Abstract
The National Cancer Institute’s (NCI) Thesaurus is a biomedical reference ontology. The NCI Thesaurus is represented using Description Logic, more specifically Ontylog, a Description logic implemented by Apelon, Inc. We are exploring the use of the DL species of the Web Ontology Language (OWL DL)—a W3C recommended standard for ontology representation—instead of Ontylog for representing the NCI Thesaurus. We have studied the requirements for knowledge representation of the NCI Thesaurus, and considered how OWL DL (and its implementation in Protégé-OWL) satisfies these requirements. In this paper, we discuss the areas where OWL DL was sufficient for representing required components, where tool support that would hide some of the complexity and extra levels of indirection would be required, and where language expressiveness is not sufficient given the representation requirements. Because many of the knowledge-representation issues that we encountered are very similar to the issues in representing other biomedical terminologies and ontologies in general, we believe that the lessons that we learned and the approaches that we developed will prove useful and informative for other researchers.
Keywords: NCI Thesaurus, OWL, biomedical ontologies
1. Introduction
The National Cancer Institute’s Thesaurus (the NCI Thesaurus) is a biomedical reference terminology with portions modeled as an ontology that covers areas of basic, translational, and clinical science. The NCI Thesaurus is a controlled terminology resource for the NCI and its collaborators in the cancer-research community intended to facilitate knowledge integration (Fragoso et al., 2004; Sioutos et al., 2007).1 The NCI Thesaurus is a product of the Enterprise Vocabulary Services (EVS) project. The EVS began publishing the NCI Thesaurus as its core reference terminology in 2000. Its goals were 1) to provide an up to date, science-based terminology for cancer research; 2) to use current terminology “best practices” to relate relevant concepts to one another in a formal structure, so that computers as well as humans can use the NCI Thesaurus for a variety of purposes; and 3) to speed the introduction of new concepts and new relationships in response to the emerging needs of basic researchers, clinical trials, information services and other users. Currently, the NCI Thesaurus is used in various capacities to index documents, to index the NCI Cancer portal cancer.gov, as the terminology source for a number of applications such as a NCI Drug Dictionary (see: http://www.cancer.gov/drugdictionary/), and for semantic annotation of metadata in the Cancer Bioinformatics Grid project caBIG™(see: http://cabig.nci.nih.gov). More recently, the NCI Center for Bioinformatics (NCICB) has launched a new terminology product, Biomedical Grid Terminology (BiomedGT)2 to support the needs of its NCICB partners, including caBIG™. This new terminology builds on the strengths of the NCI Thesaurus (concept orientation, description logic, public accessibility) and its current content, but will restructure the content to facilitate terminology federation and open content development. New tools used to facilitate collaborative terminology development as part of the BiomedGT workflow cycle will enable the wider biomedical research community to participate directly in extending and refining the terminology upon which they depend. The OWL representation discussed in this paper will be implemented, at least initially, in BiomedGT rather than the original NCI Thesaurus.
The NCI Thesaurus contains the information about diseases, their corresponding findings, associated abnormalities, and cellular origins; gene and allele information, their locations, products of gene functions, and biological processes that genes are responsible for; information on human anatomy, and anatomic sites of diseases; chemicals and drugs, and their mechanisms of actions; and other categories pertinent to the cancer domain. At the time of this writing, the NCI Thesaurus contains more than 59,000 classes, 187 roles, or properties, and 57000 asserted restrictions on properties.
The NCI Thesaurus is a description logic (DL) ontology, where classes are defined by the sets of necessary and necessary and sufficient conditions. The primary form of inference in a DL ontology is subsumption inference: a special inference engine (a DL classifier) uses the class definitions to infer additional subsumption (is-a) relationships between classes and to determine which class definitions are inconsistent.
The way one plans to use inference in a DL ontology may affect its representation. Specifically, in the NCI Thesaurus, the inference is used to ensure internal consistency of the NCI Thesaurus itself. The definitions and restrictions are not intended to be used, for instance, for making a diagnosis. For example, a definition of a disease and its necessary and sufficient conditions should ensure the correct place of the class describing the disease in the class hierarchy. The definition should not be used to make a diagnosis of a specific patient with specific conditions, for example, by classifying a patient as having a particular disease. This distinction played an important role in some of the design decisions we discuss in the paper. The goal of the NCI Thesaurus is to represent the current understanding of the defined concepts and useful information available about them. This information also makes it possible for applications to traverse relationships using the NCI Thesaurus, for example, from a disease of interest to genes that may be involved in the disease, thus facilitating knowledge discovery.
The current version of the NCI Thesaurus has been developed using the TDE environment developed by Apelon, Inc., and the ELH Ontylog description logic (Hartel et al., 2005).3 We are currently exploring the use of OWL DL, a SHOIN(D) DL ontology-representation language recommended by the World-Wide Web Consortium (Dean and Schreiber, 2004) for representing the NCI Thesaurus. We have several motivations for representing the NCI Thesaurus in OWL DL:
improve interoperability of the NCI Thesaurus with other tools that use OWL
use the greater expressive power of OWL DL;
conform to the ontology-representation standards developed by the World Wide Web Consortium;
make it easier to publish subsets or domains of the NCI Thesaurus for end users not wishing to use the entire thesaurus;
facilitate the transition to terminology federation
Golbeck and colleagues (Golbeck et al., 2003) developed the initial translation of the NCI Thesaurus into OWL DL. That translation was the first step in porting the NCI Thesaurus to OWL DL. It addressed most of the syntactic issues of the translation. However, the additional expressive power provided by OWL DL, as well as OWL’s lack of some of the features that were available in Ontylog, have led us to reexamine some of the design decisions in the structure and organization of the NCI Thesaurus. We refer the reader to a detailed comparison of Ontylog and OWL by Hartel and colleagues (Hartel et al., 2005) for more information on the similarities and differences between the two description logics.
Through extensive meetings with editors and users of the NCI Thesaurus, we have analyzed the requirements and constraints on the representation of various parts of the NCI Thesaurus. In this paper, we report our findings on the adequacy of OWL DL as the representation language for the NCI Thesaurus and similar terminologies and ontologies. We discuss the main OWL DL constructs that we used, constructs that were lacking, constructs that were not readily available, but for which we could create ontology-design patterns. We highlight components of ontology-development tools that, we believe, must be developed in order to facilitate creation of further OWL versions of the NCI Thesaurus and similar biomedical ontologies.
