Abstract
The rhesus monkey embryonic stem cell line 366.4 differentiates into serotonin neurons. We examined the genetic cascade during differentiation and compared ESC-derived serotonin neurons to adult monkey serotonin neurons. RNA was extracted from ESC colonies, embryoid bodies (EBs), neurospheres in selection (N1) and proliferation stages (N2), differentiated serotonin neurons (N3) and from laser captured (LC) serotonin neurons of spayed female macaques treated with placebo, estrogen (E), progesterone (P) or E+P. The RNA was labeled and hybridized to Rhesus Monkey Affymetrix Gene Chips (n=1 per stage and 2 per animal treatment). Gene expression was examined with GeneSifter software. 545 genes that were related to developmental processes showed a 3-fold or greater change between stages. TGFβ, Wnt, VEGF and Hedgehog signaling pathways showed the highest percent of probe set changes during differentiation. Genes in the categories (a) homeobox binding and transcription factors, (b) growth factors and receptors, (c) brain and neural specific factors and (d) serotonin specific factors are reported. Pivotal genes were confirmed with quantitative RT-PCR. In the serotonin developmental cascade, FGFR2 was robustly expressed at each stage. GATA3 was robustly expressed in EBs. Sonic hedgehog (Shh), PTCH (Shh-R), and Fev1 transcription factor expression coincided with the induction of serotonin specific marker genes during N1-selection. A majority of the examined genes were expressed in adult serotonin neurons. However, in the ESC-derived neurons, there was significant over-representation of probe sets related to cell cycle, axon guidance & dorso-ventral axis formation. This analysis suggests that the 366.4 cell line possesses cues for serotonin differentiation at early stages of differentiation, but that ESC-derived serotonin neurons are still immature.
Keywords: embryonic stem cells, development, serotonin, macaques, Affymetrix array, gene expression, dorsal raphe
Introduction
The serotonin neural system plays a crucial role in numerous autonomic and higher order functions of the central nervous system. Dysfunction of the serotonin system has been implicated in a wide array of psychopathologies including depression, anxiety, obsessive-compulsive disorder, impulse disorders, and even metabolic syndrome. As such, the serotonin neural system is a target of pharmacotherapies, steroid hormones, cytokines, neuropeptides and trophic factors, all of which impact the generation and efficacy of serotonin neurotransmission.
The proper differentiation and establishment of connections in the serotonin system during fetal development is obviously crucial for proper functioning in adult life and interference with differentiation can lead to adult psychopathologies. The developmental pathway leading to serotonin neurons has been examined in rodent knockout models and critical genes have been identified which include Shh, Nkx2.2, Lmx1b, GATA3, Mash1 and Pet1 (Cheng, et al. 2003; Hendricks, et al. 2003). Different laboratories have suggested slightly different models of the pathway employed by these genes leading to the serotonin phenotype, but it is agreed that Shh plays a very early role, potentially activating Nkx2.2, which in turn leads to activation of Lmx1b and Pet1 (Fev1 in primates). Lmx1b may act through GATA3 or GATA3 may work in concert with Pet1 to obtain terminal differentiation. There also appear to be different requirements between different populations of serotonin neurons.
Nonetheless, fetal development in rodents and primates differs in significant areas and it is unknown whether this pathway of gene expression is utilized for serotonin differentiation in higher primates. However, study of the development of the serotonin system in primates is difficult and expensive and knock out models are not available. Moreover in vivo, it has been difficult to identify markers for undifferentiated neurons, which indicate that a neuron or its progeny will become serotonergic.
Our laboratory has devoted considerable effort to the study of serotonin neural function in female macaques (Bethea, et al. 2002). We recently developed a protocol that promotes differentiation of rhesus monkey embryonic stem cells into a high percentage of serotonin neurons with immunological characteristics of adult serotonin neurons (Salli, et al. 2004). Since the stem cells are wholly undifferentiated and upon application of the protocol, become serotonergic, it is possible to examine gene expression that leads to the serotonin phenotype prior to the expression of any classical serotonin markers.
Therefore in this study, we questioned whether the identified developmental genes from rodent studies were recapitulated in the differentiating rhesus embryonic stem cells and examined the expression of genes that code for homeobox binding and transcription factors, growth factors and receptors, brain and neural specific factors and serotonin specific factors using the Rhesus Affymetrix Gene Chip. In addition, we compared the global gene expression in the ESC derived serotonin neurons to that of laser captured serotonin neurons from adult ovariectomized female monkeys treated for one month with placebo, estrogen (E), progesterone (P) or E supplemented with P for the last 14 days of treatment.
Materials and Methods
The Oregon National Primate Research Center (ONPRC) Institutional Animal Care and Use Committee approved this study.
ES Cell Culture and In Vitro Differentiation
The rhesus monkey ES cell line 366.4 was obtained from Dr. James Thomson, Wisconsin National Primate Research Center. This line, derived from an in vivo flushed preimplantation embryo, has been characterized for its pluripotency including its potential to differentiate into cells of the neural lineage (Thomson, et al. 1995). All cell culture related materials were purchased from GIBCO Invitrogen Corporation (Grand Island, NW) unless specifically indicated. The following protocol was used on the cultures generated for microarray analysis and qRT-PCR.
ES cells, starting from passage 13, were propagated and maintained as previously described(Kuo, et al. 2003). Briefly, ES cells were co-cultured with mitotically inactive (mitomycin C-treated; 1 mg/ml at 37°C for 30 min; Sigma, St Louis, MO) mouse embryonic fibroblasts (MEF) in 60 mm gelatin-coated, tissue culture dishes (Nalgen NUNC International Corp, Rochester, NY). ES cell culture medium consisted of 80% Dulbecco’s Modified Eagle Medium (DMEM; with L-glutamine, glucose and without sodium pyruvate) supplemented with 1% nonessential amino acids, 2 mM glutamine, 0.1 mM □-mercaptoethanol (Sigma, St. Louis, MO) and 20% fetal bovine serum (Hyclone, Logan, UT). ES cells and their colonies were observed daily and passaged every 6-8 days when colonies reached 1-1.5 mm in diameter. For passaging, cultures were incubated with collagenase IV (1 mg/ml) at 37°C for 20 min. Culture dishes were then rinsed with ES cell culture medium and colonies were triturated into small clumps (>15 cells/clump) and incubated at 37°C for an additional 30 min during which time MEF and differentiated cells attached to the bottom of the culture dish. Following this incubation, the medium containing unattached cells was harvested by pipette aspiration and replated into three to five dishes with fresh MEF cells.
The pluripotency of ES cells was evaluated periodically by immunocytochemistry (ICC) with antibodies to Oct-4, stage specific embryonic antigens (SSEA-3 and 4) and embryonic proteoglycans (TRA-1-60 and TRA-1-81) as described below.
A protocol consisting of multiple sequential steps was used to induce differentiation (Kuo et al. 2003; Lee, et al. 2000) as follows:
Isolation of ES cell colonies and formation of embryoid bodies (EBs): After ES cell colonies attained a 1-1.5 mm diameter, they were isolated mechanically, triturated into intermediate sized clumps (>200 cells/clump), transferred into 60 mm dishes (BD Biosciences, Bedford, MA) and cultured in ES cell medium at 37°C for 7 days. Embryoid bodies were defined as Oct-4 negative, three-dimensional structures that could potentially give rise to endo-, ecto- and mesodermal cell lineages.
Selection of Nestin-positive cells (N1 stage): After formation of EBs, ES cell medium was replaced with a selection medium composed of DMEM/Nutrient Mixture F12 (1:1) containing L-glutamine, sodium bicarbonate, pyridoxine hydrochloride, 1% ITSX (Insulin, 1 g/L; sodium selenite, 0.67 mg/L; transferrin, 0.55 g/L and ethanolamine, 0.2 g/L) and human plasma fibronectin (5 μg/ml). Embryoid bodies were cultured in selection medium for 7 days. Toward the end of this period, EBs developed into globular structures (0.5 to 1.0 mm in diameter) containing nestin- and Musashi1-positive cells. These structures have been referred to as neurospheres (Ostenfeld, et al. 2002).
Expansion of Nestin-positive cells (N2 stage): At the end of the selection period, neurospheres were cultured in expansion medium composed of DMEM/Nutrient Mixture F12 (1:1), 1% N2 supplement (Bottenstein, et al. 1980) (insulin, 500 μg/ml; transferrin, 10,000 μg/ml; progesterone, 0.63 μg/ml; putrescine, 1611 μg/ml and selenite, 0.52 μg/ml) and FGF2 (10 ng/ml) for 7-9 days changing the medium daily. FGF2, a potent mitogenic factor, promoted cell proliferation resulting in increased neurosphere diameter (0.25 to 1.25 mm in diameter).
Maturation of differentiated neural cells (N3 stage): Expanded neurospheres were chopped into small clumps of cells and then cultured on growth factor reduced (GFR)-matrigel coated coverslips or wells depending on the application. Maturation medium was the same as the expansion medium but did not contain FGF2. Dispersed cells were cultured for a further 7-9 days before use.
Animals and treatments
Eight adult female rhesus monkeys (Macaca mulatta) were ovariectomized by the surgical personnel of ONPRC between 3 and 6 months before assignment to this project according to accepted veterinary surgical protocol. All animals were born in China, were aged between 7-14 years by dental exam, weighed between 4 and 8 kg, and were in good health. Two animals each were treated with placebo (ovx-control group), or treated with estrogen (E) for 28 days (E), or treated with progesterone (P) for the last 14 days (P), or treated with E for 28 days and then supplemented with P for the final 14 of the 28 days (E+P). The treatments were obtained with Silastic capsules implanted subcutaneously in the periscapular region. The details of the treatments and the hormone levels achieved were previously published (Bethea and Reddy 2007). The monkeys were euthanized at the end of the treatment periods according to procedures recommended by the Panel on Euthanasia of the American Veterinary Association. Each animal was sedated with ketamine, given an overdose of pentobarbital (25 mg/kg, i.v.), and exsanguinated by severance of the descending aorta.
Tissue preparation and Laser Capture Dissection (n=7)
The preparation of the brain, sectioning and immunostaining for identification of serotonin neurons and the laser capture dissection were described previously (Bethea and Reddy 2007).
RNA extraction from laser captured neurons
The extraction of RNA from the laser captured serotonin neurons was described previously (Bethea and Reddy 2007).
RNA extraction from stem cell stages
RNA was obtained from cells at each stage using TriReagent and further cleaned with a Qiagen RNAeasy column (Velencia, CA). The quality of the RNA from the Qiagen column was examined on an Agilent Bioanalyzer and found acceptable and of equal quality.
Affymetrix hybridization
Labeled target cRNA was prepared from each of the stem cell stages (n=1/ESC, EB, N1, N2 and N3) and 7 pools of laser captured serotonin neurons (n=2/ovx placebo; 2/E treatment; 1/P treatment; 2/E+P treatment). The cRNA was hybridized to Rhesus Affymetrix GeneChip arrays. Microarray assays were performed in the Affymetrix Microarray Core of the OHSU Gene Microarray Shared Resource.
