Abstract
microRNAs (miRNAs) regulate gene expression post-transcriptionally by targeting mRNAs for degradation or by inhibiting translation. Dicer is an RNase III endonuclease which processes miRNA precursors into functional 21-23 nucleotide RNAs that are subsequently incorporated into the RNA-induced silencing complex. miRNA-mediated gene regulation is important for organogenesis of a variety of tissues including limb, lung and skin. To gain insight into the roles of Dicer and miRNAs in mammalian skeletal muscle development, we eliminated Dicer activity specifically in the myogenic compartment during embryogenesis. Dicer activity is essential for normal muscle development during embryogenesis and Dicer muscle mutants have reduced muscle miRNAs, die perinatally and display decreased skeletal muscle mass accompanied by abnormal myofiber morphology. Dicer mutant muscles also show increased apoptosis and Cre-mediated loss of Dicer in Myod-converted myoblasts results in enhanced cell death. These observations demonstrate key roles for Dicer in skeletal muscle and implicate miRNAs as critical components required for embryonic myogenesis.
Keywords: Dicer, miRNA, Myogenesis, Morphogenesis, Apoptosis
Introduction
Skeletal muscle development during embryogenesis is a well-defined process that requires precise regulation of a variety of factors in muscle progenitors and differentiated cell types (Parker et al., 2003; Relaix, 2006). Skeletal muscle precursors originate in the dermomyotome, a dorsal somitic compartment, during early development (Hollway and Currie, 2005). Muscle progenitors in the dermomyotome consist of a population of proliferating and undifferentiated cells that express the Pax3 and Pax7 transcription factors (Buckingham et al., 2006; Relaix et al., 2006). Precursors in the dermomyotome dorsal medial lip and ventral lateral lip establish the respective epaxial and hypaxial myotomes (Huh et al., 2005; Kalcheim et al., 1999). Epaxial myotome progenitors differentiate, elongate and fuse to give rise to deep back musculature. A sub-population of hypaxial precursors differentiate to develop trunk muscle while others remain proliferative, delaminate and migrate ventrally to form diaphragm, limb and tongue musculature.
Myofiber formation occurs during two waves of limb muscle development (Cossu and Biressi, 2005). Primary fibers, which are derived from embryonic myoblasts, form the primary myofiber scaffolding around embryonic days (E)11-14. Secondary fibers are established from fetal myoblasts around E14-16. Satellite cells also serve as precursors for the rapid muscle growth that occurs during the fetal period. Myofiber formation requires that myoblasts exit the cell cycle, fuse, elongate and remain terminally differentiated (Brand-Saberi, 2005) . Myogenic regulatory factors (MRFs) and Myocyte enhancer factor-2 (Mef2) transcription factors orchestrate a gene regulatory network which controls these processes (Berkes and Tapscott, 2005). MRFs are basic helix-loop-helix transcription factors that dimerize with E proteins to bind E boxes (CANNTG) in the promoters and enhancers of muscle-specific genes. For example, MyoD promotes myoblast determination by functioning as a master regulator of muscle-specific gene transcription and ectopic MyoD has been shown to convert non-muscle cell types to myoblasts (Weintraub et al., 1989). Another transcription factor, myogenin, plays a critical role in myoblast terminal differentiation (Venuti et al., 1995). While much is known about the transcriptional regulation that controls myogenesis, potential roles for post-transcriptional gene regulation are not as well defined.
MicroRNAs (miRNAs) are small non-coding RNA molecules that regulate gene expression post-transcriptionally (Bartel, 2004). miRNAs are transcribed as long primary transcripts that undergo a series of cleavage events to reduce them to functional 21-23 nucleotide (nt) RNAs that become incorporated into the RNA-induced silencing protein complex (RISC). The miRNA/RISC complex functions by binding to complementary sequences in the 3’-untranslated regions (UTRs) of target mRNAs to inhibit protein synthesis by preventing translation or promoting mRNA degradation.
To clarify the biological roles of miRNAs, the mRNAs they target must be identified. Bioinformatic approaches have resulted in the prediction of thousands of miRNA targets (John et al., 2006). These algorithms predict targets based on various features of the miRNA or the target, such as the 5’-end nucleotide seed sequence of the miRNA, evolutionary conservation between species, and thermodynamic free energy calculations. Although thousands of targets have been predicted in silico, few have been validated in vivo.
miRNAs were discovered in worms but have since been identified in many different organisms, including plants, flies, fish, mice and humans. Some miRNAs are ubiquitously expressed and other display tissue-specific expression patterns (Kloosterman et al., 2006; Sempere et al., 2004). It is estimated that thousands of miRNAs exist in the human genome and they may regulate 20-30% of known genes (Lewis et al., 2005). Mammalian miRNAs can exist as single-copy genes or as families of conserved and highly related sequences and multiple miRNAs have been predicted to regulate the same target. This makes it difficult to use reverse genetics to study the function of a particular miRNA because loss of a miRNA gene at one genomic locus may be compensated for by expression of other miRNAs. To determine the global functions of miRNAs during tissue development, several studies have reported the effects of mutating Dicer, the endonuclease responsible for processing pre-miRNAs into mature miRNAs. Dicer inactivation results in loss of processed miRNAs and accumulation of miRNA precursors. Mice that lack Dicer die at E7.5 (Bernstein et al., 2003). Therefore, Dicer conditional alleles were generated to inactivate Dicer in specific tissues. Dicer inactivation in limb, lung and skin results in severe morphological defects and is associated with aberrant cell death and proliferation during development (Andl et al., 2006; Harfe et al., 2005; Harris et al., 2006). In this study, we address the role of Dicer and miRNA-mediated gene silencing during skeletal muscle development by inactivating Dicer during embryonic myogenesis. Our results demonstrate that Dicer inactivation results in perinatal lethality, skeletal muscle hypoplasia, myofiber morphogenesis defects and increased apoptosis.
Materials and Methods
Mice Generation and Genotyping
To inactivate Dicer specifically in skeletal muscle, mice containing a Dicer conditional allele (Dicercond) were crossed to those expressing a MyoDcre transgene (Chen et al., 2005; Harfe et al., 2005). The resulting Dicer+/cond;MyoDcre male mice were mated to Dicercond/cond females to generate Dicer−/cond;MyoDcre mutant animals. Some females also carried the ROSA26 reporter (Soriano, 1999). It is noteworthy that males carrying a Dicercond allele and MyoDcre transgene always transmitted a Dicer deleted allele (Dicer). This is likely due to leaky MyoDcre expression in the male germline. Therefore, all mutant embryos were Dicer−/cond;MyoDcre and males were never used to transmit the ROSA26 reporter. Mice were genotyped by PCR. Adult and embryonic mouse tail snips (∼3 mm) were lysed in 200 μl 50 mM NaOH at 100°C for 30 min with vigorous vortexing every 10 min. Tail lysates were neutralized with 50 μl of 1 M Tris-Cl (pH 8.0), and 1 μl was used in a 20 μl PCR reaction containing 2.5 U Taq polymerase (New England Biolabs), 1X buffer, 0.375 mM dNTPs and the following primer sets (30 pmol of each per reaction). Dicercond: Dicer F1 (CCTGACAGTGACGGTCCAAAG) and Dicer R1 (CATGACTCTTCAACTCAAACT). MyoDcre: Cre Fwd (TGCAACGAGTGATGAGGTTCGCAAGAAC) and Cre Rev (GAACGAACCTGGTCGAAATCAGTGCGTT). ROSA26R: lacZ Fwd (TTATCGATGAGCGTGGTGGTTATGC) and lacZ Rev (GCGCGTACATCGGGCAAATAATATC). Genotyping reactions were PCR amplified through 35 cycles (95°C for 30 sec, 60°C for 30 sec and 68°C for 1 min). All experiments were performed under approved animal protocols according to the institutional guidelines established by the University of Florida IACUC committee.