While we focus on the NCI Thesaurus in our analysis, we believe that many of our findings are relevant to using OWL DL for biomedical terminologies in general, as many of the representation issues that we address are not at all unique to representing cancer-related information. These issues include representation of imprecise information (e.g., a possible outcome of a disease), exceptions (e.g., properties of genes that are changed in their alleles), role chains (e.g., representing a link from gene to gene product and then to the disease, but being able to query the link between the gene and the diseases, bypassing the gene product); reciprocals for existential restrictions (e.g., representing that gene X plays role in a process Y by using an existential restriction on the class for gene X, but being able to query which genes play role in process Y, from the process point of view); and others.
This paper makes the following contributions:
– We analyze the knowledge-representation requirements for the NCI Thesaurus and similar biomedical terminologies, based on interviews with the NCI Thesaurus developers and users.
– We identify and define ontology-design patterns that simplify modeling of role chains and reciprocal restrictions.
– We define an ontology for describing ontology-specific templates for groups of similar classes (such as Kinds in the NCI Thesaurus).
The goal of this paper is not to critique the current representational choices, but rather to discuss new possibilities for representation being opened by using a more expressive ontology language, and the hurdles that still exist.
2. The basics of representing the NCI Thesaurus in OWL DL
Like most other biomedical terminologies, the NCI Thesaurus has a class hierarchy at its core. The class hierarchy can be subdivided into 22 main subontologies, representing the major divisions of the domain (Figure 1). These categories are inter-linked, with classes from one category using classes from other categories in property restrictions.
Fig. 1. Major categories of the NCI Thesaurus.

The NCI Thesaurus is partitioned into twenty major categories that are linked with one another through properties and restrictions.
The translation of the NCI Thesaurus to OWL by Golbeck and colleagues (Golbeck et al., 2003) created the “backbone” of the OWL representation of the NCI Thesaurus. Concepts from the original Ontylog representation became classes in OWL; roles defining relations between different classes became properties in OWL; defining roles that provide local restrictions on the range of roles became existential or universal restrictions in OWL.4 Class-level information such as code, id, synonyms, preferred name, and so on became annotation properties in OWL. Relations that were not inheritable and that were not defining (associations in Ontylog) became object annotation properties in OWL.
Figure 2 shows a typical class definition after translation into OWL (in Protégé-OWL). The class in the figure, Lung_Disorder is a subclass of a Respiratory_Disorder. It has an associated anatomic site Lung (occurs in the lung) and also Thorax and Respiratory_System as associated anatomic sites inherited from Thoracic_Disorder and Respiratory_Disorder, respectively.
Fig. 2. A Protégé screenshot of a typical class definition in the NCI Thesaurus.
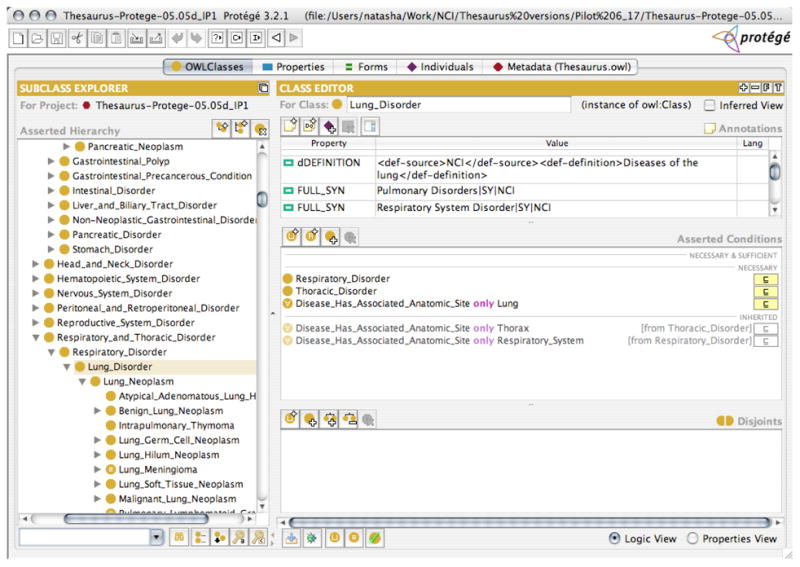
The left-hand side shows a snapshot of the class hierarchy. The right-hand side presents the definition fro the selected class, Lung_Disorder. The top part of the definition are the values for annotation properties, such as the textual definition for the class (“Diseases of the lung”), the synonyms of the class name (e.g., Pulmonary disorders, etc.), and other properties (not shown in the figure). The bottom part contains the logical definition of the class in the form of OWL restrictions.
3. The NCI Thesaurus in OWL DL: Pushing the envelope
While the basics of the translation of the NCI Thesaurus into OWL DL were relatively straightforward, many details and representation requirements proved harder to reflect in OWL DL:
– express imprecise information or information about what is typical, or common, or possible, but not always necessarily true;
– represent exceptions (such as properties of genes that are changed in alleles)
– define role chains (e.g., representing a link from gene to gene product and then to the disease, but being able to query the link between the gene and the diseases, bypassing the gene product)
– access inverses on restrictions (e.g., representing that gene X plays role in a process Y by using an existential restriction on the class for gene X, but being able to query which genes play role in process Y, from the process point of view)
– represent numeric ranges in restrictions (e.g., feature present in 20 to 50 percent of cells)
We discuss these issues in detail and recommend ways to address them in the rest of this section.
Some of our solutions suggest defining ontology design patterns (Gangemi, 2005). Design patterns have become an accepted practice in software engineering (Gamma et al., 1999) and represent an abstract representation of a common solution to a design or modeling problem. Similarly, an ontology design pattern represents an abstract representation of a common ontology-modeling solution. For example, OWL defines only binary relations between objects. If we need to represent relations of higher arity (n-ary relations), we need to use a combination of OWL statements that essentially “breaks down” an n-ary relation into a set of binary relations. An ontology-design pattern can encapsulate such detailed representation and enable a modeler to define an n-ary relation directly, by referring to the pattern (Noy and Rector, 2006). Thus, such a pattern defines, at a general level, how an n-ary relation must be translated into a set of binary OWL relations. When the pattern is used to specify a particular n-ary relation, we create an instantiation of the pattern, which is then translated into a set of specific OWL statements, based on the pattern definition.
