Abstract
The primary cilium plays a key role in the development of mammals and in the maintenance of health. Primary cilia are assembled and maintained by the process of intraflagellar transport (IFT). In this work, we characterize mouse IFT complex B by identifying all of the mammalian orthologues of complex B and B-associated proteins previously identified in Chlamydomonas and Caenorhabditis and also identify a new component (IFT25/Hspb11) of complex B by database analysis. We tagged each of these proteins with the FLAG epitope and show that all except IFT172 and IFT20 localize to cilia and the peri-basal body or centrosomal region at the base of cilia. All of the proteins except IFT172 immunoprecipitate IFT88 indicating that they are co-assembled into a complex. IFT20 is the only complex B protein that localizes to the Golgi apparatus. However, overexpression of IFT54/Traf3ip1, the mouse orthologue of Dyf-11/Elipsa, displaces IFT20 from the Golgi apparatus. IFT54 does not localize to the Golgi complex nor does it interact with GMAP210, which is the protein that anchors IFT20 to the Golgi apparatus. This suggests that IFT54s effect on IFT20 is a dominant negative phenotype caused by its overexpression.
Keywords: Golgi complex, cilia, flagella, intraflagellar transport, polycystic kidney disease, retinal degeneration
Introduction
Most types of eukaryotic cilia and flagella are assembled and maintained by the process of intraflagellar transport (IFT). During IFT, large protein complexes called IFT particles are transported along the ciliary microtubules under the ciliary membrane. It is thought that these particles carry precursors needed for ciliary assembly from the site of synthesis in the cell body to the site of assembly in the cilium. The outward movement of the particles is powered by kinesin-2 motors while inward transport is powered by dynein-2. The IFT particles themselves are composed of about 20 unique subunits that are highly conserved across the eukaryotic kingdom [Rosenbaum and Witman, 2002; Scholey, 2003 Cole, 2008].
The composition of the IFT particle is not completely known. Initial biochemical purification of the particles from Chlamydomonas showed that the proteins are organized into two distinct complexes with four proteins in the A complex (IFT144, 140, 139, 122) and 12 proteins in the B complex (IFT172, 88, 81, 80, 74/72, 57/55, 52, 46, 27, 20) [Cole et al., 1998; Piperno and Mead, 1997] (Table 1). Subsequent studies have added 2 subunits to complex A (IFT121/122b and IFT43) [Cole, 2003]. These complexes appear functionally distinct as mutational analysis in C. elegans showed that mutations in B subunit genes typically block cilia assembly while mutations in A subunit genes do not block cilia assembly but cause structural and functional defects [Qin et al., 2001]. Comparison of the phenotypes of cilia-assembly mutants and the rates of IFT in the middle and distal parts of C. elegans cilia has identified a number of IFT components and place them into functional groups [Ou et al., 2007]. Two of these functional groups correspond to the A and B complexes and contain most of the genes discussed above as well as several genes not identified by the Chlamydomonas biochemistry. New IFT proteins identified by this approach include Qilin/Dyf-3 [Ou et al., 2005b; Murayama et al., 2005], IFTA-2 [Schafer et al., 2006], Dyf-11 [Li et al., 2008; Bacaj et al., 2008; Kunitomo and Iino, 2008] in complex B and IFTA-1 [Blacque et al., 2006] in complex A. This work also defined a complex B-associated group containing Dyf-1 [Ou et al., 2005a] and Dyf-13 [Blacque et al., 2005] that is suggested to be a linker between Osm-3 kinesin and complex B. A large number of uncloned mutations remain in all of the functional groups [Ou et al., 2007] making it likely that there are new IFT components to be discovered.
Table 1.
Mouse Complex B
| Chlamydomonas | Caenorhabditis | Other | Mus Protein Accession Number | Mus Gene ID | Mus Gene Name | Mus Protein | Mus Predicted MW | FLAG Expression Clone | Cellular Distribution | |
|---|---|---|---|---|---|---|---|---|---|---|
| Complex B | IFT172 | Osm-1 | SLB Wim | NP_080574 | 67661 | Ift172 | IFT172 | 197,370 | FX24 (C) | Dispersed |
| FX20 (N) | Dispersed | |||||||||
| IFT88 | Osm-5 | Tg737 Polaris | NP_033402 | 21821 | Ift88 | IFT88 | 92,853 | JAF164 (N) | Cilia + Cent | |
| IFT81 | Ift-81 | CDV1 | NP_034009 | 12589 | Ift81 | IFT81 | 79,154 | JAF188 (N) | Cilia + Cent | |
| IFT80 | Che-2 | Wdr56 | NP_080917 | 68259 | Ift80 | IFT80 | 87,680 | JAF176 (N) | Dispersed | |
| FX58 (C) | Cilia + Cent | |||||||||
| IFT74/72/71 | Ift-74 | Cmg1 | NP_080595 | 67694 | Ift74 | IFT74 | 69,171 | FX40 (N) | Cilia + Cent | |
| IFT57/55 | Che-13 | Hippi | NP_082956 | 73716 | Ift57 | IFT57 | 48,641 | JAF156 (N) | Cilia + Cent | |
| IFT52 | Osm-6 | Ngd5 | NP_742162 | 245866 | Ift52 | IFT52 | 48,117 | FX23 (C) | Cilia + Cent | |
