Abstract
Isolated immunoglobulin CH2 domains were proposed as scaffolds for selection of binders with potential effector functions. We tested the feasibility of this approach by constructing a large (size 5×1010) library where all amino acids in two loops (BC and FG) were mutated to four residues (Y, A, D or S). Three binders were selected from this library by panning against a gp120-CD4 complex. The strongest binder, m1a1, recognized specifically a highly conserved CD4i epitope and inhibited to various extents 7 out of 9 HIV-1 isolates from different clades. The loop BC and the conformational state of the scaffold are critical for its binding. These results provide a proof of concept for the potential of CH2 as a scaffold for construction of libraries containing potentially useful binders. The newly identified HIV-1 inhibitors could be further improved to candidate therapeutics and/or used as research reagents for exploration of conserved gp120 structures.
Keywords: CH2, scaffold, antibody library, HIV, phage display, human
Introduction
Monoclonal antibodies are well-established therapeutics and invaluable tools for research. Most of these antibodies are full-size immunoglobulins (Igs) but antibody fragments of smaller size including Fabs (~60 kDa), single chain Fv fragments (scFvs) (20~30 kDa) and domain antibodies (dAbs) (12~15 kDa) are being increasingly used [1]. These antibody fragments can be selected and produced with relative ease. They penetrate solid tissues including solid tumors better and can bind to structures that are sterically constrained. A large amount of work has been aimed at developing novel human or non-human scaffolds of small size and high stability for use as therapeutics and diagnostics [2, 3].
Recently, isolated immunoglobulin constant CH2 (CH3 for IgE and IgM) domains were proposed as scaffolds for construction of libraries containing diverse binders that could confer some effector functions [4]. Isolated human CH2 is independently folded and can be expressed and purified from bacterial expression systems as a soluble, monomeric protein at a high level [4, 5]. It exhibits thermostability that is comparable to a dAb and can be further improved [6]. Importantly, as a fragment in all IgGs, which are at high concentrations in blood, γ1 CH2-based therapeutics are likely to be well tolerated in concentrations needed for achieving therapeutic efficacies. Furthermore, CH2 binders can be engineered so as to retain some of the effector functions that are possessed only by IgGs and not by other scaffolds [7]. Based on its similarities to a variable domain, one could hypothesize that the CH2 framework could sustain diversification of the loops for the development of libraries containing diverse binders. Here we provide evidence supporting this hypothesis by demonstrating that antigen specific binders based on the CH2 scaffold with synthetic loops can be obtained and that these binders are functional, i.e. capable of neutralizing HIV-1 isolates from different clades.
Materials and Methods
Primers, peptide and proteins
All the primers used in this study were purchased from Invitrogen (Carlsbad, CA). The biotin labeled peptide was from Sigma (St Louis, MO). Bal gp120-CD4 was provided by Tim Fouts (University of Maryland, Baltimore, MD) and other recombinant proteins (gp120s and gp140s) were provided by Christopher Broder (USUHS, Bethesda, MD). SCD4 was obtained through AIDS research and reagent program.
Library construction
Overlapping PCR was used to introduce mutations to the loops BC and FG to generate the library (Fig. 1). Degenerate primers containing codon KMT were utilized to introduce desired combinations of four residues to each location indicated (Fig.1). PCR fragments were subjected to SfiI digestion and ligated to the pCOM3X (provided by Dennis Burton, Scripps Institute, La Jolla, CAL). The ligated product was desalted and transformed to the electrocompetent TG1 cells using an electroporator (Bio-Rad, Hercules, CA).
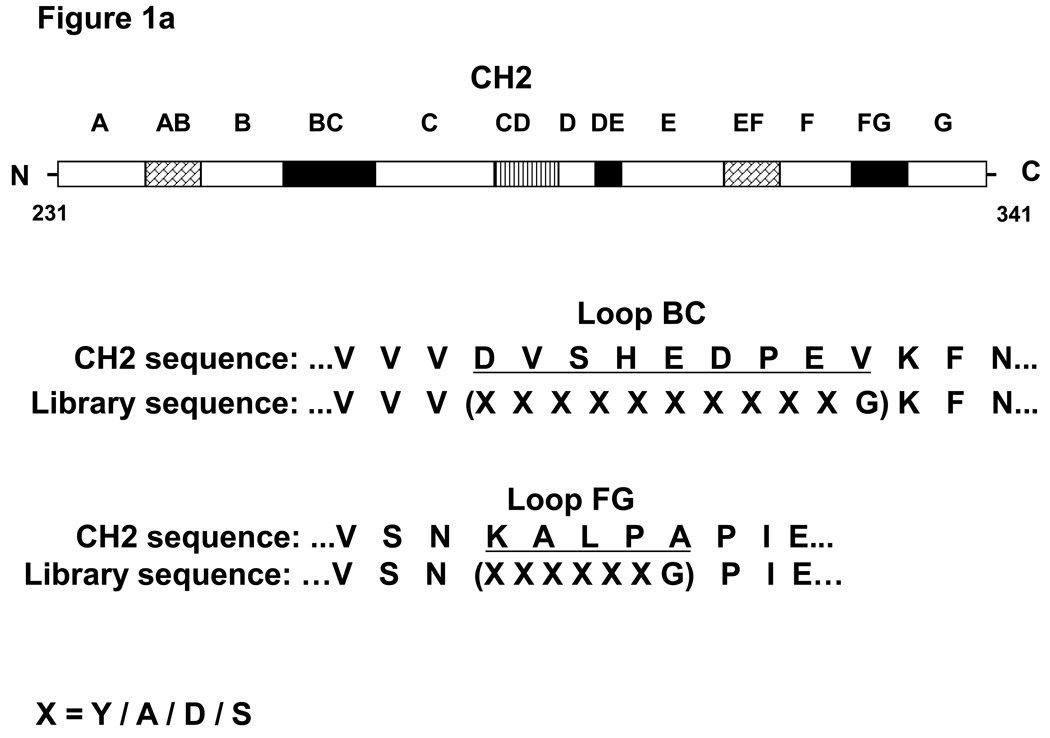
Figure 1.