3.1. Expressing and inheriting imprecise, possible information
A number of properties in the NCI Thesaurus, in particular the properties describing diseases, represent the information that is typical, or possible, for a particular disease, but is not necessary.
For example, certain features are typical characteristics of some kinds of cancers. These features often occur when the cancer is present (e.g., possible outcome), but some instances of this cancer may not have this feature. Currently, the NCI Thesaurus identifies these features as properties that have _May_Have_ in their name. For example:
– Disease_May_Have_Abnormal_Cell
– Disease_May_Have_Associated_Disease
– Disease_May_Have_Cytogenetic_Abnormality
– Disease_May_Have_Finding
Furthermore, a class high in the hierarchy may have a particular feature as a typical feature, but some of its subclasses may not exhibit this feature at all, and, in fact, explicitly exclude this feature; conversely, they may have this feature necessarily for all instances, rather than have it as a typical feature.
In general, using existential restrictions to represent these typical roles may carry incorrect semantics. For instance, suppose we say that:
Astroblastoma:
Disease_May_Have_Finding some Necrotic_Change
Such a statement implies that any instance of this disease has the instance of the finding as the value for the property Disease_May_Have_Finding. However, not all instances of this disease have this finding, and, in fact, some of the subclasses may explicitly exclude this finding. However, given that the goal of the logical constraints in the NCI Thesaurus is not to provide diagnostic classification for patients, but rather to ensure logical consistency of the terminology, using existential restrictions for these “possible” properties does not cause a problem: We do indeed want to classify a disease that may have finding subclass_of_X as a subclass of the disease that may have finding X (given that all other conditions confirm the subclass relationship).
The solution that we are proposing is to have two properties, such as Disease_has_〈X〉 and Disease_May_Have_〈X〉 (e.g., Disease_Has_Finding and Disease_May_Have_Finding). Disease_Has_〈X〉 is a subproperty of Disease_May_Have_〈X〉. This solution would handle the case where the feature is typical or optional at the higher level of the hierarchy, but is necessary at the lower levels. Given definition of subproperties, the following implication is true:
Disease_Has_Finding some Y ⇒
Disease_May_Have_Finding some Y
(because Disease_Has_Finding is a subproperty of Disease_May_Have_Finding)
If we have a class with a (possibly inherited) Disease_May_Have_〈X〉 restriction with a value Y, we can still state for this class that
not Disease_Has_〈X〉 some Y.
Thus, we will be saying that this particular class may have a particular finding (usually an inherited restriction) but does not actually have it. While possibly counter-intuitive at first, these two statements are actually logically correct, even when used together to describe the same class.
In a more common case, where in a subclass the feature becomes necessary, we can add
Disease_Has_〈X〉 some Y.
3.2. Representing Exceptions
The issue related to the problem of representing the “may” restrictions is the issue of exceptions and typical information. Describing genes and gene locations is a good example where representing exceptions would have been helpful.
The NCI Thesaurus contains classes for genes, as well as for the wild types and other allelic variant subtypes (in other contexts these subtypes of genes could be individuals or instances of the appropriate gene class). Alleles are inheritable mutations in populations (these are not mutations that occur as a result of exposure to carcinogens). As a rule, alleles inherit properties of their parent gene such as the chromosomal location, the role it plays in processes, etc. However, the really “interesting” cases are the small fraction of cases where alleles alter some of the properties of the gene. Thus, the NCI Thesaurus models only a small number of alleles—only the ones that are involved in some diseases or abnormalities.
In the NCI Thesaurus, the class Gene defines a gene by its organism, chromosomal location, and the biological processes in which it plays a role. In the hierarchy of Genes in the NCI Thesaurus, alleles are modeled as subclasses of the corresponding genes. In general, alleles inherit all the properties of the gene, but may occasionally change (override) essentially any of the gene’s properties, from chromosomal location to the process in which the gene plays a role, to roles in pathways. Ideally, we would like to specify the properties of the wild-type gene, and then represent the properties that are changed (overridden) for each allele. However, OWL DL does not support the modeling of exceptions in the class hierarchy: a subclass always inherits all properties and restrictions of its superclass.
One representation alternative is to assert that, because alleles do not inherit all the roles from the corresponding gene, there is no subclass relationship between genes and alleles. Rather, alleles are linked to the appropriate genes through a property, such as Allele_Is_Mutation_For_Gene. Thus, we will be able to link alleles and their corresponding genes. In this variant, we can also safely define restrictions on alleles that are incompatible with restrictions on the corresponding genes: the classes representing alleles are not subclasses of the gene classes and hence there is no problem with having a restriction at a class being incompatible with an inherited one. This approach has one major drawback: In most cases, alleles do inherit properties of the parent gene. If alleles are not subclasses of the parent genes, such inheritance will not be automatic and will need to be inferred by a special-purpose reasoner or a rule language. We will need to define special rules that describe how the gene properties are propagated to alleles. The rule engine will propagate the properties of the corresponding gene to the allele, unless there are conflicting properties in the allele’s definition. We can have the following rule, for example:
Allele_Is_Mutation_For_Gene (?allele, ?gene)
^ Gene_In_Chromosomal_Location (?gene, ?loc) →
Allele_In_Chromosomal_Location (?allele, ?loc)
We will need to write such rules for each property that alleles should “inherit” from genes. We will also need to add exceptions for the cases where alleles overwrite these properties. This solution is possible but it poses obvious maintenance problems.
Another alternative is to use the approach that is similar to the one we have described for diseases (Section 3.1): we can say that the roles of the wild-type gene are the typical roles for that gene. This “typicality” is still true of the allele, even though allele does something atypical. For instance, we can say the following:
Gene_X:
Gene_Typically_Associated_With_Disease some Disease_Y
Allele_Z:
subclassOf Gene_X
(inherited)
Gene_Typically_Associated_With_Disease some Disease_Y
(asserted at Allele_Z):
not ( Allele_Associated_With_Disease some Disease_Y)
Note that, in this solution, there is no subproperty relation between properties Gene_Typically_Associated_With_Disease and Allele_Associated_With_Disease. Both statements are true about Allele_Z: it is typically associated with a particular disease, but not in this specific case.