| IFT46/FAP32 | Dyf-6 | NP_076320 | 76568 | 1500035H01 Rik | IFT46 | 33,923 | JAF161 (N) | Cilia + Cent | ||
| IFT27/FAP156 | Not found | NP_080207 | 67042 | Rabl4 | IFT27 | 20,683 | JAF160 (N) | Dispersed | ||
| BK8 (C) | Cilia + Cent | |||||||||
| IFT25/FAP232 | Not found | C1orf41 HSP16.2 | NP_082670 | 72938 | 2900042B11 Rik | IFT25 | 16,149 | JAF143 (N) | Cilia + Cent | |
| IFT20 | Y110A7A.20 | NP_061342 | 55978 | Ift20 | IFT20 | 15,105 | JAF134 (N) | Golgi + Cilia + Cent | ||
| FAP22 | Dyf-3 | Qilin | NP_084014 | 76779 | Cluap1 | 47,822 | FX4 (N) | Weak Cilia + Cent | ||
| FX62 (C) | Cilia + Cent | |||||||||
| IFT22/FAP9 | IFTA-2 | NP_080349 | 67286 | Rabl5 | IFT22 | 20,702 | JAF181 (N) | Cilia + Cent | ||
| FAP116 | Dyf-11 | Elipsa | NP_082994 | 74019 | Traf3ip1 | IFT54 | 70,906 | FX34 (N) | Cilia + Cent | |
| B-Associated | FAP259 | Dyf-1 | Fleer | NP_084464 | 78802 | 4930506L13 RIK | 76,107 | FX3 (N) | Cilia + Cent | |
| DYF13 | Dyf-13 | NP_705828 | 264134 | Ttc26 | 64,020 | FX12 (N) | Cilia + Cent | |||
| Kinesin-2 | FLA | NP_032470 | 16569 | Kif3b | Kif3B | 85,288 | FX17 (N) | Cilia + Cent | ||
| Control GFP | JAF146 (N) | Dispersed |
All of the IFT particle protein genes identified in Chlamydomonas and Caenorhabditis are conserved in mammals. It appears that the mammalian proteins also co-assemble in complexes that resemble the IFT particle as IFT88, IFT52, IFT57, and IFT20 co-sediment on a sucrose gradient at ~16S [Pazour et al., 2002a; Baker et al., 2003] and mouse IFT20 [Follit et al., 2006] and IFT46 [Hou et al., 2007] can pull down other IFT complex B proteins when expressed in cultured mammalian cells. IFT particle proteins are typically found concentrated in cilia and in an intracellular pool at the base of the cilia. IFT20, a complex B protein, is localized to the Golgi complex in addition to the cilia and centrosome [Follit et al., 2006]. At the Golgi, IFT20 complexes with a protein called GMAP210 [Follit et al., 2008]. No other IFT subunit is known to localize to the Golgi apparatus but the cellular distribution of most mammalian IFTs has not been characterized. We are interested in defining the mammalian IFT complexes and determining if any other IFT complex B proteins are shared with the Golgi IFT complex. To achieve this, we FLAG-epitope tagged the mouse homologues of all the IFT complex B and complex B-associated proteins identified in Chlamydomonas and Caenorhabditis. We expressed these tagged proteins in mouse kidney epithelial cells and determined the cellular localization patterns for each and used immunoprecipitation to characterize their interactions with other IFT components.
Materials and Methods
FLAG Tagging
Each of the mouse homologues was amplified from mouse kidney, brain, or testis cDNA using primers that placed them in frame with the FLAG epitope in the p3XFLAG-myc-CMV-26 (Sigma, St. Louis, MO), pJAF113 (N-terminal tag) or p3XFLAG-myc-CMV-14 (Sigma) (C-terminal tag). pJAF113 is a modification of p3XFLAG-myc-CMV-26 made by filling in the Hind III site to shift the open reading frame. Each construct was verified by sequencing.
IFT54 Constructs
| FX34 MNAA…SRR* | N-C | 1–625 |
| FX44 MNAA…SGEISSRKLSD | N-1000 | 1–317 |
| FX45 mSGEISSRKLSD…SRR* | 1000-C | 308–625 |
| FX46 mSGEISSRKLSD…ELEDEEKH | 1000–1500 | 308–470 |
| FX47 mELEDEEKH…SRR* | 1500-C | 463–625 |
| FX48 mELEDEEKH…DYIQEDVD | 1500–1734 | 463–546 |
| FX49 DYIQEDVD…SRR* | 1734-C | 539–625 |
| FX52 DYIQEDVD…ICAVKANIL | 1734–1920 | 539–606 |
| FX53 QSITDSAV…ICAVKANIL | 1810–1920 | 570–606 |
| FX54 QSITDSAV…SRR* | 1810-C | 570–625 |
| FX60 ICAVKANIL…SRR* | 1895-C | 598–625 |
Mammalian Cell Culture
The expression plasmids were electroporated into mouse IMCD3 cells and stable lines selected with G418 using procedures described previously [Follit et al., 2006]. After drug selection, the cells were fixed with paraformaldehyde and examined by immunofluorescence microscopy as described previously [Follit et al., 2006]. If less than ~50% of the cells were FLAG positive, the cells were dilution cloned to identify a subclone that more uniformly expressed the tagged protein before further analysis.
Immunoprecipitations and Western Blotting
Cells for immunoprecipitation were rinsed once with cold PBS and lysed in Cell Lytic M + 0.1 % NP40 (Sigma), 0.1% CHAPSO (BioRad), plus Complete Protease Inhibitor (Roche) at 4° Celsius for 10 min. Lysates were centrifuged at 18,000 g for 10 minutes and clarified lysates were incubated with Agarose beads coupled with FLAG M2 antibody (Sigma) for one hour at 4° Celsius. FLAG beads were washed 3 times with Wash Buffer (50 mM Tris, 300 mM NaCl, pH 7.4, 1% NP40). Bound FLAG proteins were eluted with 200 μg/ml 3X FLAG peptide (Sigma).
Western blotting was performed as described [Follit et al., 2006] using antibodies to FLAG (Sigma), MmIFT140 (Walker and Pazour, unpublished), MmIFT88, MmIFT20 [Pazour et al., 2002a], MmIFT27 (Follit and Pazour, unpublished), MmPKD2 [Pazour et al., 2002b], and MmGMAP210 [Follit et al, 2008].