RNA Blots and RT-PCR
RNA was isolated from fresh homogenized tongue or pooled embryonic forelimbs and hindlimbs with TRI Reagent (Sigma). For small RNA blots, approximately 20 μg of total RNA was resolved on 10% urea/polyacrylamide gels and electroblotted to Hybond N+ membranes (Amersham) at 200 mA for 3 h. Blots were crosslinked using a Stratalinker (Stratagen) and prehybridized for at least 1 h at 37°C in ULTRAhyb™-Oligo (Ambion) hybridization buffer before overnight incubation at 37°C in hybridization buffer containing [32P]-end-labeled probe. Probes were generated by end-labeling 20 pmols of DNA oligonucleotide (Invitrogen) complementary to miRNA or U6 with T4 polynucleotide kinase (New England Biolabs) and 250 μCi [γ-32P] ATP (Perkin Elmer) followed by purification with MicroSpin™ G-25 columns (Amersham). Blots were washed (2X SSC, 0.1% SDS) at 37°C for 30 min followed by two 30 min room temperature washes. Blots were exposed to Kodak BioMax film and quantification was determined by densitometry of autoradiographs using ImageQuant TL v2003.02 software. For RT-PCR, 10 μg of total RNA was reverse transcribed in a 25μl reaction containing 750 ng random hexamers, 1.25μg (dT)12-18, 0.2 mM dNTPs, 1X first strand buffer, 8 mM DTT, 50 U RNasin and 250 U Superscript III reverse transcriptase (Invitrogen). PCR conditions were 25°C for 5 min, 50°C for 1 hr and 70°C for 15 min. Following PCR, reactions were treated with 1μl (2 U) RNase H at 37°C for 20 min and 2 μl of cDNA were used as template for Dicer amplification in a 50 μl reaction containing 2.5U Triplemaster Taq (Eppendorf), 1X buffer, 0.4 mM dNTPs, 30 pmol Fwd primer in exon 23 (CACACGCCTCCTACCACTACAACAC) and 30 pmol Rev primer in exon 25 (GGCTGCATCATCGGATAGTACACC). PCR conditions were 27 cycles of 95°C for 30 sec, 55°C for 30 sec and 72°C for 1 min and 15 μl of each PCR reaction was resolved on a 10% acrylamide gel and exposed to Kodak BioMax film.
Histology, X-gal Staining, Immunohistochemistry and TUNEL Assays
Mouse embryos were collected after natural overnight matings and hematoxylin and eosin (H&E) staining was performed by standard procedures. For whole mount X-gal staining, embryos were harvested in PBS and fixed in 0.4% formaldehyde overnight at 4°C. Embryos were rinsed three times for 10 min in concentrated rinse buffer (0.1 M NaH2PO4, pH 7.4, 2 mM MgCl2, 0.2% Igepal, 0.1% sodium deoxycholate) and stained with 1 mg/ml X-gal in concentrated rinse buffer, 5 mM K3Fe(CN)6 and 5 mM K4Fe(CN)6 4-16 hr at 4°C with shaking. Embryos were rinsed three times for 5 min in PBS (pH 7.4), post-fixed in 4% formaldehyde overnight at 4°C and visualized with a Leica dissecting microscope. For paraffin sectioning, embryos were fixed in 4% formaldehyde overnight at 4°C, washed in PBS three times for 10 min and stored in 70% EtOH at 4°C. Embryonic limbs or tongue were dissected and dehydrated through 10 min washes in 70% EtOH, 95% EtOH, 100% EtOH and Citrisolv (Fisher). Dehydrated tissues were incubated in paraffin three times for 1 hr under vacuum at 65°C before routine embedding. Serial sections (7 μm) were cut on a Leica rotary microtome, laid across 45°C water to remove wrinkles, positioned on Superfrost Plus microscope slides (Fisher) and melted at 65°C for at least 30 min. To compare similar planes of distal forelimbs for control and mutant littermates a reference point was established at the junction between humerus, radius and ulna bones. The first section that did not contain any portion of humerus, but showed radius and ulna was deemed the reference. Sections equidistant from the reference for control and mutant distal forelimbs were compared for all experiments. For immunohistochemistry, slides were de-paraffinized and rehydrated through 10 min washes in Citrisolv, 100% EtOH, 95% EtOH, 70% EtOH, water and PBS. Antigen retrieval was performed for all antibodies by incubating slides in 10 mM Tris (pH 9), 1 mM EDTA for 30 min at 97°C and cooling slides in the same buffer at room temperature (RT) for 20 min. For monoclonal primary antibodies, slides were treated with papain-digested mouse IgGs as described (Lu and Partridge, 1998). Slides were blocked in PBS, 1% BSA, 1% heat inactivated goat serum and 0.025% Tween-20 for 1 hr at RT. Three PBS rinses were performed and slides were treated with primary antibody (see below) diluted in blocking buffer at 4°C overnight. Slides were rinsed three times with PBS and incubated with secondary antibody diluted in blocking buffer for 1 hr at RT in the dark. To reduce autofluorescence, slides were incubated with 10 mM CuSO4, 50 mM CH3COONH4 (pH 5.5) for 30 min at RT with shaking. Samples were rinsed three times in PBS, mounted with Vectashield containing DAPI (Vector) and visualized with a Leica fluorescence or confocal microscope. Primary antibodies and dilutions were 1:100 desmin (D33, DAKO), 1:100 MHC (MY32, Sigma) and 1:200 cleaved caspase-3 (Cell Signaling). TUNEL (Roche) was performed according to the manufacturer's protocol. TUNEL combined with MyoD was performed by doing MyoD immunochemistry first as described above followed by TUNEL accordingly to manufacturer's protocol. Secondary antibodies used were 1:250 goat anti-mouse IgG1 conjugated to AlexaFluor 488 (Invitrogen). Cleaved caspase-3 was detected using the Rabbit IgG Vectastain kit in combination with Vector Red Substrate (Vector labs). Coimmunohistochemistry specific for cleaved caspase-3 and myosin heavy chain was conducted by combining primary antibodies and followed with combined secondary antibodies.
Immunoblotting
Tongues were dissected and placed in 50-100 μl of 50 mM Tris·Cl (pH = 6.8), 1 mM EDTA, 2% SDS, 0.5 mM phenylmethylsulfonyl fluoride, 5 μg/ml pepstatin A, 1 μg/ml chymostatin, 1 mM aminocaproic acid, 1 mM p-aminobenzamidine, 1 μg/ml leupeptin, 2 μg/ml aprotinin and disrupted using a miniature pestle followed by brief sonication. Extracts were centrifuged at 16,000 × g for 10 min, and supernatant proteins were fractionated on 12.5% SDS-polyacrylamide gels (40 μg protein per lane). Gels were electroblotted to nitrocellulose and blocked 30 min in 5% Nonfat dry milk, 0.05% Igepal. Blots were incubated with primary antibodies (below) diluted in 5% Nonfat dry milk, 0.05% Igepal for 1 hr at RT followed by three 5 min washes in PBS, 0.05% Igepal. Horseradish perioxidase conjugated secondary antibodies specific for mouse or rabbit IgG (Amersham) were diluted 1:5000 in 5% Nonfat dry milk, 0.05% Igepal and incubated with blot for 30 min at RT followed by three 5 min washes in PBS, 0.05% Igepal and one 5 min wash in PBS. Immunoblots were visualized by ECL (Amersham). Specific primary antibodies and dilutions were 1:1000 anti-MyoD (5.8A, Dako), 1:500 anti-myogenin (F5D, ascites fluid Developmental Studies Hybridoma), 1:1000 anti-myosin heavy chain fast isoform (MY-32, Sigma), 1:1000 anti-desmin (D33, Dako) and 1:10,000 anti-Gapdh (6C5, Abcam).