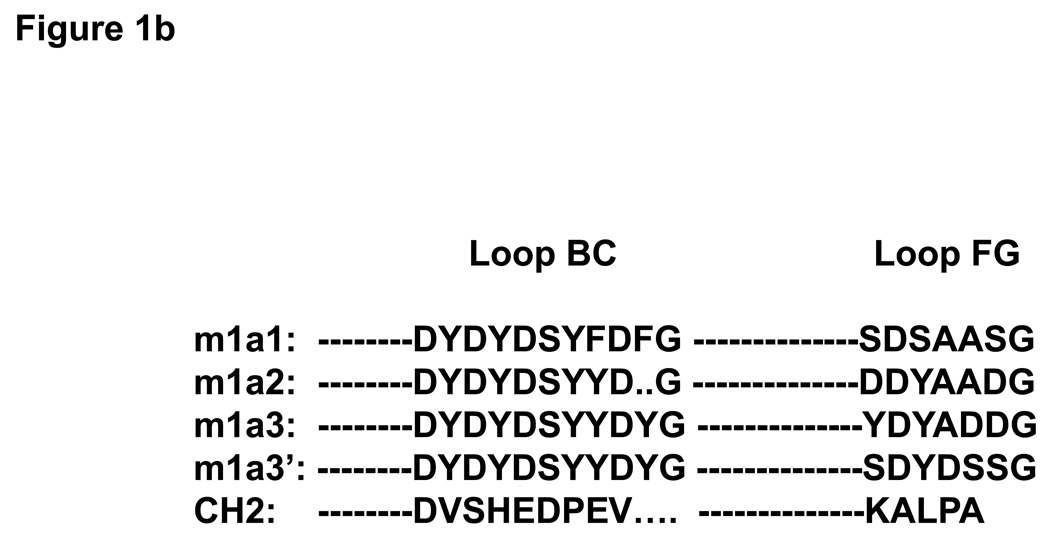
(a) Experiment design. The filled rectangles represent the loops (BC, DE and FG) which are on the same side of CH2. The shaded rectangles represent the two helices (AB and EF) and the loop (CD) which are on the opposite side of CH2. The empty rectangles labeled with letters A through G represent the seven β-strands. The numbers 231 and 341 denote the starting and ending residues corresponding to the CH2 in the context of the IgG1. Fragments containing loops BC and FG are shown below the CH2 representation where their sequences are underlined. The mutations introduced in the loops are shown in parentheses. (b) Sequence comparison among the isolated binders and WT CH2. M1a3’ has the same loop BC as m1a3 but different loop FG.
Similar overlapping PCR method was employed to graft the loop BC sequence of m1a1 to N terminus, loop DE, and loop FG of wild type CH2 respectively.
Panning
Bal gp120-CD4 was coated directly to Maxisorp plates (Nunc, Denmark) in PBS buffer at 4°C, o/n as the panning antigen. Monoclonal ELISA was then used to select for positive clones after 5 rounds of panning. Only clones displaying an ELISA signal that was at least 10 folds above the background signal were selected for further analysis.
CH2 expression and refolding
E.coli strain HB2151 was used for protein expression. Fresh transformant was inoculated into 2YT with 100 units of amp and incubated at 37°C with shaking. When the OD600 reached 0.5, IPTG was added to 1 mM and the culture was continued for another 3–5 hours. Cells were collected, lysed with polymyxin B (Sigma, St Louis) in PBS, and the supernatant was subjected to the Ni-NTA agarose bead (Qiagen, Hilden, Germany) purification. The pellet was re-suspended in a buffer containing 25 mM Tris.HCl, pH 8.0, 6 M Urea, 0.5 M NaCl, and subjected to a brief sonication. The supernatant was collected by centrifugation and subjected to the Ni-NTA agarose bead purification. CH2 obtained from the pellet was dialyzed against PBS and filtered through a 0.2 µm low protein binding filter (Pal, Ann Arbor, MI).