Results
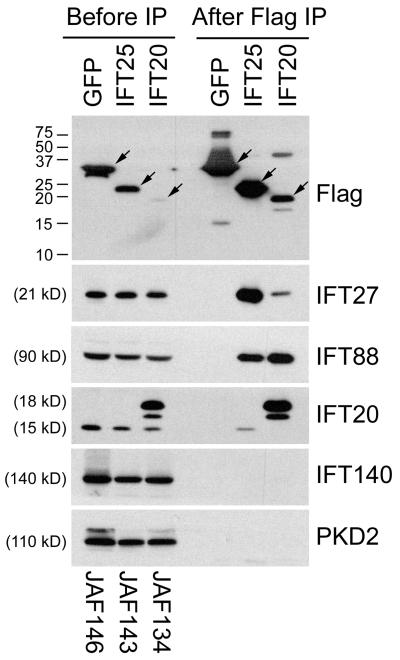
A number of large scale interactome studies have been carried out to identify protein-protein interactions in the human proteome. Some of the interactions involve known IFT particle proteins and we reasoned that some of the binding partners may be new IFT particle proteins. To test this idea, we analyzed the mammalian portion of the BioGRID database (www.thebiogrid.org), which compiles results of yeast two hybrid and other interactions from the published literature [Stark et al., 2006]. We wished to find proteins that interacted with known complex B proteins and whose Chlamydomonas orthologue was found in the membrane and matrix fraction of the flagellar proteome [Pazour et al., 2005]. Only one protein fit these criteria. C1orf41, an IFT27\Rabl4 binding protein [Rual et al., 2005], has a Chlamydomonas orthologue called Flagella Associated Protein 232 (FAP232). FAP232 was identified in the flagellar proteome by five peptides in the membrane and matrix fraction. This is the fraction where known IFT complex B subunits are found, making this a good candidate to be a new complex B subunit. To test whether C1orf41 is a component of the IFT particle, we placed a FLAG tag on the N-terminal end of the coding sequence and expressed this construct in mouse IMCD3 cells (Fig 1). The tagged protein localized to the cilium and peri-basal body region similar to other IFT particle proteins (Fig 2). To test the ability of the tagged proteins to assemble into IFT complexes, we immunoprecipitated proteins from cells stably expressing FLAG-C1orf41 (IFT25), FLAG-IFT20 or FLAG-GFP. FLAG-IFT20 and FLAG-GFP served as positive and negative controls. FLAG-C1orf41 behaved similar to FLAG-IFT20 in that it co-precipitated complex B proteins IFT27, IFT88, and IFT20, but did not bring down the complex A protein IFT140 nor the negative control PKD2. FLAG-GFP did not precipitate the complex B subunits indicating that the C1orf41 and IFT20 immunoprecipitations are specific (Fig 1). Thus we conclude that C1orf41 is a subunit of IFT complex B and name this protein IFT25 based on the size of the Chlamydomonas orthologue (FAP232) as is the standard naming convention in the field [Cole et al., 1998].
Figure 1. C1orf41/IFT25 is a subunit of IFT complex B.
Mouse IMCD3 cells stably transfected with FLAG expression constructs were lysed, immunoprecipitated with FLAG Ab and analyzed by western blotting. The FLAG-tagged bait protein is listed vertically at the top of the figure while name of the expression construct is listed vertically at the bottom. The left group is the starting material before immunoprecipitation and the right group is the precipitated material. The FLAG-tagged proteins are marked with arrows on the FLAG western. Abs used for the western blots are listed on the right side. The western blot of IFT20 shows endogenous IFT20 at 15 kD and FLAG-IFT20 at 18 kD along with a degradation product of the FLAG-IFT20 that runs slightly faster. Note that both IFT20 and C1orf41/IFT25 precipitated complex B subunits (IFT27, IFT88, IFT20) but not complex A (IFT140) or control proteins (PKD2).
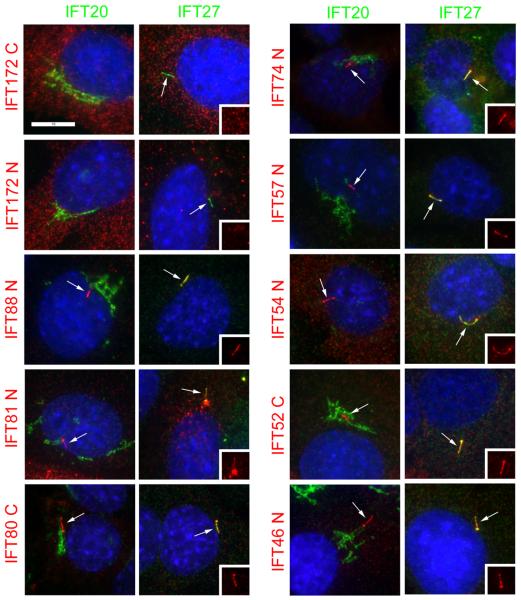
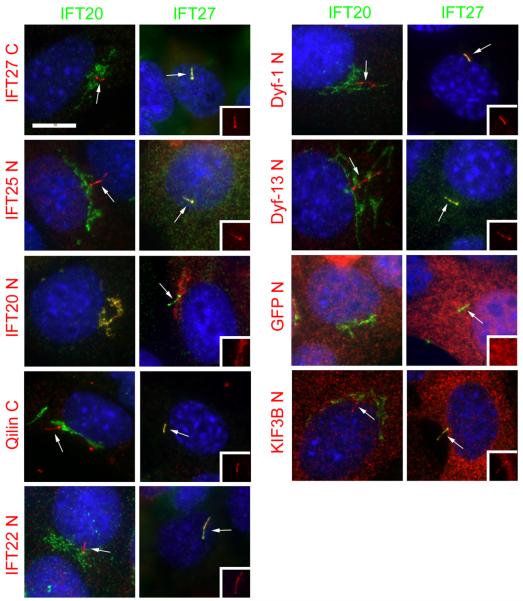
Figure 2. Localization of complex B proteins in mouse kidney cells.
Mouse IMCD3 cells stably transfected with FLAG expression constructs were serum starved for 48 hr, fixed with paraformaldehyde and stained with DAPI (blue), FLAG (red) and IFT20 (green, left panel in each pair) or IFT27 (green, right panel in each pair). The transfected FLAG construct is listed on the left side of each set of panels. N and C refer the position of the tags. Cilia are marked with arrows. Insets are the FLAG (red) channels of the cilia to more clearly document the localization of the FLAG-tagged protein to this organelle. Note that all proteins except IFT172 (with either an N- or C-terminal tag), IFT20 and GFP localize to cilia. Also note that IFT20 is the only protein localized to the Golgi complex and that IFT54 displaces native IFT20 from the Golgi complex. Size bar is 10 μm and relevant for all panels.