Morphometric and Statistical Analysis
Myofiber number and average cross-sectional area were quantified on E16.5 desmin-stained distal forelimb transverse sections. Images were captured on a Zeiss fluorescent microscope and myofibers were counted and measured using Axiovision 4 software (Zeiss). Specifically, a 60 μm2 box was arbitrarily placed in each individual muscle mass. The box was small enough so that all edges were surrounded by muscle and no region of any box fell outside the muscle mass. The number of myofibers was counted in each box and the data represented as number of myofibers per cm2 for each muscle. A total of 398 and 264 fibers were counted for control and mutant sections, respectively. To determine myofiber cross sectional area, the outline of desmin-stained fibers were traced and the area was calculated using Axiovision 4 software. Average myofiber area was calculated for fifteen randomly chosen fibers in each control and mutant muscle. Statistical differences were evaluated by Student's t-test. For TUNEL and PH3 quantification, at least three sections per animal from 2-3 sets of control and mutant littermates were examined at each developmental time point. Only stained nuclei within muscle masses were counted and statistical differences were determined using Student's t-test.
Cell Culture, Annexin-V-FITC/Propidium Iodide Staining and Cell Sorting
To create the Dicer conditional MDER cell line, mouse embryonic fibroblasts derived from embryos with two Dicer conditional alleles (see above) were infected with a retrovirus containing the pBABE vector expressing a hygromycin-resistance gene and MDER, a MyoD-estrogen receptor fusion protein (Hollenberg et al. 1993). To mutate Dicer, cells were infected at a high MOI with Ad-CMV-cre adenovirus (Vector Biolabs). Cells were maintained in growth medium (GM), which consisted of Dulbecco's Modified Eagle (DME, Gibco), without phenol red, containing 10% fetal bovine serum (Hyclone), and 1% L-glutamine. pBABE-MDER infected cells were selected in 200 μg/mL hygromycin. To induce expression of MDER, cells were switched to differentiation medium (DM), which consisted of DME containing 1% L-glutamine, 0.5% horse serum, 10 μg/mL insulin, 10 μg/ml transferrin, and 10−7 M β-estradiol. Cells were genotyped under the same conditions as the mice (see above).Phase-contrast images of unfixed, unstained Dicer conditional MDER and Cre-infected Dicer conditional MDER cells following 96 hours of treatment with DM with or without β-estradiol were captured on film on an Axiovert 10 microscope (Zeiss). Images were digitized, and if needed, minor linear adjustments to contrast were made using Adobe Photoshop software.
Dicer conditional MDER and Cre-infected Dicer conditional MDER cells were cultured in triplicate on 24 well tissue-culture treated dishes in DM with or without β-estradiol for 24 hours, as indicated. Cells were trypsinized and co-stained with Annexin V-FITC and propidium iodide using the Annexin V-FITC Apoptosis Detection Kit (BioVision) per manufacturer's instructions. Cells were sorted on a FACScan cytometer (Beckton/Dickson). As a positive control for apoptosis and to define gating parameters, cells were treated in parallel with 50 μM geldanamycin in GM for 24 hours prior to analysis, and either single-stained with Annexin V-FITC or propidium iodide, or double-stained.
Results
miRNA levels are reduced following Dicer inactivation in skeletal muscle
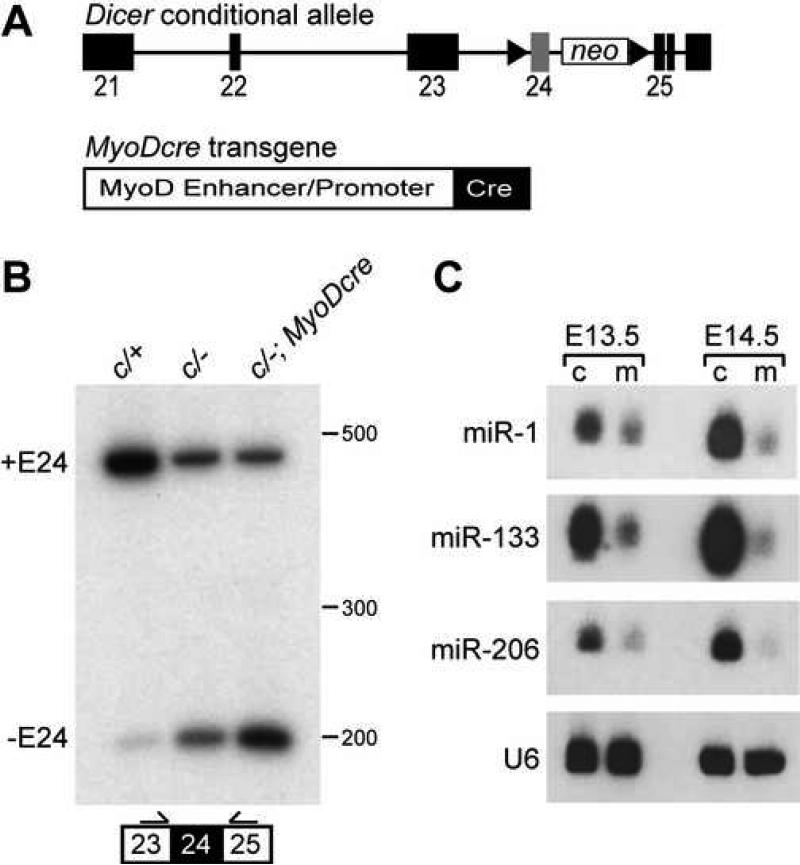
To bypass the early embryonic lethality associated with a Dicer null allele and investigate its role in skeletal muscle, we inactivated Dicer specifically in skeletal muscle using a Dicer conditional allele and a MyoDcre transgene (Chen et al., 2005; Harfe et al., 2005). The Dicer conditional allele used in this study has the exon encoding one of the RNase III domains flanked by loxP sites (Fig. 1A). Cre-mediated recombination of this exon has been shown to be effective in reducing the ability of Dicer to process miRNAs into their functional forms (Harfe et al., 2005). The MyoDcre transgene contains Cre recombinase downstream of MyoD core enhancer and promoter elements (Fig. 1A). MyoDcre activity is consistent with endogenous MyoD mRNA expression (Chen et al., 2005). It is active in the mandibular arch at E9.75, somitic myotomes at E10.5, developing limbs buds at E11.5 and all trunk musculature by E12.5. Mice containing two wild type alleles, Dicer conditional and wild type alleles (c/+), conditional and deletion alleles (c/−) or conditional and wild type alleles together with MyoDcre (c/+; MyoDcre) were phenotypically indistinguishable from one another and are referred to as controls. Mice containing Dicer conditional and deletion alleles combined with MyoDcre (c/−; MyoDcre) are designated as mutants.
Fig. 1.
Inactivation of skeletal muscle Dicer and decreased muscle-specific miRNAs. (A) Dicer conditional allele and MyoDcre transgene. Dicer conditional allele contains loxP sites (triangles) flanking the RNAse III domain encoded by exon (filled boxes) 24 (gray box). A neomycin resistance cassette (neo) is retained in intron 24. MyoDcre transgene contains Cre recombinase (filled box) downstream of the MyoD core enhancer and proximal promoter elements (open box). (B) RT-PCR on E17.5 tongue cDNA using primers flanking targeted exon 24 (filled box). Primers (arrows) were positioned in exon 23 and 25. Mice contained a combination of Dicer conditional (c), wild type (+) or Dicer deleted exon 24 (−) alleles and a MyoDcre transgene. Transcripts containing (+E24) or missing exon 24 (−E24) result in 431 and 199 bp fragments, respectively. (C) RNA blot analysis of E13.5 and E14.5 control and mutant littermate limbs to detect muscle-specific miRNAs (miR). U6 snRNA is the loading control. Direct comparison between miR levels cannot be made because probe specific activities and film exposure times varied for each miR.