Data Analysis with GeneSifter Software
The data was compressed into .cel files and converted to GS_CHP files, which were uploaded to GeneSifter (VisX Labs, Seattle, WA). Probe sets that were undetectable in all treatment groups were eliminated. The remaining probe sets were examined as follows:
Probe sets that exhibited a 3-fold change between stem cell differentiation stages were filtered for genes related to development.
Probe sets that exhibited a 5-fold change between the ESC-derived serotonin neurons and the adult laser captured neurons were subjected to KEGG Analysis. In addition, a regression analysis was performed on the expression profiles of all samples.
Quantitative (q) RT-PCR (n=3 passages/stage)
Additional RNA was extracted from each stage of stem cell differentiation from 3 different passages for qRT-PCR. Complementary DNA (cDNA) synthesis was performed using Oligo-dT 15 and Random Hexamer primers (Invitrogen, Carlsbad, CA) and Superscript III (200 U/μg of RNA, Invitrogen) at 50°C for 1 hr followed RNAse H reaction at 37°C. A pool of RNAs from different rhesus tissues was used as the standard. The pooled RNAs were extracted from 4 brain regions, testis, ovary, liver, lungs, pancreas, heart, skeletal muscle, and stem cell stages.
Quantitative polymerase chain reaction (qRT-PCR) was conducted with the Invitrogen Sybr Green qPCR Mix. This mix contains a modified DNA Taq Polymerase and Sybr Green I fluorescent dye, which is specific for double stranded DNA. The Sybr Green super mix also contains a UDG component to avoid reamplification of cDNA template. There is a linear increase in fluorescence detected as the concentration of amplified double stranded product cDNA increases during the reaction. The fluorescence was detected with the ABI 7900 thermal cycler during forty cycles. The reaction (final volume 20 μl) contained dilutions from 10 – 1000 pg of cDNA, 100 nM of forward and reverse primers and 1X Sybr Green super mix. The amount of cDNA added to the reaction mix was calculated with Nanodrop spectroscopy after the RT reaction and set to 100 ng across the samples. A standard curve was generated for all of the primer sets. The slope of the curve was used to calculate the relative pg of each transcript in the RNA extracted from the raphe blocks and all of the other tissues. Then, the ratio of each transcript to GUSB was calculated.
Primer selection
The genes of interest selected for qRT-PCR were Shh (sonic hedgehog), GATA3 (Gata binding protein isoform 3), Wnt3 (wingless type MMTV integration site family), transcription factors FEV1 (oncogene called Pet1 in rat), Lmx1b, Mash1, the receptor NTRK2 (neurotrophin tyrosine kinase type 2) and GFAP (glial marker). In addition, the specific serotonergic genes TPH2 (tryptophan hydroxylase 2), SERT (serotonin reuptake transporter) and the 5HT1A autoreceptor were examined. For most genes of interest, the probe set ID was saved as a .txt file. This file was launched to batch query at Affymetrix Netaffix program on the web. This program enabled retrieval of the annotation list of the genes of interest and provides direct access to Genebank sequence at NCBI. Thus, using Netaffix, the oligonucleotide location on each cDNA sequence was identified. Based upon the oligoset distribution, a target area of each gene of interest was selected. The target sequence was then loaded into Primer Express software, which chooses the primers for optimum qRT-PCR. Many of the probe sets on the microarray are located in the 3′ untranslated region of the mRNA. However, previously obtained primers that amplify part of the coding region were used for SERT and 5HT1A(Pecins-Thompson and Bethea 1998; Pecins-Thompson, et al. 1998). The primers were obtained from Invitrogen Life Technologies. The primers utilized for qRT-PCR on the RNA extracted from the ESC derived serotonin neurons are shown in Table 1.
Table 1.
Primer sequences utilized for qRT-PCR on stem cell RNA.
| Gene | Name | Accession ID | Primers | Tm°C | Amplicon size(bp) |
|---|---|---|---|---|---|
| SHH | Sonic hedghog | XM_001106515 | F_5′ ATCCCCAATGTGGCTGAGAAG ′3 R_3′ GGTCGGACGTGGTGATGTC ′5 |
60 58 |
317 |
| GATA3 | Rhesus Gata binding protein 3 isoform 1 | XM_001108237 | F_5′ GTGAACTGCGGAGCACCTC ′3 R_3′ TGGATGCCTTCCTTCTTCATG ′5 |
59 59 |
254 |
| NTRK2 | Neurotrophic tyrosine kinase type2 | XM_001107390 | F_5′CACACGGAGTACCACGGCT ′3 R_3′ACAGACGCAATCACCACCAC ′5 |
58 58 |
311 |
| WNT3 | Wingless type MMTV integration site family | XM_001115749 | F_5′GCGCCTCGGAGATGGTAGTAG ′3 R_3′GACGTAGCAGCACCAGTGGAA ′5 |
60 60 |
301 |
| PET1 | oncogene needed for serotonin differentiation | XM_001095962 | F_CAGAAAGGCAGCGGACAGAT ′3 R_CCTGGAAGTCGAAGCGGTAG ′5 |
59 59 |
268 |
| LMX1B | LIM homeobox transcription factor 1 | XM_001097412 | F_CTGCTACTTCCGGGATCGG(260bp-278bp) R_CCCCATCTTCATCCTCGCT(564bp_546bp) |
59 59 |
321 |
| MASH1 | Achaete-scute homolog1(Ascl1) | XM_001094978 | F_AGCGCAACCGCGTCA(397bp-411bp) R_TCGTAAGAGCCCTCGTCCG(676bp-658bp) |
58 60 |
288 |
| GFAP | Glial Fibrillary acidic protein | AL306647 | F GATTGTAAATGGAACGCCGCC(92bp-112b R TCCCAGTGACAGGAAGAG(421bp-400bp) |
62 61 |
330 |
| TPH2 | Tryptophan hydroxylase 2 brain isoform | AY_098914 | F CTGACACTGAGTACGTCGTGGA ′3 R TCCCAGTGACAGGAAGAG ′5 |
58 59 |
253 |
| SERT | SLC6A4 Serotonin transporter(human) | NM_001045 | F_TCATTTACTAACCAGCAGCATGGAC(287b R_TTCCCCTTGATGAAGCTCAGCCACTAGG |
58 58 |
258 |
| 5HT1A | Serotonin 1A auto receptor(human) | NM_000524 | F_GGCGCATATTCCGAGCTGCGC(647bp-6661 R_GCAGCCAGCAGAGGATGAAGG(1058bp-J |
59 59 |
432 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | XM_001087699 | F CTGACACTGAGTACGTCGTGGA ′3 R TGGACTGTGGTCATGAGTCCTT ′5 |
57 58 |
268 |
| PPIA | Cyclophilin A | NM_021130 | F_5′TCGTGCTCTGAGCACTGGAG′3 R_3′CATGCCTTCTTTCACTTTGCC′5 |
59 58 |
301 |
| TBP | Tata Box binding Protein | XM_001085250 | F_5′AGACCATTGCACTTCGTGCC′3 R_3′CACATCACAGCTCCCCACC ′5 |
60 58 |
253 |
| GUSB | Glucoridase ß | XM_001087699 | F_5′ACTTCGGGCACCTGGAGTT′3 R_3′TCATGAAATCGGCAAAATTCC′5 |
58 58 |
253 |
Several housekeeping genes were measured with qRT-PCR for normalization of the data. GAPDH (glyceraldehydes-3—phosphate dehydrogenase), TBP (Tata box binding protein), GUSB (glucoridase beta) and PPIA (cyclophilin A) were measured at each developmental stage. GUSB exhibited a moderate level of expression and yielded whole numbers instead of small fractions with normalization, which are easier to compile. In addition, the expression was most consistent per microgram of RNA across the stages as compared to the other housekeeping genes. Thus, we chose GUSB for normalization of the qRT-PCR data.
Results
The appearance of the ESC-derived serotonin neurons with phase contrast microscopy is shown in Figure 1. The cultures are viable in this state for several weeks. Attempts to disperse the neurospheres and replate single cells in the N3 medium did not yield viable neurons. The protein expression of Nestin and Musashi1 in the neuroprogenitor cells (N1, N2) and the expression of neuronal specific proteins MAP2 and NeuN in the differentiated neurons (N3) obtained with this protocol applied to the 366.4 cell line, was demonstrated with immunocytochemistry in two previous studies (Mitalipov, et al. 2006; Salli et al. 2004).
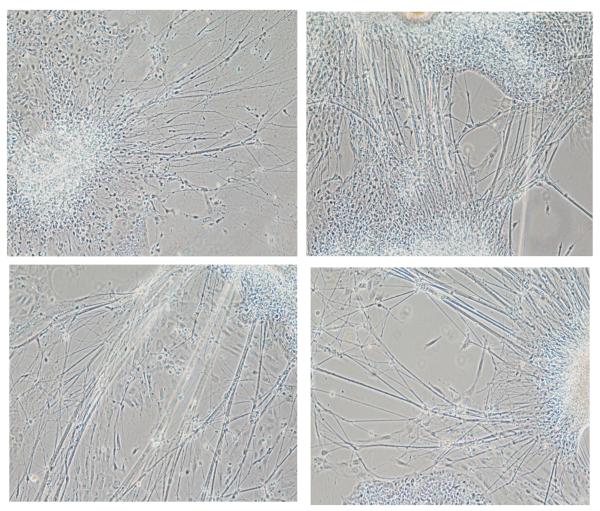
Figure 1.
Examples of ESC derived serotonin neurons after 2 weeks of culture under N3 conditions. The neurospheres send out long axonal-like processes, which form bundles.
The Affymetrix microarray data was entered into GeneSifter Software. There were 4639 genes that exhibited a present call in one stage and showed a 3-fold or greater change between the 5 stages. KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis revealed numerous categories that exhibited a significant number of 3-fold or greater changes in gene expression across the 5 stages and signaling pathways were of particular interest. In addition, for this study, we applied the gene function filter for developmental processes on the genes that showed a 3-fold or greater change and examined the 545 genes listed. We noted the pattern of their expression and then subcategorized them as homeobox binding and transcription factors, growth factors and receptors, brain and neural induction factors and serotonin specific factors. Subcategories not included in this study were related to the cytoskeleton, adhesion, metabolism and proteoglycans.