Ideally, we would also like to express that for any allele, the value for the property Allele_Not_Associated_With_Disease should be one of the values for the property Gene_Typically_Associated_With_Disease. There is no direct way to express this restriction in OWL but we can express it in a rule language, such as SWRL.5
Stevens and colleagues have also discussed the use of OWL to represent exceptions in modeling biomedical knowledge (Stevens et al., 2007). They define an ontology design pattern to address this issue. Specifically, the authors propose creating a disjoint covering partition of the class, such as the class Gene_X in our example, stating that all instances of the class must be instances of one of its two disjoint subclasses: GeneWithTypicalDiseaseAssociation_X and GeneWithAtypicalDiseaseAssociation_X. The appropriate restrictions differentiate the two subclasses. In this approach, however, we will have to create such a partition for each property that can have an atypical value for an allele—that is, for each property of the gene. Such approach results in a very large number of classes, created for the sole purpose of differentiating exceptional cases. In our approach, we focus on the cases where there are exceptions in specific alleles. Which approach works better in a particular application will depend on the specific domain being modeled, and how frequent exceptions are, and whether those exceptions are for values of the same property, or many different properties.
3.3. Role chains
A useful notion in modeling is the notion of role chains: For example, we often want to say that someone’s uncle is his father’s brother. More formally:
brother (?x, ?y) ^ father (?y, ?z) → uncle (?x, ?z)
The OWL language itself does not have role chaining (according to the language authors, introducing role chaining into the language would have made it undecidable (Horrocks et al., 2003)). However, we can use rule languages, such as SWRL, for simple role chaining. OWL 1.16 also has introduced role chaining in a decidable way (Horridge et al., 2006).
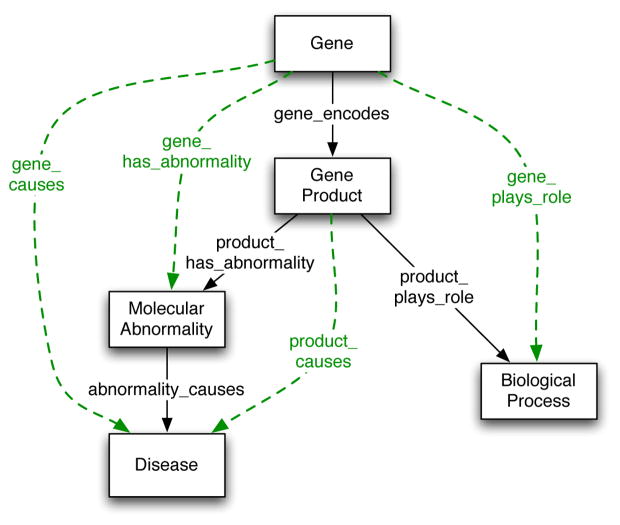
Role chaining would be extremely useful in modeling some parts of the NCI Thesaurus. In particular, the ability to chain roles addresses one of the prime concerns of the modelers for genes (Figure 3). The definitions of genes often include links from a gene to a gene product ( Gene_Encodes_Product), and then from the gene product to some molecular abnormality ( Gene_Product_Has_Abnormality). However, the users are usually interested in the direct link from gene to the abnormality or from gene to a disease.
Fig. 3. Relations between gene-related classes.
The solid black arrows indicate properties directly represented in the NCI Thesaurus. The dashed green arrows indicate properties that should be inferred.
Modeling relations between classes along the solid dark arrows in the Figure 3 would paint the most complete picture of what is going on. However, this detailed information (e.g., always going through gene products) either may not be available, or may be too tedious to enter. While technically it is the gene product rather than the gene itself that plays a role in the disease, we often want to bypass such detailed information. In fact, it is rare for people to ask what proteins (gene products) are involved in a particular disease; rather they ask about the genes themselves (that had encoded the product). Here are some typical use cases of queries of genes and gene products:
– Researchers often need a link from gene to abnormality
– Clinicians often need a link from gene to diseases: which disease is the gene associated with
– Researchers often need a link from gene to diseases to know whether there is a diseases known to be associated with a gene or gene variant
We can specify such inference as a rule in a rule language such as SWRL and then use a rule language to perform the additional inference:
Gene_Has_Product (?gene, ?product) ^
Product_Has_Abnormality (?product, ?abnormality) →
Gene_Has_Abnormality (?gene, ?abnormality)
The preceding rule is expressed in the SWRL rule language and such expression could be a solution to expressing role chaining in general. However, role chaining seems to be such a common modeling situation that it may make sense to have specific tool support both for specifying the chains and for using them in inference. In general, we can think of a role chain definition as an example of an ontology-design pattern. In this case, the user specifies the role chain at the abstract level, by stating that two (or more) properties constitute a role chain (e.g., Gene_Has_Product and Product_Has_Abnormality constitute a role chain that results in Gene_Has_Abnormality). A tool can then translate such pattern instantiation into a set of specific OWL and SWRL statements.
3.4. Reciprocal Restrictions
The OWL language has the notion of inverse properties. For example, we can declare the following two properties form the NCI Thesaurus to be inverse properties using owl:inverseOf property: Gene_Encodes_Product and Gene_Product_Encoded_by_Gene (cf. Figure 3). Having these properties defined as inverse properties, enables the following inference automatically:
Gene_Encodes_Product (?gene, ?product) ⇒
Gene_Product_Encoded_by_Gene (?product, ?gene)
However, this inference applies only to instances; in other words, if we have a statement for the property value for a specific instance of a gene, then we can fill in the gene for the corresponding product.
However, the NCI Thesaurus models classes and does not represent instances. The relationships between classes are expressed primarily through existential restrictions such as:
GeneX:
Gene_Encodes_Product some ProductY
This restriction does not imply the “inverse” restriction:
ProductY:
Gene_Product_Encoded_by_Gene some GeneX
In fact, the first restrictions says that every instance of the class GeneX must encode some instance of the class ProductY. However, this restriction says nothing that would be applicable to every instance of ProductY. It leaves open the possibility that some instances of ProductY are not encoded by any of the GeneX instances. The second restriction however states exactly that: every instance of ProductY is encoded by some instance of GeneX. So, in general, restrictions should not automatically be “inversible.”
