SUMMARY
A partial cDNA sequence was obtained from the human blood fluke, Schistosoma mansoni using a signal sequence trap approach. The full-length cDNA was cloned and termed Sm-7TM. The corresponding open reading frame had 7 membrane spanning domains and shared identity with a small, novel group of seven transmembrane (7TM) receptors from vertebrates and invertebrates, including the human ee3 receptor - a heptahelical protein implicated in neuronal signalling. Phylogenetic analysis of this novel family showed that the Sm-7TM ORF formed a clade with exclusively invertebrate sequences. Based on topology modelling with ee3, Sm-7TM was predicted to possess an intracellular C-terminal tail, which was expressed as a soluble thioredoxin fusion protein (Sm-7TMC) in Escherichia coli and purified using metal ion-affinity chromatography. A polyclonal antiserum against this domain was used to detect Sm-7TM in detergent-soluble parasite extracts and to immunolocalize the receptor to the tegument of adult S. mansoni.
Keywords: Schistosoma mansoni, seven transmembrane receptor, tegument
INTRODUCTION
Schistosomiasis of humans is caused by the infection with blood flukes of the genus Schistosoma (also called schistosomes). This disease occurs throughout the developing world and is most prevalent in the poorest populations. Schistosomes afflict more than 200 million people, with ~1 billion being at risk of schistomiasis. It is estimated that 20 million people suffer from severe consequences of this chronic and debilitating disease, responsible for hundreds of thousands of deaths per year (Ross et al. 2002; van der Werf et al. 2003; King et al. 2005). Anti-schistosome vaccines, in combination with other control strategies, including chemotherapy, are needed to make elimination of the disease possible (Todd and Colley, 2002; Capron et al. 2005; Loukas et al. 2007).
The outermost surface of the parasite, the tegument, is in intimate contact with the host immune system and presents an array of membrane-bound and membrane-associated antigens. Using signal sequence trapping, we identified novel membrane receptors (Smyth et al. 2003; Pearson et al. 2005), including a partial sequence encoding a putative seven transmembrane (7TM) receptor.
The 7TM receptors represent the largest and most ubiquitous family of membrane receptors in vertebrates (Pierce et al. 2002) and multicellular invertebrates (Mombaerts, 1999). Their ligands (where known) are as diverse as their distribution, and include hormones, neurotransmitters, chemokines, ions and even light photons. After binding to their ligands, many 7TM receptors signal by activating heterotrimeric G (guanine-nucleotide regulatory) proteins, resulting in phosphorylation of different substrates. There are 3 major families of 7TM receptors based on their sequence similarities - families A, B and C (Lefkowitz, 2004). Only 2 S. mansoni 7TM receptors have been described thus far, both from family A - a rhodopsin G Protein Coupled Receptor (GPCR) from cercariae (Hoffmann et al. 2001) and a histamine GPCR from adult worms (Hamdan et al. 2002).
Here, we describe the cloning and analysis of a full-length cDNA encoding a novel 7TM receptor from S. mansoni, Sm-7TM, which does not belong to any of the major families but shares identity with the human ee3 protein - a 7TM receptor implicated in neuronal signalling (Maurer et al. 2004). The orphan Sm-7TM receptor protein is predicted to have a long, intracellular C-terminal tail, and the protein is anchored in the tegument membrane of the adult stage.
MATERIALS AND METHODS
Nucleic acid techniques and analyses
Total RNA was extracted from mixed sex S. mansoni by homogenization in Trizol (Invitrogen), according to the manufacturer’s protocol. The full length cDNA - termed Sm-7TM - was obtained using RACE as previously described (Smyth et al. 2003), and has been deposited in the GenBank database (Accession number AY576274). Sm-7TM was amplified by PCR using oligonucleotide primers flanking the entire open reading frame (ORF) of the gene (1101 bp - excluding the stop codon), cloned into the pCR4-TOPO vector (Invitrogen) and chemically transformed into E. coli TOP10 cells (Invitrogen). The insert was sequenced and compared with those in the public databases using the BLAST algorithm (www.ncbi.nlm.nih.gov/BLAST/). Topological data were obtained using the TMPRED server (www.ch.embnet.org/software/TMPRED_form.html). N-linked glycosylation sites were predicted by the presence of NXS/T motifs and phosphorylation sites were predicted using the NetPhos (www.cbs.dtu.dk/services/NetPhos/) and NetPhosK (www.cbs.dtu.dk/services/NetPhosK/) servers. The alignment of amino acid sequences was carried out using the ClustalW 1.8 algorithm (www.searchlauncher.bcm.tmc.edu/multi-align/multi-align.html) and adjusted by eye. The phylogenetic analysis was performed with heuristic and neighbour-joining searches using PAUP beta version 8.0 for Macintosh. Bootstrap values were determined from 1000 replicates.