ELISA
Protein antigens were diluted in the PBS buffer in concentrations ranging from 1–4 µg/ml and coated to the 96 well plate at 4°C for overnight. The mouse-anti-His-HRP (Qiagen) was used to detect the His tag at the C terminus of each of the CH2 clones binding to the antigens in most of the ELISA unless indicated otherwise. ABTS (Roche, Germany) was then added to each well and OD 405 was taken 5–10 minutes afterward.
Pseudovirus neutralization assay
HIV Env pseudotyped virus preparation and neutralization was performed essentially as previously described [8].
Structure modeling
For m1a1 and germline X5 modeling, coordinates were generated by replacing and/or inserting side chain residues based on CH2 crystal structure (PDB 3DJ9) and the X5 (PDB 1RHH), respectively. Changes were made only for residues that are in the primary binding sites, loop BC and HCDR3, respectively. Swiss-model was used to generate the coordinate [9]. VMD software was used to calculate structural alignment between the CH2 and m1a1 and to present the graphics [10].
Results
Construction of a large human CH2-based library and selection of specific binders
We mutated loops BC and FG because they are the longest loops on the same side of the molecule (see (Fig. 1a)). We selected four residues, A, Y, D, and S, which frequently occur in CDRs, to randomly replace all BC and FG residues. Another residue, G, was added to the C-terminal end of each loop to increase flexibility (Fig. 1a). It has been previously observed that these four residues are sufficient to form a specific binding surface on different frameworks [11, 12]. The calculated diversity of this library is 416 = 4.3 ×109. The number of clones from the final electroporation was 5 × 1010, which would also include possible PCR mutants. More than 80% of randomly selected clones were with correct reading frames.
To test the library and select potentially useful binders we used an HIV-1 envelope glycoprotein, gp120, from the Bal isolate fused with a two-domain CD4 (gp120Bal-CD4) as a panning antigen. After five rounds of panning 200 clones were screened by phage ELISA and 15 clones that exhibited the highest level of binding to the screening antigen were isolated for further characterization. Three clones, m1a1, m1a2 and m1a3, dominated; they were represented by 7, 5 and 2 (out of 15) sequences, respectively, suggesting a specific enrichment. They have similar loop BC sequences but different loop FG (Figure 1b). The dominant clones, m1a1 and m1a2, have residues in loop BC (two Fs in loop BC, and deletion before G, respectively) which were apparently due to PCR errors.
CH2 binders recognize specifically a highly conserved CD4i epitope
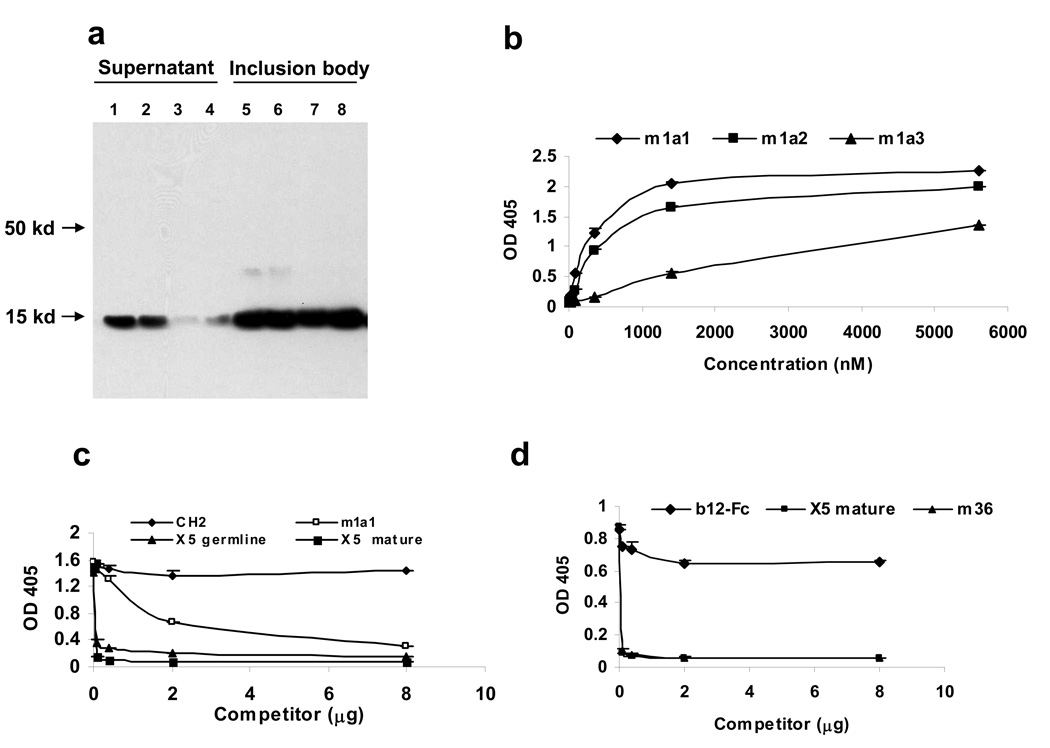
