinas of diurnal mammals provide a striking exception (Solovei et al., 2009), so in this case, we suggest that other specific forces become more significant. For example, it is quite possible that variations in the strength of the attraction between heterochromatic regions or between heterochromatin and the lamina will modulate the organization.
One of our major findings is that linear fibers do not generally form discrete territories; rather, linear fibers intermingle (Fig. 1, A, C, and D). Compact fibers are the exception (Fig. 1 B). As compaction is achieved by allowing interactions between beads in any one fiber (but not between those in different fibers) and as it is difficult to imagine what mechanism might generate such specificity, it seems unlikely that this type of compaction drives territory formation in vivo. However, looping clearly generates discrete territories (Fig. 2 C), and there is now good experimental evidence for looping (Marenduzzo et al., 2007). There are also good theoretical arguments for it. Thus, the physical separation between any two human genes in three-dimensional nuclear space (determined by fluorescence in situ hybridization) depends on the number of intervening base pairs according to a power law that is inconsistent with a random walk; instead, the fiber must fold back on itself to give the required compaction, and the best fit is given by models involving mixtures of local and giant loops (Mateos-Langerak et al., 2009). That linear and looped fibers form territories to different degrees is also in agreement with an equilibrium analysis by de Nooijer et al. (2009) and the dynamic simulations of Rosa and Everaers (2008).
Our simulations sample equilibrium conformations, and it remains possible that a true equilibrium is never reached in the lifetime of a cell. For example, Rosa and Everaers (2008) argue that entanglement and disentanglement occur so slowly that the organization is determined largely by the structure imposed by the previous mitosis. Chromosomes are initially positioned within daughter cells by the mitotic machinery, and remnants of such positioning are sometimes seen in the Rabl conformation, with centromeres at one end of the interphase nucleus and telomeres at the other (Cremer and Cremer, 2006). However, this organization is not seen in mammalian lymphocytes, so something must rearrange things. Furthermore, it would be natural to assume that the shorter chromosomes of yeast and Arabidopsis thaliana will equilibrate faster (Rosa and Everaers, 2008; de Nooijer et al., 2009), but their heterochromatin is still preferentially peripheral, as our results suggest. Note also that no interstrand crossings were allowed in the molecular dynamics simulations of Rosa and Everaers (2008), but if included (because of topoisomerase action), they would speed up disentangling (Sikorav and Jannink, 1994; Rosa and Everaers, 2008). We also believe entropic effects will still be major drivers even if dynamic (nonequilibrium) effects play some role. First, even partial equilibration still allows many configurations to be explored, and while this is happening, we would expect the system to drift toward the organization we describe. Second, starting from a mitotic chromosome (with few interchromosomal contacts) instead of intermixed fibers (in our simulations) can only facilitate the formation of interphase territories; in this case, the dynamics work with us and not against us.
We conclude that nonspecific entropic forces are major and global determinants of chromosome conformation and position within human nuclei. At first glance, this seems like an oxymoron: entropy is usually associated with disorder, but here it leads to a more ordered structure. However, there are many other such examples (e.g., the formation of a colloidal crystal from the packing of hard spheres; Chaikin and Lubensky, 1995). In passing, we note that prokaryotic genomes are significantly less confined than eukaryotic ones and so would be subject to different entropic biases acting at lower packing fractions (Fig. 1). And if evolutionary pressure acts against any of these entropic biases, the system has to expend energy to reorganize the structure. Finally, we also note that it has just become feasible to compare the contact maps for the entire human genome obtained by simulation (in the way we have done but using supercomputers) and experiment (using chromosome conformation capture allied to deep sequencing; Simonis and de Laat, 2008).