Reverse transcription PCR (RT-PCR) was used to assess the transcription profile of Sm-7TM in cercariae, 5-day in vitro-cultured schistosomula (Basch, 1981) and mixed sex adult worms. RNA was extracted from parasite tissues using Trizol (Invitrogen). First-strand cDNA was synthesized using Superscript III (Invitrogen), and RT-PCR consisted of 30 cycles of 94 °C for 1 min, 50 °C for 30 sec, 68 °C for 30 sec. Reactions were controlled by omitting reverse transcriptase from first-strand production. Triose phosphate isomerase was used as a constitutively expressed control mRNA (cf. Pearson et al. 2005).
The ORF corresponding to the intracellular C-terminal domain (Leu-252 - Glu-366) was amplified by PCR, cloned into the pBAD/Thio-TOPO vector (Invitrogen) and chemically transformed into E. coli TOP10 cells (Invitrogen). Recombinant protein was produced under non-denaturing conditions according to the manufacturer’s instructions. Supernatant from freeze-thawed cells was purified by batch binding via the 6×His tag to TALON metal affinity resin (Clontech). Fractions containing the majority of purified protein were desalted with a PD-10 desalting column (Amersham), eluted with PBS and concentrated to 1·0 mg/ml using an Amicon Ultra centrifugal filtration device (Millipore) with a 5 kDa molecular weight cut-off. The size and presence of recombinant Sm-7TMC were verified using a standard Western blotting approach using an anti-His(C-term)-HRP antibody (Invitrogen) diluted 1: 5000 in PBS/0·1% Tween 20/5% skim milk powder (PBST/SMP).
Production of specific antibody and detection/localization of Sm-7TM
An emulsion containing 100 μg of Sm-7TMC (1·0 mg/ml) and an equal volume of Freund’s Complete Adjuvant (FCA) was subcutaneously injected into a single New Zealand White rabbit. The same amount of antigen emulsified in Freund’s Incomplete Adjuvant (FIA) instead of FCA was injected 2 and 4 weeks later. The rabbit was bled 2 weeks later and the serum was collected by centrifugation. Polyclonal IgG was purified from the serum by binding to protein A-Sepharose CL-4B (Amersham). Bound antibodies were eluted with 0·1 m glycine, pH 2·8, and then equilibrated by adding 1/10 volume of 1·0 m Tris-HCl, pH 8·0. Serum collected prior to the immunization (‘pre-bleed’) was purified in the same manner.
Whole S. mansoni extracts were prepared by incubating 100 worms in 1·0 ml of PBS/1% SDS for 2 h with rotation at 4 °C. The insoluble material was pelleted by centrifugation. Ten microlitres of this extract was subjected to Western blotting using purified anti-Sm-7TMC antibody (diluted 1: 1000 in PBST/SMP) as the probe, following overnight blocking of the membrane (at 4 °C) with BALB/c mouse serum (diluted 1: 200 in PBST/SMP). The pre-bleed serum (diluted in the same manner) was used as a negative control.
Freshly perfused adult S. mansoni were embedded in OCT (Shandon) and cryostatically sectioned into 7·0 μm sections. Sections were then immunolabelled using indirect immunofluorescence as described by Li et al. (2000). Anti-Sm-7TMC serum was diluted 1: 25 in PBST/1% SMP. Serum raised against the thioredoxin tag was used as a negative control.