We characterized the three selected mutants by first expressing them as soluble proteins. Most of the expressed proteins were found in inclusion bodies (Fig. 2a) and were refolded at an average yield of 10–30 mg/L of bacterial culture. The purified CH2 binders bound to the panning antigen (gp120-CD4) specifically as measured by ELISA with EC50s ranging from 500 nM (m1a1 and m1a2) to low µM (m1a3) (Fig. 2b). Similar results were obtained for CH2 binders purified from the supernatant without refolding (data not shown). These results suggest that m1a1,2,3 retained their binding activity in soluble (not phage-displayed) form and that refolding from inclusion bodies did not affect it. To characterize the epitope of the best binder m1a1 and to confirm the specificity, we performed competition ELISA using scFv X5 and the recently reported domain antibody m36 [13]. M1a1 binding to Bal gp120-CD4 was not inhibited by high concentrations of WT CH2, inhibited modestly by unlabeled m1a1 and completely eliminated by X5 (Fig. 2c). Interestingly, germline X5, which has overlapping epitope with that of X5 (Xiao and Dimitrov, unpublished data) failed to completely eliminate m1a1 binding as did the mature X5 (Fig. 2c). M1a1 binding can also be abolished by m36, but not by the CD4bs antibody b12 (Figure 2d). Collectively, these data suggest that m1a1 binds specifically to a highly conserved CD4i epitope.
Figure 2.
Characterization of the CH2-based binders. (a) The purified CH2 binders were analyzed by Western blot. Lanes 1 through 4 show the amounts of m1a1, m1a2, m1a3 and m1a3’, respectively, purified from the supernatant; Lanes 5–8 show the same CH2 binders purified from the inclusion bodies. (b) Binding of m1a1-3 to Balgp120-CD4 as measured by ELISA. (c) Purified m1a1 was labeled with biotin and used in a competition ELISA for binding to Balgp120-CD4. Biotinylated m1a1 at 2 μg was mixed with the indicated amounts of unlabeled WT CH2, m1a1, scFv mature or germline X5 and added to each well. The bound, biotin-labeled m1a1 was detected by using streptavidin-HRP. (d) Similar competition ELISA with scFv-b12-Fc, m36 and scFv mature x5 as the competitors.
Loop BC determines the binding specificity of m1a1 and m1a2
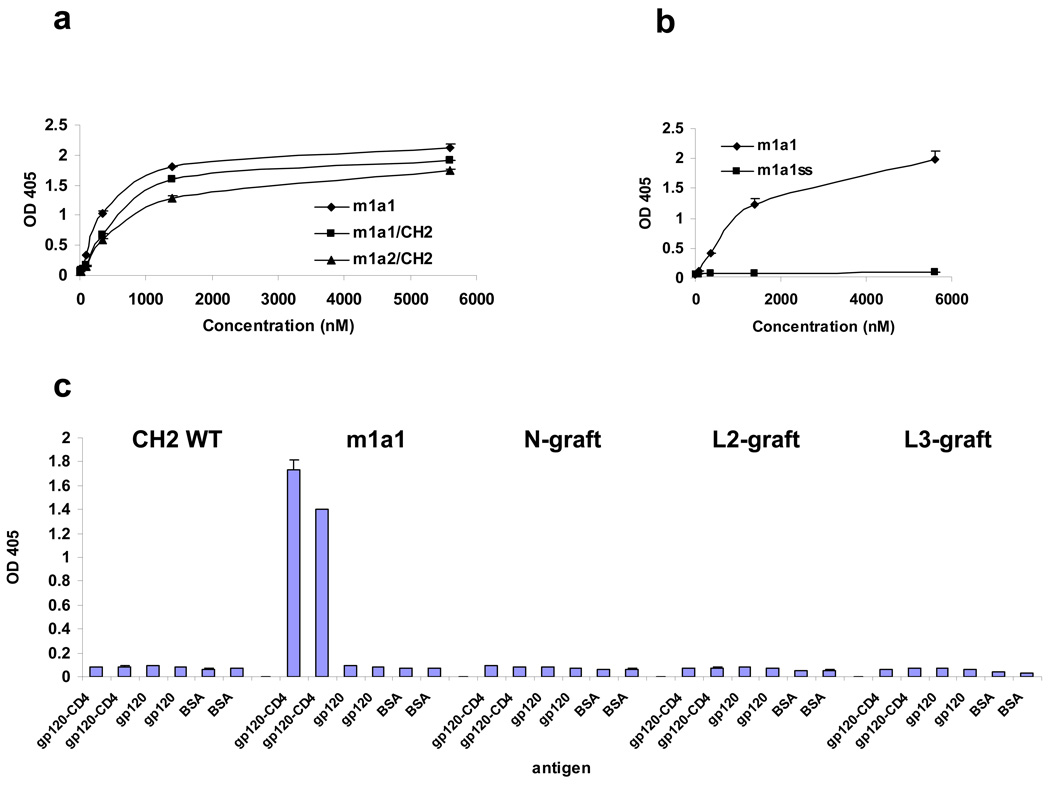
To determine the binding paratopes, we generated two hybrid clones, m1a1/CH2 and m1a2/CH2 where loops BC from m1a1 and m1a2, respectively, were grafted onto CH2 replacing its original loop BC. These hybrid antibodies expressed well and bound to the gp120-CD4 with similar affinity compared to m1a1 (Fig. 3a), indicating that loops FG are not essential for binding. To find out whether the scaffold as a whole is required for binding we tested the m1a1 loop BC in isolation – as a synthetic peptide (DYDYDSYFDFG). The biotin labeled peptide did not bind (data not shown). We also tested the effect of possible conformational changes in the scaffold on binding by creating an additional disulfide bond between strands A and G. We have shown previously [6] that such an S-S bond increases significantly the CH2 stability and does not affect significantly the mobility and the microenvironment of any CH2 residue as measured by NMR. The resulting antibody, m1a1ss, expressed well but did not bind either (Fig. 3b).