We assessed Dicer inactivation in E17.5 tongue muscle by RT-PCR with primers flanking floxed exon 24 (Fig. 1B). The neomycin resistance cassette is in intron 24 and would not have been amplified. The majority of Dicer transcripts contained exon 24 in Dicerc/+ tongue while those lacking exon 24 were present at much lower levels (lane 1). The absence of exon 24 containing mRNA may be explained by low level alternative splicing. This suggests that Dicer isoforms lacking the exon 24 encoded RNAse III domain might be required for properties other than miRNA processing. As expected, Dicerc/− tongue muscle showed relatively equal amounts of Dicer mRNA containing and missing exon 24 (lane 2). Dicerc/−; MyoDcre mutant skeletal muscle displayed higher Dicer transcript levels that lacked floxed exon 24 (lane 3). The presence of exon 24 containing transcripts in mutants could have been due to expression of Dicer in non-muscle cells of the tongue that did not express MyoDcre, such as neurons and vasculature, or was a result of inefficient MyoDcre-mediated recombination. These data revealed that MyoDcre was capable of deleting the floxed region of Dicer in skeletal muscle.
To confirm that miRNA processing was blocked in mutant embryos we performed RNA blot analysis specific for three miRNAs that are enriched in skeletal muscle, miR-1, miR-133 and miR-206, using whole limb RNA from E13.5 and E14.5 embryos (Fig. 1C). miR-1 levels were reduced by 52% at E13.5 and 76% at E14.5 in mutants compared to controls, and miR-133 and miR-206 RNAs each decreased by 74% at E13.5 and 90% at E14.5. Similar reductions were observed in mutants at later developmental stages (data not shown). Interestingly, the mature miRNA level reduction was most dramatic at E14.5, two days after MyoDcre is activated throughout E 12.5 limb muscle. This delay has been observed in other tissues and cells where Dicer was inactivated and most likely reflects the long half-life of Dicer and/or miRNAs (Andl et al., 2006). The residual amount of functional miRNA present in mutant limbs may have resulted from incomplete Cre-mediated recombination or from possible expression of these miRNAs in non-muscle cell types in limbs, including bone and cartilage. These results demonstrated that mice containing Dicer conditional alleles with a MyoDcre transgene showed an increase in Dicer exon 24 deleted transcripts which resulted in decreased skeletal muscle miRNA levels.
Dicer inactivation results in perinatal lethality and skeletal muscle hypoplasia
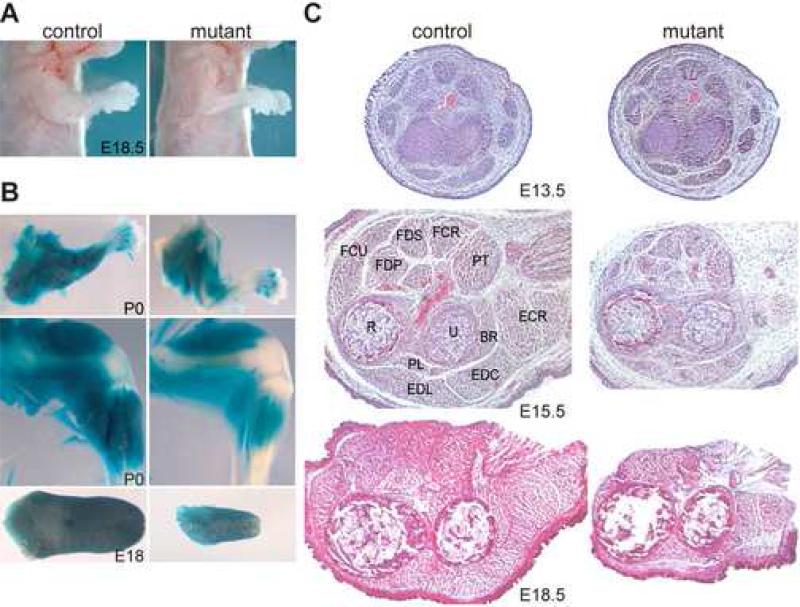
All Dicer skeletal muscle mutants died within minutes following birth. Therefore, phenotypic analysis was performed on newborns and embryos. We observed that the gross anatomy of late stage mutant embryos differed from that of control littermates. Specifically, mutant embryonic limbs appeared to be less developed (Fig. 2A). To visualize muscle morphology, we X-gal stained embryos and newborns containing ROSA26 reporters (Soriano, 1999) to specifically stain those cells in which Cre was active and lacZ was expressed (Fig. 2B). We found that mutant embryos showed a decrease in muscle mass in both limbs and tongue. Histological examination of H&E and anti-desmin, a pan myofiber marker, stained distal forelimb cross sections confirmed that there was less skeletal muscle for all muscles of mutant embryonic limbs (Fig. 2C, Fig. 3A). This decrease was first observed in E14.5 mutant embryos and became more pronounced during subsequent stages of embryogenesis. By E18.5 the amount of muscle in mutant limbs was drastically reduced compared to control littermates. Furthermore, a patterning defect was not observed in mutant embryos as each of the individual muscles was present albeit in smaller proportions.
Fig. 2.
Dicer mutants display less skeletal muscle. (A) Skinned E18.5 control and mutant littermates. Note the impaired development of mutant forelimb muscle. (B) X-gal stained P0 control and mutant littermate forelimbs (top panels), hindlimbs (middle panels) and E18 tongues (bottom panels). Mice contained the R26 reporter in which X-gal specifically stains cells that express MyoDcre. (C) H&E staining of control and mutant transverse distal forelimb sections at different developmental time points (E13.5-E18.5). Each muscle is labeled in the E15.5 control section. Muscle and bones are brachioradials (BR), extensor carpi radialis brevis (ECR), extensor digitorum commusis (EDC), extensor digitorum lateralis (EDL), flexor carpi radialis (FCR), flexor carpi ulnaris (FCU), flexor digitorum profundus caput fadiale (FDP), pollicis longus (PL), and pronator teres (PT) muscles and ulna (U) and radius (R) bones.
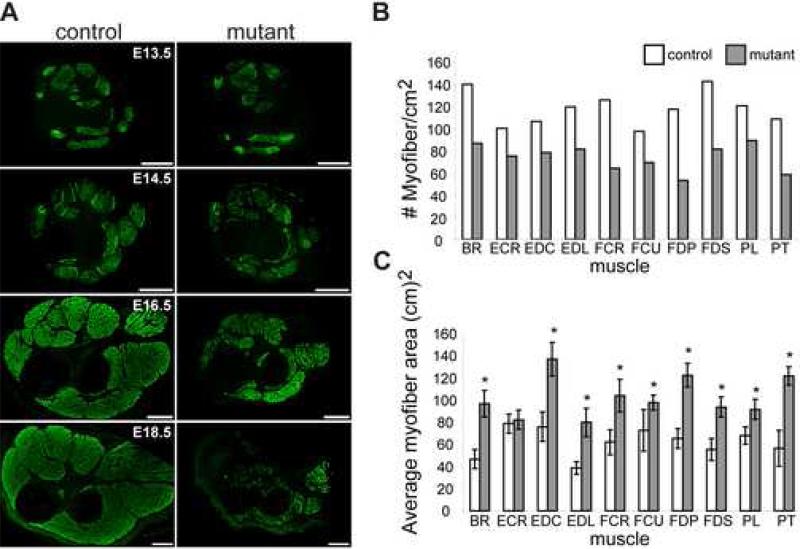
Fig.3.
Dicer mutants show skeletal muscle hypoplasia. (A) Desmin immunohistochemistry of control and mutant transverse distal forelimb sections at different developmental time points (E13.5-E18.5). Scale bars equal 250 μm. (B-C) Quantification of myofiber number (B) and cross sectional (C) area for each muscle group of E16.5 control and mutant littermate distal forelimbs. Asterisk signifies P<0.05. Muscle abbreviations are listed in Fig. 2 legend.