RESULTS AND DISCUSSION
Sm-7TM: a novel seven transmembrane receptor
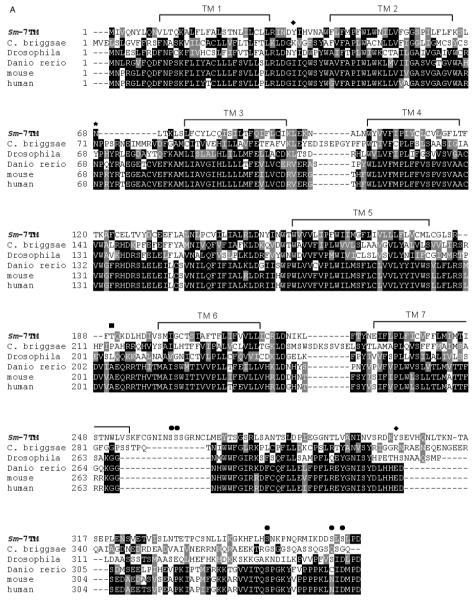
Previously, a 557 bp clone corresponding to Sm-7TM was isolated using a signal sequence trap screen of the S. mansoni cercarial transcriptome (Pearson et al. 2005). This transcript was shown to be present in males, females, eggs, miracidia and cercariae. Here, we also detected the presence of Sm-7TM mRNA in schistosomula that had been transformed and cultured for 5 days in vitro (Basch, 1981). The Sm-7TM mRNA was weakly transcribed in cercariae, but equally strongly expressed in schistosomula and adult worms relative to the triose phosphate isomerase control mRNA, further supporting our earlier findings (Pearson et al. 2005). We then used RACE to clone the full-length cDNA (Sm-7TM) from adult S. mansoni. The full-length ORF was predicted to contain 7 strong transmembrane domains. The ORF did not contain an archetypal, cleavable signal peptide, and likely undergoes secretion to the cell membrane via a mechanism similar to that used by type IIIb multiple membrane spanning proteins, where the amino terminus is exposed on the exterior surface of the membrane but does not have a cleavable signal sequence. Instead, the first hydrophobic transmembrane domain and surrounding charged residues provide the signal required to direct the protein through the secretory pathway (Spiess, 1995). While Sm-7TM exhibited no identity at the nucleotide level to any sequence deposited in GenBank, the deduced ORF (367 amino acids) shared 24-29% amino acid identity with gene products that contained 7 transmembrane domains from various metazoan organisms, including the free-living nematode Caenorhabditis briggsae (26%) and humans (26%) (Fig. 1A). Three truncated expressed sequence tags (ESTs; partial ORFs) derived from adult S. mansoni had 97-99% identity at the nucleotide level to Sm-7TM. While all of the homologues of Sm-7TM were novel 7TM receptors and did not belong to any of the 3 major families (Pierce et al. 2002), they all share identity with the human ee3 receptor, which is predicted to be a member of a new family of non-classical GPCR’s (Maurer et al. 2004). The higher order organism homologues of Sm-7TM are likely to be ee3 orthologues, as they all have >88% protein sequence identity with human ee3. Sequence identity was mostly restricted to the transmembrane domains (TMs) with conserved proline residues in TMs 2, 4, 5 and 7. Intracellular and extracellular loops were variable in length between the closest homologues of Sm-7TM (Fig. 1A); for example, Sm-7TM had a unique deletion of 11 amino acids in the loop region between transmembrane domains 2 and 3. Sm-7TM also contained a unique insertion of 17 residues in the C-terminal tail immediately after the final transmembrane domain. A common architectural feature of Sm-7TM and its homologues is the presence of a lengthy, hydrophilic C-terminal tail which, according to topology predictions, extrudes into the cytoplasm (Fig. 1B). The intracellular orientations of the C-termini of this orphan group, including Sm-7TM, is supported by the presence of multiple predicted phosphorylation sites on their C-terminal tails (for Sm-7TM, this is shown in Fig. 1A). Indeed, the C-terminus of human ee3 has been shown to immunoprecipitate with microtubule-associated protein (MAP) and co-localizes with MAP in vivo (Maurer et al. 2004). A notable feature of Sm-7TM is the prediction of a relatively large extracellular domain connecting the transmembrane segments 4 and 5. Present in the same position in the other 7TM homologues, this extracellular loop from Sm-7TM is 46 amino acids in length (13% of the coding region), is considerably longer than the equivalent loops from the homologues (which are between 9 and 11 residues long), and is the domain exhibiting the highest degree of identity (36% over 39 residues) with human ee3.
Fig. 1.