However, in modeling the NCI Thesaurus, we usually indeed want to conclude that, for many restrictions, the reciprocal restriction also holds, as in the earlier example. Indeed, it so happens that, in the specific cases encoded in the NCI Thesaurus, the “reciprocal” restriction holds in many cases. Not only that, but also many use cases require access to information from both directions: some use cases involve queries for genes given a disease and others query diseases associated with a given gene.7
Therefore, we need an ontology-design pattern or some other mechanism (as well as tool support), to specify that a particular existential restriction is reciprocal:
classX propertyP some classY ⇒
classY propertyP some classX
This expression is, in essence, a definition of another ontology-design pattern. If we have tool support for such a pattern, the user needs only to mark a restriction as reciprocal, and the tool can create the second restriction automatically.
3.5. Defining classes by numeric ranges of properties
Lack of the ability to specify numeric ranges (e.g., saying that a teenager is a person whose age is between 13 and 19) in OWL has been noted numerous times (Stevens et al., 2007). Like many other ontologies and terminologies, the NCI Thesaurus also requires the use of numeric ranges to represent some of its concepts. Consider, for example, a class such as Bone_Marrow_Dysplasia_Present_in_50_Percent_or_More_of_the_Cells_of_Two_Cell_Lines. This class inherently contains a numeric range in its definition: it is a value range for a property representing the percentage of specific cells present in cell lines. Thus, the restriction on this property must include a numeric range restriction.
Furthermore, with the NCI Thesaurus, many queries related to genes and their locations require reasoning with restrictions involving numbers or numeric ranges.
For example, genes have locations on chromosomal bands. Figure 4 shows the location definitions for a gene and for a chromosomal band. We want to be able to answer the following types of queries:
Fig. 4. Definition of a specific Gene.
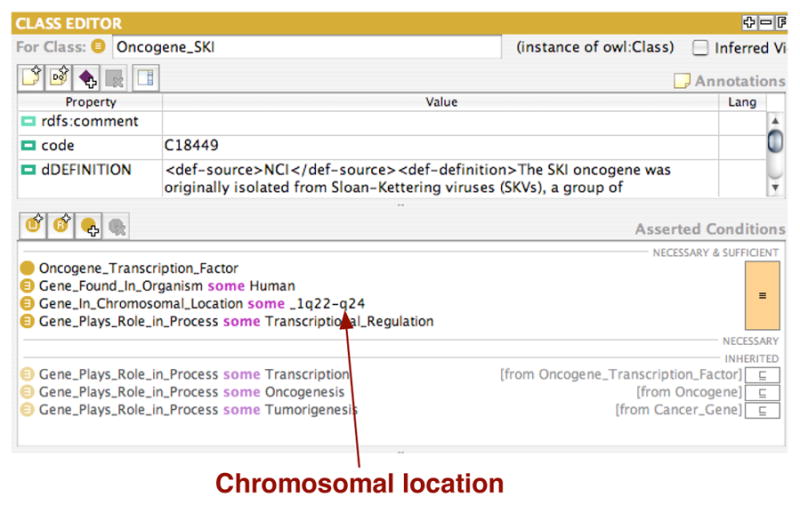
The chromosomal location is part of the definition. The location specifies a band on the chromosome and we want to be able to infer that location, such as _1q23, is part of this band.
– A person has an abnormality: they are missing a particular band on the chromosome. Which genes are affected?
– If the missing band is, for example, _1p35-p32, the result should include genes that have location chromosomal location _1p34
While the language itself does not have numeric ranges, many OWL editors, such as Protégé-OWL, enable users to represent numeric ranges by using user-defined XML schema datatypes.
In general, DL classifiers have shied away from dealing with someValuesFrom and hasValue restrictions for datatype properties in OWL. However, many queries related to genes and their locations require reasoning with these types of restrictions. Modeling and use of many ontologies and terminologies require such representation and reasoning.
4. Using Ontology Design Templates To Specify Similar Classes
The ontology-design patterns that we suggested in the previous section (such as the ones for role chains and reciprocal restrictions) deal with hiding some of the complexities of standard sets of statements, allowing users to specify these sets at a more abstract level. The ontology-design patterns are independent of a particular ontology or terminology and are defined at the level of the ontology language. Ontology templates, which we introduce in this section, are another way of modularizing ontology development and facilitating the modeling process for domain experts. One can think of an ontology template for a class as, essentially a “default” definition of a class, with some values to be filled in by the user who defines the class. We present some template examples later in this section.
As we noted earlier, the NCI Thesaurus is structured around 22 basic kinds, such as Diseases, Genes, Anatomy, and so on. Each kind has a certain set of properties that are defined for it. And for each kind, some of these properties are definitional (a set of necessary and sufficient conditions that fully defines a class) and some are necessary. In general, for each kind in the NCI Thesaurus, we can identify the set of definitional properties and restrictions and the necessary restrictions that need to be made more specific in subclasses. For example, the NCI Thesaurus developers mostly agree that the following roles are defining roles for all Genes classes (subclasses of the class Gene) and should be necessary and sufficient conditions in the definitions of genes:
– Gene_Found_In_Organism (such as Human, which is the case for 99% of the cases in the NCI Thesaurus)
– Gene_Has_Chromosomal_Location
– Gene_Plays_Role_In_Process
– Gene_In_Physical_Location (present only if the previous three characteristics are not distinguishing enough)
The rest of the roles in the Gene definitions (e.g., Gene_Is_Element_In_Pathway, Gene_Associated_With_Disease, Gene_is_Biomarker_of, etc.) should become necessary conditions. We can identify similar “templates” for other kinds. Thus, it would be very practical to enable definition of such templates for groups of classes (say, subclasses of a particular class) and to integrate their definition and use into an ontology editor.
Such templates will define, for each subtree:
– the necessary and sufficient conditions: the editors will need to make the restrictions more specific, but will have a template to start with
– the necessary conditions, giving an indication to an editor as to what conditions usually need to be filled in for a class
– correct types of restrictions (e.g., universal or existential, has Value, etc.)
– restricrtions that must be specialized for each class (e.g., gene location)
In general, such templates should be suggestive rather than prescriptive. In some cases, editors may want to deviate from these templates. For instance, for diseases, the disease finding (the clinical manifestations of the disease) is generally a necessary and sufficient condition, but may be just a necessary condition.
The editors could then be guided through the process of defining a new class in a specific hierarchy, requested to enter information that is mandatory, prompted to enter optional information, and so on.