Figure 3.
Loop BC sequence is critical for binding. (a) Two of the dominant clones, m1a1,2 as well as the two hybrids, m1a1,2/CH2, were expressed, purified and used in ELISA against Balgp120-CD4. (b) The CH2 framework provides critical structural support for loop BC. The dominant clone m1a1 and its mutant, m1a1ss, carrying an additional disulfide bond were expressed, purified and used in the ELISA. (c) Loop BC location is critical for the binding. The wild type CH2, m1a1, as well as CH2 carrying loop BC of m1a1 in different positions including N terminus (N), loop DE (DE), and loop FG (FG) were expressed, purified and used in ELISA against Bal gp120, Bal gp120-CD4, and BSA. For each clone indicated at the top, the left two bars show their respective binding at 6 (left) and 1.2 (right) µM to bal gp120-CD4, and the middle two bars show their respective binding to bal gp120, and right two bars show their respective binding to BSA at the same two concentrations.
Finally we tested if the location of the binding sequence on the CH2 is important. In one of the constructs, loop BC of m1a1 was grafted to the N terminus of the wild type CH2. The grafted m1a1 sequence was flanked by a pair of cysteines. In the other two constructs, loop DE and FG of the wild type CH2 were replaced by the loop BC sequence of m1a1, respectively. All three grafted constructs expressed well and were comparable to or better than m1a1 (data not shown). When tested in ELISA, only m1a1 showed specific binding to gp120-CD4 but not to gp120 or BSA. The three grafted clones and the wild type CH2 did not show any binding activities against any of the antigens (Fig. 3c). In addition to further confirming the highly specific binding of m1a1 to the CD4i epitope, these data suggest that the location is critical for the binding activity of m1a1.
Neutralization of HIV-1 by m1a1 and m1a2
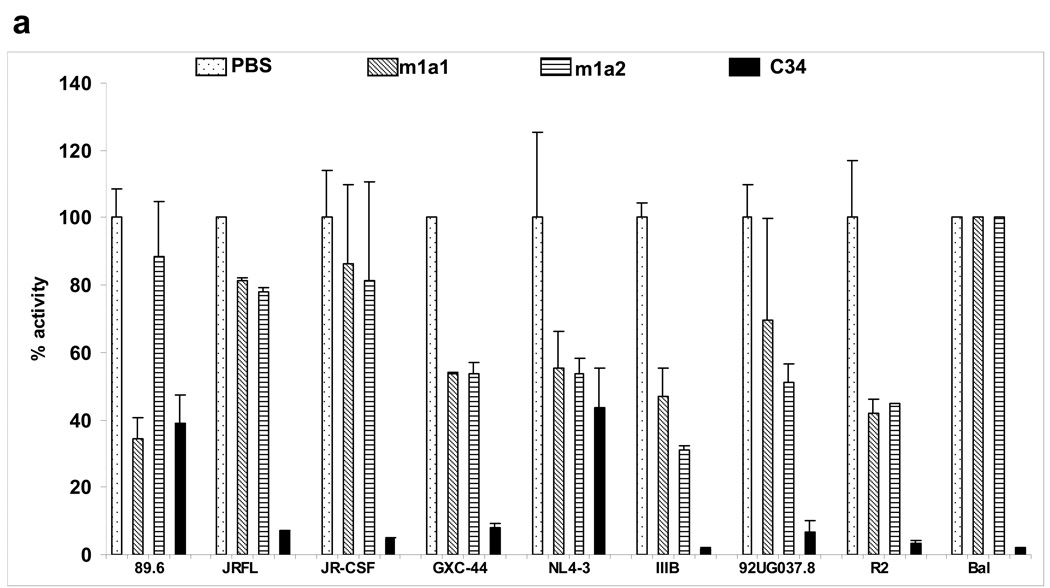
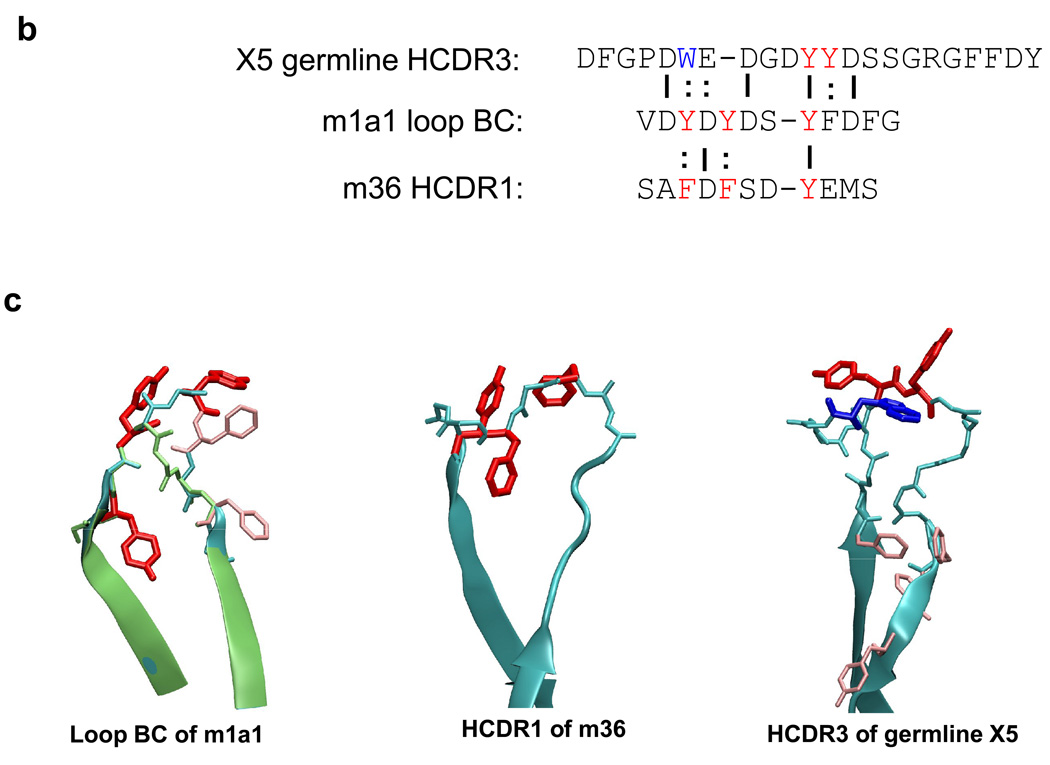
To evaluate the neutralizing activity of m1a1,2 we used a cell line based pseudovirus assay and a panel of nine HIV-1 isolates. Seven of these isolates were inhibited to certain degree by one or both antibodies (Fig. 4a). The two antibodies differentially inhibited two isolates (89.6 and IIIB) and to about the same degree five other isolates (Fig. 4a). As expected from their relatively modest binding affinity, their potency was relatively low compared to the highly potent inhibitor C34. These results provide proof of concept