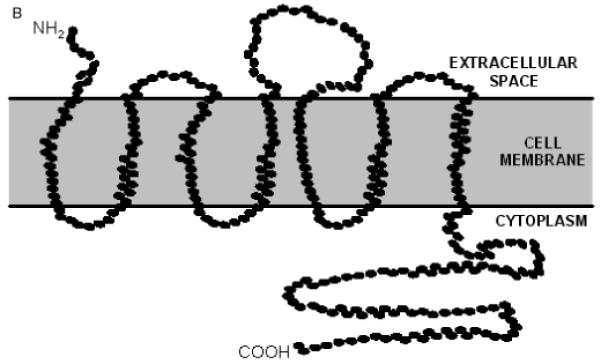
(A) Alignment of the full-length ORF of Sm-7TM with inferred gene products from Caenorhabditis briggsae, Drosophila melanogaster, Danio rerio and the mouse and human ee3 proteins. Black boxes indicate identical amino acids and grey boxes denote similarity. The predicted transmembrane domains (TM 1 - TM 7) relative to Sm-7TM are marked by a line spanning the region. The predicted N-glycosylation site is indicated by an asterisk (*). Hypothetical serine (●), threonine (■) and tyrosine (◆) phosphorylation sites are also marked. (B) Schematic representation of the predicted topology of Sm-7TM.
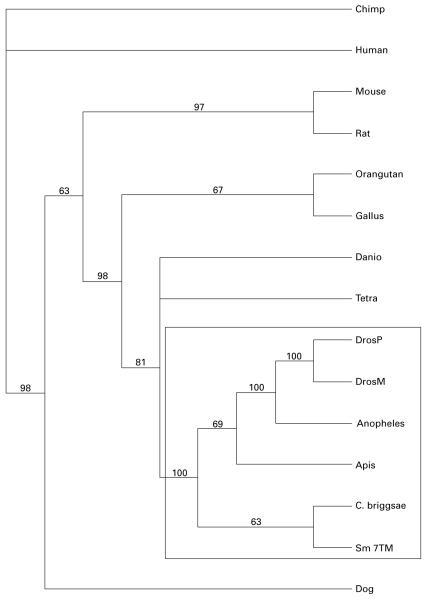
Phylogenetic analyses of data for the complete ORFs of representatives of these novel 7TM receptors (using heuristic and neighbour joining methods) revealed that Sm-7TM clustered within a clade (100% bootstrap support) of invertebrate orphan 7TM receptors (Fig. 2). By scanning the GPCR subfamily classifier website (http://www.soe.ucsc.edu/research/compbio/gpcr-subclass/), it was determined that Sm-7TM (and its homologues) did not belong to any of the known families of GPCRs.
Fig. 2.
Rooted, heuristic tree of Sm-7TM and representative homologous sequences found in GenBank. Protein sequence identifiers are as follows: chimp - XP_505903, human - AAH80607, mouse - NP_666215, rat - XP_222559, orangutan - CAH91932, Gallus - XP_420360, Danio - NP_956731, Tetra - CAF94057, DrosP - EAL32058, DrosM - NP_573360, Anopheles - XP_322017, Apis - XP_396742, C. briggsae - CAE57386, Sm-7TM - AAR84066 and dog - XP_533320. Numbers above the branches are bootstrap values calculated from 1000 replicates. The strongly supported clade of invertebrate receptors to which Sm-7TM belongs is highlighted in a box.
Recombinant expression of the C-terminal domain of Sm-7TM
Since the expression of the complete ORF of Sm-7TM was predicted to be problematic, due to the highly hydrophobic amino acid content of the transmembrane domains, we expressed the C-terminal 114 amino acids of Sm-7TM inferred to encode the large hydrophilic domain (termed Sm-7TMC). Sm-7TMC was expressed as a soluble fusion with E. coli thioredoxin (Fig. 3A) and represented a band with a molecular weight of ~29 kDa (comprising 13 kDa Sm-7TMC plus 16 kDa thioredoxin+vector tags). Once the optimal conditions for the expression were determined, a 1 l culture was induced, lysed and the soluble Sm-7TMC purified on metal affinity resin using the 6×His tag of the recombinant protein. Sm-7TMC was assessed for purity and concentration (3·0 mg/l) by SDS-PAGE (Fig. 3B) and Western blot analysis using anti-His(C-term)-HRP (Fig. 3C). The intact protein and an equal amount of degraded product (~14 kDa) of Sm-7TMC were detected. The first 10 N-terminal residues of the smaller protein were identified using Edman degradation (conducted at the Australian Proteome Analysis Facility, New South Wales) and the resultant sequence (KFRGNINSSS) matched residues 4-13 of Sm-7TMC (Lys-255 - Ser-264 of the complete ORF of Sm-7TM).
Fig. 3.