Materials and methods
Self-avoiding, semiflexible, self-interacting chromatin segments
We modeled chromosome fragments as flexible fibers with variable length, thickness, persistence length (or stiffness), and strength of (attractive) self-interactions (Bon et al., 2006; Marenduzzo et al., 2006b). Each fragment was modeled as a chain with N beads (each of diameter, σ, of 30 nm, corresponding to ∼3 kbp of DNA in chromatin) with persistence length ξ (ranging from 40 nm to 200 nm for eu- and heterochromatin, respectively). The total length varies in a range that provides a compromise between the need to maximize fragment length (to eliminate finite size effects) and minimize computational cost. Typical simulations reported in this study are for 10 0.3-Mbp fragments and a total of 1,000 beads, and we checked results using 10-fold longer fibers. Note that, for flexible polymers, our approach may be scaled: for example, each bead could represent 300 kbp (each 140 nm in diameter to maintain the same packing fraction), and then a single 1,000-bead chain could represent a 150-Mbp human chromosome.
Let us focus on one fiber, and let us identify the centers of its N beads with the set of position vectors . The associated interaction potential is then V = Vbending + Vsteric + Vself. The first term describes bending:
where (i = 1,…, N) denotes the i-th discretized tangent vector, kB the Boltzmann constant, and T the temperature. The second term stands for a two-body hard core potential between any two beads, which states that the center to center distance between any two beads cannot be less than the hard core diameter (usually 30 nm). The last term is also a sum of two-body interactions, which are attractive and promote self-aggregation of a segment. We chose the form of Vself to be the sum of square well potentials:
where V2 was taken to be equal to −kBT if is smaller than 50 nm. This interaction could be of whatsoever origin; for example, it could arise from depletion interactions acting on thicker heterochromatic beads. All segments were confined inside a nucleus, which we minimally model as a sphere of radius R.
In some cases, fibers were compacted or formed into rosettes. Compact fibers were generated by allowing a monomer in the fiber to attract other monomers in the same fiber with an energy of 1 kBT if their centers were 30–50 nm apart (the hard core diameter being 30 nm). Rosettes were generated by forcing loop formation by tethering together beads (e.g., those at positions 1, 21, 41, 61, 81, and 101 in a string to create a rosette with five loops of 60 kbp in each string) using the following interparticle potential:
which exerts a tethering force of constant modulus between the particles i and j, which we want to join to form a loop (we took f0 = 100 kBT/ σ).
Monte Carlo simulations of confined chromatin segments
The statistical properties of chromatin fragments can be sampled using Monte Carlo techniques, but conventional methods become inefficient when fragments are confined. Therefore, we generalized a Monte Carlo algorithm recently used to study the growth of biopolymers inside of a vesicle (Marenduzzo and Orlandini, 2007). To equilibrate a configuration of many confined polymers, we started with the prescribed number of shorter fragments that can readily fit inside a sphere (e.g., to equilibrate 10 fragments each with 500 beads, we started with 10 fragments each with 5 beads). In addition to crankshaft, pivot, and reptation moves, we allowed fibers to grow at a rate of kon for each fragment until the desired length was reached. Growth was then stopped, and Monte Carlo equilibration continued for many steps (typically a million). The whole process was then repeated ∼10 times to improve statistics and eliminate any bias caused by the initial conditions. Growth rendered our Monte Carlo scheme more efficient, as confinement was progressively increased so that until the last stages, the mobility of the chains was higher than in conventional fixed-length schemes. Starting from small chains also eliminated the problem of choosing an otherwise arbitrary initial condition. The volume fraction, φ, can be defined via
where Ntot is the total number of 30-nm chromatin beads in the system (the numerator in the aforementioned equation is therefore the volume of chromatin). In human lymphocyte nuclei, which are 7–8 µm in diameter and contain ∼6,000 Mbp, assuming a compaction of 3 kbp into 30 nm of a chromatin fiber, one may estimate that φ is between 10 and 15%. This value is in line with those calculated by Rosa and Everaers (2008) and by de Nooijer et al. (2009). In simulations, φ was varied by shrinking the confining sphere.
Structures were visualized using Rasmol software (Sayle and Milner-White, 1995). To characterize distributions, we recorded the time-dependent radial position of each bead inside of the sphere. The beads corresponding to each of the chromatin fragments were then separately averaged to yield the radial position. The resulting plots typically referred to normalized probability densities of beads from the middle to the periphery and could be compared directly with analogous plots obtained by chromosome painting. Note that in all plots, the probability density at the center (position 0) is zero as the volume there is zero.