The expression changes exhibited a variety of patterns. In the simplest terms these patterns were as follows:
High expression in ESC and then decreased markedly
Low in ESC, increase markedly in EBs and stay markedly expressed
Gradually increase from ESC through N3
Low in ESC, increase markedly in EBs and then gradually decline in N1, N2, N3
Low in ESC, increase markedly in EBs and N1, then decline in N2 and N3
Low in ESC and EBs, increased markedly in N1 and then decline in N2 and N3
Low in ESC and EBS, increased markedly in N1 and N2, then decline in N3
Increase only in N2
Increase only in N3
Increase in N2 and N3
Microarray Gene Expression
Shown in Tables 2, 3, 4 and 5 are genes in the four subcategories of interest that exhibited relatively robust expression and 3-fold or greater changes in expression across the 5 stages. The signal intensity (arbitrary units) reported on the microarray is shown for each gene. Robust and peak expression values are highlighted in red. The genes are arranged from top to bottom depending on the stage of peak expression with early genes at the top and later genes at the bottom. For example, in Table 1 in the homeobox and transcription factor category, NANOG (homeobox transcription factor) was robustly expressed in the ESC and then was not detected in any other stage. NPM1 (nucleophosmin, nucleolar protein) was robustly expressed in the ESC and relatively high expression continued through all of the stages. SOX9 (regulates transcription of the anti-Muellerian hormone gene) was detected in EBs and no other stage. SOX11 (may function in the developing nervous system) turned on at the N1-selection stage and increased to robust signal intensity at the N3-differentiation stage. A number of genes including PAX6 (paired box 6 transcription factor expressed in developing nervous system), DKK3 (Dickkopf family, Wnt antagonist) and DACH1 (dachshund 1, eye development) were elevated during N2-proliferation and N3-differentiation. Genes that were only expressed in N3-differentiation were rare in this category.
Table 2.
The expression patterns of genes encoding homebox binding and transcription factors during differentiation of ESC into serotonin neurons. The signal intensity on the microarray is reported. Robust and peak expressions are highlighted in red.
| Homeobox Binding and Transcription Factors | |||||
|---|---|---|---|---|---|
| ESC | EB | N1 | N2 | N3 | |
| ID1 | 3858 | 1511 | 2153 | 982 | 1287 |
| NPM1 | 5098 | 4625 | 2506 | 1907 | 1149 |
| ZIC2 | 5521 | 1433 | 860 | 2267 | 2484 |
| NANOG | 3693 | 54 | 43 | 85 | 68 |
| TNFRSF12A | 2015 | 86 | 283 | 171 | 295 |
| OTX | 1737 | 44 | 50 | 453 | 359 |
| PHC1 | 2183 | 543 | 542 | 799 | 760 |
| FLI 1 | 549 | 266 | 173 | 166 | 253 |
| HUS1 | 342 | 174 | 128 | 79 | 95 |
| MEF2B | 406 | 300 | 269 | 112 | 87 |
| HOXC6 | 140 | 1294 | 961 | 962 | 1136 |
| PBX1 | 606 | 1116 | 1563 | 1982 | 1781 |
| ID2 | 353 | 1348 | 3041 | 1649 | 2904 |
| SALL1 | 421 | 2054 | 394 | 698 | 970 |
| SOX9 | 85 | 2554 | 149 | 361 | 360 |
| HOXA3 | 37 | 696 | 372 | 509 | 491 |
| MITF | 57 | 459 | 355 | 206 | 116 |
| BLZF1 | 158 | 497 | 457 | 465 | 350 |
| SOX11 | 623 | 1269 | 3133 | 6045 | 8079 |
| TCF12 | 1153 | 1743 | 3045 | 3915 | 3352 |
| NCOA6 | 242 | 794 | 966 | 1197 | 1337 |
| HOXD10 | 54 | 335 | 652 | 1197 | 1303 |
| PAX6 | 299 | 166 | 122 | 1315 | 1324 |
| MNT | 296 | 477 | 429 | 1033 | 856 |
| DKK3 | 322 | 783 | 735 | 1487 | 2260 |
| PHC3 | 188 | 572 | 708 | 1189 | 998 |
| DACH1 | 48 | 209 | 624 | 1087 | 1390 |
| CUTL1 | 390 | 431 | 808 | 1174 | 768 |
| DKK1 | 77 | 617 | 272 | 793 | 556 |
| FOXF2 | 32 | 247 | 313 | 420 | 157 |
| DVL3 | 216 | 382 | 445 | 781 | 623 |
| FMR2 | 58 | 81 | 142 | 308 | 560 |
Table 3.
The expression patterns of genes encoding growth factors and growth factor receptors during differentiation of ESC into serotonin neurons. The signal intensity on the microarray is reported. Robust and peak expressions are highlighted in red.
| Growth Factors & Receptors | |||||
|---|---|---|---|---|---|
| ESC | EB | N1 | N2 | N3 | |
| FGFR2 | 1099 | 1585 | 2458 | 4398 | 2777 |
| FGFR1 | 759 | 409 | 250 | 358 | 428 |
| FGF2 | 619 | 94 | 92 | 129 | 101 |
| BAX | 1198 | 343 | 342 | 244 | 250 |
| FST | 3094 | 974 | 288 | 272 | 346 |
| BST2 | 1060 | 637 | 756 | 234 | 245 |
| TGIF | 2938 | 953 | 767 | 948 | 701 |
| IGFBP3 | 537 | 6147 | 1742 | 5775 | 5116 |
| SMAD5 | 944 | 3190 | 3103 | 3216 | 2706 |
| BMPR2 | 501 | 1730 | 1740 | 2480 | 2809 |
| VEGF | 1073 | 3533 | 2299 | 5169 | 6003 |
| ING3 | 147 | 586 | 660 | 595 | 402 |
| MTSS1 | 56 | 696 | 588 | 1055 | 1279 |
| TGFb R3 | 185 | 1130 | 1480 | 838 | 491 |
| ANGPT1 | 28 | 1295 | 1240 | 651 | 260 |
| VEGF-R (KDR) | 156 | 1144 | 555 | 303 | 202 |
| CSF1 | 133 | 653 | 296 | 223 | 192 |
| FZD4 | 148 | 638 | 295 | 253 | 295 |
| NRP2 | 185 | 953 | 675 | 551 | 607 |
| NRP1 | 185 | 953 | 675 | 551 | 607 |
| FZD4 | 148 | 638 | 295 | 253 | 295 |
| IGFBP5 | 1713 | 813 | 3409 | 6805 | 6370 |
| TGFb2 | 149 | 291 | 908 | 1483 | 1032 |
| SFRP2 | 669 | 538 | 1482 | 2972 | 2979 |
| LPIN1 | 272 | 687 | 1001 | 1350 | 987 |
| CRABP1 | 2018 | 1518 | 4753 | 6574 | 8449 |
| IGFBP7 | 161 | 134 | 196 | 835 | 994 |
Table 4.
The expression patterns of genes encoding brain and neural specific proteins during differentiation of ESC into serotonin neurons. The signal intensity on the microarray is reported. Robust and peak expressions are highlighted in red.
| Brain & Neural Specific | |||||
|---|---|---|---|---|---|
| ESC | EB | N1 | N2 | N3 | |
| ADAM10 | 2604 | 1324 | 1119 | 770 | 1067 |
| PCDH1 | 341 | 72 | 68 | 80 | 10 |
| PES1 | 449 | 266 | 252 | 100 | 239 |
| CHRDL1 | 696 | 43 | 87 | 226 | 161 |
| MDK | 3125 | 2205 | 2863 | 722 | 521 |
| PCDHB9 | 1203 | 2342 | 3096 | 3903 | 3876 |
| PCDH18 | 554 | 1265 | 2120 | 2751 | 1659 |
| SLIT2 | 158 | 1440 | 2220 | 2080 | 2502 |
| SEMA6D | 293 | 1107 | 951 | 1686 | 1395 |
| SERPINI-1 | 134 | 424 | 458 | 419 | 597 |
| PLXNA2 | 49 | 318 | 361 | 442 | 686 |
| GAS7 | 107 | 549 | 1007 | 627 | 691 |
| STMN2 | 372 | 791 | 5021 | 5577 | 9892 |
| ROBO2 | 300 | 451 | 1269 | 2214 | 2431 |
| PAFAH1B1 | 386 | 919 | 1198 | 1247 | 1373 |
| NEGF1 | 516 | 232 | 1126 | 1591 | 3724 |
| ROBO1 | 528 | 996 | 1134 | 2177 | 1698 |
| DCX | 73 | 213 | 886 | 2585 | 4228 |
| CRIM1 | 461 | 684 | 670 | 1585 | 1548 |
| ENC1 | 772 | 857 | 995 | 1585 | 3058 |
| WHSCI | 567 | 530 | 1044 | 903 | 1937 |
| PALM | 686 | 1236 | 965 | 2390 | 2730 |
| CHL1 | 42 | 27 | 310 | 1179 | 2701 |
| NTRK2 | 94 | 294 | 775 | 1824 | 3540 |
| OLFM1 | 501 | 506 | 684 | 1829 | 3482 |
| NCAM1 | 172 | 463 | 918 | 2001 | 2933 |
| NRXN1 | 421 | 149 | 282 | 842 | 1572 |
| NRXN3 | 36 | 132 | 232 | 748 | 1102 |
| CTNND2 | 105 | 250 | 256 | 582 | 767 |
| INA | 1194 | 556 | 779 | 2615 | 5784 |
| FEZ1 | 732 | 106 | 305 | 729 | 2418 |
| CDKSR1 | 79 | 88 | 216 | 571 | 1246 |
| GAP43 | 168 | 143 | 382 | 411 | 1378 |
Table 5.
The expression patterns of genes encoding proteins believed to promote the serotonin phenotype during differentiation of ESC into serotonin neurons. The signal intensity on the microarray is reported. Robust and peak expressions are highlighted in red.