Our identification of IFT20 as a Golgi-associated protein [Follit et al., 2006] prompted us to question whether any of the other mouse IFT particle proteins showed a similar distribution. The mouse homologues of each of the complex B and complex B-associated proteins that had been identified in Chlamydomonas or Caenorhabditis were amplified, cloned into mammalian expression vectors as fusions to the FLAG epitope tag, and expressed in mammalian cells (Table 1). We initially cloned each of the proteins except IFT52 with an N-terminal tag. We were unable to clone IFT52 as an N-terminal fusion in bacteria. All clones were either in the reverse orientation or in the rare cases that they were in the correct orientation they contained frame shift mutations near the start of the gene. The cause of this lethality is unknown, but a C-terminal fusion was easily obtained. These constructs were expressed in mouse IMCD3 cells and stable lines were selected with G418. The majority of the FLAG-tagged proteins localized to the cilia and centrosomes as predicted (Table 1, Fig 2). As we previously reported, IFT20 is found at the Golgi complex and cilia plus centrosome but only the Golgi pool can be observed in paraformaldehyde fixed material due to epitope blocking [Follit et al., 2006]. FLAG-IFT81 localized to cilia and centrosomes but also localized to large cytoplasmic puncta of unknown origin. N-terminal tagged IFT172, IFT80, IFT27, failed to localize to cilia and centrosomes as expected and were recloned with the tags at the C-terminal ends. N-terminal tagged Qilin showed weak localization to cilia and centrosomes and also was cloned with a C-terminal tag. The C-terminal tagged versions of IFT80, IFT27, and Qilin localized to cilia and centrosomes. C-terminal tagged IFT172, like the N-terminal tagged version, did not localize to cilia or centrosomes. As controls, we examined a kinesin-2 subunit (KIF3B) and a completely unrelated protein (GFP).
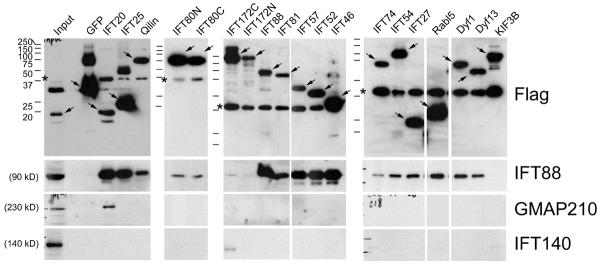
To more directly test the assignment of these proteins as IFT complex B subunits, we immunoprecipitated FLAG-tagged proteins from each of the cell lines and examined the precipitates by western blotting with antibodies to IFT88 (complex B), IFT140 (complex A), and GMAP210 (Golgi IFT complex) (Fig 3). All of the potential complex B proteins except for IFT172 co-precipitated IFT88 indicating that they are associated with complex B. The failure of both N- and C-terminally tagged IFT172 to immunoprecipitate IFT88 is consistent with the localization studies and likely indicates that both the N- and C-terminal ends of this protein are important for assembly into the particle. The behavior of IFT80 was unusual as the N-terminally tagged version did not localize to cilia but did precipitate IFT88. This suggests that the tag on the N-terminal end of the protein differentially disrupt interactions with other components of the particle. None of the proteins brought down IFT140 except for a small amount precipitated by the N-terminal tagged IFT172. The inability of complex B proteins to precipitate complex A proteins is expected based on work in Chlamydomonas where complexes A and B are not tightly associated with each other [Cole et al., 1998]. The reason for the small amount of IFT140 being brought down by N-terminally tagged IFT172 is not clear. It is possible that this indicates that IFT172 can at least partially interact with the A complex or it could be an artifact since the proteins are not properly localized in the cell. None of the proteins except IFT20 immunoprecipitated GMAP210, indicating that IFT20 is uniquely associated with the Golgi IFT complex. This is consistent with the immunolocalization, which did not detect any other complex B proteins at the Golgi complex (Fig 2).
Figure 3. Immunoprecipitation of FLAG-tagged proteins.
Mouse IMCD3 cells stably transfected with FLAG expression constructs were lysed, immunoprecipitated with FLAG Ab and analyzed by western blotting. The FLAG-tagged bait protein is listed at the top of the figure. Abs used for the western blots are listed on the right side. Arrows indicate the FLAG-tagged protein. * at ~37 kD marks a non-specific protein that is immunoprecipitated by the FLAG antibody. Ladders are shown to the left of each of the gels. The sizes of the markers are the same for all and shown on the left most ladder. Input (left most lane of the first gel) is starting material from the IFT20 immunoprecipitation equivalent to 1/6 of the amount in the precipitated lane. Note that GFP-FLAG was much more abundant than the other proteins and 1/5 as much of this immunoprecipitation was loaded on the gel for the FLAG western. The amount was not altered in the gels for the IFT88, GMAP210 or IFT140 westerns.
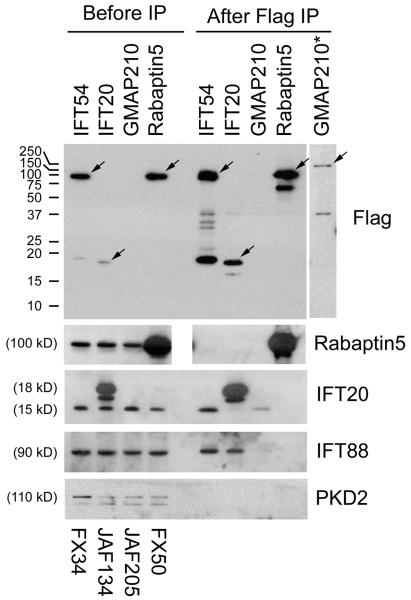
Although none of the complex B proteins except IFT20 localize to the Golgi apparatus, the mouse homologue of Caenorhabditis Dyf-11 and Chlamydomonas FAP116, which we named IFT54, showed the unique property of displacing IFT20 from the Golgi complex (Fig 2). Golgi structure is not disrupted by the expression of FLAG-IFT54 (data not shown). We were never able to observe any association of this protein with the Golgi complex. In low expressing cells, IFT54 associated uniquely with the cilium and centrosome. In high expressing cells, it spread throughout the cytoplasm in addition to the cilium and centrosome but did not concentrate at the Golgi structure. This suggests that the ability of IFT54 to displace IFT20 is a dominant negative effect caused by its over expression. While this work was in progress, it was reported that the Danio orthologue of IFT54/Dyf-11/FAP116 is encoded by the elipsa gene. In this work, it was demonstrated that immunoprecipitates of Elipsa brought down complex B proteins and yeast two-hybrid analysis with Elipsa as bait identified IFT20 and Rabaptin5 as interacting proteins. From this work, it was concluded that Elipsa and IFT20 interact within the IFT particle. In addition, the authors proposed that Elipsa has the additional function of linking the IFT particle to membrane trafficking by coupling Rabaptin5 to Rab8 [Omori et al., 2008]. To further characterize the relationship of IFT20 and GMAP210 to IFT54 and Rabaptin5, we carried out immunoprecipitations from cells expressing FLAG-tagged IFT20, GMAP210, IFT54 and Rabaptin5 (Fig 4). IFT20 and IFT54 were able to co-precipitate complex B proteins consistent with both being associated with complex B. In addition, GMAP210 precipitated IFT20 but not other complex B proteins, consistent with GMAP210 associating with IFT20 in a complex different from complex B. We were not able to immunoprecipitate Rabaptin5 with GMAP210, IFT20, or IFT54 nor was an immunoprecipitation with FLAG-Rabaptin5 able to precipitate any of the IFT components. This indicates that in mouse cells there is not a strong association between Rabaptin5 and the IFT complex.