Additional simulations supported results obtained in Fig. 1. Thus, fibers with different lengths and/or persistence lengths gave qualitatively similar trends as those seen in Fig. 1 A. For example, with five flexible and five stiff fibers of 1.5 Mbp (ξ = 40 and 200 nm) and at φ = 8%, mean radial positions (when scaled by radius) were 0.76 (root mean square deviation [rmsd] = 0.04) for flexible fibers and 0.66 (rmsd = 0.04) for stiff ones. At φ = 13.5%, averaged scaled positions were 0.71 (rmsd = 0.03) for flexible fibers and 0.77 (rmsd = 0.01) for stiff ones. We also confirmed that flexible polymers close to the sphere surface lose more entropy than stiff ones as follows. For example, in a Monte Carlo run with four stiff or four flexible fibers in the outermost concentric shell of a sphere (0.3 Mbp; ξ = 40 or 200 nm; φ = 26%; radius of shell = 192–333 nm), 75% of Monte Carlo moves made in flexible fibers were rejected as they lay outside of the confining surface compared with 55% for stiff ones. This confirms qualitatively that flexible fibers lose more entropy. We also performed additional simulations in which a set of flexible and stiff polymers were confined in a spherical shell close to the surface; to apply the same pressure on the surface, stiff fibers needed to be packed more tightly than flexible ones. For example, a pressure of 10 kBT/σ3 (where σ is bead diameter) is exerted by stiff and flexible polymers when φ is 36% and 33%, respectively, and a pressure of 20 kBT/σ3 is exerted when φ is 62% and 47%. In other words, flexible polymers exert a larger force on the surface because of their larger entropy, and this makes it more difficult to position them peripherally.
Acknowledgments
This work has made use of the resources provided by the University of Edinburgh Compute and Data Facility, which is partially supported by the e-Science Data, Information, and Knowledge Transformation initiative.
Footnotes
Abbreviation used in this paper:
- rmsd
- root mean square deviation
References
- Asakura S., Oosawa F. 1958. Interactions between particles suspended in solutions of macromolecules.Journal of Polymer Science. 33:183–192 doi:10.1002/pol.1958.1203312618 [Google Scholar]
- Berg H.C. 1993. Random Walks in Biology. Princeton University Press, Princeton, NJ: 152 pp [Google Scholar]
- Berger A.B., Cabal G.G., Fabre E., Duong T., Buc H., Nehrbass U., Olivo-Marin J.C., Gadal O., Zimmer C. 2008. High-resolution statistical mapping reveals gene territories in live yeast.Nat. Methods. 5:1031–1037 doi:10.1038/nmeth.1266 [DOI] [PubMed] [Google Scholar]
- Bickmore W.A., Teague P. 2002. Influences of chromosome size, gene density and nuclear position on the frequency of constitutional translocations in the human population.Chromosome Res. 10:707–715 doi:10.1023/A:1021589031769 [DOI] [PubMed] [Google Scholar]
- Bolzer A., Kreth G., Solovei I., Koehler D., Saracoglu K., Fauth C., Müller S., Eils R., Cremer C., Speicher M.R., Cremer T. 2005. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes.PLoS Biol. 3:e157 doi:10.1371/journal.pbio.0030157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bon M., Marenduzzo D., Cook P.R. 2006. Modeling a self-avoiding chromatin loop: relation to the packing problem, action-at-a-distance, and nuclear context.Structure. 14:197–204 doi:10.1016/j.str.2005.10.016 [DOI] [PubMed] [Google Scholar]
- Bonthuis D.J., Meyer C., Stein D., Dekker C. 2008. Conformation and dynamics of DNA confined in slitlike nanofluidic channels.Phys. Rev. Lett. 101:108303 doi:10.1103/PhysRevLett.101.108303 [DOI] [PubMed] [Google Scholar]
- Branco M.R., Pombo A. 2007. Chromosome organization: new facts, new models.Trends Cell Biol. 17:127–134 doi:10.1016/j.tcb.2006.12.006 [DOI] [PubMed] [Google Scholar]