We next sought to determine if the reduction in skeletal muscle was due to skeletal muscle hypoplasia (fewer fibers) and/or hypotrophy (smaller fibers). To distinguish between these two possibilities, we counted the number of myofibers per given area and measured myofiber cross-sectional area for each distal limb muscle of E16.5 embryos (Fig. 3B, C). These measurements showed that all mutant limb muscles contained fewer fibers and that the mutant fibers were larger than the controls. An overall increase in average myofiber size was observed because mutant muscles contained fewer smaller fibers compared to controls. We also examined fiber type distribution in control and mutant limbs by immunohistochemistry with antibodies specific for myosin isoforms present in fast or slow fibers. We observed the presence of both fast and slow fibers and similar staining patterns in distal forelimbs of control and mutant embryos at E13.5-E16.5 (data not shown). These data suggested that the reduction in muscle mass observed in Dicer mutant embryos resulted from skeletal muscle hypoplasia, rather than an inability to pattern skeletal muscle or specify myofiber type.
Defective morphogenesis and maintenance of Dicer mutant myofibers
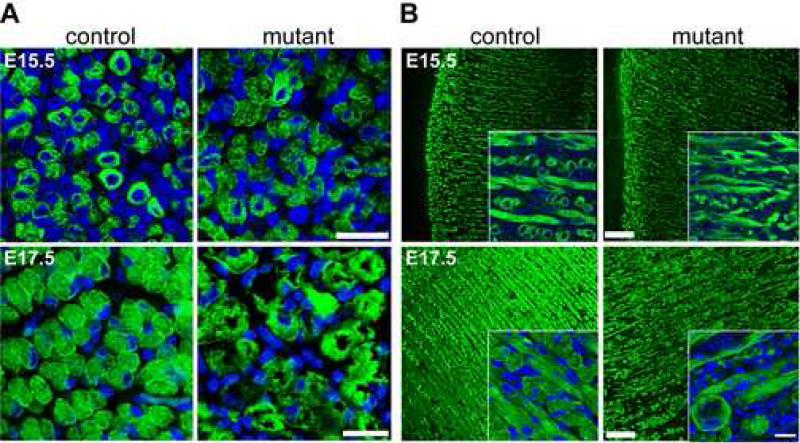
To address if Dicer is important for myofiber formation or maturation we examined myofiber development in limb and tongue musculature using immunohistochemistry with antibodies specific for desmin, a pan myofiber marker (Fig. 4). Initiation of myofiber formation at E15.5 in limb (Fig. 4A) and tongue (Fig. 4B) appeared similar in mutant embryos compared to control. We observed small circular desmin-positive myotubes with centralized nuclei that are indicative of immature fibers. By E16.5, early maturation was mildly affected in mutant fibers in limb and tongue (data not shown). Control and mutant fibers had increased in size and the majority of nuclei migrated to the periphery. However, mutant fibers were slightly disorganized compared to control. The E16.5 control fibers began to become organized in rows while mutant fibers showed less organization and this continued in E17.5 embryos and was especially noticeable in tongue (Fig. 4B). The E17.5 control tongues displayed a regular lattice of longitudinal and transverse myofibers that was not observed in mutants. Furthermore, the morphology of mutant fibers was considerably different than control at later developmental time points. Mutant fibers lacked desmin staining in the center, varied in size and were irregularly shaped. These results indicated that Dicer activity was dispensable for initiating myofiber formation, but was required for myofiber morphogenesis.
Fig. 4.
Abnormal myofiber morphology and organization in Dicer mutants. (A-B) Control and mutant littermate extensor carpi radialis transverse sections (A) or tongue sagittal sections (B) stained with antibody specific for desmin at E15.5 and E17.5. DAPI (blue) stains nuclei. Insets show representative higher magnification images of tongues. All scale bars equal 25 μm.
Increased apoptosis in Dicer mutant muscle
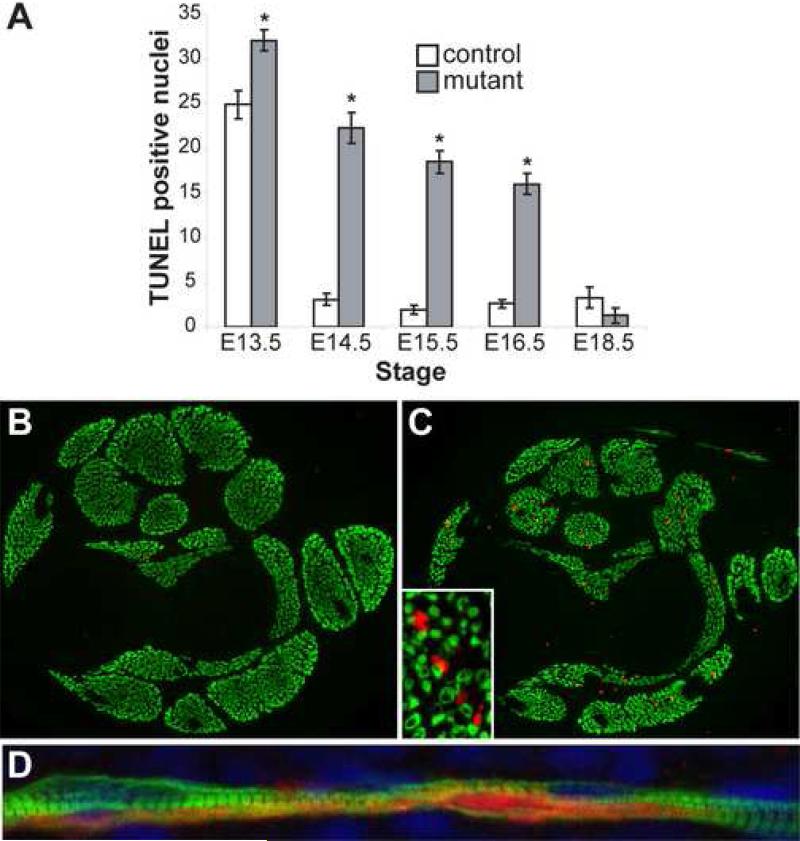
Increased cell death has been observed when Dicer was inactivated in limb mesoderm, lung epithelium and cultured cells (Fukagawa et al., 2004; Harfe et al., 2005; Harris et al., 2006). Therefore, we hypothesized that increased cell death of myogenic cells contributed to the decreased skeletal muscle in mutant embryos. To test this hypothesis, we performed TUNEL, a specific marker of DNA fragmentation associated with apoptotic nuclei, on control and mutant forelimb sections at different developmental stages (Fig. 5 and Supplemental Fig. S1). At E13.5, both control and mutant limb muscles were observed to have an appreciable number of apoptotic nuclei although the number was slightly higher in mutant skeletal muscle (Fig. 5A). By E14.5, the number of TUNEL positive nuclei diminished drastically in control embryos but remained high in mutants (P<0.001). We observed a similar significant increase in apoptosis in mutant skeletal muscle compared to control at E15.5 and E16.5. By E18.5, the number of apoptotic nuclei was similarly low in control and mutant skeletal muscle. This is most likely because the majority of mutant muscle cells had already died.
Fig. 5.
Increased apoptosis in Dicer mutant skeletal muscle. (A) Quantification of TUNEL positive nuclei in control and mutant littermate distal forelimb muscle masses at different developmental stages. At least three sections from each of two control and mutant embryos were counted for each time point. Please refer to Supplemental Fig. S2 for images. Asterisk indicates P<0.05. (B-C) Cleaved caspase-3 (red) and myosin heavy chain (green) coimmunohistochemistry on E15.5 control (B) and mutant littermate (C) transverse distal forelimb sections (10X). Inset (40X) shows higher magnification of a mutant section. (D) Cleaved caspase-3 (red) and myosin heavy chain (green) coimmunohistochemistry on E16.5 mutant sagittal distal forelimb section. Nuclei are stained with DAPI (blue).