that functional binders can be selected from libraries based on the CH2 scaffold. To explore the mechanistic basis for the similar epitope between m1a1 and the two CD4i antibodies and its neutralizing abilities, we first compared the sequence of loop BC of m1a1 with the sequences of the HCDR3 of X5 and HCDR1 of m36. Tyrosine residues flanked by acidic residues have been found to be directly involved in interacting with residues from gp120 CD4i epitopes. This prominent feature is evident in the loop BC of m1a1 (Fig. 4b). In addition, the positioning of the aromatic residues in the loop BC of m1a1 is quite similar to that of the m36 HCDR1. The third tyrosine in the loop BC is conserved in both m36 and germline X5, suggesting significant contribution of this tyrosine in gp120 interaction. The germline X5 has a slightly increased affinity to bal gp120-CD4 than the mature X5 that has a serine residue instead of tyrosine in this position (Xiao and Dimitrov, unpublished dada), supporting the above observation. Structure modeling based on the crystal structure of wild type CH2 demonstrated that the loop BC of m1a1 is larger than that of the wild type and has the aromatic residues at the tip of the loop (Fig. 4c).
Figure 4.
Inhibition of HIV-1 pseudovirus infection by CH2-based binders. (a) The two binders, m1a1 and m1a2, were used at a fixed concentration of 100 µg/ml against a panel of nine viruses pseudotyped with envelope glycoproteins of HIV-1 isolates from different clades. The peptide C34 at concentration of 6 µg/ml was used as a positive control. (b) Sequence comparison between m1a1 loop BC, HCDR3 of X5, and HCDR1 of m36. (c) Structure modeling of loop BC of m1a1, HCDR1 of m36, and HCDR3 of germline X5. In (c), red or blue highlighted-aromatic residues are those high-lighted in the same color in (b), and other aromatics in sequence (b) are shown by pink
Discussion
In this study we evaluated the feasibility of using CH2 as a novel scaffold for the development of antigen specific binders that could be useful as therapeutics. The successful construction of a large size (5×1010) library and selection of a number of binders to gp120 that could neutralize HIV-1 albeit at relatively modest potency suggests that CH2 is a promising scaffold for constructions of libraries of binders. The antigen specific binders described here also provide hope that a new class of synthetic therapeutics may be developed in the fight against AIDS and other diseases.