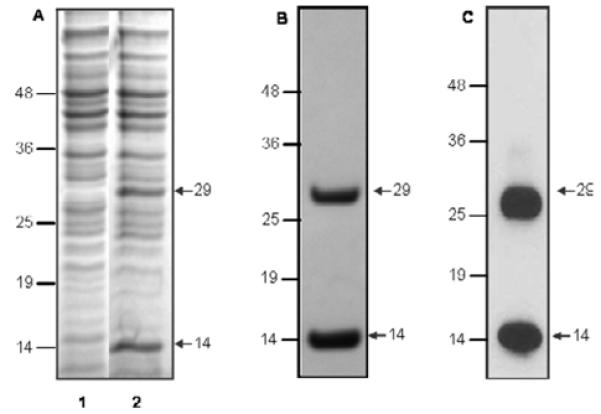
Expression of the C-terminal domain of Sm-7TM (Sm-7TMC). (A) SDS-polyacrylamide gel stained with Coomassie Brilliant Blue, showing soluble protein profiles from Escherichia coli before (lane 1) and 4 h after (lane 2) induction with 0·2% arabinose. Sm-7TMC was subsequently purified by metal affinity resin and analysed by (B) SDS-PAGE and (C) Western blotting using anti-His(C-term)-HRP. Molecular weight markers are shown in kDa.
Anti-Sm-7TMC antibody recognizes recombinant Sm-7TMC and native Sm-7TM
The ability of the antiserum to recognize both recombinant and native protein was tested by probing purified, recombinant Sm-7TMC and a SDS-solubilized S. mansoni extract. Western blot analysis (Fig. 4A) showed that the anti-Sm-7TMC antibody recognized the recombinant Sm-7TMC (~29 kDa) as well as its degradation product (~14 kDa; without the tag). Anti-Sm-7TMC serum strongly identified a 39 kDa protein (the approximate predicted molecular weight of full-length Sm-7TM) by Western blotting of solubilized parasite extracts (Fig. 4B), and the negative (pre-bleed) serum did not recognize the native or recombinant Sm-7TMC. A protein of ~80 kDa was recognized weakly in the S. mansoni extract by both the antiserum and negative control serum (Fig. 4B). This protein was not recognized by the conjugated secondary antibody alone (in the absence of rabbit primary antibody), and may represent an uncharacterized protein with sequence identity to Sm-7TM.
Fig. 4.
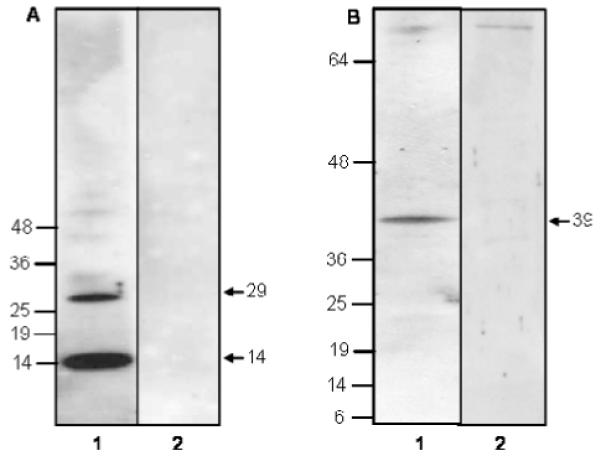
Western blots showing antigen detection by anti-Sm-7TMC rabbit serum. (A) Blot contains recombinant Sm-7TMC. Lane 1 - anti-Sm-7TMC rabbit serum, lane 2 - pre-bleed serum. (B) Blot contains SDS-solubilized adult Schistosoma mansoni extract. Lane 1 - anti-Sm-7TMC rabbit serum, lane 2 - pre-bleed serum. Molecular weight markers are shown in kDa.
Sm-7TM is localized to the tegument of adult Schistosoma mansoni
Fluorescence was observed along the outer tegument in adult worm sections probed with the specific antibody to Sm-7TM, including the tubercles of male worms (Fig. 5A and 5B), but was not detected in the other epithelial surfaces, such as the gastrodermis (Fig. 5B). The tegument of female worms also fluoresced (not shown), but no fluorescence was detected for sections probed with sera raised against the thioredoxin tag alone (Fig. 5C).
Fig. 5.