Although the apoptotic nuclei were present in skeletal muscle masses, we wanted to determine if the TUNEL positive nuclei were indeed myonuclei. Unfortunately, we were unable to detect MyoD or myogenin in TUNEL-positive nuclei probably due to apoptosis-associated protein degradation which precedes DNA fragmentation. As an alternative, we examined the level of cleaved caspase-3, an earlier marker of cell apoptosis. Dicer mutants showed increased cleaved caspase-3 levels compared to controls (Fig. 5B,C). Importantly, the majority of cleaved caspase-3 positive cells were present within skeletal muscles although the cleaved caspase-3 staining did not appear to completely overlap with MHC on E15.5 mutant transverse sections (Fig. 5C inset). However, sagittal limb sections revealed that mutant myofibers expressed both cleaved caspase-3 and MHC and that the cleaved caspase-3 signal was not homogeneously distributed throughout individual myofibers (Fig. 5D). This result confirmed that Dicer mutant skeletal muscle cells were indeed dying.
Loss of Dicer in myoblasts also leads to enhanced cell death
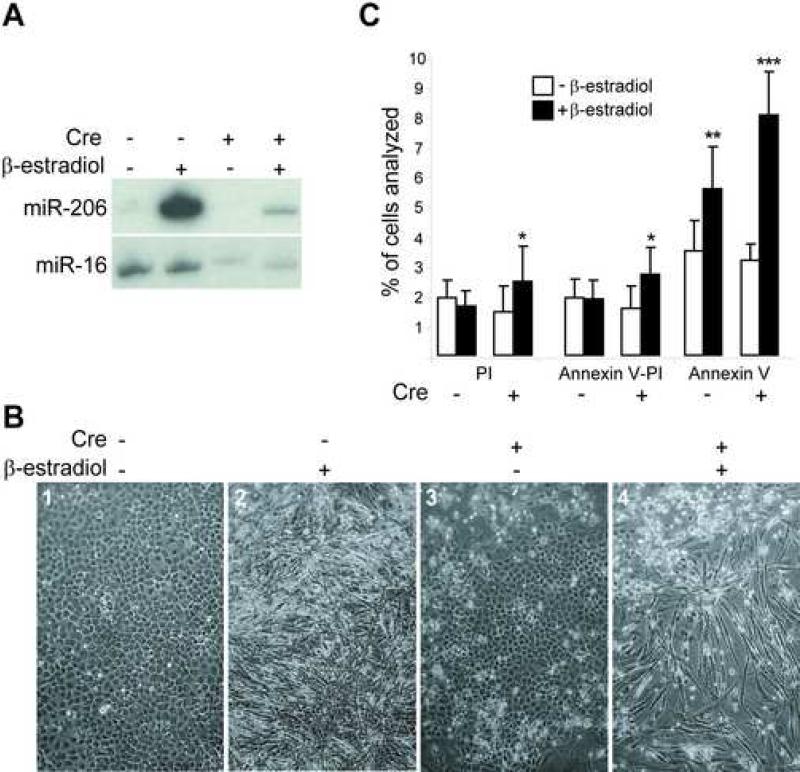
We also investigated the effect of Dicer inactivation on myogenic differentiation in vitro. We were unable to derive myoblasts from mutant embryos, although we successfully cultured control myoblasts. This was likely due to impaired growth and/or increased apoptosis of mutant myoblasts. To circumvent this problem, we converted Dicer conditional mouse embryonic fibroblasts (MEFs) to myoblasts by Myod overexpression and mutated Dicer by ectopically expressing Cre to yield a relatively pure population of Dicer mutant myoblasts. More specifically, we infected spontaneously immortalized Dicer conditional fibroblasts with a retroviral construct that constitutively expresses a fusion protein between mouse Myod and the hormone binding domain of the estrogen receptor (Hollenberg et al., 1993). Under conditions of low serum and in the presence of β-estradiol, the transcriptional activity of the Myod-ER fusion (MDER) is induced and is sufficient to convert fibroblasts to the muscle phenotype (Bergstrom et al., 2002). To inactivate Dicer in conditional MDER MEFs, we infected cells with an adenovirus harboring the Cre recombinase gene regulated by the CMV promoter (Ad-CMV-Cre). We confirmed Dicer exon 24 recombination in Ad-CMV-Cre cultures by genomic PCR analysis (data not shown). We also determined if muscle miRNA levels decreased in Cre-infected Dicer conditional MDER by RNA blotting (Fig. 6A). As previously reported (Rosenberg et al., 2006), we observed increased levels of muscle-specific miR-206 in β-estradiol induced Dicer conditional MDER that have Myod activated. Importantly, miR-206 levels decreased when these cells were infected with Cre. miR-16, which is not sensitive to Myod activation, was not affected by β-estradiol and decreased upon Cre infection. Therefore, muscle miRNAs increased upon Myod activation and decreased when Cre was expressed.
Fig. 6.
Decreased muscle miRNAs and increased apoptosis of Dicer mutant muscle cells in vitro. (A) RNA blot analysis of Dicer condtional MDER (− Cre) and Cre-infected Dicer conditional MDER (+ Cre) fibroblast (− β-estradiol) and muscle cell (+ β-estradiol, 24 hrs.) cultures. miR-206 (which is induced by Myod-ER expression) and miR-16 (which is constitutively expressed in these cells and can be used as a loading control for first two lanes) were analyzed by stripping and sequential hybridization of the same blot. (B) Phase-contrast images of unfixed Dicer conditional MDER (− Cre) and Cre-infected Dicer conditional MDER (+ Cre) cells, following 96 hours in DM with or without β-estradiol, as indicated. 10X magnification. (C) Flow cytometric analysis of Dicer conditional MDER (− Cre) and Cre-infected Dicer conditional MDER (+ Cre) cells. Cells were cultured in DM with (+) or without β-estradiol (−) for 24 hours, as indicated. Cells were trypsinized, stained with Annexin V-FITC and propidium and sorted and quantified by FACS. Shown are summary data of the percentage of positively-stained cells, representing three independent trials. *: P<0.05, **: P<0.01 and ***: P<0.001
We then examined the ability of the Dicer conditional MDER and Cre-infected Dicer conditional MDER cells to differentiate into myotubes in vitro. Strikingly, we found that while the Dicer conditional MDER cells were competent to form myotubes, the Cre-infected cultures exhibited a marked level of cell death during differentiation, as revealed by phase-contrast microscopy (Fig. 6B, panels 2 and 4). Importantly, this increased cell death was dependent on the induction of MDER activity, as Cre-infected cells that were cultured in DM without β-estradiol did not exhibit the same level of cell death (Fig. 6B, panels 3 and 4). To quantify this apparent increase in cell death and to determine whether it reflected increased apoptosis in differentiating Cre-infected Dicer conditional MDER cultures, we performed Annexin V-FITC/propidium iodide staining of the cultures and quantified the positively-stained cells by FACS analysis (Fig. 6 and Supplemental Fig. S3). AnnexinV-FITC staining identifies apoptotic cells by specifically detecting the translocation of phosphatidylserine from the inner face of the plasma membrane to the cell surface that occurs during the early stages of apoptosis. To permit quantitative cell sorting by FACS, we chose to examine an early stage of myogenic differentiation (24 hours) when we expected that apoptosis may be initiated, but before the cells had fused to become multinucleated myotubes. All samples were analyzed in the presence of both propidium iodide and Annexin V-FITC, and cell sorting revealed the percentage of cells that were viable, nonviable (either apoptotic or necrotic; propidium iodide positive), or apoptotic (Annexin V-FITC positive) (Fig. 6C). Consistent with the increased cell death observed in Cre-infected Dicer conditional MDER cells in the presence of DM plus β-estradiol, we observed a greater than two-fold increase in Annexin V-positive, apoptotic cells when Myod activity was induced in the absence of Dicer function, compared to uninduced Cre-infected Dicer conditional MDER cells (Fig. 6C). Although there was also an increase in the percentage of Dicer conditional MDER cells that were Annexin V positive when Myod activity was induced compared to uninduced Dicer conditional MDER cells, we note that the magnitude of increased apoptosis was significantly greater in differentiating cells that lack Dicer function. Importantly, not all of the Cre-infected Dicer conditional MDER cells underwent apoptosis in the presence of DM with β-estradiol, as evidenced by the development of myotubes at later stages of Myod-induced differentiation (Fig. 6B, panel 4).