Figure 4. Interaction between IFT54 and the IFT complex.
Mouse IMCD3 cells transiently transfected with FLAG expression constructs were lysed 48 hrs after electroporation, immunoprecipitated with FLAG Ab and analyzed by western blotting. The FLAG-tagged bait protein is listed vertically at the top of the figure while name of the expression construct is listed vertically at the bottom. The left group is the starting material before immunoprecipitation and the right group is the precipitated material. The FLAG-tagged proteins are marked with arrows on the FLAG western. Note that GMAP210 is less abundant than the other proteins and is not visible at exposures reasonable for them. The right most lane (*) shows a longer exposure of immunoprecipitated GMAP210 from a different gel. Abs used for the western blots are listed on the right side. The western blot of IFT20 shows endogenous IFT20 at 15 kD and FLAG-IFT20 at 18 kD along with a degradation product of the FLAG-IFT20 that runs slightly faster. Note that the blank lane was inadvertently left out of the Rabaptin5 gel. To allow the lanes to line up, the image was split in the middle.
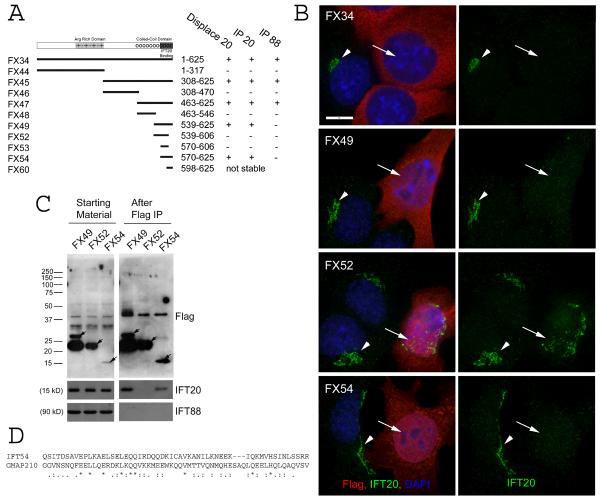
The yeast two hybrid analysis of Danio Elipsa and our observation that IFT54 can displace IFT20 from the Golgi complex suggests that these two proteins directly interact. We recently mapped the IFT20 binding site on GMAP210 [Follit et al., 2008] and we were interested to determine if there are similarities between the IFT20 binding sites in the two proteins. To do this, we progressively split IFT54 into smaller fragments and tested the effect of these fragments on the localization of IFT20 to the Golgi complex and for the ability of the fragments to immunoprecipitate IFT20 (Fig 5). This analysis indicates that the IFT20 binding site is located at the very C-terminal end of IFT54 and that residues 570–625 are sufficient to both displace IFT20 from the Golgi complex and to immunoprecipitate IFT20. It is possible that the binding site is smaller but fragments smaller than these were not stable in mammalian cells. Comparison of this motif to the IFT20 binding motif in GMAP210 revealed very little commonality between the two motifs aside from a pair of glutamines in a stretch of weak similarity (Fig 5D).
Figure 5. Characterization of the IFT20 binding site on IFT54.
A. Diagram summarizing data from experiments to map the IFT20 binding site on IFT54. Map illustrating the motif structure of IFT54 is shown on the top. Expression construct names are on the left. Exact amino acids are listed in the materials and methods. Arg rich domain is an unusual stretch of alternating positive and negatively charged residues. The coiled-coil domain is marked by series of circles. The IFT20 binding domain is marked by a solid dark bar. Summary of the activity of each of the constructs is shown on the right. B. IF of selected constructs to illustrate the ability of the C-terminal end of IFT54 (Red) to displace native IFT20 (green) from the Golgi complex. Left panel is a composite of red and green channels while the right panel shows only IFT20 staining in green. Arrows mark transfected cells. Arrowheads mark untransfected cells. Scale bar is 10 μm and relevant for all panels. C. Immunoprecipitation of selected constructs to illustrate the ability of C-terminal fragments of IFT54 to precipitate IFT20 but not IFT88. Mouse IMCD3 cells transiently transfected with FLAG expression constructs were lysed, immunoprecipitated with FLAG Ab and analyzed by western blotting. The FLAG-tagged expression construct is listed at the top of the figure. The left group is the starting material before immunoprecipitation and the right group is the precipitated material. The FLAG-tagged proteins are marked with arrows on the FLAG western. Antibodies used for western blotting are listed on the right. D. ClustalW alignment of the IFT20 binding site from IFT54 to a portion of the IFT20 binding site from GMAP210. * indicate identity, : and . mark similarity.
Discussion
IFT particle proteins organize into at least three distinct complexes called A, B and the Golgi IFT complex. We hypothesized that IFT20 and the Golgi IFT complex play a role in sorting or transport of ciliary membrane proteins [Follit et al. 2006] (Fig 6). The direct interaction between IFT20 and IFT54 [this work and Omori et al. 2008] suggests that IFT54 provides a bridge between the Golgi IFT complex and IFT complex B. In this model, IFT54 in the complex B pool at the basal body “catches” IFT20 on the surface of Golgi derived vesicles to complete the assembly of complex B. This could allow the contents of IFT20 marked vesicles to fuse with the plasma membrane in the periciliary region, providing a link between ciliary membrane cargo and the IFT particle. Compared to complex A and B, little is known about the composition of the Golgi IFT complex. At this point, two subunits, GMAP210 and IFT20, have been identified, but it is likely that additional proteins are present in the complex. Since IFT20 is also part of complex B, we questioned whether other complex B proteins may be associated with the Golgi IFT complex. To test this, we tagged each of the known complex B proteins with the FLAG epitope and expressed them in mammalian cells. When expressed in mouse cells, all of the proteins except for IFT172 localized to cilia/centrosomes and immunoprecipitated endogenous IFT88. The ability of these proteins to localize to the cilium and to immunoprecipitate IFT88 is good evidence that they are part of IFT complex B and provides strong support for the assignment of these proteins as orthologues of the Chlamydomonas and Caenorhabditis proteins. The failure of IFT172 to localize and interact as expected may indicate that the mouse protein proposed to be IFT172 is not a true orthologue or that the tags interfere with the localization and interaction with other IFT proteins. The latter explanation is more likely as there are several lines of evidence suggesting that the mouse protein is a true orthologue. These include a high level of sequence conservation between the mouse protein and homologues in Chlamydomonas, Caenorhabditis and Tetrahymena. In these organisms, IFT172 has been extensively characterized as an IFT component [Pedersen et al., 2005; Bell et al., 2006; Cole et al., 1998; Tsao and Gorovsky, 2008]. In addition, mouse mutations in IFT172 have phenotypes similar to mutations in other IFT complex B-encoding genes [Huangfu et al., 2003; Gorivodsky et al., 2008]. Furthermore, we immunoprecipitated endogenous IFT172 from mouse cells with the complex B subunit IFT20 [Follit et al. 2008]. Thus, it is more likely that both ends of IFT172 are important for localization to the cilium and interaction with other IFT components and that the failure to localize is an artifact of the tags.