A naïve library based on Fab scaffold should contain typically more than 1010 clones in order to select binders against a broad spectrum of antigens with good affinity [14]. The CH2-based library described here has a calculated diversity of 109 and is appropriate for an initial test. Indeed, we selected several binders specific for HIV. We anticipate that binders against some antigens but not others may be obtained from this library. This has been demonstrated for Fab as a scaffold where certain combinations of amino acids mutated during the library construction resulted in binders to some antigens but not to others [12]. The affinities of the selected binders are modest but typical of those selected from the first generation of scaffolds without further improvement [11].
The relatively simple structures and the consequent ease of manipulation and productions are often cited as major reasons for the development of libraries based on small scaffolds [2]. CH2 meets such criteria. Even though the three binders selected against HIV have poor soluble expression, similar phenomenon was observed for the first generation of monobody binders [11]. Additionally, the recovery of the CH2 binders was very efficient from the inclusion bodies. Many therapeutics and vaccines in clinical trials have been produced from inclusion bodies after fermentation and CH2 binders are not expected to face any significant challenges in this aspect [15, 16].
Our data indicate that loop BC is critical for binding. The lack of participation of other loops could reflect the nature of the structure recognized by these clones. It has been demonstrated that antibodies recognizing epitopes defined by X5 or 17b on the HIV-1 gp120 often have a long, protruding CDR3s of the heavy chain extending into a sterically restrained crevice [17–19]. Other CDRs play limited roles in the direct contact with the gp120. The binding potentials of loop DE and FG are to be further explored as they are involved in interactions with multiple molecules in the context of Ig to facilitate various effector functions [20].
To characterize the epitope of the best binder, m1a1, we found that it competed with two other antibodies, X5 and m36, which are representatives of so-called CD4 induced (CD4i) antibodies [21, 22]. CD4i epitopes, especially those of X5 and m36, are highly conserved among various HIV-1 isolates, transiently exposed only upon CD4 engagement and are better accessed by antibody fragments as scFv or Fabs than by full-size antibodies. The transient nature of this epitope requires that CD4i antibodies have high affinity in order to exhibit significant neutralizing activity because of the short half-life of the virus entry intermediates. This might explain the lack of neutralizing ability of CH2 binders for the Bal isolate. Even though they bound Bal gp120-CD4, their affinity was too low. However, in some isolates whose cellular entry is CD4 independent, this epitope is already exposed. Among these are IIIBx and R2 [23, 24]. Indeed R2 was inhibited in the pseudovirus assay (Fig. 4a). The difference in neutralization against various isolates by CH2 binders is not unusual as X5 also neutralized some but not others [25]. The conserved nature of this epitope either as induced by CD4 or in the absence of CD4 for some isolates indicates its potential as a target for therapeutics and as a potentially helpful tool for vaccine design. The newly identified CH2 binders provide a unique platform for studying the detailed information of the paratopes making efficient contact with the epitope and helping to design new classes of therapeutics and vaccines.
Acknowledgements
We thank Christopher Broder, Gerald Quinnan, Dennis Burton and Tim Fouts for providing reagents, and the members of our group Weizao Chen, Zhongyu Zhu, Rui Gong, John Owens and Meiyun Zhang for help. This work was supported by the Intramural AIDS Targeted Antiviral Program (IATAP), National Institutes of Health (NIH) and by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dimitrov DS, Marks JD. Therapeutic antibodies: Current state and future trends – is a paradigm change coming soon? Meth Mol Biol. 2009;525:1–27. doi: 10.1007/978-1-59745-554-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skerra A. Alternative non-antibody scaffolds for molecular recognition. Curr.Opin.Biotechnol. 2007;18:295–304. doi: 10.1016/j.copbio.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat.Biotechnol. 2005;23:1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- 4.Dimitrov DS. Engineered CH2 domains (nanoantibodies) mAbs. 2009;1:26–28. doi: 10.4161/mabs.1.1.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vu B, Walsh J, Dimitrov DS, Ishima R. Dynamics of antibody domains studied by solution NMR. Meth Mol Biol. 2009;525:533–544. doi: 10.1007/978-1-59745-554-1_29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gong R, Vu BK, Feng Y, Prieto DA, Dyba MA, Walsh JD, Prabakaran P, Veenstra TD, Tarasov SG, Ishima R, Dimitrov DS. Engineered human antibody constant domains with increased stability. J.Biol Chem. 2009 Mar 23; doi: 10.1074/jbc.M900769200. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duncan AR, Winter G. The Binding-Site for Clq on Igg. Nature. 1988;332:738–740. doi: 10.1038/332738a0. [DOI] [PubMed] [Google Scholar]