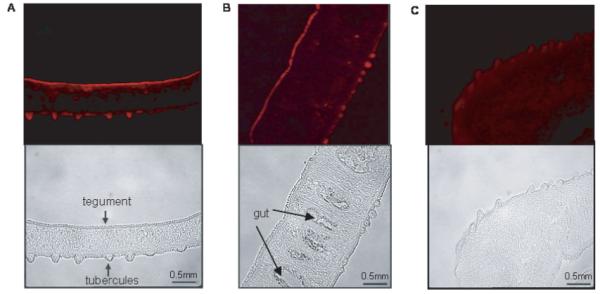
Immunostaining of unfixed, male Schistosoma mansoni sections with anti-Sm-7TMC rabbit serum and sheep anti-rabbit IgG-Cy3. All images are shown at×40 magnification with corresponding bright-field images shown beneath fluorescent images; (A) and (B) sections were probed with anti-Sm-7TMC rabbit serum; (C) section was probed with anti-thioredoxin serum.
Implications and concluding remarks
The 7TM receptors comprise the largest, most versatile family of membrane proteins known. As well as mediating a vast array of cell signalling and signal transduction processes, they are targeted by ~90% of all chemotherapeutic drugs (Pierce et al. 2002). Many of the well-characterized 7TM receptors are GPCRs, but the novel family of orphan receptors to which Sm-7TM belongs does not contain the required motifs to bind to G-proteins (Karchin et al. 2002) and is proposed to initiate signal transduction in a non-classical way.
Sequencing of 163 000 S. mansoni ESTs (Verjovski-Almeida et al. 2003) yielded partial sequences encoding probable 7TM receptors. Of 611 randomly sequenced, full-length cDNAs from S. japonicum, only 1 encoded a 7TM receptor (Hu et al. 2003), implying that this family of proteins is less common in schistosomes than in other organisms (Pierce et al. 2002), at least at the transcriptional level. An orthologue of Sm-7TM has not been identified from the S. japonicum ESTs (Hu et al. 2003).
Sm-7TM shared identity with a number of evolutionarily diverse homologues, including those from humans, Xenopus, zebrafish and fruit flies. Classification of 7TM receptors to known families usually requires at least 25% sequence identity (Pierce et al. 2002), implying that Sm-7TM and its homologues (Fig. 2) constitute a novel family of orphan receptors. All homologues possessed a long, hydrophilic C-terminal tail (predicted) that shared little to no identity between the invertebrate species - interestingly, this region most likely contains at least part of the ligand binding pocket, suggesting that this orphan family of receptors likely binds diverse ligands. Conversely, the relative homology observed between membrane spanning regions could be explained by the fact that certain aspects of signal transduction, such as receptor phosphorylation and membrane translocation, are relatively conserved (Lefkowitz, 2004).
Given its tegumental location, particular motifs of Sm-7TM are likely to be exposed to the host environment, making this receptor an attractive candidate for intervention strategies against schistosomiasis. Indeed, it has been shown that protective immunity can be induced in mice by immunization with purified adult worm surface membranes (Smithers et al. 1989) and, more recently, recombinant forms of the membrane and/or membrane-associated proteins, Sm-TSP-2 (Tran et al. 2006), Sm14 (Tendler et al. 1996), Sm23 (Da’dara et al. 2001) and Sm25 (Suri et al. 1997). Recent studies have explored the tegument proteome of S. mansoni (see Braschi et al. 2006; Braschi and Wilson, 2006), but Sm-7TM had not been identified using this approach. Its relatively low expression at the mRNA level (3 ESTs identified) might equate to low-level protein expression in the tegument, accounting for its absence from these reports in which the detection of proteins is limited to the sensitivity of the approach used for proteomic analysis.
Acknowledgments
This work was funded by a project grant (A.L.) and a block grant (D.P.M.) from the National Health and Medical Research Council of Australia (NHMRC). M.S.P. was supported by an Australian Postgraduate Award. A.L. was supported by a Career Development Award from the NHMRC. We gratefully acknowledge Ms Mary Duke for maintaining the life-cycle of S. mansoni.