| Serotonin Specific | |||||
|---|---|---|---|---|---|
| ESC | EB | N1 | N2 | N3 | |
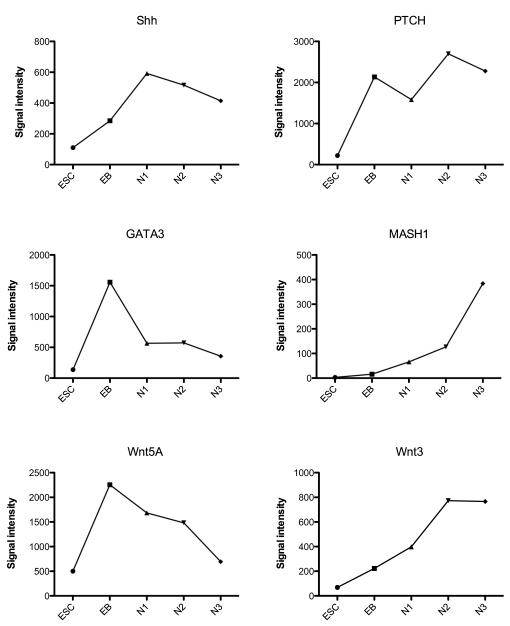
| GATA3 | 138 | 1558 | 565 | 572 | 356 |
| Wnt5A | 502 | 2256 | 1684 | 1483 | 692 |
| PTCH (shhR) | 220 | 2137 | 1584 | 2699 | 2279 |
| Nkx2.3 | 24 | 151 | 176 | 120 | 48 |
| BDNF | 41 | 94 | 211 | 268 | 107 |
| Shh | 110 | 285 | 592 | 516 | 414 |
| En1 | 29 | 35 | 112 | 112 | 197 |
| ISL2 | 51 | 42 | 92 | 216 | 177 |
| WNT9A | 96 | 205 | 214 | 448 | 264 |
| Wnt3 | 69 | 223 | 398 | 773 | 766 |
| PHOX2b | 5 | 26 | 94 | 415 | 490 |
| MASH1 | 3 | 16 | 66 | 127 | 384 |
| Nkx2.2 | 11 | 16 | 36 | 81 | 129 |
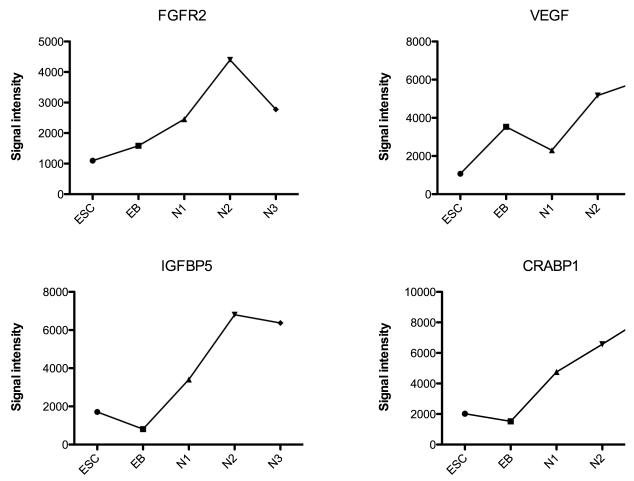
In Table 2, genes of note in the growth factor and receptor category include FGFR2 (fibroblastic growth factor receptor 2), which was robustly expressed in each stage. FGF2 (fibroblastic growth factor 2) was expressed by ESCs, but not other stages. A number of genes increased expression during the EB stage and remained relatively highly expressed such as IGFBP3 (insulin like growth factor binding protein 3), SMAD5 (signal transduction for transforming growth factor), and VEGF (vascular endothelial growth factor). Other genes were modestly induced during the EB stage and then declined such as KDR (VEGF receptor), NRP1 and 2 (neuropilins, tyrosine kinase coreceptor for VEGF and semaphorin). Several genes increased expression during N1-selection and remained relatively highly expressed such as IGFBP5 (insulin like growth factor binding protein 5), TGFβ2 (transforming growth factor, beta 2), and CRABP1 (cellular retinoic acid binding protein). Genes that were induced only during N3-differentiation were also rare in this category.
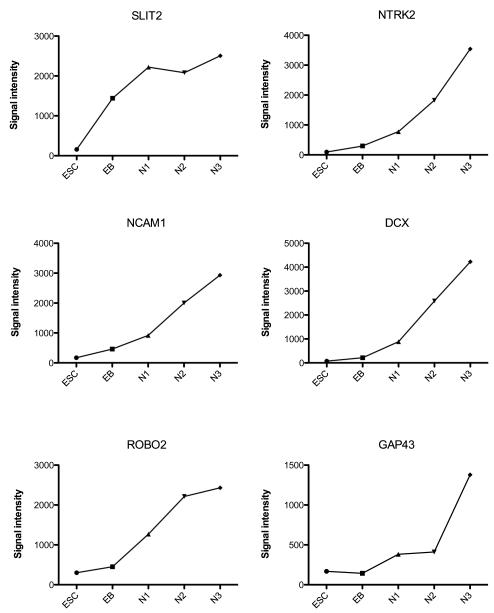
In Table 3, in the neural category, Adam10 (cell surface metalloproteinase) and MDK (midkine, neurite growth-promoting factor 2) were the only genes that were robustly expressed in ESCs. However, from the EB stage through terminal differentiation a number of neuronal genes are robustly expressed, which likely play a role in the neuronal lineage development. Several genes were induced during the EB stage and remained highly expressed in all stages such as PCDHB9 (protocadherin beta 9, neural adhesion protein) and SLIT2 (axonogenesis). Then, ROBO2 (roundabout, axon guidance receptor) and others were induced during N1-selection and remained highly expressed in the remaining stages. A significant number of genes were up-regulated during N2-proliferation and remained expressed in N3-differentiation. Finally, several genes were up-regulated only in N3-differentiation such as FEZ1 (axonal outgrowth, bundling and elongation) and GAP43 (neuronal growth cone protein).
Studies in rodent species have provided insight into genes that are pivotal for attaining the serotonin phenotype. Based upon those studies, we constructed Table 5 to show the expression of implicated genes in serotonin development as reported by the microarray during differentiation in vitro. Many of the implicated genes in serotonin development were detected on the microarray. GATA3 (transcription factor) was induced during the EB stage and declined thereafter. Wnt5a (signal transduction with a role in axis induction) was induced in EBs, and declined thereafter. This overlapped with the expression of Wnt3 (sonic hedgehog signaling), which was slightly elevated during N2-proliferation and N3-differentiation. Shh (sonic hedgehog) has been highly implicated in serotonin development and it was modestly increased during N1-selection and thereafter. However, the Shh receptor, PTCH, was robustly expressed at the EB stage and thereafter. Nkx2.2 (homeobox transcription factor) was generally below the level of detection of the chip, and Nkx2.3 was barely detectable. Mash1 (or Ascl1, transcription factor in neuronal commitment) showed a modest up-regulation at the N2-proliferation stage. Two pivotal transcription factor genes, Fev1 and Lmx1b were undetectable on the chip. When this was checked with qRT-PCR as described below, it was found that these genes were in fact expressed, suggesting that the probe sets on the chip were not optimal. Phox2b (paired-like homeobox 2b, role in neurotransmitter phenotype) increased modestly during N2-proliferation and N3-differentiation.
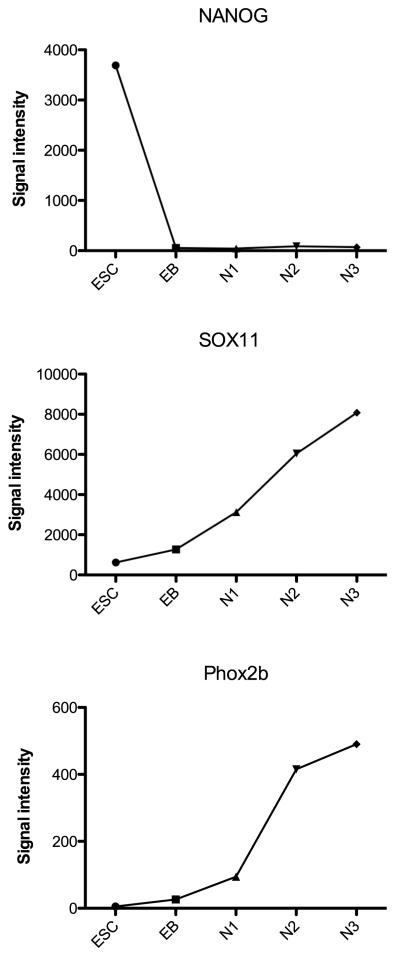
To better illustrate examples of signal intensity on the microarray, several genes with different patterns of expression were selected from each category and their expression levels are shown in Figures 2, 3, 4 and 5. The signal intensity in arbitrary units is plotted on the Y-axis. (There are no error bars, because the graphs illustrate the expression with one chip per stage of differentiation. Shown below, portions this data are confirmed with qRT-PCR on 3 preparations of each stage.) In Figure 2, NANOG is robustly expressed in the ESC and then completely suppressed upon initiation of differentiation. SOX11 and Phox2b increased throughout differentiation. In Figure 3, FGFR2 was robustly expressed at each stage with peak expression during N2-proliferation. VEGF was robustly expressed at each stage, but the peak expression occurred at N3-differentiation. IGFBP5 was also robustly expressed at each stage with peak expression during N2-proliferation stage. CRABP1 increased throughout differentiation. In Figure 4, 6 pivotal genes in neuronal differentiation are illustrated which have different patterns of expression. SLIT2 increased 1000 fold between ESC and EB stages and remained robustly expressed thereafter. NTRK2 (nerve growth factor tyrosine kinase receptor), and NCAM (neural cell adhesion molecule) increased gradually across the stages and peaked during the final N3-differentiation stage. ROBO2 and DCX (doublecortex regulates stability of microtubules) were markedly up-regulated and increased from N1-selection to N3-differentiation stages. Finally, GAP43 showed significant induction during N3-differentiation. In Figure 5, the microarray results were graphed for 6 genes that have been implicated in serotonin differentiation. Shh and its receptor, PTCH, were expressed in a complimentary fashion starting at the EB stage and thereafter. GATA3 was markedly up-regulated only during the EB stage, whereas Wnt5a was induced during the EB stage and gradually declined in the succeeding stages. Wnt3 increased in parallel with the decline in Wnt5a. Mash1 was modestly up-regulated during N3-differentiation.
Figure 2.
Illustration of the signal intensity on the microarray of NANOG, SOX11 and Phox2b from the homeobox or transcription factor category at the end of each developmental stage on the microarray (n=1 chip/stage).
Figure 3.
Illustration of the signal intensity on the microarray of FGFR2, VEGF, IGFBP5 and CRABP1 from the growth factor/receptor category at the end of each developmental stage on the microarray (n=1 chip/stage).
Figure 4.
Illustration of the signal intensity on the microarray of SLIT2, NTRK2, NCAM, ROBO2, DCX and GAP43 from the brain specific category at the end of each developmental stage on the microarray (n=1 chip/stage).
Figure 5.
Illustration of the signal intensity on the microarray of Shh, PTCH, GATA3, MASH1, Wnt5A and Wnt3 from the serotonin genetic hierarchy, at the end of each developmental stage on the microarray (n=1 chip/stage).