Examination of the propidium iodide-positive population and the dually positive Annexin V-FITC and propidium iodide positive cells, revealed an approximate 1.7-fold increase in cell death in the Dicer−/− MDER cells at 24 hours following Myod induction (Fig. 6C). We cannot attribute these changes directly to apoptosis, but because propidium iodide can enter cells in the later stages of apoptosis, it is likely that the doubly positive cells represent late stages of apoptosis rather than necrosis. As this effect was not observed in Dicer conditional MDER cells following the induction of Myod activity, and as the percentage of propidium iodide-positive cells was comparable in both Dicer conditonal MDER and Cre-infected Dicer conditional MDER cells in the absence of Myod activity, we attribute this effect specifically to Myod activation in the absence of Dicer function.
Overall, these results are consistent with both the increased cell death that we observed in late-stage differentiated Cre-infected Dicer conditional MDER cultures in vitro (Fig. 6B, panel 4) and with the increased apoptosis seen in Dicer mutant muscle in vivo. Therefore, we conclude that the loss of Dicer function in skeletal muscle promoted increased apoptosis in both myofibers and myoblasts .
Discussion
Dicer and miRNAs are important for a variety of developmental processes (Hornstein et al., 2005; Lee et al., 2006; O'Rourke et al., 2006). The objective of this study was to determine the role of Dicer and miRNA-mediated gene regulation in skeletal muscle. Our analysis demonstrates that Dicer activity is essential for skeletal muscle development during embryogenesis and post-natal life. Dicer inactivation in skeletal muscle results in lower levels of muscle-specific miRNAs and Dicer muscle mutants die perinatally and are characterized by skeletal muscle hypoplasia. Reduced skeletal muscle is accompanied by abnormal myofiber morphology. The skeletal muscle defects associated with loss of Dicer function may be explained by increased apoptosis. This study is the first, to our knowledge, to provide in vivo evidence of the importance of Dicer function and miRNA-mediated gene silencing during mammalian skeletal muscle development.
Dicer is essential for skeletal muscle morphogenesis
The phenotype associated with Dicer inactivation and reduced muscle miRNAs in mice is similar to that observed in Drosophila miR-1 (dmir-1) mutants (Kwon et al., 2005; Sokol and Ambros, 2005). Knockout dmir-1 flies arrest during embryogenesis or larval development, display disorganized muscle morphology and have aberrant expression of muscle-specific genes. Further, dmir-1 targets Delta, a membrane-bound ligand for Notch and dmir-1 mutants have disrupted Delta-Notch signaling. Notch signaling also plays an important role in mammalian myogenesis (Luo et al., 2005). Therefore, potential defects in Notch signaling in mouse Dicer mutant embryos may contribute to abnormal muscle morphogenesis. These observations argue that miR-1 plays an essential role in embryonic muscle development.
Interestingly, Dicer skeletal muscle mutants are observed to have reduced skeletal muscle in the presence of increased levels of MRFs (see Supplemental Fig. S3). MyoD, myogenin and Mef2 promote miR-1, miR-133 and miR-206 expression by binding to upstream cis elements (Rao et al., 2006). This raises the possibility that MRFs are upregulated to compensate for reduced miRNA levels in Dicer mutants. However, this does not explain why increased MRFs fail to promote myogenesis in Dicer mutant skeletal muscle. One possibility is that myogenic inhibitory factors that antagonize MRFs are upregulated in Dicer mutants. Examples of such factors are MyoR and Id family members (Benezra et al., 1990; Lu et al., 1999). These proteins function as myogenic antagonists by inhibiting binding of MRFs to E boxes in the promoters of muscle-specific genes. Indeed, overexpression of miR-206 in cultured myoblasts results in decreased expression of MyoR, Id1, Id2 and Id3 (Kim et al., 2006). Further, preliminary results indicate that Id2 mRNA is elevated 1.5 fold in Dicer mutant tongues (data not shown). Thus, muscle miRNAs may promote myogenesis by downregulating the expression of inhibitory factors.
The decrease in muscle mass in Dicer mutants is strikingly similar to phenotypes associated with muscular dystrophies and aged skeletal muscle. It will be interesting to see if Dicer mutations, or disrupted miRNA-mediated gene regulation, contribute to skeletal muscle myopathy and age-related sarcopenia.
Dicer is required for muscle cell survival during embryogenesis
Increased apoptosis in Dicer mutant skeletal muscle provides one explanation why mutant embryos are characterized by skeletal muscle hypoplasia and abnormal myofiber morphology. However, the relationship between Dicer, miRNAs and apoptosis is unclear. Programmed cell death occurs in a variety of cell types during embryogenesis and is thought to be required for rapid tissue remodeling (Huppertz and Herrler, 2005). Productive tissue growth during development requires the rate of cell proliferation to exceed that of cell death and miRNA-mediated gene silencing may function to restrict or inhibit apoptosis during embryonic development by targeting pro-apoptotic factors. miR-21 has anti-apoptotic activity in glioblastoma cells and several other miRNAs are implicated in tumor suppression (Chan 2005, Chen 2006). It is possible that muscle miRNAs have an anti-apoptotic activity that is required to ensure that the apoptosis required at early developmental time points does not persist during later periods of rapid proliferation and growth. For example, muscle miRNA(s) may function in the decline of apoptosis observed in limb skeletal muscle between E13.4 and E14.5 (Fig 5) by inhibiting the expression of pro-apoptotic factors. Loss of this regulation may then result in increased apoptosis and decreased skeletal muscle. It is also possible that the apoptosis observed in mutant skeletal muscle is associated with non-muscle cell types.
Alternatively, increased apoptosis in Dicer mutant muscle may be independent of miRNAs and result from uncharacterized Dicer functions. Loss of Dicer in limb and lung also results in increased cell death and morphological defects (Harfe et al., 2005; Harris et al., 2006). In fission yeast, Dicer and RNA interference are important for regulating chromosome segregation and heterochromatic gene silencing (Hall et al., 2003; Provost et al., 2002). Therefore, Dicer inactivation might lead to perturbations in chromosome segregation and condensation which trigger cell death to avoid aberrant cell division and gene expression.
Dicer's miRNA processing ability has been shown to be essential for limb, lung, skin and hair development. Our results demonstrate that Dicer and miRNAs are also indispensable for embryonic skeletal muscle development. It will be interesting to see if Dicer and miRNAs function during post-natal myogenesis and myofiber regeneration.
Supplementary Material
Fig. S1. Increased apoptosis in Dicer mutant skeletal muscle. (A) TUNEL analysis of E13.5 and 14.5 control and mutant littermate distal forelimb cross sections. Arrowheads point to TUNEL positive nuclei within skeletal muscles. TUNEL positive nuclei without arrowheads are positioned outside of skeletal muscle masses and were not included in the quantification in Fig.6. Inset shows blowup of one arrowhead and a TUNEL positive nucleus. Scale bars equal 100 μm.