Such templates can be stored as a set of metadata (class-level properties) at the root of the subtree. We have developed a small ontology for defining such a templates. This ontology should be imported during editing when a tool can use the information for presenting the templates. Since these metadata are not part of the domain description itself, ontology developers may often choose to remove the import before distributing the ontology. Figures 5, 6, and 7 present main components of the template ontology and its use in the NCI Thesaurus in Protégé. Figure 5 shows a metaclass that is added as a type for any subtree root that defines a new template (e.g., the class Gene in the NCI Thesaurus). This metaclass adds two new class-level properties to such subtree root: a set of necessary and sufficient restrictions that its subclasses should define and a set of necessary conditions for them. Each restriction is specified as a template (Figure 6) that defines the type of the restrictions, the property being restricted, the value for that property, and whether or not this restriction must be specialized at each subclass. The last component essentially tells the tool whether an ontology developer must enter some new value for this restriction at each new subclass he creates.
Fig. 5. Definition of the metaclass, Subtree_Template defining a template for restrictions in a class.
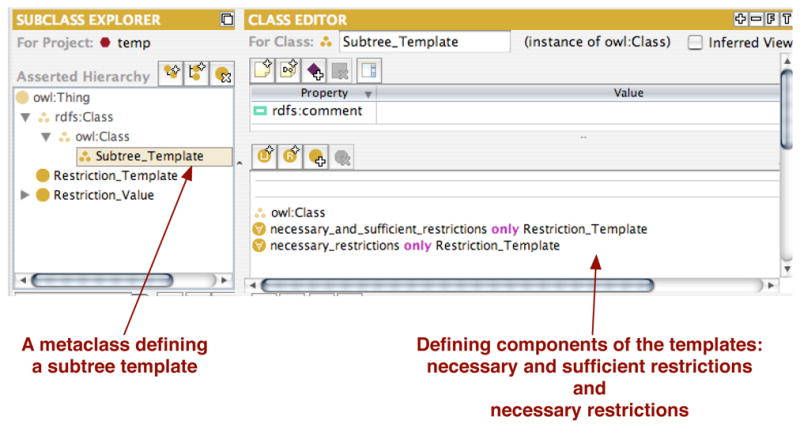
Domain classes that are roots of the subtrees for which there is a template, will have Subtree_Template as an additional type.
Fig. 6. Defining a template for a particular restriction.
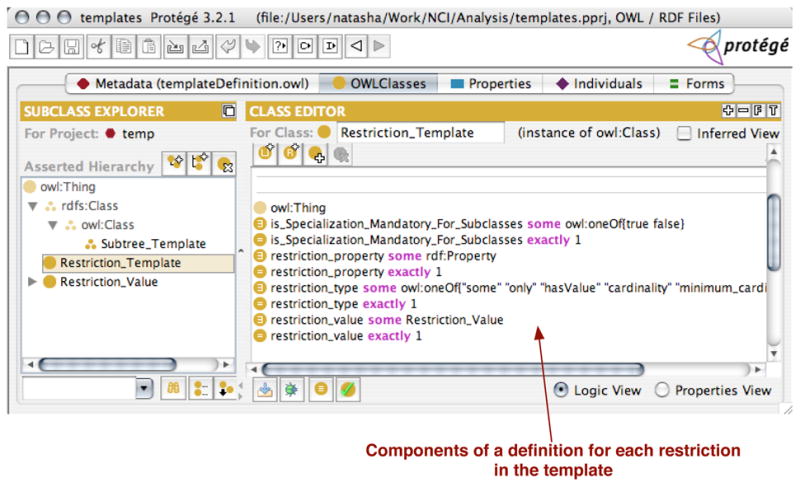
Instances of this class are entries in a subtree template, one per each restriction in the subtree template. Each instance describes a restriction to be included in the template: the property in the restriction ( restriction_property), the type of the restriction, such as some or only ( restriction_type), and the value of the restriction, such as a class in a some restriction ( restriction_value)
Fig. 7. Adding a subtree template to a class.
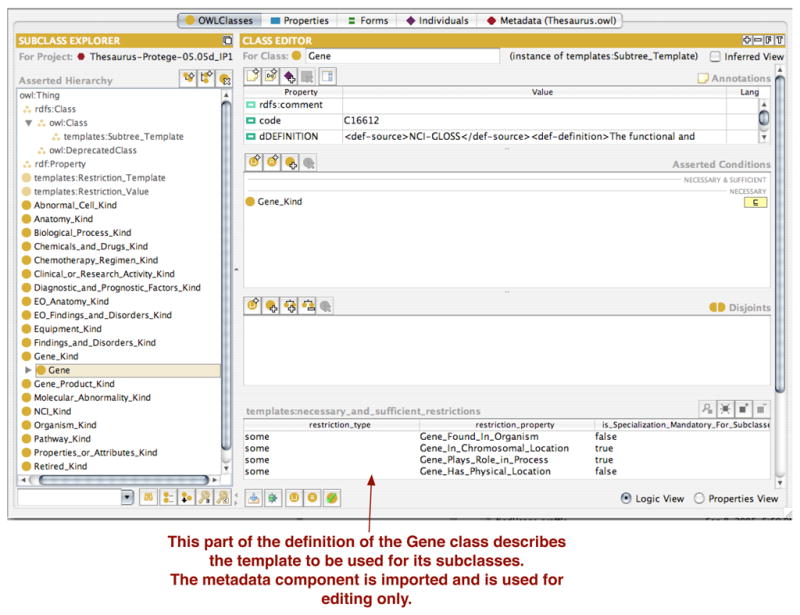
Gene. The template defines several necessary and sufficient conditions that subclasses of the class Gene must specify. Two of these conditions, Gene_Has_Chromosomal_Location and Gene_Plays_Role_In_Process, must be specialized at each of the subclasses, that is there are must be non-inherited values for these restrictions at each of the subclasses.
Figure 7 shows the use of the subtree template to define a template for subclasses of the class Gene. There are four necessary and sufficient restrictions that developers are expected to enter for each subclass of Gene.8 Two of these restrictions must be specialized at each subclass.
Note that the template ontology is in OWL Full, and hence importing it makes the domain ontology itself an OWL Full ontology. However, we expect that the templates will be used only at development time and hence the ontology developers can simply remove the import statement (and, therefore, all the template information) when distributing the ontology. Thus the original OWL species of the domain ontology is preserved.