Two important factors in serotonin differentiation, FEV1 (or Pet1) and Lmx1b were not detected on the microarray. In addition, other serotonin related genes were poorly represented. Hence, FEV1, Lmx1b, tryptophan hydroxylase 2 (TPH2), the serotonin reuptake transporter (SERT), and the 5HT1A autoreceptor were examined with qRT-PCR. Other pivotal genes and GFAP, a marker for glia, were also examined.
qRT-PCR Confirmation
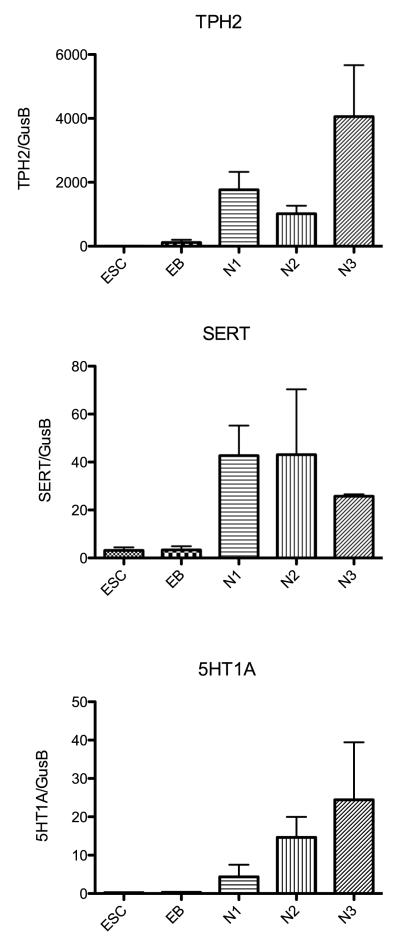
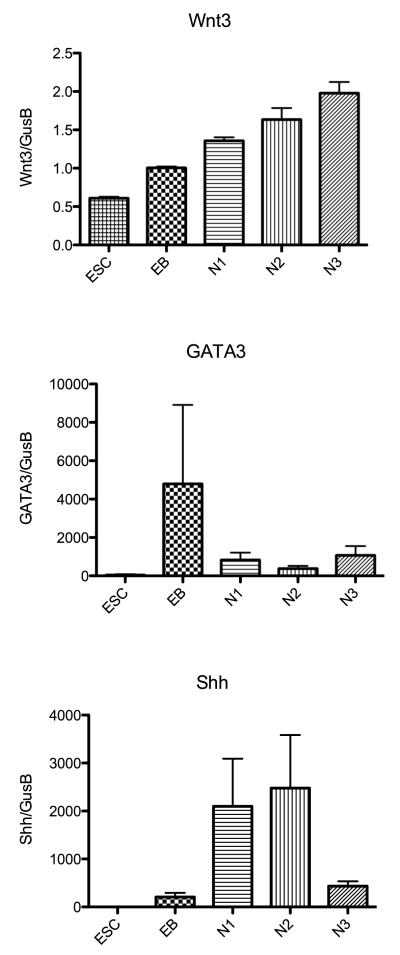
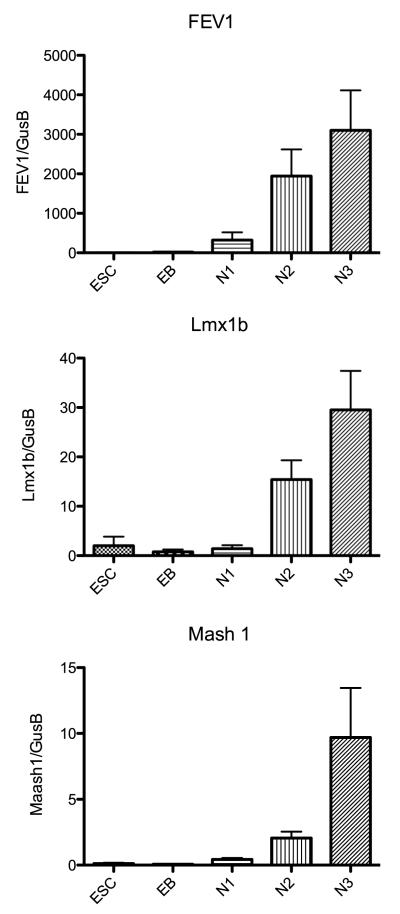
Therefore, to verify and add to the array results, pivotal genes from each category that have been implicated in the serotonergic genetic hierarchy were selected and subjected to quantitative (q) RT-PCR on 3 different passages of each stage of development. The genes selected were TPH2, SERT, 5HT1A, GATA3, Shh, Wnt3, FEV1, Lmx1b, Mash1, NTRK2, and GFAP. The mean (± sem) relative expression (n=3 passages per stage) of each of these genes is illustrated in Figures 6, 7, 8 and 9. In Figure 6, 3 genes that are markers for serotonin neurons are illustrated. TPH2 codes for the rate-limiting enzyme in serotonin synthesis; SERT codes for the serotonin reuptake transporter and 5HT1A codes for the autoreceptor located in the dendrite region of serotonin neurons. TPH2 was an abundant transcript. The expression of TPH2 was highest in differentiated neuronal cultures, but it was expressed during the selection and proliferation stages as well. SERT was a modestly expressed transcript that exhibited a peak in expression during the selection and proliferation stages. The slightly lower expression at N3-differentiation stage is not significantly different from N1-selection and N2-proliferation expression. 5HT1A gene expression was also modest, but it peaked during the terminal differentiation stage. Together, the data confirm our earlier report that the 366.4 ESC line differentiates into serotonin neurons. In Figure 7, the expression pattern of Wnt3 and GATA3 was similar to the expression reported on the microarray. Wnt3 increased gradually through each stage, but it was not a prevalent RNA species. GATA3 expression peaked at the EB stage and then declined markedly. Shh expression was highest at the N1 and N2 stages but declined at N3 more so than depicted by the microarray. Figure 8 illustrates the expression of FEV1 and LMX1b, which were not detected by the microarray, and Mash1. FEV1 and Lmx1b each increased markedly during proliferation and peaked during differentiation. FEV1 was a more abundant transcript than Lmx1b. Mash1 expression increased gradually throughout differentiation as reported by the microarray but it was not a prevalent transcript.
Figure 6.
Histograms illustrating the relative expression of 3 pivotal genes that define serotonin neurons at each developmental stage as determined with qRT-PCR (n=3 passages/stage). Expression was normalized to GUSB. The genes code for TPH2, the rate limiting enzyme in serotonin synthesis; SERT, the serotonin reuptake transporter; and 5HT1A serotonin autoreceptor.
Figure 7.
Histograms illustrating the relative expression of 3 pivotal genes in serotonin development at the end of each developmental stage as determined with qRT-PCR (n=3 passages/stage). Expression was normalized to GUSB. The genes code for Wnt3, GATA3 and Shh.
Figure 8.
Histograms illustrating the relative expression of an additional 3 pivotal genes in serotonin development at the end of each developmental stage as determined with qRT-PCR(n=3 passages/stage). Expression was normalized to GUSB. The genes code for FEV1, Lmx1b and Mash1.
Figure 9.
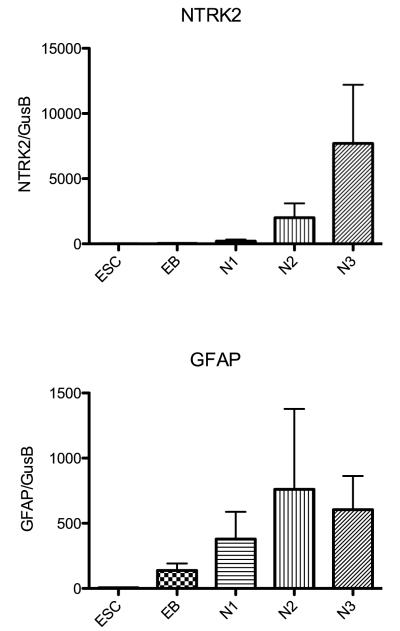
Histograms illustrating the relative expression of a gene that is localized to neurons, NTRK2, and a gene localized to glia, GFAP, at the end of each developmental stage as determined with qRT-PCR and normalized to GUSB (n=3 passages/stage). NTRK2 was 10-fold more prevalent than GFAP throughout development.
In Figure 9, we compared the relative expression of NTRK2, a nerve growth factor receptor tyrosine kinase, to GFAP, a marker for glia. The expression of NTRK2 is ten-fold higher than GFAP indicating that the cultures are enriched for neuronal cells.
Signaling Pathway Analysis
To corroborate our interpretation of pivotal signaling genes, a pathway analysis was conducted based upon the KEGG database and the results are shown in Table 6. The total number of probe sets on the array that are involved in a particular pathway were compared with the number of those probe sets that were altered 3-fold or greater during differentiation within the pathway. The results are shown in Table 6. It is important to note that the analysis detects changes in probe sets and there may be multiple probe sets for a single gene. Moreover, there is significant intersection between the pathways and so there is redundancy in the probe sets between the pathways. Nonetheless, the greatest percent of gene regulation occurred in the TGFβ pathway (which intersects with the MAPK signaling pathway), followed by VEGF, Wnt and Hedgehog. The MAPK pathway, which utilizes the FGFs and their receptors, showed moderate gene regulation. Gene expression in Jak-STAT, PPAR, Notch and Toll-like signaling pathways showed minor regulation during differentiation.
Table 6.
Number of probe sets that were over background and changed 3 fold or greater (increase or decrease) during differentiation relative to designated probe sets in different signaling pathways according to KEGG analysis. It is important to note that there may be multiple probe sets for the same gene and there was significant redundancy in the probe sets between pathways.
| *Pathway | Total on Microarray | #Changed >3 fold | Percent |
|---|---|---|---|
| TGF beta | 58 | 14 | 24 |
| VEGF | 51 | 12 | 23 |
| Wnt | 110 | 55 | 22 |
| Hedgehog | 43 | 8 | 19 |
| Insulin | 107 | 20 | 19 |
| MAPK | 184 | 33 | 18 |
| ErbB (EGF) | 70 | 11 | 16 |
| Calcium | 143 | 22 | 15 |
| Jak-STAT | 104 | 14 | 13 |
| PPAR | 52 | 5 | 10 |
| Notch | 35 | 3 | 8 |
| Toll-like | 55 | 3 | 5 |
mTOR is omitted due to its role in the cell cycle which is still very active in these cultures.
ESC and Adult Serotonin Neuron Gene Expression Compared
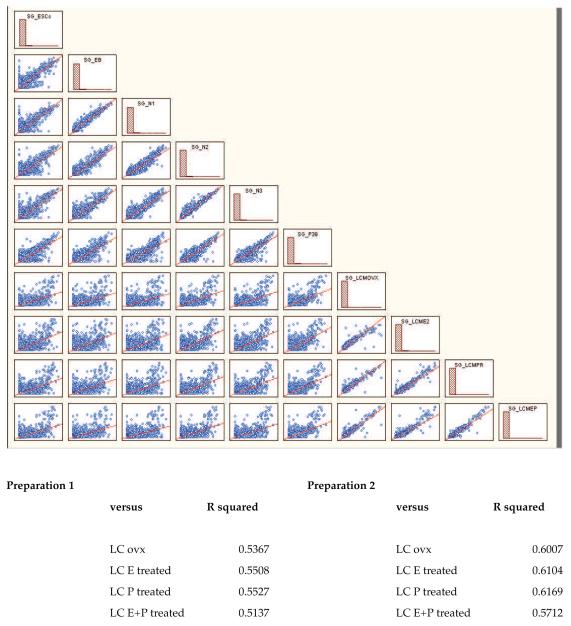
Global gene expression was compared between all of the developmental stages (including an extra preparation of terminal neurons called p39-N3) and laser captured serotonin neurons from ovariectomized monkeys treated with placebo, E, P or E+P for one month (n=1 chip each for this comparison). Regression analysis was then performed and the results are shown in Figure 10. The highest correlation coefficients (R squared) were between sequential stages of development and between the laser-captured pools. The 2 preparations of terminal neurons exhibited a correlation coefficient of 0.8475. The best correlations between the two terminal ESC-derived serotonin neuron preparations and the laser-captured pools were between the N3-differentiation stages and the P-treated monkey (0.5527 for preparation 1, called N3 and 0.6169 for preparation 2, called p39-N3).
Figure 10.