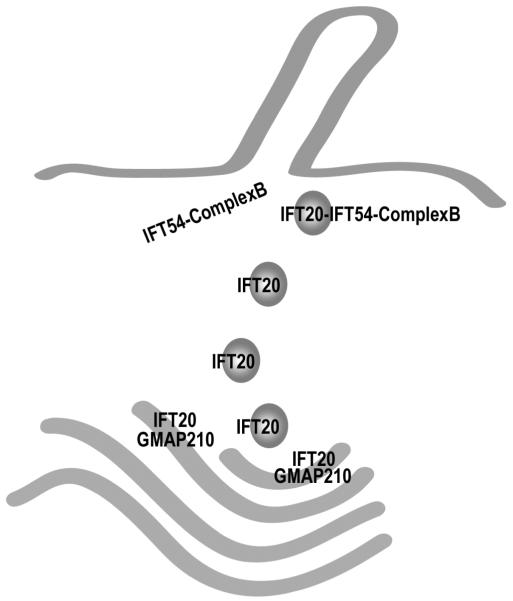
Figure 6. Model for the coordination of IFT20 in the Golgi IFT complex and complex B.
The finding that IFT20 is part of the Golgi IFT complex as well as complex B presents interesting possibilities for its function. This speculative model presents our view of how this may be occurring. IFT20 is anchored to the Golgi membrane by the golgin protein GMAP210 [Follit et al., 2008]. IFT20 is dynamic and moves from the Golgi complex to the cilium [Follit et al., 2006] but GMAP210 appears to remain at the Golgi. IFT20 binds directly to the complex B subunit IFT54 [this work and Omori et al. 2008] but IFT54 does not appear to localize to the Golgi complex and it is not part of the Golgi IFT complex [this work]. Thus, we hypothesize that IFT20 and GMAP210 function at the Golgi complex to sort proteins into vesicles destined for the ciliary membrane. These vesicles leave the Golgi with IFT20 associated with them. Once the vesicles arrive at the base of the cilium, IFT20 on the vesicles interacts with the IFT54 subunit of complex B to form the complete IFT complex. The vesicles fuse with the plasma membrane at the base of the cilium and the IFT complex with attached cargos is carried into the cilium by kinesin-2. By having the IFT complex assembled on the surface of the vesicle, the transport of membrane and non-membranous cargos could be coordinated.
No complex B protein, besides IFT20, localized to the Golgi complex or was able to immunoprecipitate GMAP210 indicating that IFT20 is the only known complex B protein also present in the Golgi IFT complex. However, we discovered the Elipsa/Dyf-11 orthologue, which we named IFT54, is able to displace IFT20 from the Golgi complex. Since IFT54 did not localize to the Golgi complex and did not immunoprecipitate GMAP210, it is likely that IFT54 is a subunit of IFT complex B and the ability to displace IFT20 is a dominant negative effect caused by its overexpression (Fig 6). This suggests that GMAP210 and IFT54 bind to a common site on IFT20. The small size of IFT20 has precluded us from making progress in determining the IFT54 binding site on IFT20 however we were able to map the IFT20 binding site in IFT54. IFT54 consists of an Arg-rich motif in the center of the protein that contains a very unusual stretch of alternating positive and negatively charged residues and a region of coiled-coil at the C-terminal end (Fig 5A). The IFT20 binding site was mapped to the coiled-coil region at the C-terminal end. Comparison of this sequence with the IFT20 binding site in GMAP210 [Follit et al. 2008] does not reveal strong similarity although both binding sites are located within regions of predicted coil-coil. IFT54 was originally identified as MIP-T3/Traf3ip1 and suggested to link the tumor necrosis factor receptor-associated factor 3 (TRAF3) to microtubules. In this work, IFT54/MIP-T3/Traf3ip1 was found to associate with microtubules in a pull down assay and N-terminally FLAG tagged IFT54/MIP-T3/Traf3ip1 co-localized with microtubules when expressed in cultured cells [Ling and Goeddel, 2000]. In our hands, N-terminally FLAG tagged IFT54 localized to cilia and centrosomes when expressed at low levels (Fig 2) while at high expression levels, it is evenly distributed throughout the cytoplasm without any notable association with microtubules (Fig 5B). IFT54/MIP-T3/Trafip1 is also reported to bind to Disrupted-In-Schizophrenia 1 (DISC1) [Morris et al., 2003]. GFP-tagged DISC1 localizes to large puncta in the cell that are concentrated around the centrosome [Morris et al., 2003]. The nature of these puncta is unknown but appears similar to what we observed with FLAG-tagged IFT81 (Fig 2). The relationship between DISC1 and IFT has not been examined. However, DISC1 appears to be a vertebrate-specific protein and so is not likely to be a critical component of the IFT system.