Neither Protégé, nor other ontology editors currently use such template specifications to facilitate editing. However, we believe that such a facility would be extremely useful for developers of biomedical terminologies, such as the NCI Thesaurus.
5. Discussion
Our analysis of the NCI Thesaurus shows that the expressive power of OWL DL enables us to specify quite precisely the distinctions that we want to make explicit in the NCI Thesaurus. We can define classes by their sets of necessary and sufficient conditions; we can identify templates for classes belonging to each sub-hierarchy (Kind) in the NCI Thesaurus;9 we use the full gamut of OWL DL constructs from object-type and datatype properties, to various types of restrictions.
We have also identified a number of representational requirements that OWL DL does not readily address. Most of these requirements, however, can be solved by instrumenting our tools to be more custom-tailored to the requirements of ontology developers, without having to change the language itself. The tools can then (1) hide the complexity of some constructs; (2) support design patterns; (3) support definition of ontology-specific templates for classes in a subtree; and (4) support some of the things that could not be directly expressed.
We are implementing the OWL representations discussed in this paper, at least initially, in BiomedGT, the open-content–development version of the NCI Thesaurus. We are also working on support for ontology templates in the Protégé environment.
The NCI Thesaurus has been the subject of scholarly analysis before. For example, Ceusters and colleagues (Ceusters et al., 2005) studied the definitions and the terms used to name entities in the NCI Thesaurus and pointed out inconsistencies, missing definitions, and the imprecise use of terminology in some places. They also noted missing relationships between concepts, and a lack of unifying principles in creating the classification itself. The authors of this study also have pointed out problems with the initial OWL representation of the NCI Thesaurus. The the NCI Thesaurus authors have since addressed many of those problems, and the OWL representation that we discuss here goes further along the path of principled and precise modeling of knowledge in the NCI Thesaurus. Kumar and Smith (Kumar and Smith, 2005) looked at the modeling of concepts that are relevant for one particular cancer—colon carcinoma—and noted some inaccuracies there. The analysis that we have presented in this paper takes a largely different course, looking at the formal structure and representation, rather than at the content of specific definitions. The two types of analyses are complementary. Furthermore, a more precise modeling of the domain might by itself eliminate or highlight the content problems that other authors have noted.
Other studies–most notably, a study by Stevens and colleagues (Stevens et al., 2007)—have looked at the representational requirements of biomedical ontologies in general and how well OWL addresses these requirements. Some of the problems that this study highlighted are the same ones we found when representing the NCI Thesaurus in OWL, such as representing exceptions, numeric ranges, and imprecise information. The authors also pointed to the requirements for representing n-ary relations, lists, qualified cardinality restrictions, and other complex property restrictions. Researchers have also published papers discussing their experiences in using OWL for representing such biomedical ontologies and terminologies as the Foundational Model of Anatomy, which is a large reference ontology of human anatomy (Golbreich et al., 2006; Dameron et al., 2005; Noy and Rubin, 2008), BioPAX, which is a data exchange format for biological pathway data (Ruttenberg et al., 2006), representation of phenotypes (Mungall et al., 2007), and many others. All these studies pointed to some problems in using OWL for representing the complexities of biomedical knowledge, but suggested workarounds and pragmatic solutions for most of the problems.
It is important to view the representation of the NCI Thesaurus in OWL DL in the larger context of the collaborative development of large medical terminologies. In addition to the purely knowledge-representation issues that we have discussed, the development of the NCI Thesaurus brought to the fore other related problems, such as ontology modularization, ontology maintenance, ontology evolution, change management, and scalability of tools and languages for open terminology development.
For example, the NCI Thesaurus is currently one monolithic OWL ontology. We are considering modularization approaches that will provide several advantages (see(Seidenberg and Rector, 2006)). First, many users don’t need all of the components of the the NCI Thesaurus, and would like to reuse only some of them in their applications or ontologies. The breakdown into separate modules that import one another provides a natural division of the NCI Thesaurus, enabling users to import only the components that they need. Second, the sheer size of the NCI Thesaurus poses scalability problems to classification using Description Logic classifiers. Classifying the NCI Thesaurus from within an editing environment such as Protégé currently requires dedicated hardware. In a multi-CPU 64-bit linux server with sufficient memory, it takes approximately 5–20 minutes depending on the classifier (Racer, FaCT++, or pellet 1.4+ with performance enhancements). Hence editors cannot practically conduct classification on demand as they are making changes. Instead, a lead editor must perform scheduled classifications as part of the workflow. Researchers are currently discussing ways to classify portions of a large OWL ontology rather than have the whole ontology classified (Stuckenschmidt and Klein, 2007). Incremental classification (Parsia et al., 2006) is another approach that would allow editors to classify the NCI Thesaurus on demand.
With BiomedGT, the content development becomes a community-based process. Tool developers are currently working on several approaches to support such open community-based development of ontologies and terminologies. And the developers of both the NCI Thesaurus and BiomedGT are using several tools to support collaborative development, track issues and problems and discuss modeling issues. First, the GForge site10 provides support for issue tracking, mostly for issues with the software itself. The LexWiki environment, currently at the core of the BiomedGT platform, supports community-based development of BiomedGT, enabling the wider community to comment on the definitions of classes and property values, suggest new values, and corrections to the current ones. Finally, Collaborative Protégé (Tudorache and Noy, 2007) is an extension of the Protégé environment that integrates ontology-development process with the process of reaching consensus, carrying out discussions on the ontology, adding new development tasks, and so on. Each of the tools addresses a particular set of users and modalities of development. Our discussion in this paper, however, has demonstrated that the tools that support ontology development itself must be extensible to support ontology patterns and ontology templates to facilitate the development and address the problems where the native OWL constructs are not appropriate or sufficient for modeling biomedical ontologies.
The NCI Thesaurus has an enviable distinction of being actively used by many users while still being very actively under development. This situation poses problems with ontology maintenance and evolution (Noy et al., 2006). Not only the NCI Thesaurus is being edited simultaneously by many editors–and their edits need to be synchronized and approved–but also the users need to have explicit and machine-processable information of what has changed from one baseline version to the next. We are currently considering using Protégé and a suite of its plugins for change management, conflict identification, and quality control.