Regression analysis of global gene expression between the different developmental stages and laser captured (LC) serotonin neurons from ovariectomized female monkeys treated with placebo, estrogen, progesterone or estrogen plus progesterone for 1 month. This analysis is done with a chip to chip comparison, hence there is 1 chip/stage or treatment) The culture stages and the monkeys are listed along the right side of the graph in red. The regression comparison of two groups is located by finding the intersection graph. For example, to see the correlation between EB and N3 gene expression, locate the EB group (second from top) and drop down to the N3 row. It is apparent that there is more deviation from the regression line in the comparison of EB and N3 gene expression than in the comparison of EB and N1 gene expression. Similarly, there is little deviation from the regression line in comparisons of the LC neurons from adult monkeys (LCM-OVX, LCM-E2, LCM-PR, LCM-EP). However, when N3 (preparation 1) or p39-N3 (preparation 2) are compared to any of the LC preparations, there is marked deviation from the regression line. The comparison between preparation 2 and the progesterone treated monkey (LCM-PR) gave the best correlation coefficient of 0.6169.
Further comparison of gene expression in the p39-N3 preparation and the P-treated monkey indicated that 4115 genes showed a 5-fold difference in expression. Of these, 1969 were 5-fold or more lower in the ESC-derived serotonin neurons than the laser captured serotonin neurons and 2146 were 5-fold or more higher in the ESC-derived serotonin neurons than the laser captured serotonin neurons. Z scores of +2.5 or greater in the KEGG Analysis indicated that higher gene expression was over-represented in the subcategories of neuroactive ligands and receptors, calcium signaling, long term potentiation, long term depression, and gap junctions in the laser captured serotonin neurons compared to the ESC-derived serotonin neurons (Table 7). In contrast, elevated expression of genes was over-represented in the subcategories of cell cycle, cell proliferation, axon guidance, dorso-ventral axis formation and development, in the ESC-derived serotonin neurons compared to the laser captured serotonin neurons (Table 8).
Table 7.
KEGG analysis of genes with 5-fold greater expression in adult serotonin neurons than in ESC-derived serotonin neurons. These categories were significantly over-represented in adult neurons.
| Gene categories | z-score |
|---|---|
| Response to stimuli | 3.59 |
| Neuroactive ligand/receptor | 4.21 |
| Long term potentiation | 5.46 |
| Long term depression | 2.96 |
| Gap junction | 3.93 |
Table 8.
KEGG analysis of genes with 5-fold greater expression in ESC-derived serotonin neurons than in adult serotonin neurons. These categories were significantly over-represented in ESC neurons.
| Gene categories | z-score |
|---|---|
| Development | 2.36 |
| Cell cycle | 6.21 |
| Axon guidance | 3.56 |
| Dorso-ventral axis formation | 3.29 |
The pivotal genes that increased or decreased during differentiation in vitro were examined in the laser captured neurons from adult monkeys and the results are shown in Table 9. In this analysis, the average signal intensity on 6 microarray chips hybridized with RNA from 6 ovariectomized animals treated with either placebo, E or E+P (n=2/treatment) was obtained. The different treatments contributed to the variation around the mean, but the goal was to examine general expression levels. A majority of the selected genes that peaked during N3 were expressed at moderate to robust levels in the adult neurons. Moreover, NANOG and Shh, which declined by N3 stage, exhibited low to undectable gene expression in the adult neurons. However, CRABP1 was robustly expressed in the N3 neurons, but it was very low in adult neurons. Lmx1b and Fev1 were detectable in the adult neurons, but we contend that these probe sets are not optimal.
Table 9.
Microarray signal intensity of pivotal developmental genes in laser captured serotonin neurons. The average represents the mean of 6 animals or chips. The animals were ovariectomized and treated with placebo, estrogen or estrogen plus progesterone (n=2 chips/treatment) for one month.
| Gene | Average Signal Intensity ± sem (n=6 chips) |
|---|---|
| NANOG | 98±24 |
| SOX11 | 150±40 |
| Phox2b | 158±87 |
| FGFR2 | 1884±667 |
| VEGF | 753±221 |
| IGFBP5 | 337±63 |
| CRABP1 | 90±50 |
| SLIT2 | 753±76 |
| NTRK2 | 5033±2400 |
| NCAM1 | 1053±344 |
| DCX | 455±75 |
| Robo2 | 488±111 |
| GAP43 | 573±211 |
| Shh | ND |
| PTCH | 663±239 |
| GATA3 | 268±57 |
| MASH1 | 528±123 |
| Wnt5a | 280±14 |
| Lmx1b | 132±86 |
| Fev1 | 225±71 |
Discussion
Mental illnesses involving the serotonin system are common. Dysfunction of the serotonin neural system plays a role in various mental illnesses, and pharmacotherapies that alleviate their symptoms target various aspects of serotonin function or serotonin receptors. However, current pharmacotherapies do not cure and they are usually needed for life. With the advent of stem cell technology, the potential to replace injured or degenerative neurons needs to be explored. Several recent successes have encouraged further effort (Cunningham, et al. 2007; Ren-Patterson, et al. 2005; Vazey, et al. 2006).
This laboratory sought to culture primate serotonin neurons to probe cellular and molecular questions that are not possible with whole monkeys. The protocol used in this study was initially shown to promote the differentiation of monkey 366.4 ES cells into neural lineages (Kuo et al. 2003). We then found that the generated neurons were almost entirely serotonergic (Salli et al. 2004). These 2 studies exhaustively demonstrated the expression of neural markers during differentiation of 366.4 ES cells. In particular, Nestin and Musashi1 were expressed in neuroprogenitor cells and MAP2 , NeuN and Tuj III were expressed in the differentiated neurons. We did not repeat these observations again. Rather, we applied the identical derivation protocol to the same cell line and based upon scientific premise, the same results were ensured. Subsequently, Mitalopov and colleagues have generated new rhesus ES cell lines, called ORMES, and found that the same protocol produces serotonin neurons from the ORMES lines as well (Mitalipov et al. 2006). Hence, the ability of primate ESCs to differentiate into serotonin neurons has been demonstrated in 3 independent studies with different cell lines.
Homeobox genes
We confirmed that NANOG is a robust marker of pluripotent ES cells (Chen and Khillan 2008) and its expression became undetectable immediately upon starting a differentiation protocol. In this case, NANOG expression disappeared with the formation of the EBs. We previously confirmed that the ES cells robustly express Oct 4 and alkaline phosphatase (Salli et al. 2004). It is clear from Table 2 that various homeobox and transcription factor genes are induced at various stages and then decline or remain elevated through differentiation. SOX11 showed the greatest fold change of all the homeobox genes with the peak occurring during N3-differentiation. This intronless gene encodes a member of the SOX (SRY-related HMG-box) family of transcription factors involved in the regulation of embryonic development and in the determination of the cell fate. The encoded protein may act as a transcriptional regulator after forming a protein complex with other proteins and function in the developing nervous system. SOX family members were altered between fetal neural precursors and mature fetal neurons (Yu, et al. 2006). The expression of SOX11 upon differentiation of stem cells into serotonin neurons supports it role in nervous system development. Recently, Pax6 was shown to increase during in vitro generation of neurons from striatal neural stem cells (Kallur, et al. 2008), which is consistent with the increased expression we observed at N2-proliferation and N3-differentiation stages.
Pathway analysis and similarities to in vivo studies
Our pathway analysis of gene expression changes in signaling pathways was informative. By expressing the number of probe set changes as a percent of the total number of probe sets in the pathway, we found that genes in the TGFβ pathway were prominently regulated during differentiation. Indeed, TGFβ2 and TGFβ receptor 3 were markedly increased on the microarray during differentiation. This is consistent with a number of other studies that have shown the importance of TGFβ in attainment of a neuronal phenotype (Aigner and Bogdahn 2008; Cai, et al. 2006; Falk, et al. 2008; Park, et al. 2008).
Alternations in gene expression in the VEGF pathway during differentiation were numerous. Moreover, a marked increase in VEGF per se was observed on the microarray and confirmed with qRT-PCR. VEGF has not received the attention of TGFβ or FGF, however increasing evidence points to a pivotal role in neural differentiation (Le Bras, et al. 2006). Consistent with this study, VEGF and its receptor KDR, were increased in a previous microarray study of ESC differentiation induced by retinoic acid (Kelly and Rizzino 2000).
Not surprisingly, there were significant changes in gene expression in the Wnt and Hedgehog pathways. These pathways play a prominent role in neural and serotonergic differentiation. In mouse, serotonin neurons develop from the most ventral hindbrain near the floorplate (Lidov and Molliver 1982; Wallace and Lauder 1983). Sonic hedgehog (Shh) derived from the floor plate initiates serotonin neuron formation (Goridis and Rohrer 2002). The appearance of Shh in the EBs and its subsequent increase during N1-selection and N2-proliferation is consistent with reports from mice indicating that early expression of Shh in the floorplate is essential for the birth of serotonin neurons (Ye, et al. 1998). Moreover, PTCH, the sonic hedgehog receptor (Stone, et al. 1996), increased markedly at the EB stage and remained robustly expressed throughout differentiation.
The intracellular signaling pathway utilized by Shh involves the Wnt proteins (Viti, et al. 2003). The Wnt family proteins play a pivotal role in neural development and in vitro neural differentiation across species (Ciani and Salinas 2005). Wnt proteins have been implicated in forebrain and spinal cord patterning, hippocampal development and neural crest specification (Ciani and Salinas 2005). In vitro, Wnt signaling was required for the maintenance, proliferation and survival of neuroprogenitor cells cultured in neurosphere conditions (Davidson, et al. 2007). With respect to specific genes, Wnt5 was strongly expressed in the EBs and Wnt3 was induced later. Indeed, Wnt5a and Wnt3 are inversely expressed after formation of EBs. It is also noteworthy that the Wnt antagonist, Dickkopf (DKK3) peaked during N3-differentiation when the neurons extrude axons and begin migration out of the neurospheres.
The insulin and MAPK pathways demonstrated a moderate level of expression changes during development. Insulin is a critical requirement for these cultures and it is provided exogenously. Moreover, IGFBP3 and 5 were markedly increased during differentiation. Our data is consistent with the observation that IGFBP family members were altered between fetal neural precursors and mature fetal neurons (Yu et al. 2006). Fibroblastic growth factors, FGF4 and FGF8, are members of the MAPK pathway and they play a pivotal role in neural differentiation (Tarasenko, et al. 2004; Viti et al. 2003; Ye et al. 1998), but their gene expression was not detected on the microarray. FGF2 gene expression was detectable only in ESCs. This may underlie the need to provide exogenous FGF to these cultures. Indeed, exogenous FGF was proposed to play a central role in the expression of c-Myc and maintenance of the cell cycle in neuroprogenitor cells (Karsten et al., 2003). FGF addition was also necessary for the survival of embryonic callosal projection neurons in culture (Catapano et al., 2004). Nonetheless, the FGF receptor-2 was robustly expressed during each stage of differentiation, consistent with an important role of the FGFs for the neuronal phenotype. Thus, there are several pivotal signals in the genetic hierarchy construed from mice for a neural and serotonin phenotype that are expressed early during in vitro differentiation of embryonic stem cells into serotonin neurons such as TGFβ2, TGFβR3, FGFR2, Shh, PTCH, and Wnt5A.