In this work we also identified a new IFT complex B subunit. This protein, IFT25, was initially found in a large scale yeast two hybrid analysis to bind IFT27 [Rual et al., 2005]. We verified that IFT25 can interact with the IFT particle by co-immunoprecipitation assays and showed that IFT25 localizes to cilia and centrosomes like other IFT components. IFT25 is conserved in many but not all ciliated eukaryotes. It is well conserved throughout the vertebrate kingdom, in some invertebrates such urchins (XP_0012014989), anemones (XP_001640906), and Trichoplax (XP_002114382). It is not found in plants, Saccharomyces, Caenorhabditis, Drosophila, or Plasmodium, but is conserved in Chlamydomonas (XP_001698507.1) and there are distant homologues in the parasitic protozoa Leishmania (XP_001463577.1) and Trypanosoma (XP_829387). Even though the Blast scores are low for the parasitic homologues, it is likely they are significant as reciprocal Blast searches identify IFT25 homologues as the top matches. In Chlamydomonas, IFT25 was found in the flagellar proteome (FAP232) where it was identified by 5 peptides in the membrane and matrix fraction, which is consistent with it being an IFT particle protein [Pazour et al., 2005]. Accompanying work indicates that the Chlamydomonas orthologue is a component of the IFT particle [Lechtreck et al. 2009]. The phylogenetic distribution of IFT25 closely resembles the distribution of other IFT proteins, with the exception that most IFT proteins are conserved in Drosophila and Caenorhabditis. However, IFT27 is likewise not conserved in Drosophila and Caenorhabditis and IFT25 is likely to interact directly with IFT27. The reason for the lack of conservation of these proteins in Drosophila and Caenorhabditis is not known but it has been suggested that IFT27 plays a role in coupling the assembly of cilia to the cell cycle and cilia are assembled only on non-cycling cells in these two species [Qin et al., 2007]. Perhaps IFT25 is functioning with IFT27 in this process. IFT25 is relatively uncharacterized in any species. The protein contains a galactose-binding like domain (IPR008979), which in other proteins binds various ligands such as carbohydrates, phospholipids and nucleic acids. It will be interesting to determine to how this domain is functioning in IFT25. IFT25 was first identified as a salt extractable protein from placenta and named pp25 [Bohn and Winckler, 1991]. It was later described as a small heat shock protein [Bellyei et al., 2007a; Bellyei et al., 2007b] and called Hspb11, Hsp16.2 and HSPCO34. The percent identity between IFT25/Hsp16.2 and small heat shock proteins is low (16%) but IFT25/Hsp16.2 levels are elevated in cultured mammalian cells by exposure to 42°C and overexpression of the protein protects against oxidative stress [Bellyei et al., 2007a]. Interestingly, levels of IFT25/Hsp16.2 were dramatically elevated in aggressive forms of brain tumors as compared to normal tissues or less aggressive tumors [Pozsgai et al., 2007]. It will be important to understand the function of IFT25 to determine if the over expression of IFT25 plays a role in the transformation of these tumor cells.
Acknowledgements
This work was supported by the National Institutes of Health (GM060992). Core resources supported by the Diabetes Endocrinology Research Center grant DK32520 were also used.
Abbreviations
- IFT
Intraflagellar Transport
- CrFP
Chlamydomonas Flagellar Proteome
- FAP
Flagella Associated Protein
- IP
immunoprecipitate
- IF
immunofluorescence
- Ab
antibody
- Mm
Mus musculus
References
- Bacaj T, Lu Y, Shaham S. The conserved proteins CHE-12 and DYF-11 are required for sensory cilium function in Caenorhabditis elegans. Genetics. 2008;178:989–1002. doi: 10.1534/genetics.107.082453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SA, Freeman K, Luby-Phelps K, Pazour GJ, Besharse JC. IFT20 links kinesin II with a mammalian intraflagellar transport complex that is conserved in motile flagella and sensory cilia. J Biol Chem. 2003;278:34211–34218. doi: 10.1074/jbc.M300156200. [DOI] [PubMed] [Google Scholar]
- Bell LR, Stone S, Yochem J, Shaw JE, Herman RK. The molecular identities of the Caenorhabditis elegans intraflagellar transport genes dyf-6, daf-10 and osm-1. Genetics. 2006;173:1275–1286. doi: 10.1534/genetics.106.056721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellyei S, Szigeti A, Boronkai A, Pozsgai E, Gomori E, Melegh B, Janaky T, Bognar Z, Hocsak E, Sumegi B, Gallyas F., Jr. Inhibition of cell death by a novel 16.2 kD heat shock protein predominantly via Hsp90 mediated lipid rafts stabilization and Akt activation pathway. Apoptosis. 2007a;12:97–112. doi: 10.1007/s10495-006-0486-x. [DOI] [PubMed] [Google Scholar]
- Bellyei S, Szigeti A, Pozsgai E, Boronkai A, Gomori E, Hocsak E, Farkas R, Sumegi B, Gallyas F., Jr. Preventing apoptotic cell death by a novel small heat shock protein. Eur J Cell Biol. 2007b;86:161–171. doi: 10.1016/j.ejcb.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Blacque OE, Li C, Inglis PN, Esmail MA, Ou G, Mah AK, Baillie DL, Scholey JM, Leroux MR. The WD repeat-containing protein IFTA-1 is required for retrograde intraflagellar transport. Mol Biol Cell. 2006;17:5053–5062. doi: 10.1091/mbc.E06-06-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque OE, Perens EA, Boroevich KA, Inglis PN, Li C, Warner A, Khattra J, Holt RA, Ou G, Mah AK, McKay SJ, Huang P, Swoboda P, Jones SJ, Marra MA, Baillie DL, Moerman DG, Shaham S, Leroux MR. Functional genomics of the cilium, a sensory organelle. Curr Biol. 2005;15:935–941. doi: 10.1016/j.cub.2005.04.059. [DOI] [PubMed] [Google Scholar]
- Bohn H, Winckler W. Isolation and characterization of five new soluble placental tissue proteins (PP22, PP23, PP24, PP25, PP26) Arch Gynecol Obstet. 1991;248:111–115. doi: 10.1007/BF02390087. [DOI] [PubMed] [Google Scholar]
- Cole DG. The intraflagellar transport machinery of Chlamydomonas reinhardtii. Traffic. 2003;4:435–442. doi: 10.1034/j.1600-0854.2003.t01-1-00103.x. [DOI] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DG. Intraflagellar Transport. In: Witman GB, editor. The Chlamydomonas Sourcebook, Second Edition. Vol. 3. Cell Motility and Behavior. Elsevier; New York, NY: 2008. pp. 71–113. [Google Scholar]