REFERENCES
- Basch PF. Cultivation of Schsitosoma mansoni in vitro. I. Establishment of cultures from cercariae and development until pairing. Journal of Parasitology. 1981;67:179–185. [PubMed] [Google Scholar]
- Braschi S, Wilson RA. Proteins exposed at the adult schistosome surface revealed by biotinylation. Molecular and Cellular Proteomics. 2006;5:347–356. doi: 10.1074/mcp.M500287-MCP200. [DOI] [PubMed] [Google Scholar]
- Braschi S, Curwen RS, Ashton PD, Verjovski-Almeida S, Wilson A. The tegument surface membranes of the human blood parasite Schistosoma mansoni: a proteomic analysis after differential extraction. Proteomics. 2006;6:1471–1482. doi: 10.1002/pmic.200500368. [DOI] [PubMed] [Google Scholar]
- Capron A, Riveau G, Capron M, Trottein F. Schistosomes: the road from host-parasite interactions to vaccines in clinical trials. Trends in Parasitology. 2005;21:143–149. doi: 10.1016/j.pt.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Da’dara AA, Skelly PJ, Wang MM, Harn DA. Immunization with plasmid DNA encoding the integral membrane protein, Sm23, elicits a protective immune response against schistosome infection in mice. Vaccine. 2001;20:359–369. doi: 10.1016/s0264-410x(01)00374-7. [DOI] [PubMed] [Google Scholar]
- Hamdan FF, Abramovitz M, Mousa A, Xie J, Durocher Y, Ribeiro P. A novel Schistosoma mansoni G protein-coupled receptor is responsive to histamine. Molecular and Biochemical Parasitology. 2002;119:75–86. doi: 10.1016/s0166-6851(01)00400-5. [DOI] [PubMed] [Google Scholar]
- Hoffmann KF, Davis EM, Fischer ER, Wynn TA. The guanine protein coupled receptor rhodopsin is developmentally regulated in the free-living stages of Schistosoma mansoni. Molecular and Biochemical Parasitology. 2001;112:113–123. doi: 10.1016/s0166-6851(00)00352-2. [DOI] [PubMed] [Google Scholar]
- Hu W, Yan Q, Shen DK, Liu F, Zhu ZD, Song HD, Xu XR, Wang ZJ, Rong YP, Zeng LC, Wu J, Zhang X, Wang JJ, Xu XN, Wang SY, Fu G, Zhang XL, Wang ZQ, Brindley PJ, McManus DP, Xue CL, Feng Z, Chen Z, Han ZG. Evolutionary and biomedical implications of a Schistosoma japonicum complementary DNA resource. Nature Genetics. 2003;35:139–147. doi: 10.1038/ng1236. [DOI] [PubMed] [Google Scholar]
- Karchin R, Karplus K, Haussler D. Classifying G-protein coupled receptors with support vector machines. Bioinformatics. 2002;18:147–159. doi: 10.1093/bioinformatics/18.1.147. [DOI] [PubMed] [Google Scholar]
- King CH, Dickman K, Tisch DJ. Reassessment of the cost of chronic helmintic infection: a meta-analysis of disability-related outcomes in endemic schistosomiasis. Lancet. 2005;365:1561–1569. doi: 10.1016/S0140-6736(05)66457-4. [DOI] [PubMed] [Google Scholar]
- Lefkowitz RJ. Historical review: a brief history and personal retrospective of seven-transmembrane receptors. Trends in Pharmacological Sciences. 2004;25:413–422. doi: 10.1016/j.tips.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Li Y, Auliff A, Jones MK, Yi X, McManus DP. Immunogenicity and immunolocalization of the 22·6 kDa antigen of Schistosoma japonicum. Parasite Immunology. 2000;22:415–424. doi: 10.1046/j.1365-3024.2000.00319.x. [DOI] [PubMed] [Google Scholar]
- Loukas A, Tran M, Pearson MS. Schistosome membrane proteins as vaccines. International Journal for Parasitology. 2007;37:257–263. doi: 10.1016/j.ijpara.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Maurer MH, Grunewald S, Gassler N, Rossner M, Propst F, Wurz R, Weber D, Kuner T, Kuschinsky W, Schneider A. Cloning of a novel neuronally expressed orphan G-protein-coupled receptor which is up-regulated by erythropoietin, interacts with microtubule-associated protein 1b and colocalizes with the 5-hydroxytryptamine 2a receptor. Journal of Neurochemistry. 2004;91:1007–1017. doi: 10.1111/j.1471-4159.2004.02799.x. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Seven-transmembrane proteins as odorant and chemosensory receptors. Science. 1999;286:707–711. doi: 10.1126/science.286.5440.707. [DOI] [PubMed] [Google Scholar]