Differences in genetic hierarchy between mice and ESC derived serotonin neurons
Differences were observed in the ontogeny of gene expression across the differentiation stages from that proposed for obtaining the serotonin phenotype in mice. This was deduced by comparing the timing of expression of serotonin specific genes to genes previously described as essential for the serotonin phenotype in mice. The serotonin specific genes were (1) TPH2, the rate-limiting enzyme in serotonin synthesis; (2) SERT, the serotonin reuptake transporter; and (3) 5HT1A, the serotonin autoreceptor. Each gene was induced during N1-proliferation and increased to peak values during N3-differentiation. TPH2 was the most prevalent of the three marker genes.
Nkx2.2 is a transcription factor involved in morphogenesis of the nervous system. Pivotal experiments demonstrated that early Nkx2.2 expression was required for serotonin neuron production and serotonin neurons were lost in Nkx2.2 knock out mice except in the dorsal raphe nucleus (Briscoe, et al. 1999; Pattyn, et al. 2003). However, during in vitro differentiation Nkx2.2 was poorly expressed. Nkx2.3 was modestly induced early and may subserve a similar purpose. Moreover, a recent study found populations of serotonin progenitor cells in mice without Nkx2.2, which is consistent with our microarray data (Jensen, et al. 2008). Down-regulation of Phox2b was required for serotonin neuron production in mice (Briscoe et al. 1999; Pattyn et al. 2003). However, in vitro, the Phox2b gene increased during differentiation, coincident with the serotonin specific genes, to maximal expression in the terminally differentiated serotonin neurons with moderate signal intensity. Thus, downregulation of Phox2b was not necessary in vitro.
The Ascl1 or Mash1 gene plays a role in the neuronal commitment and differentiation and in the generation of olfactory and autonomic neurons and it is considered essential for the birth of serotonin neurons in mice (Pattyn, et al. 2004). It was proposed that the Mash1 gene is activated by Shh in parallel with Nkx2.2 and together they induce Lmx1b and Pet 1 (Fev1 in primates) (Cheng et al. 2003; Pattyn et al. 2004). Mash1 was induced during N2-proliferation, after the large increase in Shh and after induction of the serotonin marker genes in N1-selection stage. Moreover, it peaked during N3-differentiation when Shh declined. Hence, the expression of Mash1 in vitro was delayed compared to in vivo reports and did not correlate with Shh expression, but it may contribute to maintenance of the serotonin phenotype.
Several reports have provided compelling evidence that Lmx1b, a LIM homeodomain-containing gene, operates either concurrently or downstream of Nkx2.2 and is essential for the expression of serotonin (Cheng et al. 2003; Ding, et al. 2003). Lmx1b either activates, or is expressed concurrently with, Pet-1 in mice. In ES cell stages. Lmx1b was induced during N2-proliferaton and peaks during N3-differentiation so it was also delayed relative to the expression of serotonin specific genes. Another gene, Fev1 (Pet1) appears essential for the final production of the serotonin phenotype in mice (Hendricks, et al. 1999; Hendricks et al. 2003). Fev1 was induced at N1 stage and robustly expressed at the N2-proliferation and N3-differentiation stages as determined by qRT-PCR. Thus, Fev1 induction appears to precede Lmx1b induction, which differs from the proposed genetic hierarchy in mice. Moreover, it is important that Fev1 expression coincided with the induction of TPH2, SERT and 5HT1A genes.
The GATA-3 gene product, expressed in parallel with Lmx1b and Pet-1 in mice, plays a role in the organization of the dorsal raphe nucleus although knockouts still had numerous serotonin neurons (van Doorninck, et al. 1999). In vitro, GATA-3 was markedly up-regulated in the EBs in the absence of significant Mash1 or Nkx2.2 expression. EBs slightly resemble early embryos and may be initiating axis formation. However, GATA-3 declined as the EBs were dissociated and differentiation proceeded.
Thus, it appears that the Shh and Wnt pathways were operational very early, thereby committing the stem cells to the serotonergic phenotype. Fev1 was expressed in N1-selection coincident with the emergence of the serotonin specific genes as would be predicted from in vivo studies. Thus, we conclude that Fev1 upon a background of Shh and Wnt pathway activation may be sufficient to produce the serotonin phenotype in culture. However, Mash1 and Nkx2.2 thought to be early in the genetic cascade in vivo, are not expressed significantly until the terminal differentiation stage. Lmx1b was present, but did not achieve significant expression until N2-proliferation, which was after the appearance of the serotonin marker genes. GATA-3, important for organization of the raphe, was robustly induced in EBs and then declined, although the signal intensity remained quite high. Hence, there appear to be some differences in the genetic hierarchy between in vivo and in vitro serotonin differentiation. This is not entirely surprising since the ESC-derived serotonin neurons have no dimensional organization whatsoever.
It is should be noted that we confirmed 5 false negative reports from the microarray. Fev1, Lmx1b, TPH2, SERT and 5HT1 genes were undetectable. Thus, the lack of detection of FGF4, FGF8 and Nkx2.2 must be received with skepticism. In contrast, no false positives were found. If a gene was robustly expressed on the microarray, then it was universally confirmed with qRT-PCR. The probe sets utilized by Affymetrix are usually located in the 3′ UTR. In the qRT-PCR reaction, primer sets generated against the 3′UTR region yielded no amplification of SERT or 5H1A genes (data not shown) whereas primer sets against the coding region detected significant expression (reported).
Comparison to Adult Serotonin Neurons
To determine whether in vitro derived serotonin neurons resembled adult serotonin neurons, a regression analysis was performed on global gene expression of each culture stage versus laser captured serotonin neurons from adult female monkeys. The pools of laser captured serotonin neurons were collected and hybridized to the Rhesus Affymetrix chip in a previous study in which the effect of ovarian hormones on gene expression was examined (Bethea and Reddy 2007). Thus, once the ESC stages were hybridized, it was straightforward to perform the regression analysis. Global gene expression in the two terminal preparations of ESC-derived serotonin neurons correlated best with the adult neurons from the animal that had been ovariectomized and then treated for one month with progesterone. This is notable because progesterone is the only steroid component of the supplement that is used for neuronal differentiation of stem cells (Bottenstein et al. 1980). We intend to manipulate this variable in future experiments.
When we compared gene expression between the progesterone-treated animal and the second preparation of ESC-derived serotonin neurons (passage 39-N3), there were 4115 genes that exhibited a 5-fold or greater difference in expression. KEGG analysis of these genes revealed that gene expression associated with immature and mitotically active neural precursors were over-represented in the ESC-derived serotonin neurons suggesting that these neurons need a longer developmental period in a different, perhaps 3 dimensional environment to achieve a mature serotonin phenotype. Different substrata for culture are under consideration.
Of import was our observation that a majority of developmental genes that peaked in the N3 serotonin neurons were also expressed in laser captured neurons from adult monkeys. For example, FGFR2, PTCH, VEGF, NTRK2 and NCAM were robustly expressed in adult serotonin neurons in a manner similar to the ESC-derived serotonin neurons. Moreover, 2 genes that declined by the N3 stage, namely NANOG and Shh, were low or undetectable in adult serotonin neurons. The exception was CRABP1, or cellular retinoic acid binding protein. This gene was highly expressed in the N3 neurons but very low in adult neurons. The ES cells are highly responsive to retinoic acid, which has been used to obtain neural progeny (Sarkar and Sharma 2002). However, retinoic acid is teratogenic and highly regulated in animals (Sasai 2002). Nonetheless, these data lend credence to the notion developmental gene expression in ESC-derived serotonin neurons was similar to adult serotonin neurons.
In conclusion, expression profiles of serotonin neurons derived from rhesus embryonic stem cells under current laboratory conditions suggest that the serotonin phenotype appears in vitro after induction of GATA3 and coincident with expression of Shh, its receptor (PTCH) and Fev1. Although ESC-derived serotonin neurons are immature in some regards as compared to adult serotonin neurons, there are a number of genes that are consistently expressed in both. Efforts are ongoing to improve the yield and viability of these cultures and to demonstrate physiological characteristics that define serotonin neurons.
Acknowledgements
We are deeply grateful to the dedicated staff of the Division of Animal Resources including the staff of the Departments of Surgery and Pathology for their expertise and helpfulness in all aspects of monkey management and to the dedicated staff of the Assisted Reproductive Technology Core for the generation of the MEF and embryonic stem cell cultures. The staff of the OHSU Gene Microarray Shared Resource was essential for this study.