- Chaikin P.M., Lubensky T.C. 1995. Principles of Condensed Matter Physics. Cambridge University Press, Cambridge: 699 pp [Google Scholar]
- Chandrasekhar S. 1943. Stochastic problems in physics and astronomy.Reviews of Modern Physics. 15:1–89 doi:10.1103/RevModPhys.15.1 [Google Scholar]
- Cohen A.E., Moerner W.E. 2007. Principal-components analysis of shape fluctuations of single DNA molecules.Proc. Natl. Acad. Sci. USA. 104:12622–12627 doi:10.1073/pnas.0610396104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P.R. 2002. Predicting three-dimensional genome structure from transcriptional activity.Nat. Genet. 32:347–352 doi:10.1038/ng1102-347 [DOI] [PubMed] [Google Scholar]
- Cremer M., von Hase J., Volm T., Brero A., Kreth G., Walter J., Fischer C., Solovei I., Cremer C., Cremer T. 2001. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells.Chromosome Res. 9:541–567 doi:10.1023/A:1012495201697 [DOI] [PubMed] [Google Scholar]
- Cremer T., Cremer C. 2006. Rise, fall and resurrection of chromosome territories: a historical perspective. Part II. Fall and resurrection of chromosome territories during the 1950s to 1980s. Part III. Chromosome territories and the functional nuclear architecture: experiments and models from the 1990s to the present.Eur. J. Histochem. 50:223–272 [PubMed] [Google Scholar]
- de Nooijer S., Wellink J., Mulder B., Bisseling T. 2009. Non-specific interactions are sufficient to explain the position of heterochromatic chromocenters and nucleoli in interphase nuclei.Nucleic Acids Res. 37:3558–3568 doi:10.1093/nar/gkp219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehghani H., Dellaire G., Bazett-Jones D.P. 2005. Organization of chromatin in the interphase mammalian cell.Micron. 36:95–108 doi:10.1016/j.micron.2004.10.003 [DOI] [PubMed] [Google Scholar]
- Dinsmore A.D., Wong D.T., Nelson P., Yodh A.G. 1998. Hard spheres in vesicles: curvature-induced forces and particle-induced curvature.Phys. Rev. Lett. 80:409–412 doi:10.1103/PhysRevLett.80.409 [Google Scholar]
- Donev A., Cisse I., Sachs D., Variano E.A., Stillinger F.H., Connelly R., Torquato S., Chaikin P.M. 2004. Improving the density of jammed disordered packings using ellipsoids.Science. 303:990–993 doi:10.1126/science.1093010 [DOI] [PubMed] [Google Scholar]
- Faro-Trindade I., Cook P.R. 2006. A conserved organization of transcription during embryonic stem cell differentiation and in cells with high C value.Mol. Biol. Cell. 17:2910–2920 doi:10.1091/mbc.E05-11-1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert N., Gilchrist S., Bickmore W.A. 2005. Chromatin organization in the mammalian nucleus.Int. Rev. Cytol. 242:283–336 doi:10.1016/S0074-7696(04)42007-5 [DOI] [PubMed] [Google Scholar]
- Haber C., Ruiz S.A., Wirtz D. 2000. Shape anisotropy of a single random-walk polymer.Proc. Natl. Acad. Sci. USA. 97:10792–10795 doi:10.1073/pnas.190320097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann F.A., Cremer M., Walter J., Kreth G., von Hase J., Bauer K., Wienberg J., Cremer C., Cremer T., Solovei I. 2001. Arrangements of macro- and microchromosomes in chicken cells.Chromosome Res. 9:569–584 doi:10.1023/A:1012447318535 [DOI] [PubMed] [Google Scholar]
- Jhunjhunwala S., van Zelm M.C., Peak M.M., Cutchin S., Riblet R., van Dongen J.J., Grosveld F.G., Knoch T.A., Murre C. 2008. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions.Cell. 133:265–279 doi:10.1016/j.cell.2008.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun S. 2008. Can entropy save bacteria? Quantitative Biology. arXiv.org, Cornell University Library. Available at: http://arxiv.org/abs/0808.2646(accessed September 15, 2008)