Finally, our analysis has shown that when we consider representational requirements of a domain, we must consider not only the ontology language but also the tool support for ontology development. It is unlikely that there is an ontology language that suits all domains and and addresses all requirements perfectly. However, flexible tool support can often mitigate whatever shortcomings a formalism might have in addressing representational requirements of a subject domain.
6. Conclusions
As we have studied and developed the representation of the NCI Thesaurus in OWL DL, we have encountered many representation problems that were not readily addressed in the current literature and by the traditional use of OWL DL constructs. We have presented a real-life challenge to the DL representation and discussed the ways to resolve the many issues. We believe that developers of large medical terminologies (and ontologies and terminologies in other domains) will be faced with similar issues and we hope our experiences and lessons learned will help in resolving them.
Acknowledgments
We are very grateful to Alan Rector for his comments on an earlier draft of this paper. We thank the developers and editors of the NCI Thesaurus for their help in understanding the representation requirements in the cancer informatics domain. This work was supported in part by a contract from the U.S. National Cancer Institute. Protégé is a national resource supported by grant LM007885 from the United States National Library of Medicine.
Footnotes
Universal and existential restrictions were not used consistently in the NCI Thesaurus. As the result, the use of allValuesFrom and someValuesFrom restrictions in OWL was also not always correct.
Ontology editors can provide access to such information. For instance, in the Protégé user interface provides modelers with access to this information in both direction through the “find usage” button
We used a special widget in Protégé that allows display of a set of instances—here instance of the class Restriction_Template—as rows in a table.
We use OWL Full to define the templates themselves
https://gforge.nci.nih.gov/projects/protegegui/
References
- Ceusters W, Smith B, Goldberg L. A terminological and ontological analysis of the NCI Thesaurus. Methods of Information in Medicine. 2005;44(4):498–507. [PubMed] [Google Scholar]
- Dameron O, Rubin DL, Musen MA. Challenges in converting frame-based ontology into OWL: the Foundational Model of Anatomy case-study. AMIA Annual Symposium; 2005. [PMC free article] [PubMed] [Google Scholar]
- Dean M, Schreiber G. Web ontology language (OWL) reference. 2004 http://www.w3.org/tr/owl-ref/
- Fragoso G, de Coronado S, Haber M, Hartel F, Wright L. Overview and utilization of the NCI Thesaurus. Comparative and Functional Genomics. 2004;5(8):648–654. doi: 10.1002/cfg.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamma E, Helm R, Johnson R, Vlissides J. Design Patterns: Elements of Reusable Object Oriented Software. Addison-Wesley Co; Reading, MA: 1999. [Google Scholar]
- Gangemi A. Ontology design patterns for semantic web content. 4th International Semantic Web Conference (ISWC2005); Galway, Ireland: Springer; 2005. pp. 262–276. [Google Scholar]
- Golbeck J, Fragoso G, Hartel F, Hendler J, Parsia B, Oberthaler J. The National Cancer Institute’s Thesaurus and Ontology. Journal of Web Semantics. 2003;1(1) [Google Scholar]
- Golbreich C, Zhang S, Bodenreider O. The Foundational Model of Anatomy in OWL: Experience and perspectives. Journal of Web Semantics. 2006;4(3):181–195. doi: 10.1016/j.websem.2006.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartel FW, Coronado Sd, Dionne R, Fragoso G, Golbeck J. Modeling a description logic vocabulary for cancer research. Journal of Biomedical Informatics. 2005;38(2):114–129. doi: 10.1016/j.jbi.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Horridge M, Tsarkov D, Redmond T. Supporting early adoption of OWL 1.1 with Protege-OWL and FaCT++ Second OWL Experiences and Directions Workshop (OWLED); 2006. [Google Scholar]
- Horrocks I, Patel-Schneider P, van Harmelen F. From SHIQ and RDF to OWL: The making of a web ontology language. Journal of Web Semantics. 2003;1(1):7–26. [Google Scholar]
- Kumar A, Smith B. Oncology ontology in the NCI Thesaurus. 10th Conference on Artificial Intelligence in Medicine, AIME 2005; Aberdeen, UK. 2005. pp. 213–220. [Google Scholar]
- Mungall C, Gkoutos G, Washington N, Lewis S. OWL: Experiences and Directions (OWLED 2007) Innsbruk; Austria: 2007. Representing phenotypes in OWL. [Google Scholar]
- Noy NF, Rector A. Defining N-ary relations on the Semantic Web. Technical report, W3C Working Group Note 2006 [Google Scholar]
- Noy NF, Chugh A, Liu W, Musen MA. A framework for ontology evolution in collaborative environments. Fifth International Semantic Web Conference, ISWC, volume LNCS 4273; Athens, GA. Springer; 2006. [Google Scholar]
- Noy NF, Rubin DL. Translating the Foundational Model of Anatomy into OWL. Journal of Web Semantics. 2008 doi: 10.1016/j.websem.2007.12.001. to appear. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsia B, Halaschek-Wiener C, Sirin E. Towards incremental reasoning through updates in OWL-DL. Reasoning on the Web Workshop-WWW2006 2006 [Google Scholar]
- Ruttenberg A, Rees J, Zucker J. OWL: Experiences and Directions (OWLED 2006) Athens; Georgia: 2006. What BioPAX communicates and how to extend OWL to help it. [Google Scholar]
- Seidenberg J, Rector A. Web ontology segmentation: Analysis, classification and use. 15th International World Wide Web Conference; Edinburgh, Scotland. 2006. [Google Scholar]
- Sioutos N, de Coronado S, Haber M, Hartel F, Shaiu W, Wright L. NCI Thesaurus: A semantic model integrating cancer-related clinical and molecular information. Journal of Biomedical Informatics. 2007;40(1):30–43. doi: 10.1016/j.jbi.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Stevens R, Egaña Aranguren M, Wolstencroft K, Sattler U, Drummond N, Horridge M, Rector A. Using OWL to model biological knowledge. International Journal of Human-Computer Studies. 2007;65(7):583–594. [Google Scholar]
- Stuckenschmidt H, Klein M. Reasoning and change management in modular ontologies. Data & Knowledge Engineering. 2007;63(2):200–223. [Google Scholar]
- Tudorache T, Noy N. Collaborative Protégé. Workshop on Social and Collaborative Construction of Structured Knowledge (CKC 2007) at the 16th International World Wide Web Conference (WWW2007); Banff, Canada: CEUR; 2007. [Google Scholar]