- 8.Choudhry V, Zhang MY, Sidorov IA, Louis JM, Harris I, Dimitrov AS, Bouma P, Cham F, Choudhary A, Rybak SM, Fouts T, Montefiori DC, Broder CC, Quinnan GV, Dimitrov DS. Cross-reactive HIV-1 neutralizing monoclonal antibodies selected by screening of an immune human phage library against an envelope glycoprotein (gp140) isolated from a patient (R2) with broadly HIV-1 neutralizing antibodies (vol 363, pg 79, 2007) Virology. 2007;368:431. doi: 10.1016/j.virol.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 10.Hsin J, Arkhipov A, Yin Y, Stone JE, Schulten K. Using VMD: an introductory tutorial Chapter 5. Curr.Protoc.Bioinformatics. 2008 doi: 10.1002/0471250953.bi0507s24. Unit 5.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koide A, Bailey CW, Huang X, Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J.Mol Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 12.Fellouse FA, Wiesmann C, Sidhu SS. Synthetic antibodies from a four-aminoacid code: a dominant role for tyrosine in antigen recognition. Proc.Natl.Acad.Sci.U.S.A. 2004;101:12467–12472. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W, Zhu Z, Feng Y, Xiao X, Dimitrov DS. Construction of a large phage-displayed human antibody domain library with a scaffold based on a newly identified highly soluble, stable heavy chain variable domain. J.Mol.Biol. 2008;382:779–789. doi: 10.1016/j.jmb.2008.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Haard HJ, van Neer N, Reurs A, Hufton SE, Roovers RC, Henderikx P, de Bruine AP, Arends JW, Hoogenboom HR. A large non-immunized human Fab fragment phage library that permits rapid isolation and kinetic analysis of high affinity antibodies. Journal of Biological Chemistry. 1999;274:18218–18230. doi: 10.1074/jbc.274.26.18218. [DOI] [PubMed] [Google Scholar]
- 15.Beers R, Chowdhury P, Bigner D, Pastan I. Immunotoxins with increased activity against epidermal growth factor receptor vIII-expressing cells produced by antibody phage display. Clinical Cancer Research. 2000;6:2835–2843. [PubMed] [Google Scholar]
- 16.Goodin JL, Nellis DF, Powell BS, Vyas VV, Enama JT, Wang LC, Clark PK, Giardina SL, Adamovicz JJ, Michiel DF. Purification and protective efficacy of monomeric and modified Yersinia pestis capsular F1-V antigen fusion proteins for vaccination against plague. Protein Expression and Purification. 2007;53:63–79. doi: 10.1016/j.pep.2006.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darbha R, Phogat S, Labrijn AF, Shu Y, Gu YJ, Andrykovitch M, Zhang MY, Pantophlet R, Martin L, Vita C, Burton DR, Dimitrov DS, Ji XH. Crystal structure of the broadly cross-reactive HIV-1-neutralizing Fab X5 and fine mapping of its epitope. Biochemistry. 2004;43:1410–1417. doi: 10.1021/bi035323x. [DOI] [PubMed] [Google Scholar]
- 19.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Structure of a V3-containing HIV-1 gp120 core. Science. 2005;310:1025–1028. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Radaev S, Motyka S, Fridman WH, Sautes-Fridman C, Sun PD. The structure of a human type III Fc gamma receptor in complex with Fc. Journal of Biological Chemistry. 2001;276:16469–16477. doi: 10.1074/jbc.M100350200. [DOI] [PubMed] [Google Scholar]
- 21.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. Characterization of Conserved Human-Immunodeficiency-Virus Type-1 Gp120 Neutralization Epitopes Exposed Upon Gp120-Cd4 Binding. Journal of Virology. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moulard M, Phogat SK, Shu Y, Labrijn AF, Xiao XD, Binley JM, Zhang MY, Sidorov IA, Broder CC, Robinson J, Parren PWHI, Burton DR, Dimitrov DS. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6913–6918. doi: 10.1073/pnas.102562599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoffman TL, LaBranche CC, Zhang WT, Canziani G, Robinson J, Chaiken I, Hoxie JA, Doms RW. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang PF, Bouma P, Park EJ, Margolick JB, Robinson JE, Zolla-Pazner S, Flora MN, Quinnan GV. A variable region 3 (V3) mutation determines a global neutralization phenotype and CD4-independent infectivity of a human immunodeficiency virus type 1 envelope associated with a broadly cross-reactive, primary virus-neutralizing antibody response. Journal of Virology. 2002;76:644–655. doi: 10.1128/JVI.76.2.644-655.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K, Franti M, Binley J, Vivona V, Grundner C, Huang CC, Venturi M, Petropoulos CJ, Wrin T, Dimitrov DS, Robinson J, Kwong PD, Wyatt RT, Sodroski J, Burton DR. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gpl120 is sterically restricted on primary human immunodeficiency virus type 1. Journal of Virology. 2003;77:10557–10565. doi: 10.1128/JVI.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]