Fig. S2. Flow cytometric analysis of Dicer conditional MDER (− Cre) and Cre-infected Dicer conditional MDER (+ Cre) cells. Cells were cultured in DM with or without β-estradiol for 24 hours, as indicated. Cells were trypsinized and stained with Annexin VFITC and propidium iodide using the Annexin V-FITC Apoptosis Detection Kit (BioVision) per manufacturer's instructions. As a positive control for apoptosis and to define gating parameters, cells were treated in parallel with 50 μM geldanamycin in GM for 24 hours prior to analysis. Cells were sorted and quantified by FACS. Shown are representative panels from one of three independent trials.
Fig. S3. Dicer mutants show aberrant expression of myogenic factors. Immunoblot analysis of E18.5 control (n=3) and mutant (n=3) tongue extracts with antibodies specific for MyoD, Myogenin, myosin heavy chain fast isoform and desmin. Gapdh was the loading control.
Acknowledgements
We thank Lauren McIntyre (University of Florida) for help with statistical analysis and Xin Sun (University of Wisconsin, Madison) for helpful comments on the manuscript. This study was supported by grants from the Muscular Dystrophy Association and the National Institutes of Health to B.D.H, D.J.G. (AG20911), S.A.G. (AR052581) and M.S.S. (AR46799).
Appendix A. Supplementary data
Supplementary data associated with this article can be found in the online version at
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andl T, et al. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16:1041–9. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–63. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Benezra R, et al. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Bergstrom DA, et al. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell. 2002;9:587–600. doi: 10.1016/s1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–95. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Bernstein E, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–7. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Brand-Saberi B. Genetic and epigenetic control of skeletal muscle development. Ann Anat. 2005;187:199–207. doi: 10.1016/j.aanat.2004.12.018. [DOI] [PubMed] [Google Scholar]
- Buckingham M, et al. Myogenic progenitor cells in the mouse embryo are marked by the expression of Pax3/7 genes that regulate their survival and myogenic potential. Anat Embryol (Berl) 2006;211(Suppl 7):51–6. doi: 10.1007/s00429-006-0122-0. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–4. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- Chen JC, et al. MyoD-cre transgenic mice: a model for conditional mutagenesis and lineage tracing of skeletal muscle. Genesis. 2005;41:116–21. doi: 10.1002/gene.20104. [DOI] [PubMed] [Google Scholar]
- Chen JF, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–33. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossu G, Biressi S. Satellite cells, myoblasts and other occasional myogenic progenitors: possible origin, phenotypic features and role in muscle regeneration. Semin Cell Dev Biol. 2005;16:623–31. doi: 10.1016/j.semcdb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Fukagawa T, et al. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol. 2004;6:784–91. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- Hall IM, et al. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci U S A. 2003;100:193–8. doi: 10.1073/pnas.232688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, et al. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KS, et al. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci U S A. 2006;103:2208–13. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg SM, et al. Use of a conditional MyoD transcription factor in studies of MyoD trans-activation and muscle determination. Proc Natl Acad Sci U S A. 1993;90:8028–32. doi: 10.1073/pnas.90.17.8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollway G, Currie P. Vertebrate myotome development. Birth Defects Res C Embryo Today. 2005;75:172–9. doi: 10.1002/bdrc.20046. [DOI] [PubMed] [Google Scholar]
- Hornstein E, et al. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–4. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- Huh MS, et al. Muscle function and dysfunction in health and disease. Birth Defects Res C Embryo Today. 2005;75:180–92. doi: 10.1002/bdrc.20045. [DOI] [PubMed] [Google Scholar]
- Huppertz B, Herrler A. Regulation of proliferation and apoptosis during development of the preimplantation embryo and the placenta. Birth Defects Res C Embryo Today. 2005;75:249–61. doi: 10.1002/bdrc.20056. [DOI] [PubMed] [Google Scholar]
- John B, et al. Prediction of human microRNA targets. Methods Mol Biol. 2006;342:101–13. doi: 10.1385/1-59745-123-1:101. [DOI] [PubMed] [Google Scholar]
- Kalcheim C, et al. Myotome formation: a multistage process. Cell Tissue Res. 1999;296:161–73. doi: 10.1007/s004410051277. [DOI] [PubMed] [Google Scholar]
- Kim HK, et al. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol. 2006;174:677–87. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, et al. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–9. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- Kwon C, et al. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci U S A. 2005;102:18986–91. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CT, et al. MicroRNAs in mammalian development. Birth Defects Res C Embryo Today. 2006;78:129–39. doi: 10.1002/bdrc.20072. [DOI] [PubMed] [Google Scholar]
- Lewis BP, et al. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lu J, et al. MyoR: a muscle-restricted basic helix-loop-helix transcription factor that antagonizes the actions of MyoD. Proc Natl Acad Sci U S A. 1999;96:552–7. doi: 10.1073/pnas.96.2.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QL, Partridge TA. A new blocking method for application of murine monoclonal antibody to mouse tissue sections. J Histochem Cytochem. 1998;46:977–84. doi: 10.1177/002215549804600813. [DOI] [PubMed] [Google Scholar]
- Luo D, et al. The regulation of Notch signaling in muscle stem cell activation and postnatal myogenesis. Semin Cell Dev Biol. 2005;16:612–22. doi: 10.1016/j.semcdb.2005.07.002. [DOI] [PubMed] [Google Scholar]
- O'Rourke JR, et al. MicroRNAs in mammalian development and tumorigenesis. Birth Defects Res C Embryo Today. 2006;78:172–9. doi: 10.1002/bdrc.20071. [DOI] [PubMed] [Google Scholar]
- Parker MH, et al. Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nat Rev Genet. 2003;4:497–507. doi: 10.1038/nrg1109. [DOI] [PubMed] [Google Scholar]
- Provost P, et al. Dicer is required for chromosome segregation and gene silencing in fission yeast cells. Proc Natl Acad Sci U S A. 2002;99:16648–53. doi: 10.1073/pnas.212633199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao PK, et al. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A. 2006;103:8721–6. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F. Skeletal muscle progenitor cells: from embryo to adult. Cell Mol Life Sci. 2006;63:1221–5. doi: 10.1007/s00018-006-6015-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relaix F, et al. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol. 2006;172:91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg MI, et al. MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol. 2006;175:77–85. doi: 10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sempere LF, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol NS, Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 2005;19:2343–54. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–1. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Venuti JM, et al. Myogenin is required for late but not early aspects of myogenesis during mouse development. J Cell Biol. 1995;128:563–76. doi: 10.1083/jcb.128.4.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, et al. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci U S A. 1989;86:5434–8. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, et al. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–20. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Increased apoptosis in Dicer mutant skeletal muscle. (A) TUNEL analysis of E13.5 and 14.5 control and mutant littermate distal forelimb cross sections. Arrowheads point to TUNEL positive nuclei within skeletal muscles. TUNEL positive nuclei without arrowheads are positioned outside of skeletal muscle masses and were not included in the quantification in Fig.6. Inset shows blowup of one arrowhead and a TUNEL positive nucleus. Scale bars equal 100 μm.
Fig. S2. Flow cytometric analysis of Dicer conditional MDER (− Cre) and Cre-infected Dicer conditional MDER (+ Cre) cells. Cells were cultured in DM with or without β-estradiol for 24 hours, as indicated. Cells were trypsinized and stained with Annexin VFITC and propidium iodide using the Annexin V-FITC Apoptosis Detection Kit (BioVision) per manufacturer's instructions. As a positive control for apoptosis and to define gating parameters, cells were treated in parallel with 50 μM geldanamycin in GM for 24 hours prior to analysis. Cells were sorted and quantified by FACS. Shown are representative panels from one of three independent trials.
Fig. S3. Dicer mutants show aberrant expression of myogenic factors. Immunoblot analysis of E18.5 control (n=3) and mutant (n=3) tongue extracts with antibodies specific for MyoD, Myogenin, myosin heavy chain fast isoform and desmin. Gapdh was the loading control.