- Follit JA, Tuft RA, Fogarty KE, Pazour GJ. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell. 2006;17:3781–3792. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follit JA, SanAgustin JT, Xu F, Jonassen J, Pazour GJ. The golgin GMAP210/TRIP11 anchors IFT20 to the Golgi complex. PLoS Genetics. 2008 doi: 10.1371/journal.pgen.1000315. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorivodsky M, Mukhopadhyay M, Wilsch-Braeuninger M, Phillips M, Teufel A, Kim C, Malik N, Huttner W, Westphal H. Intraflagellar transport protein 172 is essential for primary cilia formation and plays a vital role in patterning the mammalian brain. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Qin H, Follit JA, Pazour GJ, Rosenbaum JL, Witman GB. Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella. J Cell Biol. 2007;176:653–665. doi: 10.1083/jcb.200608041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- Kunitomo H, Iino Y. Caenorhabditis elegans DYF-11, an orthologue of mammalian Traf3ip1/MIP-T3, is required for sensory cilia formation. Genes Cells. 2008;13:13–25. doi: 10.1111/j.1365-2443.2007.01147.x. [DOI] [PubMed] [Google Scholar]
- Lechtreck KF, Luro S, Witman GB. HA-tagging of putative flagellar proteins in Chlamydomonas identifies a novel component of IFT complex B. Cell Motil Cytoskeleton. 2009 doi: 10.1002/cm.20369. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Inglis PN, Leitch CC, Efimenko E, Zaghloul NA, Mok CA, Davis EE, Bialas NJ, Healey MP, Heon E, Zhen M, Swoboda P, Katsanis N, Leroux MR. An essential role for DYF-11/MIP-T3 in assembling functional intraflagellar transport complexes. PLoS Genet. 2008;4:e1000044. doi: 10.1371/journal.pgen.1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling L, Goeddel DV. MIP-T3, a novel protein linking tumor necrosis factor receptor-associated factor 3 to the microtubule network. J Biol Chem. 2000;275:23852–23860. doi: 10.1074/jbc.M001095200. [DOI] [PubMed] [Google Scholar]
- Morris JA, Kandpal G, Ma L, Austin CP. DISC1 (Disrupted-In-Schizophrenia 1) is a centrosome-associated protein that interacts with MAP1A, MIPT3, ATF4/5 and NUDEL: regulation and loss of interaction with mutation. Hum Mol Genet. 2003;12:1591–1608. doi: 10.1093/hmg/ddg162. [DOI] [PubMed] [Google Scholar]
- Murayama T, Toh Y, Ohshima Y, Koga M. The dyf-3 gene encodes a novel protein required for sensory cilium formation in Caenorhabditis elegans. J Mol Biol. 2005;346:677–687. doi: 10.1016/j.jmb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Omori Y, Zhao C, Saras A, Mukhopadhyay S, Kim W, Furukawa T, Sengupta P, Veraksa A, Malicki J. Elipsa is an early determinant of ciliogenesis that links the IFT particle to membrane-associated small GTPase Rab8. Nat Cell Biol. 2008;10:437–444. doi: 10.1038/ncb1706. [DOI] [PubMed] [Google Scholar]
- Ou G, Blacque OE, Snow JJ, Leroux MR, Scholey JM. Functional coordination of intraflagellar transport motors. Nature. 2005a;436:583–587. doi: 10.1038/nature03818. [DOI] [PubMed] [Google Scholar]
- Ou G, Koga M, Blacque OE, Murayama T, Ohshima Y, Schafer JC, Li C, Yoder BK, Leroux MR, Scholey JM. Sensory ciliogenesis in Caenorhabditis elegans: assignment of IFT components into distinct modules based on transport and phenotypic profiles. Mol Biol Cell. 2007;18:1554–1569. doi: 10.1091/mbc.E06-09-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G, Qin H, Rosenbaum JL, Scholey JM. The PKD protein qilin undergoes intraflagellar transport. Curr Biol. 2005b;15:R410–R411. doi: 10.1016/j.cub.2005.05.044. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Baker SA, Deane JA, Cole DG, Dickert BL, Rosenbaum JL, Witman GB, Besharse JC. The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J Cell Biol. 2002a;157:103–113. doi: 10.1083/jcb.200107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, San Agustin JT, Follit JA, Rosenbaum JL, Witman GB. Polycystin-2 localizes to kidney cilia and the ciliary level is elevated in orpk mice with polycystic kidney disease. Curr Biol. 2002b;12:R378–R380. doi: 10.1016/s0960-9822(02)00877-1. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Miller MS, Geimer S, Leitch JM, Rosenbaum JL, Cole DG. Chlamydomonas IFT172 is encoded by FLA11, interacts with CrEB1, and regulates IFT at the flagellar tip. Curr Biol. 2005;15:262–266. doi: 10.1016/j.cub.2005.01.037. [DOI] [PubMed] [Google Scholar]
- Piperno G, Mead K. Transport of a novel complex in the cytoplasmic matrix of Chlamydomonas flagella. Proc Nat Acad Sci USA. 1997;94:4457–4462. doi: 10.1073/pnas.94.9.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozsgai E, Gomori E, Szigeti A, Boronkai A, Gallyas F, Jr., Sumegi B, Bellyei S. Correlation between the progressive cytoplasmic expression of a novel small heat shock protein (Hsp16.2) and malignancy in brain tumors. BMC Cancer. 2007;7:233. doi: 10.1186/1471-2407-7-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Rosenbaum JL, Barr MM. An autosomal recessive polycystic kidney disease gene homolog is involved in intraflagellar transport in C. elegans ciliated sensory neurons. Curr Biol. 2001;11:457–461. doi: 10.1016/s0960-9822(01)00122-1. [DOI] [PubMed] [Google Scholar]
- Qin H, Wang Z, Diener D, Rosenbaum J. Intraflagellar transport protein 27 is a small G protein involved in cell-cycle control. Curr Biol. 2007;17:193–202. doi: 10.1016/j.cub.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Schafer JC, Winkelbauer ME, Williams CL, Haycraft CJ, Desmond RA, Yoder BK. IFTA-2 is a conserved cilia protein involved in pathways regulating longevity and dauer formation in Caenorhabditis elegans. J Cell Sci. 2006;119:4088–4100. doi: 10.1242/jcs.03187. [DOI] [PubMed] [Google Scholar]
- Scholey JM. Intraflagellar transport. Annu Rev Cell Dev Biol. 2003;19:423–443. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao CC, Gorovsky MA. Different effects of Tetrahymena IFT172 domains on anterograde and retrograde intraflagellar transport. Mol Biol Cell. 2008;19:1450–1461. doi: 10.1091/mbc.E07-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]