- Pearson MS, McManus DP, Smyth DJ, Lewis FA, Loukas A. In vitro and in silico analysis of signal peptides from the human blood fluke, Schistosoma mansoni. FEMS Immunology and Medical Microbiology. 2005;45:201–211. doi: 10.1016/j.femsim.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nature Reviews. Molecular Cell Biology. 2002;3:639–650. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- Ross AG, Bartley PB, Sleigh AC, Olds GR, Li Y, Williams GM, McManus DP. Schistosomiasis. New England Journal of Medicine. 2002;346:1212–1220. doi: 10.1056/NEJMra012396. [DOI] [PubMed] [Google Scholar]
- Smithers SR, Hackett F, Ali PO, Simpson AJ. Protective immunization of mice against Schistosoma mansoni with purified adult worm surface membranes. Parasite Immunology. 1989;11:301–318. doi: 10.1111/j.1365-3024.1989.tb00669.x. [DOI] [PubMed] [Google Scholar]
- Smyth D, McManus DP, Smout MJ, Laha T, Zhang W, Loukas A. Isolation of cDNAs encoding secreted and transmembrane proteins from Schistosoma mansoni by a signal sequence trap method. Infection and Immunity. 2003;71:2548–2554. doi: 10.1128/IAI.71.5.2548-2554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess M. Heads or tails - what determines the orientation of proteins in the membranes? FEBS Letters. 1995;369:76–79. doi: 10.1016/0014-5793(95)00551-j. [DOI] [PubMed] [Google Scholar]
- Suri PK, Goldberg M, Madikizela M, Petzke MM, Bungiro RD, Jr., Davies SJ, Chakraborty B, Nguyen KB, McCray JW, Jr., Knopf PM. Evaluation of recombinant protein r140, a polypeptide segment of tegumental glycoprotein Sm25, as a defined antigen vaccine against Schistosoma mansoni. Parasite Immunology. 1997;19:515–529. doi: 10.1046/j.1365-3024.1997.d01-160.x. [DOI] [PubMed] [Google Scholar]
- Tendler M, Brito CA, Vilar MM, Serra-Freire N, Diogo CM, Almeida MS, Delbem AC, Da Silva JF, Savino W, Garratt RC, Katz N, Simpson AS. A Schistosoma mansoni fatty acid-binding protein, Sm14, is the potential basis of a dual-purpose anti-helminth vaccine. Proceedings of the National Academy of Sciences, USA. 1996;93:269–273. doi: 10.1073/pnas.93.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd CW, Colley DG. Practical and ethical issues in the development of a vaccine against schistosomiasis mansoni. American Journal of Tropical Medicine and Hygiene. 2002;66:348–358. doi: 10.4269/ajtmh.2002.66.348. [DOI] [PubMed] [Google Scholar]
- Tran MH, Pearson MS, Bethony JM, Smyth DJ, Jones MK, Duke M, Don TA, McManus DP, Correa-Oliveira R, Loukas A. Tetraspanins on the surface of Schistosoma mansoni are protective antigens against schistosomiasis. Nature Medicine. 2006;12:835–840. doi: 10.1038/nm1430. [DOI] [PubMed] [Google Scholar]
- van der Werf MJ, de Vlas SJ, Brooker S, Looman CW, Nagelkerke NJ, Habbema JD, Engels D. Quantification of clinical morbidity associated with schistosome infection in sub-Saharan Africa. Acta Tropica. 2003;86:125–139. doi: 10.1016/s0001-706x(03)00029-9. [DOI] [PubMed] [Google Scholar]
- Verjovski-Almeida S, DeMarco R, Martins EA, Guimaraes PE, Ojopi EP, Paquola AC, Piazza JP, Nishiyama MY, Jr., Kitajima JP, Adamson RE, Ashton PD, Bonaldo MF, Coulson PS, Dillon GP, Farias LP, Gregorio SP, Ho PL, Leite RA, Malaquias LC, Marques RC, Miyasato PA, Nascimento AL, Ohlweiler FP, Reis EM, Ribeiro MA, Sa RG, Stukart GC, Soares MB, Gargioni C, Kawano T, Rodrigues V, Madeira AM, Wilson RA, Menck CF, Setubal JC, Leite LC, Dias-Neto E. Transcriptome analysis of the acoelomate human parasite Schistosoma mansoni. Nature Genetics. 2003;35:148–157. doi: 10.1038/ng1237. [DOI] [PubMed] [Google Scholar]