- Jun S., Mulder B. 2006. Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome.Proc. Natl. Acad. Sci. USA. 103:12388–12393 doi:10.1073/pnas.0605305103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser T.E., Intine R.V., Dundr M. 2008. De novo formation of a subnuclear body.Science. 322:1713–1717 doi:10.1126/science.1165216 [DOI] [PubMed] [Google Scholar]
- Khalil A., Grant J.L., Caddle L.B., Atzema E., Mills K.D., Arneodo A. 2007. Chromosome territories have a highly nonspherical morphology and nonrandom positioning.Chromosome Res. 15:899–916 doi:10.1007/s10577-007-1172-8 [DOI] [PubMed] [Google Scholar]
- Küpper K., Kölbl A., Biener D., Dittrich S., von Hase J., Thormeyer T., Fiegler H., Carter N.P., Speicher M.R., Cremer T., Cremer M. 2007. Radial chromatin positioning is shaped by local gene density, not by gene expression.Chromosoma. 116:285–306 doi:10.1007/s00412-007-0098-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langowski J. 2006. Polymer chain models of DNA and chromatin.Eur Phys J E Soft Matter. 19:241–249 doi:10.1140/epje/i2005-10067-9 [DOI] [PubMed] [Google Scholar]
- Man W., Donev A., Stillinger F.H., Sullivan M.T., Russel W.B., Heeger D., Inati S., Torquato S., Chaikin P.M. 2005. Experiments on random packings of ellipsoids.Phys. Rev. Lett. 94:198001 doi:10.1103/PhysRevLett.94.198001 [DOI] [PubMed] [Google Scholar]
- Marenduzzo D., Orlandini E. 2007. Dynamics of fibers growing inside soft vesicles.Europhys. Lett. 80:48004 doi:10.1209/0295-5075/80/48004 [Google Scholar]
- Marenduzzo D., Finan K., Cook P.R. 2006a. The depletion attraction: an underappreciated force driving cellular organization.J. Cell Biol. 175:681–686 doi:10.1083/jcb.200609066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenduzzo D., Micheletti C., Cook P.R. 2006b. Entropy-driven genome organization.Biophys. J. 90:3712–3721 doi:10.1529/biophysj.105.077685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marenduzzo D., Faro-Trindade I., Cook P.R. 2007. What are the molecular ties that maintain genomic loops? Trends Genet. 23:126–133 doi:10.1016/j.tig.2007.01.007 [DOI] [PubMed] [Google Scholar]
- Marko J.F., Siggia E.D. 1997. Polymer models of meiotic and mitotic chromosomes.Mol. Biol. Cell. 8:2217–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateos-Langerak J., Bohn M., de Leeuw W., Giromus O., Manders E.M., Verschure P.J., Indemans M.H., Gierman H.J., Heermann D.W., van Driel R., Goetze S. 2009. Spatially confined folding of chromatin in the interphase nucleus.Proc. Natl. Acad. Sci. USA. 106:3812–3817 doi:10.1073/pnas.0809501106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer R., Brero A., von Hase J., Schroeder T., Cremer T., Dietzel S. 2005. Common themes and cell type specific variations of higher order chromatin arrangements in the mouse.BMC Cell Biol. 6:44 doi:10.1186/1471-2121-6-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. 2001. The concept of self-organization in cellular architecture.J. Cell Biol. 155:181–185 doi:10.1083/jcb.200108110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neusser M., Schubel V., Koch A., Cremer T., Müller S. 2007. Evolutionarily conserved, cell type and species-specific higher order chromatin arrangements in interphase nuclei of primates.Chromosoma. 116:307–320 doi:10.1007/s00412-007-0099-3 [DOI] [PubMed] [Google Scholar]