Supported by NIH grants: MH73564 and MH62677 to CLB, U54 contraceptive Center Grant HD 18185, and RR000163 for the operation of ONPRC
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Aigner L, Bogdahn U. TGF-beta in neural stem cells and in tumors of the central nervous system. Cell Tissue Res. 2008;331:225–241. doi: 10.1007/s00441-007-0466-7. [DOI] [PubMed] [Google Scholar]
- Bethea C, Reddy A. Effect of ovarian hormones on survival genes in laser captured serotonin neurons from macaques. J Neurochem. 2007;105:1129–1143. doi: 10.1111/j.1471-4159.2008.05213.x. [DOI] [PubMed] [Google Scholar]
- Bethea CL, Lu NZ, Gundlah C, Streicher JM. Diverse actions of ovarian steroids in the serotonin neural system. Frontiers in Neuroendocrinology. 2002;23:41–100. doi: 10.1006/frne.2001.0225. [DOI] [PubMed] [Google Scholar]
- Bottenstein JE, Skaper SD, Varon SS, Sato GH. Selective survival of neurons from chick embryo sensory ganglionic dissociates utilizing serum-free supplemented medium. Exp Cell Res. 1980;125:183–190. doi: 10.1016/0014-4827(80)90202-5. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Sussel L, Serup P, Hartigan-O’Connor D, Jessell TM, Rubenstein JL, Ericson J. Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature. 1999;398:622–627. doi: 10.1038/19315. [DOI] [PubMed] [Google Scholar]
- Cai Y, Wu P, Ozen M, Yu Y, Wang J, Ittmann M, Liu M. Gene expression profiling and analysis of signaling pathways involved in priming and differentiation of human neural stem cells. Neuroscience. 2006;138:133–148. doi: 10.1016/j.neuroscience.2005.11.041. [DOI] [PubMed] [Google Scholar]
- Catapano LA, Arlotta P, Cage TA, Macklis JD. Stage-specific and opposing roles of BDNF, NT-3 and bFGF in differentiation of purified callosal projection neurons toward cellular repair of complex circuitry. Eur J Neurosci. 2004;19:2421–2434. doi: 10.1111/j.0953-816X.2004.03303.x. [DOI] [PubMed] [Google Scholar]
- Chen L, Khillan JS. Promotion of feeder independent self-renewal of embryonic stem cells by retinol (Vitamin A) Stem Cells. 2008;26:1858–1864. doi: 10.1634/stemcells.2008-0050. [DOI] [PubMed] [Google Scholar]
- Cheng L, Chen CL, Luo P, Tan M, Qiu M, Johnson R, Ma Q. Lmx1b, Pet-1, and Nkx2.2 coordinately specify serotonergic neurotransmitter phenotype. J Neuroscience. 2003;23:9961–9967. doi: 10.1523/JNEUROSCI.23-31-09961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciani L, Salinas PC. WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nat Rev Neurosci. 2005;6:351–362. doi: 10.1038/nrn1665. [DOI] [PubMed] [Google Scholar]
- Cunningham MG, Donalds RA, Carlezon WA, Jr., Hong S, Kim DS, Kim DW, Kim KS. Antidepressant effect of stem cell-derived monoaminergic grafts. NeuroReport. 2007;18:1663–1667. doi: 10.1097/WNR.0b013e3282f0eb1c. [DOI] [PubMed] [Google Scholar]
- Davidson KC, Jamshidi P, Daly R, Hearn MT, Pera MF, Dottori M. Wnt3a regulates survival, expansion, and maintenance of neural progenitors derived from human embryonic stem cells. Mol Cell Neurosci. 2007;36:408–415. doi: 10.1016/j.mcn.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Ding YQ, Marklund U, Yuan W, Yin J, Wegman L, Ericson J, Deneris E, Johnson RL, Chen ZF. Lmx1b is essential for the development of serotonergic neurons. Nat Neurosci. 2003;6:933–938. doi: 10.1038/nn1104. [DOI] [PubMed] [Google Scholar]
- Falk S, Wurdak H, Ittner LM, Ille F, Sumara G, Schmid MT, Draganova K, Lang KS, Paratore C, Leveen P, et al. Brain area-specific effect of TGF-beta signaling on Wnt-dependent neural stem cell expansion. Cell Stem Cell. 2008;2:472–483. doi: 10.1016/j.stem.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Goridis C, Rohrer H. Specification of catecholaminergic and serotonergic neurons. Nat Rev Neurosci. 2002;3:531–541. doi: 10.1038/nrn871. [DOI] [PubMed] [Google Scholar]
- Hendricks T, Francis N, Fyodorov D, Deneris ES. The ETS domain factor Pet-1 is an early and precise marker of central serotonin neurons and interacts with a conserved element in serotonergic genes. J Neuroscience. 1999;19:10348–10356. doi: 10.1523/JNEUROSCI.19-23-10348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks TJ, Fyodorov DV, Wegman LJ, Lelutiu NB, Pehek EA, Yamamoto B, Silver J, Weeber EJ, Sweatt JD, Deneris ES. Pet-1 ETS gene plays a critical role in 5-HT neuron development and is required for normal anxiety-like and aggressive behavior. Neuron. 2003;37:233–247. doi: 10.1016/s0896-6273(02)01167-4. [DOI] [PubMed] [Google Scholar]
- Jensen P, Farago AF, Awatramani RB, Scott MM, Deneris ES, Dymecki SM. Redefining the serotonergic system by genetic lineage. Nat Neurosci. 2008;11:417–419. doi: 10.1038/nn2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallur T, Gisler R, Lindvall O, Kokaia Z. Pax6 promotes neurogenesis in human neural stem cells. Mol Cell Neurosci. 2008;38:616–628. doi: 10.1016/j.mcn.2008.05.010. [DOI] [PubMed] [Google Scholar]
- Karsten SL, Kudo LC, Jackson R, Sabatti C, Kornblum HI, Geschwind DH. Global analysis of gene expression in neural progenitors reveals specific cell cycle, signaling, and metabolic networks. Dev Biol. 2003;261:165–182. doi: 10.1016/s0012-1606(03)00274-4. [DOI] [PubMed] [Google Scholar]
- Kelly DL, Rizzino A. DNA microarray analyses of genes regulated during the differentiation of embryonic stem cells. Mol Reprod Develop. 2000;56:113–123. doi: 10.1002/(SICI)1098-2795(200006)56:2<113::AID-MRD1>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Kuo HC, Pau KY, Yeoman RR, Mitalipov SM, Okano H, Wolf DP. Differentiation of monkey embryonic stem cells into neural lineages. Biol Reproduction. 2003;68:1727–1735. doi: 10.1095/biolreprod.102.012195. [DOI] [PubMed] [Google Scholar]
- Le Bras B, Barallobre MJ, Homman-Ludiye J, Ny A, Wyns S, Tammela T, Haiko P, Karkkainen MJ, Yuan L, Muriel MP, et al. VEGF-C is a trophic factor for neural progenitors in the vertebrate embryonic brain. Nat Neurosci. 2006;9:340–348. doi: 10.1038/nn1646. [DOI] [PubMed] [Google Scholar]
- Lee SH, Lumelsky N, Studer L, Auerbach JM, McKay RD. Efficient generation of midbrain and hindbrain neurons from mouse embryonic stem cells. Nat Biotechnol. 2000;18(6):675–679. doi: 10.1038/76536. [DOI] [PubMed] [Google Scholar]
- Lidov HG, Molliver ME. Immunohistochemical study of the development of serotonergic neurons in the rat CNS. Brain Res Bull. 1982;9:559–604. doi: 10.1016/0361-9230(82)90164-2. [DOI] [PubMed] [Google Scholar]
- Mitalipov S, Kuo HC, Byrne J, Clepper L, Meisner L, Johnson J, Zeier R, Wolf D. Isolation and characterization of novel rhesus monkey embryonic stem cell lines. Stem Cells. 2006;24:2177–2186. doi: 10.1634/stemcells.2006-0125. [DOI] [PubMed] [Google Scholar]
- Ostenfeld T, Joly E, Tai YT, Peters A, Caldwell M, Jauniaux E, Svendsen CN. Regional specification of rodent and human neurospheres. Dev Brain Res. 2002;134:43–55. doi: 10.1016/s0165-3806(01)00291-7. [DOI] [PubMed] [Google Scholar]
- Park SM, Jung JS, Jang MS, Kang KS, Kang SK. Transforming growth factor-beta1 regulates the fate of cultured spinal cord-derived neural progenitor cells. Cell Prolif. 2008;41:248–264. doi: 10.1111/j.1365-2184.2008.00514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn A, Simplicio N, van Doorninck JH, Goridis C, Guillemot F, Brunet JF. Ascl1/Mash1 is required for the development of central serotonergic neurons. Nat Neurosci. 2004;7:589–595. doi: 10.1038/nn1247. [DOI] [PubMed] [Google Scholar]
- Pattyn A, Vallstedt A, Dias JM, Samad OA, Krumlauf R, Rijli FM, Brunet JF, Ericson J. Coordinated temporal and spatial control of motor neuron and serotonergic neuron generation from a common pool of CNS progenitors. Genes and Development. 2003;17:729–737. doi: 10.1101/gad.255803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecins-Thompson M, Bethea CL. Ovarian steroid regulation of 5HT1A autoreceptor messenger ribonucleic acid expression in the dorsal raphe of rhesus macaques. Neuroscience. 1998;89:267–277. doi: 10.1016/s0306-4522(98)00326-1. [DOI] [PubMed] [Google Scholar]
- Pecins-Thompson M, Brown NA, Bethea CL. Regulation of serotonin re-uptake transporter mRNA expression by ovarian steroids in rhesus macaques. Mol Brain Res. 1998;53:120–129. doi: 10.1016/s0169-328x(97)00286-6. [DOI] [PubMed] [Google Scholar]
- Ren-Patterson RF, Kim DK, Zheng X, Sherrill S, Huang SJ, Tolliver T, Murphy DL. Serotonergic-like progenitor cells propagated from neural stem cells in vitro: survival with SERT protein expression following implantation into brains of mice lacking SERT. Faseb J. 2005;19:1537–1539. doi: 10.1096/fj.04-3657fje. [DOI] [PubMed] [Google Scholar]
- Salli U, Reddy AP, Salli N, Lu NZ, Kuo H-C, Pau FK-Y, Wolf DP, Bethea CL. Serotonin neurons derived from rhesus monkey embryonic stem cells: similarities to CNS serotonin neurons. Exper Neurology. 2004;188:351–364. doi: 10.1016/j.expneurol.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Sarkar SA, Sharma RP. Modulation of c-myc, max, and mad gene expression during neural differentiation of embryonic stems cells by all-trans-retinoic acid. Gene Expression. 2002;10(3):125–135. [PMC free article] [PubMed] [Google Scholar]
- Sasai Y. Generation of dopaminergic neurons from embryonic stem cells. J Neurol. 2002;249(Suppl 2):S41–S44. doi: 10.1007/s00415-002-1208-0. [DOI] [PubMed] [Google Scholar]
- Stone DM, Hynes M, Armanini M, Swanson TA, Gu Q, Johnson RL, Scott MP, Pennica D, Goddard A, Phillips H, et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- Tarasenko YI, Yu Y, Jordan PM, Bottenstein J, Wu P. Effect of growth factors on proliferation and phenotypic differentiation of human fetal neural stem cells. J Neurosci Res. 2004;78:625–636. doi: 10.1002/jnr.20316. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Kalishman J, Golos TG, Durning M, Harris CP, Becker RA, Hearn JP. Isolation of a primate embryonic stem cell line. Proc Natl Acad Sci U S A. 1995;92:7844–7848. doi: 10.1073/pnas.92.17.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorninck JH, van Der Wees J, Karis A, Goedknegt E, Engel JD, Coesmans M, Rutteman M, Grosveld F, De Zeeuw CI. GATA-3 is involved in the development of serotonergic neurons in the caudal raphe nuclei. J Neuroscience. 1999;19:RC12. doi: 10.1523/JNEUROSCI.19-12-j0002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazey EM, Chen K, Hughes SM, Connor B. Transplanted adult neural progenitor cells survive, differentiate and reduce motor function impairment in a rodent model of Huntington’s disease. Exp Neurol. 2006;199:384–396. doi: 10.1016/j.expneurol.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Viti J, Gulacsi A, Lillien L. Wnt regulation of progenitor maturation in the cortex depends on Shh or fibroblast growth factor 2. J Neuroscience. 2003;23:5919–5927. doi: 10.1523/JNEUROSCI.23-13-05919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace JA, Lauder JM. Development of the serotonergic system in the rat embryo: an immunocytochemical study. Brain Res Bull. 1983;10:459–479. doi: 10.1016/0361-9230(83)90144-2. [DOI] [PubMed] [Google Scholar]
- Ye W, Shimamura K, Rubenstein JL, Hynes MA, Rosenthal A. FGF and Shh signals control dopaminergic and serotonergic cell fate in the anterior neural plate. Cell. 1998;93:755–766. doi: 10.1016/s0092-8674(00)81437-3. [DOI] [PubMed] [Google Scholar]
- Yu S, Zhang JZ, Xu Q. Genes associated with neuronal differentiation of precursors from human brain. Neuroscience. 2006;141:817–825. doi: 10.1016/j.neuroscience.2006.02.080. [DOI] [PubMed] [Google Scholar]