- Nicodemi M., Panning B., Prisco A. 2008. A thermodynamic switch for chromosome colocalization.Genetics. 179:717–721 doi:10.1534/genetics.107.083154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenheimer J., Kreth G., Heermann D.W. 2005. Dynamic simulation of active/inactive chromatin domains.J. Biol. Phys. 31:351–363 doi:10.1007/s10867-005-7286-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polioudaki H., Kourmouli N., Drosou V., Bakou A., Theodoropoulos P.A., Singh P.B., Giannakouros T., Georgatos S.D. 2001. Histones H3/H4 form a tight complex with the inner nuclear membrane protein LBR and heterochromatin protein 1.EMBO Rep. 2:920–925 doi:10.1093/embo-reports/kve199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse I., Perlman M.D., Scalzo D., Kooperberg C., Groudine M., Kosak S.T. 2009. The emergence of lineage-specific chromosomal topologies from coordinate gene regulation.Proc. Natl. Acad. Sci. USA. 106:6679–6684 doi:10.1073/pnas.0900986106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roix J.J., McQueen P.G., Munson P.J., Parada L.A., Misteli T., Misteli T. 2003. Spatial proximity of translocation-prone gene loci in human lymphomas.Nat. Genet. 34:287–291 doi:10.1038/ng1177 [DOI] [PubMed] [Google Scholar]
- Rosa A., Everaers R. 2008. Structure and dynamics of interphase chromosomes.PLOS Comput. Biol. 4:e1000153 doi:10.1371/journal.pcbi.1000153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnick J., Gaspari G. 1987. The shapes of random walks.Science. 237:384–389 doi:10.1126/science.237.4813.384 [DOI] [PubMed] [Google Scholar]
- Sayle R.A., Milner-White E.J. 1995. RASMOL: biomolecular graphics for all.Trends Biochem. Sci. 20:374–376 doi:10.1016/S0968-0004(00)89080-5 [DOI] [PubMed] [Google Scholar]
- Shopland L.S., Lynch C.R., Peterson K.A., Thornton K., Kepper N., Hase J., Stein S., Vincent S., Molloy K.R., Kreth G., et al. 2006. Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence.J. Cell Biol. 174:27–38 doi:10.1083/jcb.200603083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorav J.L., Jannink G. 1994. Kinetics of chromosome condensation in the presence of topoisomerases: a phantom chain model.Biophys. J. 66:827–837 doi:10.1016/S0006-3495(94)80859-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M., de Laat W. 2008. FISH-eyed and genome-wide views on the spatial organisation of gene expression.Biochim. Biophys. Acta. 1783:2052–2060 doi:10.1016/j.bbamcr.2008.07.020 [DOI] [PubMed] [Google Scholar]
- Solovei I., Kreysing M., Lanctôt C., Kösem S., Peichl L., Cremer T., Guck J., Joffe B. 2009. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution.Cell. 137:356–368 doi:10.1016/j.cell.2009.01.052 [DOI] [PubMed] [Google Scholar]
- Takizawa T., Meaburn K.J., Misteli T. 2008. The meaning of gene positioning.Cell. 135:9–13 doi:10.1016/j.cell.2008.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegenfeldt J.O., Prinz C., Cao H., Chou S., Reisner W.W., Riehn R., Wang Y.M., Cox E.C., Sturm J.C., Silberzan P., Austin R.H. 2004. From the Cover: The dynamics of genomic-length DNA molecules in 100-nm channels.Proc. Natl. Acad. Sci. USA. 101:10979–10983 doi:10.1073/pnas.0403849101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet J.H., Luyten M.C., ten Brinke G. 1992. Scaling behavior of dilute polymer solutions confined between parallel plates.Macromolecules. 25:3802–3806 doi:10.1021/ma00040a029 [Google Scholar]
- Versteeg R., van Schaik B.D., van Batenburg M.F., Roos M., Monajemi R., Caron H., Bussemaker H.J., van Kampen A.H. 2003. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes.Genome Res. 13:1998–2004 doi:10.1101/gr.1649303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidemann T., Wachsmuth M., Knoch T.A., Müller G., Waldeck W., Langowski J. 2003. Counting nucleosomes in living cells with a combination of fluorescence correlation spectroscopy and confocal imaging.J. Mol. Biol. 334:229–240 doi:10.1016/j.jmb.2003.08.063 [DOI] [PubMed] [Google Scholar]
- Weierich C., Brero A., Stein S., von Hase J., Cremer C., Cremer T., Solovei I. 2003. Three-dimensional arrangements of centromeres and telomeres in nuclei of human and murine lymphocytes.Chromosome Res. 11:485–502 doi:10.1023/A:1025016828544 [DOI] [PubMed] [Google Scholar]